Abstract
Viral replication relies on the host to supply nucleosides. Host enzymes involved in nucleoside biosynthesis are potential targets for antiviral development. Ribavirin (a known antiviral drug) is such an inhibitor that suppresses guanine biosynthesis; depletion of the intracellular GTP pool was shown to be the major mechanism to inhibit flavivirus. Along similar lines, inhibitors of the pyrimidine biosynthesis pathway could be targeted for potential antiviral development. Here we report on a novel antiviral compound (NITD-982) that inhibits host dihydroorotate dehydrogenase (DHODH), an enzyme required for pyrimidine biosynthesis. The inhibitor was identified through screening 1.8 million compounds using a dengue virus (DENV) infection assay. The compound contains an isoxazole-pyrazole core structure, and it inhibited DENV with a 50% effective concentration (EC50) of 2.4 nM and a 50% cytotoxic concentration (CC50) of >5 μM. NITD-982 has a broad antiviral spectrum, inhibiting both flaviviruses and nonflaviviruses with nanomolar EC90s. We also show that (i) the compound inhibited the enzymatic activity of recombinant DHODH, (ii) an NITD-982 analogue directly bound to the DHODH protein, (iii) supplementing the culture medium with uridine reversed the compound-mediated antiviral activity, and (iv) DENV type 2 (DENV-2) variants resistant to brequinar (a known DHODH inhibitor) were cross resistant to NITD-982. Collectively, the results demonstrate that the compound inhibits DENV through depleting the intracellular pyrimidine pool. In contrast to the in vitro potency, the compound did not show any efficacy in the DENV-AG129 mouse model. The lack of in vivo efficacy is likely due to the exogenous uptake of pyrimidine from the diet or to a high plasma protein-binding activity of the current compound.
INTRODUCTION
Dengue is an increasingly prevalent mosquito-borne viral disease affecting human beings. The etiological agents involved are four serotypes of dengue virus (DENV), which belongs to the genus Flavivirus in the family Flaviviridae. An estimated 50 to 100 million dengue infections occur annually, with 500,000 cases of dengue hemorrhagic fever and 22,000 deaths, mainly among children (http://www.who.int/csr/disease/dengue/impact/en/index.html). Besides DENV, pathogenic flaviviruses also include yellow fever virus (YFV), West Nile virus (WNV), Japanese encephalitis virus (JEV), and tick-borne encephalitis virus (TBEV). No clinically approved antiviral therapy is currently available for the treatment of flavivirus infections. Human vaccines for flaviviruses are available only for YFV, JEV, and TBEV (16). Developing a dengue vaccine is challenging because a safe and efficacious dengue vaccine should provide durable protection against all four serotypes of DENV simultaneously to eliminate the potential pathological consequences of nonneutralizing, serotype-cross-reactive immune responses (31). With the incidence of dengue growing dramatically around the world, there is an urgent need for therapies.
The flaviviral genome is a single-stranded, positive-sense RNA of about 11 kb in length. The single open reading frame of the viral genome encodes three structural proteins (capsid [C], premembrane [prM], and envelope [Env]) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The structural proteins form the viral particle. The nonstructural proteins participate in the replication of the RNA genome, virion assembly (20, 27), and attenuation of host antiviral responses. For viral replication, NS1 participates in replication complex formation (24). NS3 acts as a protease (with NS2B as a cofactor) (14, 23), a nucleotide triphosphatase (NTPase) (48, 49), an RNA triphosphatase (4), and a helicase (7). NS4A induces membrane rearrangements in infected cells (32, 41), whereas NS4B, along with NS2A, colocalizes with replication complexes (28, 32). NS5 functions as a methyltransferase (MTase) (12, 40, 51) and an RNA-dependent RNA polymerase (RdRp) (1, 18). Both the structural and NS proteins are potential targets to block the viral life cycle. For attenuation of host antiviral responses, NS1 antagonizes complement activation (9). NS4B, and to a lesser extent NS2A and NS4A, blocks type I interferon signaling (17, 26, 33, 34). NS5 also antagonizes type I interferon signaling (5, 21) and mediates STAT2 degradation (2). Pharmacological blocking of viral protein-mediated antagonism of the host immune response may also bring clinical benefits to infected patients.
Besides viral targets, host proteins could also be targeted for potential antiviral development (37). A number of host proteins or cellular pathways have been reported to be critical for flavivirus replication. Furin cleaves the flavivirus prM protein on the virus surface into the mature M protein (13, 45). Glucosidase is responsible for the proper folding and glycosylation of the flaviviral prM, E, and NS1 (11). Host kinases have been shown to be involved in DENV and WNV assembly and secretion (8, 19). The cholesterol biosynthesis pathway has been linked to DENV entry and replication as well as to the host immune response (29, 42). We recently showed that brequinar inhibits DENV through depletion of the intracellular level of pyrimidine (39). Brequinar was originally developed as an antimetabolite in cancer and immunosuppression therapies through inhibition of dihydroorotate dehydrogenase (DHODH), the fourth enzyme in the de novo pyrimidine biosynthesis pathway (25, 30). Our results suggested that inhibitors of the pyrimidine synthesis pathway could potentially be developed for DENV therapy. Along similar lines, ribavirin, a drug for treatment of hepatitis C virus (HCV) infection, inhibits cellular IMP dehydrogenase (IMPDH), an enzyme that is essential for de novo biosynthesis of guanine nucleotides (15). Ribavirin was previously shown to inhibit flaviviruses, primarily through depletion of the intracellular GTP pool (22).
In this study, we report on a novel inhibitor of host DHODH. The compound was identified through a cell-based high-throughput screening (HTS). It has a broad spectrum of antiviral activities. To analyze the mechanism of inhibition, we show that the compound directly binds and inhibits recombinant DHODH. Addition of uridine to the culture medium reverses the compound-mediated antiviral activity. Furthermore, we show that DENV type 2 (DENV-2) variants resistant to brequinar were cross resistant to the newly identified compound. Overall, the results have strengthened the concept that targeting pyrimidine biosynthesis is a potential approach for antiviral development.
MATERIALS AND METHODS
Viruses, compounds, and antibodies.
