Abstract
Efficient horizontal transmission is a signature trait of chronic wasting disease (CWD) in cervids. Infectious prions shed into excreta appear to play a key role in this facile transmission, as has been demonstrated by bioassays of cervid and transgenic species and serial protein misfolding cyclic amplification (sPMCA). However, the source(s) of infectious prions in these body fluids has yet to be identified. In the present study, we analyzed tissues proximate to saliva, urine, and fecal production by sPMCA in an attempt to elucidate this unique aspect of CWD pathogenesis. Oropharyngeal, urogenital, and gastrointestinal tissues along with blood and obex from CWD-exposed cervids (comprising 27 animals and >350 individual samples) were analyzed and scored based on the apparent relative CWD burden. PrPCWD-generating activity was detected in a range of tissues and was highest in the salivary gland, urinary bladder, and distal intestinal tract. In the same assays, blood from the same animals and unseeded normal brain homogenate controls (n = 116 of 117) remained negative. The PrP-converting activity in peripheral tissues varied from 10−11- to 100-fold of that found in brain of the same animal. Deer with highest levels of PrPCWD amplification in the brain had higher and more widely disseminated prion amplification in excretory tissues. Interestingly, PrPCWD was not demonstrable in these excretory tissues by conventional Western blotting, suggesting a low prion burden or the presence of protease-sensitive infectious prions destroyed by harsh proteolytic treatments. These findings offer unique insights into the transmission of CWD in particular and prion infection and trafficking overall.
INTRODUCTION
Chronic wasting disease (CWD) is an efficiently transmitted prion disease of cervids (e.g., deer, elk, and moose) and is the only known prion disease affecting free-ranging, nondomestic animals. While the origins of CWD are uncertain, the disease has been present in wild cervid populations of northern Colorado and southern Wyoming for over 40 years (56, 57) and has now been identified in both captive and free-ranging cervids in 17 states, 2 Canadian provinces, and the Republic of Korea (44). With intensified national surveillance efforts, CWD continues to be identified in areas previously thought to be free of infection, including recent discoveries in Virginia and Missouri (20, 21). The prevalence of CWD varies across North America but can be as high as 30% in some areas of Colorado or up to 100% in captive populations (23).
While much remains to be learned about the mechanisms of horizontal CWD transmission, the environment and environmental fomites were previously shown to be capable of harboring infectious prions in both outdoor and, later, indoor cervid housing facilities (31, 33). Our previous work demonstrated that both saliva and blood from CWD-positive (CWD+) animals contained infectious prions able to transmit infection to naïve white-tailed deer (32). Subsequently, the application of a transgenic mouse bioassay and serial protein misfolding cyclic amplification (sPMCA) to saliva, urine, and fecal samples from infected cervids and to deer inoculated with urine and feces has demonstrated lower levels of infectious prions in these excreta (17, 18, 50).
The identification of infectious prions in excreta has shed light on the transmission and pathogenesis of CWD; however, the proximal source of the excreted prions remains elusive, i.e., whether they are produced in organs of excretion or, alternatively, are derived from central nervous or lymphoid tissues and excreted into bodily fluids and feces, questions pertinent to our understanding of prion transmission and pathogenesis in nature.
Limited evidence exists for protease-resistant, disease-associated forms of the prion protein (PrPRES/Sc/d) in glandular epithelial and excretory tissues. However, researchers have reported the presence of PrPRES by immunohistochemistry (IHC), paraffin-embedded tissue (PET) blotting, or Western blotting (WB) in the kidneys of scrapie-affected sheep (28, 35, 46, 47) and CWD+ deer (19), the lingual tissues of hamsters inoculated with the HY (hyper) strain of transmissible mink encephalopathy (10, 34), and the salivary glands of scrapie-affected sheep (55) and CWD-infected cervidized transgenic mice (43). While the identification of protease-resistant prion proteins in cervid excretory tissues using conventional assays has proven difficult (2, 13, 14, 24, 49), bioassay studies identifying genuine prion infectivity in excretory tissues, albeit limited, date to the 1960s (16) and implicate both the salivary glands of Creutzfeldt-Jakob disease (CJD)-infected mice (41) and kidneys of scrapie-infected mice (12). sPMCA (38, 48) now offers a more time- and animal-efficient means to detect low levels of infectious prions and thereby reexamine questions and mechanisms of prion excretion and transmission.
In the present study, we have applied standardized sPMCA to examine tissues associated with saliva, urine, and fecal excretion (e.g., salivary glands, tongue, nasal mucosa, kidney, ureter, urinary bladder, and intestinal tract) from 27 CWD-exposed versus naïve white-tailed deer to gain insight into the potential peripheral mechanisms underlying the facile horizontal transmission of chronic wasting disease.
MATERIALS AND METHODS
Infected cervids.