We used the following viruses: WNV (strain 3356), YFV (17D vaccine strain), DENV-2 (strains New Guinea C and TSV01), Western equine encephalitis virus (WEEV) (strain Cova 746), and vesicular stomatitis virus (VSV) (New Jersey serotype). The sources of these viruses were reported previously (50). Vero, A549, and Huh-7 cells were used for virus infection as specified for individual experiments. All compounds used in this study were synthesized in house and were dissolved in 90% dimethyl sulfoxide (DMSO) for in vitro experiments. DENV-specific mouse monoclonal antibody 4G2 against envelope protein was prepared from a hybridoma cell line purchased from the American Type Culture Collection (ATCC).
CFI assay.
The cell-based flavivirus immunodetection (CFI) assay was performed as described previously (47). Briefly, A549 cells were infected with DENV-2 (strain New Guinea C; multiplicity of infection [MOI] of 0.3) in the presence of 2-fold serial dilutions of compounds. After incubation at 37°C for 48 h, viral antigen production was quantified by immunodetection using the 4G2 antibody and goat anti-mouse IgG conjugated with horseradish peroxidase as primary and secondary antibodies, respectively. The concentrations of compounds that decreased the envelope protein production by 50% (50% effective concentrations [EC50s]) were calculated by nonlinear regression analysis.
Viral titer inhibition assay.
The following viruses were used in the viral titer inhibition assay: DENV-2 (strain New Guinea C), WNV (strain 3356), YFV (17D vaccine strain), WEEV (strain Cova 746), and VSV (New Jersey serotype). Plaque assay was performed as reported previously (38). Briefly, Vero cells (4 × 105 cells/well) were seeded in a 12-well plate. At 24 h postseeding, the cells were infected with the indicated viruses at an MOI of 0.1. For DENV, WNV, YFV, and WEEV infections, samples were collected at 42 h postinfection (p.i.). For VSV, samples of culture medium were collected at 16 h postinfection.
Cell viability assay.
Cell viability was measured using the CellTiter 96 Aqueous One Solution cell proliferation assay (Promega) according to the manufacturer's protocol. Approximately 1 × 104 Vero or A549 cells in 100 μl medium were seeded in a 96-well plate. After 16 h of incubation, the cells were treated with test compound. After another 48 h of incubation, 20 μl of CellTiter 96 solution was added to 100 μl of medium. After 2 h of incubation at 37°C with 5% CO2, the absorbance was measured at 490 nm in a SaffireII microplate reader (Tecan, Austria).
Replicon assay.
The replicon assay was performed as described previously (35). Briefly, A549 dengue virus replicon cells or Huh-7 HCV replicon cells were treated with 2-fold serial dilutions of the test compound. After incubation at 37°C for 48 h, luciferase substrate (EnduRen [Promega] for dengue virus replicon and Britelite [Perkin-Elmer] for HCV replicon) was added according to the manufacturer's protocols. Luminescence was measured in Clarity luminometer (Biotek). The concentrations of compounds that decreased the luciferase expression by 50% (EC50s) were calculated by nonlinear regression analysis.
Cloning, expression, and purification of human DHODH protein.
The cDNA encoding an N-terminally truncated human DHODH was synthesized and cloned into the pET-19b expression vector. The human DHODH protein was expressed in Escherichia coli strain BL21(DE3) as a six-histidine fusion protein. Expression of the fusion protein was induced with 1 mM isopropyl-β-d-thiogalactoside at an optical density at 600 nm of 0.6. Cells were grown for another 3 h at 37°C in Terrific broth medium. The expressed fusion protein was then purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography (Qiagen), cleaved with Tev protease (500 units/mg fusion protein, overnight at 4°C), and further purified on a SuperDex75 size exclusion column (GE Healthcare).
Chromogen reduction assay.
A chromogen reduction assay was performed to measure the DHODH activity as described previously with modifications (3, 10). A100-μl reaction mixture was used, and the reaction was carried out in a 96-half-well plate (Costar) at room temperature. The oxidation of l-dihydroorotate (Sigma) with cosubstrate ubiquinone-10 (CoQ10) (Sigma) was coupled to 2,6-dichlorophenolindophenol (DCIP) (Fluka) reduction. l-Dihydroorotate (200 μM) was preincubated with CoQ10 (100 μM) and DCIP (60 μM) in assay buffer (0.1% Triton X-100, 100 mM KCl, and 100 mM Tris-Cl, pH 8.0) for 20 min at room temperature before addition of the enzyme to initiate the reaction. Changes in absorbance at 610 nm due to DCIP reduction were monitored using a SaffireII microplate reader.
ITC.
Isothermal titration calorimetry (ITC) experiments were performed using an Auto ITC titration calorimetric system (MicroCal Inc.). For the direct determination of the binding of the compound NITD-102 and brequinar to DHODH, the calorimetric cell, which contained purified DHODH protein dissolved in 20 mM HEPES (pH 8.0), 10% glycerol, 150 mM KCl, and 5% DMSO, was titrated with NITD-102 or brequinar dissolved in the same buffer. The concentration of DHODH was 23 μM, and the concentration of inhibitor in the injection syringe was 233 μM for NITD-102 and 200 μM for brequinar. The titration experiment was performed by adding the titrant in steps of 10 μl at 25°C.
De novo RdRp assay.
The de novo RdRp assay was performed as described previously (36). The reaction mixture (25 μl) contained 50 mM HEPES (pH 8.0), 10 mM KCl, 5 mM MgCl2, 2 mM MnCl2, 10 mM dithiothreitol (DTT), 0.5 mM ATP, 0.5 mM UTP, 0.5 mM CTP, 2.5 μCi [33P]GTP (10 μCi/μl, 3,000 Ci/mmol; Perkin-Elmer), 0.25 μg NS5, 0.5 μg RNA template, and the indicated concentrations of compound NITD-982. The de novo RdRp reaction mixtures were incubated at 23°C for 30 min, and the enzymatic reaction products were analyzed on a 10% denaturing polyacrylamide gel and quantified with a Typhoon 9410 variable-mode imager (GE Healthcare).
Pharmacokinetics and in vivo efficacy analyses.
The pharmacokinetic profiles of NITD-982 were determined in cotton rats following oral (p.o.) and subcutaneous (s.c.) administration of 10 mg/kg of body weight. Blood samples were collected at 0.25, 0.5, 1, 2, 4, 8, and 24 h after dosing. There were three cotton rats per time point. Plasma concentrations of NITD-982 were measured using liquid chromatography-tandem mass spectrometry (LC–MS-MS). Pharmacokinetic parameters were calculated by a noncompartmental approach using the WinNonLin software (Pharsight).
The extent of plasma protein binding of NITD-982 was determined by equilibrium dialysis using the Pierce RED device from Thermo Scientific (Rockford, IL) according to the manufacturer's protocol. In brief, CD-1 mouse plasma (Innovative Research) was spiked with 5 μg/ml of compound and transferred to the red chamber, while phosphate-buffered saline (PBS) was added to the white chamber. To determine nonspecific binding, PBS containing compound was added to a separate insert. The RED device was incubated at ∼37°C while shaking at 100 rpm for 4 h. Afterwards, an aliquot was removed from each chamber, and equal amounts of fresh plasma and PBS were added to the respective incubated aliquots. The plasma-PBS mixtures were precipitated using acetonitrile. After mixing and centrifugation, the supernatant was transferred to a clean 96-well plate for LC–MS-MS analysis.
The in vivo efficacy of NITD-982 was evaluated in a dengue virus viremia model in mice (43). AG129 mice (with knockout alpha/beta interferon and gamma interferon receptors), purchased from B & K Universal, were injected intraperitoneally with 0.4 ml of RPMI 1640 medium containing 5 × 106 PFU/ml of DENV-2 (strain TSV01). The infected mice were then dosed with NITD-982 or vehicle (100% corn oil) by s.c. injection. The mice (n = 6 per group) were monitored twice a day; blood samples were collected on day 3 p.i. for viral titer determination by plaque assay, and plasma uridine concentrations were quantitatively analyzed by LC–MS-MS on a Sciex Applied Biosystems 4000QTRAP coupled to an Agilent 1100 high-pressure liquid chromatography (HPLC) system. Statistical analysis was performed with Student's t test using SigmaPlot/SigmaStat software (Systat Software Inc.).
RESULTS
Cell-based CPE inhibition assay.
We developed a high-throughput screening (HTS) assay to monitor compound-mediated inhibition of viral infection-induced cytopathic effect (CPE). Huh-7 cells (a human hepatocyte-derived carcinoma cell line; 750 cells per well) were infected with DENV-2 (New Guinea C strain) at an MOI of 1 in a 1,536-well format. After incubation of the infected cells with compounds for 3 days, compound-mediated inhibition of CPE was determined by measuring the intracellular level of ATP using the CellTiter-Glo luminescent-cell viability assay (Promega). The assay had a signal-to-noise ratio of 15 and a Z′ value of 0.67. The CPE assay measures a gain of signal. Therefore, the assay reduces false-positive hits that are cytotoxic.
Identification of NITD-982.
The CPE-based HTS assay was used to screen the Novartis compound library of about 1.8 million compounds at a single concentration of 5 μM. The overall hit rate of the screen was 0.14% using a cutoff of 25% inhibition. All hits were reordered for retesting of their antiviral activities, with a reconfirmation rate of 33%. The screening effort led to identification of a class of compounds with an isoxazole-pyrazole core with anti-DENV activity. One such compound, NITD-982 (Fig. 1A), inhibited the virus-induced CPE by 71% at 5 μM; as a positive control, treatment of the infected cell with NITD-008, a known nucleoside inhibitor of DENV (50), gave an EC50 of 1.6 μM (data not shown). To validate the antiviral activity, we analyzed the compound using a CFI assay. The CFI assay is an enzyme-linked immunosorbent assay (ELISA)-based assay that measures the amount of envelope protein in cells infected with DENV (47). The compound reduced viral E protein production in a dose-dependent manner, with an estimated EC50 of 2.4 nM (Fig. 1B). Next, the antiviral activity of NITD-982 was further confirmed by a viral titer reduction assay. As shown in Fig. 1C, the compound inhibited virus production with an EC90 of 5.2 nM; the control compound NITD-008 showed an EC90 of 2.8 μM (data not shown). Treatment with 50 nM NITD-982 suppressed the viral titer to an undetectable level. Furthermore, NITD-982 inhibited a DENV-2 luciferase-reporting replicon (Fig. 1D), suggesting that it suppresses viral translation and/or RNA synthesis. A cell proliferation-based 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay (Promega) showed that the compound was not cytotoxic in A549 (a human alveolar basal epithelial cell line) and Vero (a kidney epithelial cell line from African green monkey) cells at up to 5 μM (Fig. 1E), suggesting that the observed antiviral activity was not due to compound-mediated toxicity.
Fig. 1.
(A) Structure of NITD-982. (B) Effect of NITD-982 on expression of viral envelope protein. A549 cells were infected with DENV-2 (MOI of 0.3) in the presence of 2-fold serial dilutions of NITD-982. After incubation at 37°C for 48 h, the expression of viral envelope protein was quantified by CFI assay. (C) Effect of NITD-982 on the growth of DENV-2. Vero cells were infected with DENV-2 (MOI of 0.1). After incubation at 37°C for 48 h, cell culture supernatants were harvested for plaque assay in BHK-21 cells. The limit of quantification (L.O.Q.) (indicated by a dotted line) for the plaque assay is around 100 PFU/ml. (D) Effect of NITD-982 on DENV-2 replicon. A549 cells containing a luciferase replicon of DENV-2 were treated with NITD-982 at the indicated concentrations for 48 h. The inhibition of viral replication by the compound was measured by the luciferase activity. (E) Cytotoxicity of NITD-982 in Vero and A549 cells. Cytotoxicity was examined by incubation of Vero and A549 cells for 2 days with the indicated concentrations of NITD-982. Cell viability was measured by an MTS assay. Average results and standard deviations (n = 3) are presented.
Time-of-addition analysis.