Twenty-seven white-tailed deer (Odocoileus virginianus) were exposed to CWD from positive and negative sources in various forms (e.g., urine and feces, saliva, environmental fomites, blood, or brain tissue) and by various routes (e.g., orally [p.o.], intravenously [i.v.], intraperitoneally [i.p.], intracranially [i.c.], or through environmental exposure) (30–32). All animals were inoculated and maintained in dedicated, restricted-access, indoor CWD research facilities in accordance with Colorado State University IACUC guidelines. The sources of inoculum included terminally ill mule deer (Odocoileus hemionus) of an unknown PrP genotype (courtesy of Michael Miller, Colorado Division of Wildlife) or white-tailed deer utilized in previous and concurrent studies of either of two genotypes: homozygous for glycine (i.e., G/G) at cervid PrP position 96 or heterozygous at that position, with alleles for both glycine and serine (G/S). The deer were monitored for 16 to 27 months postinoculation (mpi), during which time the majority became tonsil biopsy positive for CWD by immunohistochemistry (IHC). The duration of clinical disease prior to necropsy ranged from 0 weeks (i.e., no clinical signs) to 72 weeks. At scheduled necropsy dates, or when exhibiting signs of terminal disease, deer were euthanized and subjected to an extensive necropsy using fresh instruments and collection vessels for each individual, at which time bodily fluids and an array of tissues were collected and frozen at −80°C. Animals that were not tonsil biopsy positive for CWD prior to euthanasia were thoroughly evaluated for CWD infection after necropsy, including an evaluation of neural and lymphoid tissues, which have been shown to be early diagnostic sites for CWD (e.g., obex, retropharyngeal lymph nodes, and tonsils) by IHC and Western blotting (WB). A complete description of animals, inoculation routes and sources, CWD status, time of first tonsil biopsy specimen positivity in mpi, incubation period at necropsy (in mpi), and duration of clinical signs (in weeks) is shown in Table 1.
Table 1.
Summary of source animals with tissues evaluated by sPMCA
| Animal ID | Inoculum | Route of exposure | PrP genotype (position 96) | CWD tonsil biopsy status | Time to first tonsil biopsy-positive result (mpi) | Incubation period at necropsy (mpi) | Duration of clinical signs (wk)a |
|---|---|---|---|---|---|---|---|
| 144 | Saliva | p.o. | G/G | + | 12 | 26 | 6 |
| 113 | Saliva | p.o. | G/G | + | 12 | 19 | NA |
| 132 | Saliva | p.o. | G/G | + | 19 | 19 | NA |
| 122 | Saliva | p.o. | G/G | + | 12 | 19 | NA |
| 147 | Saliva | p.o. | G/G | − | NA | 16 | NA |
| 107 | Saliva | p.o. | G/G | + | 19 | 19 | NA |
| 4119 | Blood | i.v. | G/S | + | 12 | 24 | 38 |
| 347 | Blood | i.v. | G/G | + | 6 | 27 | 44 |
| 114 | Blood | i.v. | G/G | + | 19 | 19 | NA |
| 108 | Blood | i.p. | G/G | + | 12 | 19 | NA |
| 110 | Blood | i.v. | G/G | + | 12 | 19 | NA |
| 133 | Blood | i.v. | G/G | + | 12 | 26 | 50 |
| 137 | Blood | i.v. | G/S | + | 19 | 19 | NA |
| 146 | Blood | i.v. | G/G | + | 12 | 29 | 72 |
| 141 | Urine/feces | p.o. | G/G | − | NA | 19 | NA |
| 150 | Urine/feces | p.o. | G/G | − | NA | 19 | NA |
| 111 | Urine/feces | p.o. | G/S | − | NA | 19 | NA |
| 134 | Urine/feces | p.o. | G/G | − | NA | 19 | NA |
| 124 | Urine/feces | p.o. | G/S | − | NA | 19 | NA |
| 4461 | Environment | Unknown | G/G | + | 15 | 19 | NA |
| 4129 | Environment | Unknown | G/S | + | 19 | 19 | NA |
| 106 | CWD-positive brain | i.c. | G/S | + | 12 | 22 | 12 |
| 121 | CWD-positive brain | i.c. | G/S | + | 6 | 26 | 24 |
| 123 | CWD-negative brain | i.c./p.o. | G/G | − | Negative | 19 | NA |
| 103 | CWD-negative brain | i.c./p.o. | G/G | − | Negative | 19 | NA |
| 4488 | CWD-negative brain | i.c./p.o. | G/S | − | Negative | 19 | NA |
| 4516 | CWD-negative brain | i.c./p.o. | G/G | − | Negative | 19 | NA |
NA, not applicable.
Study samples and preparation.