A time-of-addition experiment was performed to estimate the inhibitory step of NITD-982 (Fig. 2). Vero cells were infected with DENV-2 (strain New Guinea C) containing a Renilla luciferase reporter. The luciferase gene was inserted to the capsid gene of DENV-2, as previously reported (44). NITD-982 (25 nM) was added to the infected cells at various time points after infection. Luciferase activities were measured at 24 h after infection. As a negative control, DMSO (0.45%) was added to the infected cells at various time points for estimation of its effects on viral replication. The results showed that the inhibition of viral replication by NITD-982 gradually diminished when the compound was added at late time points (up to 16 h p.i.), suggesting that the compound blocks a late stage of the viral life cycle. The time-of-addition result agrees with the replicon result (Fig. 1D), arguing that NITD-982 likely inhibits viral RNA replication.
Fig. 2.
Time-of-addition analysis. DENV-2 (strain New Guinea C) containing a Renilla luciferase reporter was used to infect Vero cells in a 96-well plate. The luciferase gene was engineered at the capsid gene of the DENV-2, as previously reported by Shustov and colleagues (44). Approximately 2 × 104 Vero cells were seeded per well. After an overnight incubation, the cells were infected with the reporter virus at an MOI of 1. NITD-982 was then added to the infected cells at a final concentration of 25 nM at the indicated time points. At 24 h p.i., the cells were assayed for luciferase activities. As negative controls, an equivalent concentration of DMSO (0.45%) was included at various time points. Average results and standard deviations (n = 4) are presented.
Broad antiviral spectrum.
We examined the antiviral spectrum of NITD-982 against other flaviviruses (yellow fever virus [YFV] and West Nile virus [WNV]), a plus-stranded RNA alphavirus (Western equine encephalitis virus [WEEV]), and a negative-strand RNA rhabodovirus (vesicular stomatitis virus [VSV]). Vero cells were infected with YFV, WNV, WEEV, or VSV at an MOI of 0.1. The infected cells were treated with various concentrations of NITD-982. Culture medium was collected for plaque assays at 42 h p.i. (YFV, WNV, and WEEV) and 16 h p.i. (VSV). As shown in Fig. 3A, NITD-982 potently inhibited YFV, WNV, WEEV, and VSV, with EC90 values of 12.8 nM, 0.31 nM, 33.8 nM, and 38.7 nM, respectively. In addition, we found that the compound also inhibited an HCV replicon, with an EC50 of 1.5 nM; cell viability assay showed that the anti-HCV activity was not due to compound-mediated cytotoxicity (Fig. 3B). These results demonstrate that NITD-982 has a broad spectrum of antiviral activity. The results also suggest that the compound inhibits a host pathway or a cellular factor that is required for the replication of these evolutionarily distant viruses. In agreement with this notion, the compound did not show any activity when tested against DENV protease, NTPase, MTase, and RdRp (see below and data not shown).
Fig. 3.
Antiviral spectrum of NITD-982. (A) Vero cells were infected with indicated viruses at an MOI of 0.1; the infected cells were immediately treated with NITD-982. Virus production was quantified using plaque assays. The detection limit of the plaque assay is about 100 PFU/ml for all tested viruses. (B) Huh-7 cells containing a luciferase replicon of HCV were treated with NITD-982 at the indicated concentrations for 48 h. The inhibition of viral replication was measured by luciferase activity (left panel). Replicon cell viability (right panel) was determined by CellTiter-Glo luminescent-cell viability assay (Promega). Average results and standard deviations (n = 3) are presented.
DHODH as the target of NITD-982.
Using a chemoproteomics approach, Bonavia and colleagues recently reported that an NITD-982 analogue binds specifically to the host enzyme dihydroorotate dehydrogenase (DHODH) (6). The details of chemoproteomics-based target deconvolution have been reported elsewhere by Bonavia and coworkers (6). DHODH is a mitochondrial protein that catalyzes the oxidation of dihydroorotate to orotate, the fourth enzymatic step of the de novo pyrimidine biosynthesis. To validate DHODH as the target of NITD-982, we prepared recombinant human DHODH (with a deletion of the N-terminal 29-amino-acid signal peptide and transmembrane domain; GenBank accession no. NC_000016) using an E. coli expression system (Fig. 4A). A chromogen reduction assay was used to monitor the activity of DHODH by measuring the reduction of 2,6-dichlorophenolindophenol (DCIP), which is stoichiometrically equivalent to the oxidation of dihydroorotate to orotate; the reaction was quantified with a spectrophotometer at a wavelength of 610 nm (3, 10). As shown in Fig. 4B, NITD-982 inhibited DHODH activity with a 50% inhibitory concentration (IC50) of 103 nM. As a positive control, brequinar, a known DHODH inhibitor (46), exhibited an IC50 of 2.1 nM.
Fig. 4.
DHODH is the target of NITD-982. (A) Recombinant human DHODH protein made in E. coli. Human DHODH containing an N-terminal His tag was overexpressed in an E. coli expression system. The cell lysates were purified through an affinity nickel column. Samples from total cell lysate, soluble cell lysate (soluble fraction), column flowthrough, washings, and elution were analyzed by SDS-PAGE with Coomassie blue staining. (B) NITD-982 and its analogue NITD-102 inhibit DHODH activity in vitro. DHODH activity was measured with a chromogen reduction assay as described in Materials and Methods. Brequinar was included as a positive control. Average results and standard deviations (n = 3) are presented. (C) Calorimetric titration of DHODH with NITD-102 and brequinar. The concentration of DHODH was 23 μM, and the syringe contained NITD-102 (233 μM) or brequinar (200 μM). The structures of NITD-102 and brequinar are shown in the bottom panels. The ITC titration curves and the calculated Kd values are shown in the top panels.
Next, we measured the compound binding to DHODH by isothermal titration calorimetry (ITC). ITC is a physical technique to determine the thermodynamic parameters of small molecules binding to larger macromolecules. Since NITD-982 has a low aqueous solubility (5 μM) that limits its use in the ITC assay, we synthesized an analogue, NITD-102 (Fig. 4C), with a solubility of >100 μM to determine the compound-DHODH interaction. NITD-102 inhibited DHODH at an IC50 of 18 nM (Fig. 4B). NITD-102 (at 233 μM; the compound was completely dissolved in solution at this concentration) was added in 10-μl increments up to a molar ratio of about 2:1 for DHODH protein (23 μM). As shown in Fig. 4C, the binding of NITD-102 to DHODH protein gave a measureable heat change with an apparent dissociation constant (Kd) of 33 nM at 25°C and a binding stoichiometry close to 1. Compound binding is driven mainly by enthalpy, with ΔH° being −9.9 kcal/mol out of a ΔG° value of −10.2 kcal/mol. As a positive control, brequinar was also shown to bind to DHODH protein with a Kd value of 8 nM, although the protein-compound stoichiometry was shifted to 2:1 (Fig. 4C). The shift in stoichiometry most likely came from the uncertainty in protein concentration determination due to the presence of cofactors. Nevertheless, the results demonstrate that the compound directly binds to DHODH and inhibits its enzymatic activity.
Pyrimidine reverses the antiviral effect of NITD-982.
We examined the ability of uridine to counteract the inhibition of DENV replication in cells treated with NITD-982. If DHODH is the cellular target of NITD-982, then supplementing uridine in the culture medium would allow pyrimidine synthesis to proceed via the salvage pathway and reverse the antiviral effect of NITD-982. Indeed, addition of uridine rescued DENV replication in the NITD-982-treated cells in a dose-dependent manner (Fig. 5). Supplementing with 5 μM uridine partially restored viral replication; supplementing with 25 or 50 μM uridine completely rescued viral replication. This result supports that DHODH is the target of NITD-982.
Fig. 5.
Uridine reverses the antiviral effect of NITD-982. A549 cells were infected with DENV-2 and treated with different doses of NITD-982 and uridine. At 48 h p.i., viral envelope protein production was quantified by the CFI assay (see experimental details in Materials and Methods). Average results and standard deviations (n = 3) are presented.
Brequinar-resistant viruses are cross resistant to NITD-982.
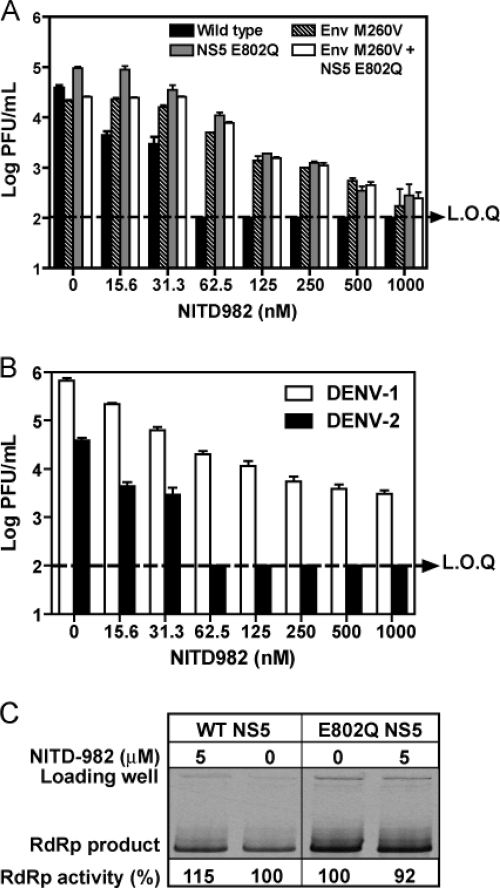
We previously selected brequinar-resistant viruses by serial passaging of DENV-2 in the presence of increasing concentrations of brequinar. We found that mutation M260V in Env alone, E802Q in NS5 alone, or Env M260V plus NS5 E802Q conferred resistance of DENV-2 to brequinar (39). Given the fact that DHODH is the target of both brequinar and NITD-982, we hypothesized that the brequinar-resistant viruses would be insensitive to the inhibition of NITD-982. As expected, a viral titer reduction assay showed that all three brequinar-resistant viruses (generated from the corresponding infectious cDNA clones) were cross resistant to NITD-982 (Fig. 6A). At a concentration of 62.5 nM, NITD-982 suppressed the wild-type (WT) virus by >300-fold to an undetectable level; in contrast, the titers of three brequinar-resistant viruses were suppressed by <10-fold. Similar to the brequinar resistance profile (39), the double mutant DENV-2 (Env M260V plus NS5 E802Q) did not exhibit more resistance than the single mutant viruses (Env M260V or NS5 E802Q). It is currently not known why the Env M260V and NS5 E802Q mutations are not additive.
Fig. 6.
(A) Brequinar-resistant viruses are cross resistant to NITD-982. Vero cells were infected with wild-type or recombinant DENV-2 (strain TSV01; MOI of 0.1) in the presence or absence of NITD-982. At 48 h p.i., viral titers were quantified by plaque assays. A dotted line indicates the limit of quantification (L.O.Q.) of the plaque assay. (B) Resistance analyses of WT DENV-1 (strain Western Pacific) and DENV-2 (strain TSV01). Vero cells were infected with WT DENV-1 and DENV-2 at an MOI of 0.1 in the presence of NITD-982. Viral titers were quantified by plaque assays at 48 h postinfection. (C) Inhibition of de novo RNA synthesis by NITD-982. The RdRp assay was performed as described in Materials and Methods. Average results and standard deviations (n = 3) are presented.
The WT sequences of DENV-1, -3, and -4 bear a Gln residue at position 802 of NS5. The fact that DENV-2 containing the E802Q substitution was resistant to NITD-982 suggests that the WT DENV-1 is resistant to the compound. As shown in Fig. 6B, a viral titer reduction assay showed that DENV-1 was much less sensitive to NITD-982 inhibition than DENV-2, with EC90 values of 30 nM and 5.2 nM, respectively. Similarly to DENV-1, DENV-3 and DENV-4 were also less sensitive to NITD-982 inhibition, with EC90 values of 27 nM and 97 nM, respectively (data not shown).