Obex, parotid salivary gland, tongue, nasal respiratory epithelium, kidney, ureter, urinary bladder, proximal and distal intestinal tract, and blood were collected at necropsy and frozen at −80°C. Tissue samples were thawed briefly, and 10 to 50 mg of each sample was trimmed individually and homogenized as a 1% (wt/vol) solution in PMCA buffer (1% [vol/vol] Triton X-100, 5 mM EDTA, and 150 mM NaCl in phosphate-buffered saline [PBS] adjusted to a pH of 7.2) by using a FastPrep tissue homogenizer for 60 s at power setting 6.5. Whole-blood samples remained suspended in 5% EDTA until sPMCA evaluation. All tissues were prepared in individual microcentrifuge tubes and homogenized in parallel in the same machine concurrent with both positive and negative controls. Prepared homogenates were coded to allow for blinded evaluation by sPMCA.
Western blotting of peripheral tissues.
Peripheral tissue homogenates, prepared as described above, were blindly analyzed for the presence of PrPCWD using conventional Western blotting. Fifteen microliters of the 1% sample homogenate was mixed with 7 μl of sample buffer (0.1% [vol/vol] Triton X-100 and 4% [wt/vol] SDS in PBS); pretreated with 2 μl collagenase A, 2 μl dispase II, and 1 μl DNase I (Boehringer-Mannheim, Germany) (final concentrations, 1 mg/ml, 1 mg/ml, and 0.05 mg/ml, respectively) for 30 min at 37°C; and ultimately digested with 3 μl proteinase K at 500 μg/ml (Invitrogen) (final concentration, 60 μg/ml) for 30 min at 37°C. Ten microliters of 4× running buffer was then added to the sample, followed by denaturation for 3 min at 95°C. Twenty microliters of this preparation was run on a precast 12% SDS-PAGE gel (Invitrogen) in a Bio-Rad electrophoresis apparatus for 1 h at 160 mV. Samples were then transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore) for 1 h at 115 mV in a Bio-Rad transfer apparatus. PVDF membranes were subsequently probed with a PrP-specific monoclonal antibody (BAR224) conjugated to horseradish peroxidase, diluted 1:20,000 in 5% (wt/vol) powdered milk in 0.2% Tween 20 in Tris-buffered saline (TBST) for 1 h. Following washing with TBST, immunoreactivity was detected by using an enhanced chemiluminescence detection system (ECL-Plus; Amersham Biosciences) with an LAS 3000 imaging system (Fuji Inc., Valhalla, NY).
Preparation of normal brain homogenate for sPMCA.
Normal brain homogenate (NBH), the substrate for prion conversion in vitro, was prepared from Tg(CerPrP)5037 mice in a room that had not previously been used for prion research (18, 26). Following euthanasia and perfusion with 5 mM EDTA in PBS, whole brain was collected from naïve Tg(CerPrP)5037 mice and placed on ice. Brain homogenates were prepared as a 10% (wt/vol) solution in PMCA buffer with the addition of Complete protease inhibitors (Roche Pharmaceuticals, Indianapolis, IN) using a Dounce homogenizer. Homogenates were then centrifuged for 1 min at 2,000 rpm, and the supernatant was frozen in single-experiment aliquots at −80°C in a “prion-free” room until use for sPMCA. Each preparation was composed of brains from 4 to 6 mice to minimize the potential influence of expression variation in CerPrP or other cofactors (1, 7); multiple batches of NBH were utilized over the course of the experiments.
sPMCA of tissues.
Tissue homogenates from CWD-exposed deer were blindly assayed for PrPCWD, in duplicate, by sPMCA. All test samples were prepared in parallel with tissue-matched positive and negative controls as a 1% homogenate in PMCA buffer, as described above, and subsequently spiked into normal brain homogenate for amplification (17, 18, 25, 26). Ten microliters of test or control tissue homogenate was added to 50 μl of NBH and assayed, in parallel and in adjacent wells of a 96-well plate (USA Scientific, Ocala, FL), along with normal brain homogenate prepared from unexposed Tg(CerPrP)5037 mice as additional, unseeded negative controls. Plates were then sonicated by using an ultrasonic processor (Misonix, Farmingdale, NY) and incubated at 37°C. Sonication parameters were set at 40-s bursts at power level 7.0, followed by 30 min of incubation. Ninety-two cycles of sonication were performed over 48 h, with a 10-μl aliquot of sonicated material transferred into 50 μl of fresh NBH for serial amplification. Following each round of amplification, samples were evaluated by Western blotting, as described below, for the presence of PrPCWD. Each sample received a score based on the number of rounds in which that particular sample was positive (i.e., a maximum of 3 in a three-round experiment), and the scores for each duplicate run were totaled to arrive at a final score, with “0” being the lowest and “6” being the highest score that a given sample could receive. In each experimental run, between 25 and 50% of samples evaluated were tissue-matched negative controls. Additionally, 10 to 20% of the samples in a run were unspiked, NBH negative controls.
sPMCA of blood.