The requirement of Gln at amino acid 802 of NS5 for viral resistance to NITD-982 raised the possibility that NS5 was an alternative target of the inhibitor. To examine this possibility, we performed de novo RdRp assays using recombinant DENV-2 NS5 proteins with or without the E802Q mutation. The mutant E802Q NS5 protein generated 2.3-fold more RNA product than the WT NS5 (Fig. 6C). NITD-982 at 5 μM (the maximal aqueous solubility) did not suppress the RdRp activity of either WT or E802Q NS5. Overall, the results demonstrate that (i) brequinar-resistant viruses are cross resistant to NITD-982 and (ii) viral NS5 is not the direct target of NITD-982, but the E802Q mutation in NS5 is able to confer resistance, probably through enhancement of polymerase activity.
In vivo pharmacokinetics and efficacy of NITD-982.
We determined the pharmacokinetic parameters of NITD-982 in cotton rats. Due to the low solubility of the compound, corn oil was selected as the delivery vehicle. Cotton rats were dosed with NITD-982 at 10 mg/kg of body weight. As shown in Fig. 7A, after subcutaneous (s.c.) injection, the compound was slowly absorbed, as indicated by a Tmax (time required to reach Cmax [maximum plasma concentration] after dosing) of 8 h, reaching a Cmax of 528 nM. After oral (p.o.) dosing, the compound exhibited a Cmax of 260 nM and a Tmax of 2 h. As indicated by the area under the curve (AUC), the total exposure of the s.c. dosing was much higher than that of the p.o. dosing. We also measured the plasma protein-binding activity for NITD-982. The in vitro binding assay showed that the compound had a mouse plasma protein-binding activity of 99.76%.
Fig. 7.
In vivo testing of NITD-982. (A) Plasma concentrations of NITD-982 after oral and subcutaneous (s.c.) administration in cotton rats at a dosage of 10 mg/kg of body weight (n = 3 per time point). A dotted line indicates the limit of quantification (L.O.Q.) by LC–MS-MS. (B) In vivo efficacy of NITD-982. AG129 mice were intraperitoneally inoculated with 2 × 106 PFU of DENV-2 (strain TSV01) on day 0. The mice (6 animals per group) were subcutaneously dosed with NITD-982 twice a day. The peak viremia on day 3 p.i. was quantified by plaque assay. A dotted line indicates the L.O.Q. of the plaque assay. (C) Plasma uridine level in NITD-982-treated mice. Uridine levels in plasma samples were quantitatively analyzed by LC–MS-MS. The asterisk denotes a Student t test P value of < 0.05. A dotted line indicates the L.O.Q.
Next, we examined the in vivo efficacy of NITD-982 in a dengue virus viremia mouse model (43). AG129 mice (lacking alpha/beta interferon and gamma interferon receptors) were infected with DENV-2 (strain TSVO1), treated immediately with NITD-982 (1, 3, 10, or 30 mg/kg of weight in mice twice per day), and measured for peak viremia on day 3 postinfection. Based on the above pharmacokinetic results that s.c. dosing yielded a better exposure than p.o. dosing, we chose to dose the animals through the s.c. route. No reduction in viremia was observed, even when the animals were treated with 30 mg/kg (Fig. 7B). To explore the reason for the lack of in vivo efficacy, we measured the uridine concentrations in plasma collected on day 3 postinfection. The average plasma concentrations of uridine ranged from 6.6 to 8.2 μM in the compound-treated mice (Fig. 7C). Compared with the vehicle-treated group, only the 30-mg/kg-treated group showed significantly lower uridine concentration in plasma (P < 0.05 by Student's t test). Collectively, the results suggest that the compound was not able to suppress the uridine concentration to a level low enough to achieve in vivo efficacy.
DISCUSSION
Host factors are potential targets for antiviral development. The targets could be cellular proteins that are required for viral replication. Alternatively, host factors that are involved in pathogenesis could be targeted to mitigate disease manifestations. For development of dengue therapeutics, the latter strategy requires blocking the host pathway that leads to plasma leakage, the hallmark of the severe disease form; it is currently challenging to pursue this strategy because the molecular understanding of dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) is lacking. In this study, we explored the feasibility of the former strategy for anti-DENV drug discovery. A novel inhibitor of host DHODH was identified through a CPE-based HTS. Since DHODH is involved in the biosynthesis of pyrimidine, the compound exhibited a broad spectrum of antiviral activity in cell culture. Besides inhibiting flavivirus, alphavirus, and rhabodovirus (Fig. 3), the compound also suppresses HIV and respiratory syncytial virus (RSV) in cell culture (data not shown).
NITD-982 inhibits DENV replication through suppression of host DHODH. This mode of action is supported by four lines of evidence in the current study. (i) The compound inhibits the enzymatic activity of DHODH. (ii) An analogue of NITD-982 directly binds to the recombinant DHODH. (iii) The compound-mediated inhibition of viral replication could be reversed by supplementing the culture medium with uridine. (iv) DENV-2 variants resistant to brequinar (a known DHODH inhibitor) are cross resistant to NITD-982. However, these results do not exclude the possibility that besides inhibiting DHODH, the compound may exert its antiviral activities through another mechanism(s). In line with this notion, we found that DENV-2 containing an M260V substitution in the viral envelope protein (located at a helix in domain II) was cross resistant to both brequinar and NITD-982. The M260V envelope mutation was previously shown to reduce the efficiency of virion assembly/release, but the mutant virus became less sensitive to brequinar inhibition at the step of virion assembly/release (39). The exact mechanism of the envelope M260V mutation in conferring resistance remains to be defined.
Cross-resistance analysis showed that besides the M260V mutation in envelope protein, an E802Q substitution in the viral RdRp domain could also confer partial resistance to both brequinar and NITD-982 (Fig. 6A). The E802Q mutation is located at the priming loop of the NS5 polymerase. This mutation was previously shown to enhance the de novo RNA synthesis of recombinant NS5 protein, as well as the efficiency of viral RNA synthesis of a DENV-2 replicon (39). However, neither brequinar nor NITD-982 inhibits the RdRp activity. These results indicate that although RdRp is not the target of these inhibitors, DENV-2 could develop resistance through enhancement of polymerase activity. Sequence alignment indicates that the wild-type amino acid at position 802 of DENV-1, -3, and -4 NS5 is Gln, suggesting that viruses from these three serotypes would be partially resistant to NITD-982. Indeed, DENV-1, -3, and -4 were less sensitive to NITD-982 inhibition than DENV-2. The variation in sensitivity among different serotypes of DENV represents a major weakness of the DHODH inhibitors for further antiviral development.