EDTA-preserved whole-blood samples from CWD-exposed deer were blindly analyzed in duplicate as described above for tissues, with some modifications. Using a protocol initially described by Tattum and colleagues (51), we spiked 1 μl of whole blood into 100 μl of normal brain homogenate. Sixty microliters of this preparation was added to a 96-well plate and analyzed by sPMCA. Sonication parameters were set at 20-s bursts at power level 7.5, followed by 30 min of incubation at 37°C. One hundred forty-four cycles of sonication were performed over 72 h, with a 10-μl aliquot of sonicated material transferred into 50 μl of fresh NBH for serial amplification. Each sample was evaluated by WB as described below for PrPCWD signals following each round of amplification.
sPMCA of obex dilution series and approximation of tissue scores.
Obex tissue dilutions from each deer, collected and processed as described above, were made by diluting each of the initial 1% obex homogenates into 10% normal Tg(CerPrP)5037 brain homogenate (NBH) at 1:10 dilutions ranging from 100 to 10−14 (i.e., final dilutions of 10−2 to 10−16). Prior to dilution, the initial 1% obex homogenates were sonicated for 20 s at power setting 7.0 in an ultrasonic processor. Ten microliters of each dilution was added to 50 μl of NBH and analyzed for three rounds of sPMCA as described above for tissues. Each dilution then received an sPMCA score as described above for peripheral tissues; these scores were then used to estimate the relative log obex dilution equivalent (LODE) scores for individual tissues within animals. Tissue LODE scores were conservatively assigned based on the lowest obex dilution yielding a given score.
WB of sPMCA-amplified tissues.
Following each round of sPMCA, an aliquot of each sonicated sample was subjected to Western blotting for the evaluation of the PrPCWD signal. Fifteen microliters of sample homogenate was mixed with 7 μl of sample buffer (0.1% [vol/vol] Triton X-100 and 4% [wt/vol] SDS in PBS), digested with 3 μl proteinase K at 500 μg/ml (final concentration, 60 μg/ml) for 20 min at 37°C followed by 10 min at 45°C, and analyzed for PrPCWD using the Western blotting protocol described above.
Evaluation of intrarun variability in duplicate experiments.
As all samples were run in duplicate on two different sonicating machines, we sought to compare the intrarun variabilities in experimental results. A commercially available calculator for the categorical Cohen's kappa value was used to define the agreement of results between the two different experiments (28a). Because scores progressed linearly in the sPMCA evaluation, a linear weighted approach was used to calculate Cohen's kappa. The significance of the correlation was determined by using a method described previously by Landis and Koch (27).
Statistical analysis of data.
To broadly analyze the factors affecting the peripheral PrPCWD distribution and intensity, we used a permutational-based multivariate analysis of variance (PERMANOVA). We used this test to evaluate (i) the effect of the obex score on the PrPCWD distribution and intensity among peripheral tissues and (ii) the relationship between the source and route of inoculum on the PrPCWD distribution and intensity among peripheral tissues. We conducted these analyses on two presentations of our data: a semiquantitative scoring measure (raw scoring of sPMCA intensity) and a more quantitative measurement of sPMCA intensity relative to the amount of amplifiable PrPCWD found in the brain of that animal (LODE score or relative PrPCWD burden). Post hoc analyses were used to evaluate pairwise comparisons of treatment groups within each PERMANOVA. As the distribution of PrPCWD intensity (measured by sPMCA scores) across tissues was fairly limited (ranging from 0 to 6 for the raw sPMCA score and 0 to 10 for the LODE score, i.e., relative PrPCWD burden), we did not transform the data prior to PERMANOVAs. PERMANOVAs were based on Bray-Curtis similarity matrices and were graphically represented using nonmetric multidimensional scaling (NMDS) plots.
RESULTS
In an effort to determine the proximal source of CWD prions in saliva, urine, and feces, and to add to our understanding of CWD pathogenesis, we blindly evaluated, in duplicate, a range of organs and tissues associated with saliva, urine, and fecal production and excretion for PrPCWD amplification using a standardized sPMCA assay. Tissues were collected at necropsy from 27 deer experimentally exposed to various sources of CWD by several different routes (Table 1). These tissues were then scored based on the number of rounds demonstrating amplification; individual tissue scores were then grouped according to the source of inoculum and correlated with intra-animal obex dilution scores.
Substantial CWD prion/PrPCWD-amplifying activity detected in excretory tissues by sPMCA.
Salivary glands, tongue, nasal mucosa, kidney, ureter, urinary bladder, and both proximal and distal intestinal tract were assayed for evidence of prion amplification activity by a standard three-round sPMCA protocol. Tissues were then assigned a score based on the total number of positive rounds in each of two replicates. In total, over 300 individual samples were evaluated, including 118 negative-control samples. Animals were initially grouped based on inoculum source (e.g., saliva, blood, urine, and feces); specific tissue scores for each of these groups were averaged to determine the mean tissue score for each group. In exposed animals, PrPCWD was amplified to various degrees in all tissues evaluated but was most predominant in salivary gland, urinary bladder, and distal intestinal tract (Table 2 and Fig. 1; see also Fig. 4). Amplification scores ranged from 1 to 6; animals inoculated i.v. or i.p. with whole blood appeared to exhibit the most widespread distribution of PrPCWD-amplifying activity, while those inoculated orally with either saliva or urine and feces demonstrated a more limited distribution. Interestingly, the amplification activity from some peripheral tissues, such as salivary gland and urinary bladder, rivaled that observed for the obex of the same animal (e.g., salivary gland and urinary bladder from deer 144 and urinary bladder from deer 4461, etc.) (Table 2). Among 118 negative-control samples, a single false-positive result was identified for a kidney section from deer 103. This particular sample was evaluated, in duplicate, in four separate experiments and received a total score of “1” in a single sPMCA experiment; no amplification was observed in the three remaining experiments.
Table 2.
Summary of sPMCA results for blood and tissues associated with production and excretion of saliva, urine, and feces in CWD-exposed deera
| Animal ID | Inoculum | Route of exposure | sPMCA score |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Obex | Salivary gland | Tongue | Kidney | Ureter | Urinary bladder | Proximal intestine | Distal intestine | Nasal epithelium | Blood | |||
| 144 | Saliva | p.o. | 6 | 6 | 2 | 0 | 2 | 6 | 2 | 5 | 3 | 0 |
| 113 | Saliva | p.o. | 6 | 6 | 0 | 0 | 0 | 5 | 2 | 3 | 0 | 0 |
| 132 | Saliva | p.o. | 6 | 0 | 0 | 0 | 0 | 3 | 2 | 3 | 1 | 0 |
| 122 | Saliva | p.o. | 6 | 0 | 0 | 5 | 0 | 1 | 1 | 3 | 0 | 0 |
| 147 | Saliva | p.o. | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 107 | Saliva | p.o. | 4 | 3 | 1 | 0 | 0 | 0 | 1 | 1 | 3 | 0 |
| 4119 | Blood | i.v. | 6 | 6 | 2 | 0 | 5 | 4 | 2 | 6 | NA | 0 |
| 347 | Blood | i.v. | 4 | 2 | 0 | 2 | 0 | 0 | 1 | 1 | NA | 0 |
| 114 | Blood | i.v. | 6 | 5 | 0 | 4 | 2 | 6 | 1 | 2 | 2 | 0 |
| 108 | Blood | i.p. | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 |
| 110 | Blood | i.v. | 6 | 6 | 3 | 5 | 3 | 6 | 1 | 2 | 3 | 0 |
| 133 | Blood | i.v. | 6 | 6 | 6 | 5 | 4 | 1 | 2 | 4 | 1 | 0 |
| 137 | Blood | i.v. | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 |
| 146 | Blood | i.v. | 6 | NA | 1 | 5 | 2 | 6 | 1 | 4 | 2 | 0 |
| 141 | Urine/feces | p.o. | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 150 | Urine/feces | p.o. | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 111 | Urine/feces | p.o. | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 |
| 134 | Urine/feces | p.o. | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 124 | Urine/feces | p.o. | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| 4461 | Environment | Unknown | 6 | 0 | 4 | 3 | 4 | 6 | 3 | 5 | NA | 0 |
| 4129 | Environment | Unknown | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | NA | 0 |
| 106 | CWD-positive brain | i.c. | 6 | 5 | 0 | 0 | 0 | 4 | 3 | 4 | 5 | 0 |
| 121 | CWD-positive brain | i.c. | 6 | 6 | 0 | 0 | 1 | 6 | 2 | 4 | NA | 0 |
| 123 | CWD-negative brain | i.c./p.o. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 103 | CWD-negative brain | i.c./p.o. | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4488 | CWD-negative brain | i.c./p.o. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4516 | CWD-negative brain | i.c./p.o. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Across individuals, all tissues were variously positive, with scores ranging from 0 to 6. Surprisingly, scores for peripheral tissues occasionally rivaled or exceeded those of the obex (i.e., salivary gland and urinary bladder of deer 144 and tongue and ureter of deer 4461). NA, sample not available.