A discrepancy was observed between brequinar and NITD-982 with regard to their relative potencies in enzyme assay versus in cell culture antiviral assay. In the DHODH enzyme assay, brequinar and NITD-982 showed IC50s of 2.1 nM and 103 nM (Fig. 4B), respectively, whereas in the cell culture-based CFI antiviral assay, brequinar and NITD-982 showed EC50s of 78 nM (39) and 2.4 nM (Fig. 1B), respectively. Since both compounds need to enter cells before exerting their antiviral functions, one explanation for the observed discrepancy is that the two compounds have different efficiencies in penetrating the cell membrane. Although brequinar is more potent in inhibiting DHOHD enzyme than NITD-982, brequinar may be less efficient in penetrating the cell membrane, resulting in a lower potency than NITD-982 in antiviral activity.
NITD-982 exhibited a potent anti-DENV-2 activity in cell culture, with an EC90 of 5.2 nM (Fig. 1C). Pharmacokinetic analysis showed that the compound reached a Cmax of 528 nM after dosing at 10 mg/kg in cotton rats (Fig. 7A); the Cmax of 528 nM was about 100-fold above the EC90. However, the compound did not achieve any in vivo efficacy, even when the infected mice were treated at 30 mg/kg (Fig. 7B). Similar to the results for DENV, NITD-982 inhibited RSV with a single-digit nanomolar EC50; however, no in vivo efficacy was observed in an RSV cotton rat model (unpublished results). The discrepancy between the in vitro and in vivo efficacies is most likely due to high concentrations of plasma uridine in the treated animals. The uridine concentration in the compound-treated animal remained at 6.6 to 8.2 μM (Fig. 7C); uridine at these concentrations could reverse the compound-mediated antiviral activity in cell culture (Fig. 5). Two parameters could contribute to the lack of reduction of the uridine concentration in the treated animals. The first parameter is the plasma protein-binding activity of the compound. Computational analysis using the software GastroPlus ADMET Predictor (SimulationsPlus, Inc.) suggests that NITD-982 has a high plasma protein-binding activity of 99%. In agreement with this prediction, the in vitro binding assay showed that the compound had a plasma protein-binding activity of 99.76%. Given this protein-binding activity, the true exposure of free NITD-982 in plasma was estimated to be 1.3 nM after the animals were dosed with 10 mg/kg (528 nM × 0.24% = 1.3 nM), which did not reach the EC90. The second parameter is the uridine uptake from diet. Even when de novo synthesis of pyrimidine was inhibited by the DHODH inhibitors, the external source of uridine could replenish/maintain a high concentration of pyrimidine in the plasma. Therefore, under physiological conditions, the uridine uptake from diet could preclude in vivo efficacy. To differentiate the effects of the two parameters on the in vivo efficacy of DENV inhibition, one needs to reduce the plasma protein-binding activity of the inhibitor through chemical modifications; this should concomitantly increase the free plasma compound concentration to levels higher than the EC90. Such a compound will allow us to examine whether the inhibitor is able to suppress the plasma concentration of pyrimidine to a level low enough to suppress viral replication. If in vivo efficacy cannot be achieved with such inhibitors, the feasibility of targeting the pyrimidine biosynthesis pathway for antiviral development has to be questioned.
In summary, we have identified an inhibitor that has a broad spectrum of antiviral activity in cell culture. Mode-of-action studies demonstrated that the compound inhibits host DHODH, an enzyme involved in the cellular pyrimidine biosynthesis. Inhibiting DHODH activity can be considered to deplete intracellular pyrimidine pools, leading to suppression of viral RNA synthesis. Our findings suggest that host proteins could be targeted for development of DENV therapeutics. However, translation of in vitro efficacy to in vivo efficacy of the host inhibitors remains to be achieved. One challenge to target host proteins for antiviral development is potential toxicity. Toward this end, it is critical to design compounds that specifically block the function essential for viral infection without affecting normal functions of the host protein. The issues discussed in this study should be applicable to other host pathway-related antiviral projects.
ACKNOWLEDGMENTS
We thank Gang Zou, Christian Noble, Andy Yip, Lee Kok Sin, and other colleagues at Novartis Institute for Tropical Diseases and colleagues from Novartis Institutes for BioMedical Research for helpful discussions and support during the course of this study.
Footnotes
Published ahead of print on 20 April 2011.