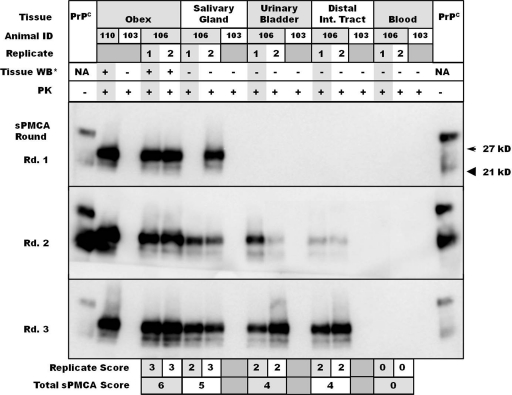
Fig. 1.
Representative Western blots of replicate salivary gland, urinary bladder, distal intestinal tract, and blood samples from study deer. Obex samples from deer 110 and 103 are used as positive and negative controls, respectively, while replicate tissues are those of deer 106. Tissue-matched negative controls from deer 103 were included. Tissue replicate scores appear below the Western blots, with cumulative scores summed to derive sPMCA scores (e.g., deer 106 obex, “6”; salivary gland, “6”). *Tissues were Western blotted prior to amplification to determine whether PrPCWD could be identified in unamplified homogenates. PK, proteinase K.
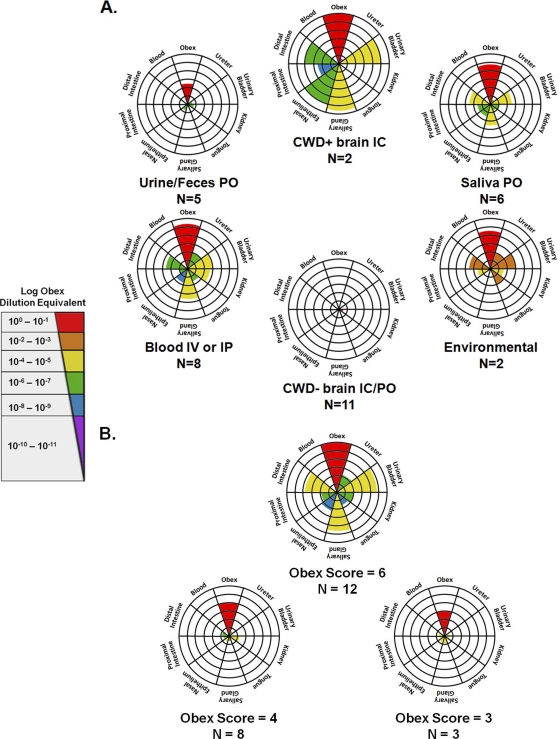
Fig. 4.
Radar plot summary of tissue sPMCA and LODE results. Animals were grouped according to the source of inoculum (blood and saliva, etc.) (A) and obex PMCA score (B). Central concentric rings represent a score of “1,” with scores progressing radially outward up to a score of “6.” For each radar plot, colored wedges indicate the mean tissue sPMCA score for that particular group, while wedge colors indicate the mean LODE score. Patterns of distribution varied with both the source of inoculum and obex burden, with the most widely distributed patterns identified for animals inoculated with blood either i.v. (IV) or i.p. (IP) and animals with an obex score of “6.” IC, intracranially; PO, orally.
Despite clear evidence that blood and its components harbor infectious CWD prions (30, 32), an effective sPMCA protocol has not yet been identified for amplifying PrPCWD from blood. sPMCA protocols have been described for the amplification of scrapie from the blood of hamsters and mice (e.g., see references 7a and 34a); however, for the present study we chose a protocol that was recently shown to be effective for sheep scrapie (51). In three rounds of sPMCA, all 27 whole-blood samples from both CWD-exposed and -naïve deer remained negative for PrPCWD amplification (Fig. 1 and Table 2) despite the amplification of concurrent CWD+ brain positive controls. Thus, we surmised that it was unlikely that blood contamination was responsible for the positive data obtained for the tissue homogenates described above. These findings do not, however, rule out the feasibility of sPMCA to detect PrPCWD in further rounds of sPMCA or using protocol modifications not employed in the present studies.
Individual intrarun sPMCA scores show a high level of agreement.
To determine the level of agreement between scores assigned for each of two sPMCA replicate experiments, we used a commercially available calculator to determine the categorical Cohen's kappa value. The sum of results available from blood and tissue sPMCA experiments was analyzed, and a summary of the categories and scores achieved in duplicate runs on sonicators “A” and “B” are presented in Table 3. Because the progression of scores for each experiment is a linear one (i.e., from 0 to 3), the kappa value was weighted linearly. The observed kappa value for the sum of sPMCA experiments was 0.80 (standard error [SE], 0.031; 95% confidence interval [CI], 0.74 to 0.87), which is considered substantial to near-perfect agreement according to guidelines proposed by Landis and Koch (27).
Table 3.