REFERENCES
- 1. Ackermann M., Padmanabhan R. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926–39937 [DOI] [PubMed] [Google Scholar]
- 2. Ashour J., Laurent-Rolle M., Shi P. Y., Garcia-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 83:5408–5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bader B., Knecht W., Fries M., Loffler M. 1998. Expression, purification, and characterization of histidine-tagged rat and human flavoenzyme dihydroorotate dehydrogenase. Protein Expr. Purif 13:414–422 [DOI] [PubMed] [Google Scholar]
- 4. Bartelma G., Padmanabhan R. 2002. Expression, purification, and characterization of the RNA 5′-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology 299:122–132 [DOI] [PubMed] [Google Scholar]
- 5. Best S. M., et al. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828–12839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonavia A., et al. 2011. Organic synthesis toward small-molecule probes and drugs special feature: identification of broad-spectrum antiviral compounds and assessment of the druggability of their target for efficacy against respiratory syncytial virus (RSV). Proc. Natl. Acad. Sci. U.S.A. 108:6739–6744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borowski P., et al. 2001. Purification and characterization of West Nile virus nucleoside triphosphatase (NTPase)/helicase: evidence for dissociation of the NTPase and helicase activities of the enzyme. J. Virol. 75:3220–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chu J. J., Yang P. L. 2007. c-Src protein kinase inhibitors block assembly and maturation of dengue virus. Proc. Natl. Acad. Sci. U. S. A. 104:3520–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chung K. M., et al. 2006. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc. Natl. Acad. Sci. U. S. A. 103:19111–19116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Copeland R. A., et al. 1995. Recombinant human dihydroorotate dehydrogenase: expression, purification, and characterization of a catalytically functional truncated enzyme. Arch. Biochem. Biophys. 323:79–86 [DOI] [PubMed] [Google Scholar]
- 11. Courageot M. P., Frenkiel M. P., Dos Santos C. D., Deubel V., Despres P. 2000. Alpha-glucosidase inhibitors reduce dengue virus production by affecting the initial steps of virion morphogenesis in the endoplasmic reticulum. J. Virol. 74:564–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Egloff M. P., Benarroch D., Selisko B., Romette J. L., Canard B. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elshuber S., Allison S. L., Heinz F. X., Mandl C. W. 2003. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 84:183–191 [DOI] [PubMed] [Google Scholar]
- 14. Falgout B., Miller R. H., Lai C. J. 1993. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B-NS3 protease activity. J. Virol. 67:2034–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Foster G., Mathurin P. 2008. Hepatitis C virus therapy to date. Antivir. Ther. 13(Suppl. 1):3–8 [PubMed] [Google Scholar]
- 16. Gubler D., Kuno G., Markoff L. 2007. Flaviviruses, p. 1153–1253 In Knipe D. M., Howley P. M. (ed.), Fields virology, 5th ed., vol. 1 Lippincott William & Wilkins, Philadelphia, PA [Google Scholar]
- 17. Guo J., Hayashi J., Seeger C. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guyatt K. J., Westaway E. G., Khromykh A. A. 2001. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J. Virol. Methods 92:37–44 [DOI] [PubMed] [Google Scholar]
- 19. Hirsch A. J., et al. 2005. The Src family kinase c-Yes is required for maturation of West Nile virus particles. J. Virol. 79:11943–11951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kummerer B. M., Rice C. M. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laurent-Rolle M., et al. 2010. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J. Virol. 84:3503–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leyssen P., Balzarini J., De Clercq E., Neyts J. 2005. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J. Virol. 79:1943–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H., Clum S., You S., Ebner K. E., Padmanabhan R. 1999. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 73:3108–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindenbach B., Rice C. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu S., Neidhardt E. A., Grossman T. H., Ocain T., Clardy J. 2000. Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. Structure 8:25–33 [DOI] [PubMed] [Google Scholar]
- 26. Liu W., et al. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79:1934–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu W. J., Chen H. B., Khromykh A. A. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 77:7804–7813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mackenzie J. M., Khromykh A. A., Jones M. K., Westaway E. G. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203–215 [DOI] [PubMed] [Google Scholar]
- 29. Mackenzie J. M., Khromykh A. A., Parton R. G. 2007. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2:229–239 [DOI] [PubMed] [Google Scholar]
- 30. McLean J. E., Neidhardt E. A., Grossman T. H., Hedstrom L. 2001. Multiple inhibitor analysis of the brequinar and leflunomide binding sites on human dihydroorotate dehydrogenase. Biochemistry 40:2194–2200 [DOI] [PubMed] [Google Scholar]
- 31. Miller N. 2010. Recent progress in dengue vaccine research and development. Curr. Opin. Mol. Ther. 12:31–38 [PubMed] [Google Scholar]
- 32. Miller S., Kastner S., Krijnse-Locker J., Buhler S., Bartenschlager R. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 282:8873–8882 [DOI] [PubMed] [Google Scholar]
- 33. Munoz-Jordan J. L., et al. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004–8013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Munoz-Jordan J. L., Sanchez-Burgos G. G., Laurent-Rolle M., Garcia-Sastre A. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U. S. A. 100:14333–14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ng C. Y., et al. 2007. Construction and characterization of a stable subgenomic dengue virus type 2 replicon system for antiviral compound and siRNA testing. Antiviral Res. 76:222–231 [DOI] [PubMed] [Google Scholar]
- 36. Niyomrattanakit P., et al. 2010. Inhibition of dengue virus polymerase by blocking of the RNA tunnel. J. Virol. 84:5678–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Noble C. G., et al. 2010. Strategies for development of dengue virus inhibitors. Antiviral Res. 85:450–462 [DOI] [PubMed] [Google Scholar]
- 38. Puig-Basagoiti F., et al. 2006. Triaryl pyrazoline compound inhibits flavivirus RNA replication. Antimicrob. Agents Chemother. 50:1320–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qing M., et al. 2010. Characterization of dengue virus resistance to brequinar in cell culture. Antimicrob. Agents Chemother. 54:3686–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ray D., et al. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 80:8362–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roosendaal J., Westaway E. G., Khromykh A., Mackenzie J. M. 2006. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 80:4623–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rothwell C., et al. 2009. Cholesterol biosynthesis modulation regulates dengue viral replication. Virology 389:8–19 [DOI] [PubMed] [Google Scholar]
- 43. Schul W., Liu W., Xu H. Y., Flamand M., Vasudevan S. G. 2007. A dengue fever viremia model in mice shows reduction in viral replication and suppression of the inflammatory response after treatment with antiviral drugs. J. Infect. Dis. 195:665–674 [DOI] [PubMed] [Google Scholar]
- 44. Shustov A., Mason P., Frolov I. 2007. Production of pseudoinfectious yellow fever virus with a two-component genome. J. Virol. 81:11737–11748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stadler K., Allison S. L., Schalich J., Heinz F. X. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ullrich A., Knecht W., Fries M., Loffler M. 2001. Recombinant expression of N-terminal truncated mutants of the membrane bound mouse, rat and human flavoenzyme dihydroorotate dehydrogenase. A versatile tool to rate inhibitor effects? Eur. J. Biochem. 268:1861–1868 [PubMed] [Google Scholar]
- 47. Wang Q. Y., et al. 2009. A small-molecule dengue virus entry inhibitor. Antimicrob. Agents Chemother. 53:1823–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Warrener P., Tamura J. K., Collett M. S. 1993. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J. Virol. 67:989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wengler G., Wengler G. 1991. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology 184:707–715 [DOI] [PubMed] [Google Scholar]
- 50. Yin Z., et al. 2009. An adenosine nucleoside inhibitor of dengue virus. Proc. Natl. Acad. Sci. U. S. A. 106:20435–20439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou Y., et al. 2007. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 81:3891–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]