Summary of scores achieved in duplicate sPMCA experimentsa
| Sonicator A score | No. of samples with sonicator B score of: |
|||
|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |
| 0 | 220 | 13 | 0 | 1 |
| 1 | 9 | 9 | 3 | 1 |
| 2 | 3 | 3 | 9 | 3 |
| 3 | 0 | 0 | 5 | 24 |
Tissues were evaluated in duplicate on two different sonicators, sonicators “A” and “B,” receiving a score ranging from 0 to 3 in each replicate. Corresponding scores from duplicated experiments were tallied and evaluated categorically to derive Cohen's kappa value (0.80 ± 0.031).
Peripheral versus central nervous system (brain) prion amplification in individual animals.
Serial dilutions of 1% brain (obex) homogenates, ranging from 100 to 10−14 (i.e., final dilutions of 10−2 to 10−16), from each study deer were evaluated in the standardized three-round PMCA assay, with each dilution assigned a score as described above for tissues. Individual peripheral/excretory tissues were then correlated with the lowest obex dilution receiving that particular score and assigned a log obex dilution equivalent (LODE) score (Fig. 2 and Table 4). Tissues that failed to amplify PrPCWD in three rounds of sPMCA were assigned a LODE score less than or equal to the first obex dilution that failed to amplify CWD prions by sPMCA. The apparent peripheral level and distribution were dependent primarily on the central nervous system burden (Fig. 3a and c and 4), while the inoculum source and route of inoculation did not significantly correlate with the peripheral burden (Fig. 3b and d and 4).
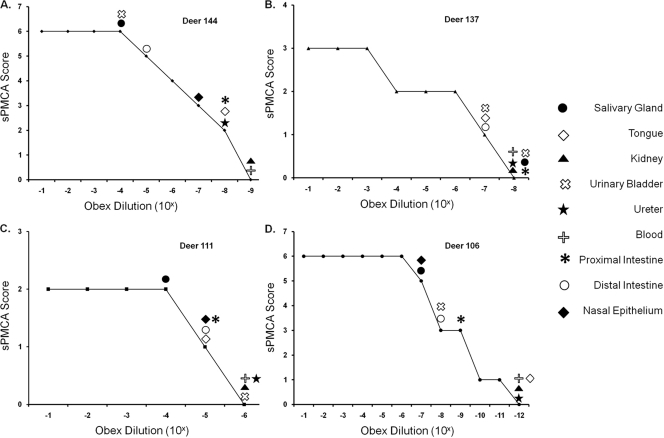
Fig. 2.
Representative LODE scores for deer inoculated with saliva orally (deer 144) (A), blood intravenously (deer 137) (B), urine and feces orally (deer 111) (C), and brain intracranially (deer 106) (D). Peripheral excretory tissue sPMCA scores of individual deer were conservatively translated into LODE scores based on the lowest equivalent obex dilution (from the source deer) yielding that particular sPMCA score.
Table 4.
Summary of individual tissue LODE scores for CWD-infected deera
| Animal ID | Inoculum | Route of exposure | LODE score |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Salivary gland | Tongue | Kidney | Ureter | Urinary bladder | Proximal intestinal tract | Distal intestinal tract | Nasal epithelium | |||
| 144 | Saliva | p.o. | 10−4 | 10−8 | ≤10−9 | 10−8 | 10−4 | 10−8 | 10−5 | 10−7 |
| 113 | Saliva | p.o. | 10−1 | ≤10−9 | ≤10−9 | ≤10−9 | 10−3 | 10−8 | 10−6 | ≤10−9 |
| 122 | Saliva | p.o. | ≤10−10 | ≤10−10 | 10−1 | ≤10−10 | 10−9 | 10−9 | 10−7 | ≤10−10 |
| 107 | Saliva | p.o. | 10−2 | 10−3 | ≤10−4 | ≤10−4 | ≤10−4 | 10−3 | 10−3 | 10−2 |
| 132 | Saliva | p.o. | ≤10−11 | ≤10−11 | ≤10−11 | ≤10−11 | 10−7 | 10−10 | 10−7 | 10−11 |
| 147 | Saliva | p.o. | ≤10−7 | ≤10−7 | 10−6 | ≤10−7 | ≤10−7 | ≤10−7 | ≤10−7 | ≤10−7 |
| 4119 | Blood | i.v. | 10−1 | 10−6 | ≤10−9 | 10−2 | 10−4 | 10−6 | 10−1 | NA |
| 110 | Blood | i.v. | 10−3 | 10−9 | 10−4 | 10−9 | 10−3 | 10−9 | ≤10−10 | 10−9 |
| 347 | Blood | i.v. | 10−3 | ≤10−7 | 10−3 | ≤10−7 | ≤10−7 | 10−6 | 10−6 | NA |
| 146 | Blood | i.v. | NA | 10−9 | 10−4 | 10−8 | 10−2 | 10−9 | 10−5 | 10−8 |
| 133 | Blood | i.v. | 10−5 | 10−5 | 10−6 | 10−6 | 10−11 | 10−8 | 10−6 | 10−11 |
| 114 | Blood | i.v. | 10−5 | ≤10−10 | 10−8 | 10−9 | 10−3 | 10−10 | 10−9 | 10−9 |
| 137 | Blood | i.v. | ≤10−8 | ≤10−8 | 10−7 | ≤10−8 | 10−7 | ≤10−8 | 10−7 | 10−7 |
| 108 | Blood | i.p. | ≤10−5 | ≤10−5 | ≤10−5 | ≤10−5 | ≤10−5 | ≤10−5 | ≤10−5 | 10−5 |
| 111 | Urine/feces | p.o. | ≤10−6 | ≤10−6 | ≤10−6 | ≤10−6 | ≤10−5 | ≤10−6 | ≤10−6 | 10−10 |
| 134 | Urine/feces | p.o. | ≤10−7 | ≤10−7 | 10−6 | ≤10−7 | ≤10−7 | ≤10−7 | ≤10−7 | ≤10−7 |
| 124 | Urine/feces | p.o. | ≤10−8 | ≤10−8 | 10−7 | ≤10−8 | ≤10−8 | ≤10−8 | ≤10−8 | 10−7 |
| 141 | Urine/feces | p.o. | ≤10−6 | ≤10−6 | ≤10−6 | ≤10−6 | ≤10−6 | ≤10−6 | ≤10−6 | 10−6 |
| 150 | Urine/feces | p.o. | ≤10−5 | ≤10−5 | ≤10−5 | ≤10−5 | ≤10−5 | ≤10−5 | ≤10−5 | ≤10−5 |
| 4461 | Environment | Unknown | ≤10−7 | 10−2 | 10−3 | 10−2 | 100 | 10−3 | 10−1 | NA |
| 4129 | Environment | Unknown | ≤10−5 | ≤10−5 | ≤10−5 | ≤10−5 | ≤10−5 | ≤10−5 | 10−4 | NA |
| 121 | CWD-positive brain | i.c. | 10−4 | ≤10−10 | ≤10−10 | 10−9 | 10−4 | 10−9 | 10−6 | NA |
| 106 | CWD-positive brain | i.c. | 10−6 | ≤10−11 | ≤10−11 | ≤10−11 | 10−7 | 10−8 | 10−7 | 10−6 |
Serial PMCA scores were initially determined for obex dilutions from individual animals. Tissues from the respective animals were then conservatively assigned a score based on the equivalent log obex dilution equivalent (LODE). Tissues that failed to amplify PrPCWD in three rounds received a LODE score equal to or less than the first obex dilution of the source animal scoring a “0.” These findings suggest that peripheral levels vary widely compared to PrPCWD levels in the central nervous system but on occasion may approach levels seen for the obex of infected animals.
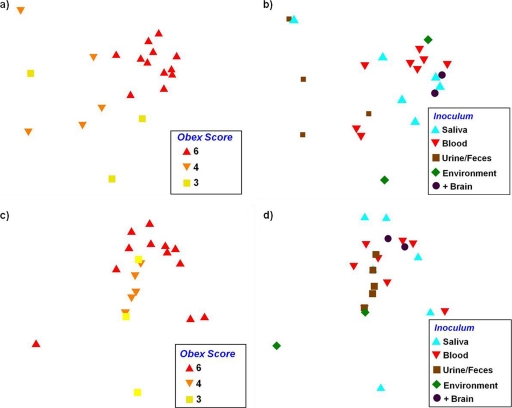
Fig. 3.
NMDS ordination plots showing the relationship between peripheral PrPCWD distribution and intensity based on the raw sPMCA score (a and b) and LODE score/relative PrPCWD burden (c and d). The obex score (a and c) significantly correlated with peripheral accumulation (P = 0.0001 and 0.042), demonstrating that as central nervous system levels increased, peripheral PrPCWD showed greater distribution and intensity and less variability. While the correlation between the inoculum source and peripheral accumulation did not correlate significantly (b and d), a loose association was observed (P = 0.072 and 0.185).
Peripheral PrPCWD intensity and distribution correlate with central nervous system burden.
The distribution and intensity of PrPCWD were significantly related to the obex score for both presentations of our data (F2,19 = 6.2 and P = 0.0001 for the raw sPMCA score; F2,19 = 2.3 and P = 0.042 for the relative PrPCWD burden [determined by PERMANOVA]) (Fig. 3a and c). Individuals with an obex score of 6 differed from individuals with scores of 3 or 4 (P value of <0.001 in both cases for the scoring measure; P = 0.05 and 0.08, respectively, for the relative PrPCWD burden [determined by a post hoc test]), but the distribution and intensity of PrPCWD among the latter two groups did not differ from one another (P = 0.44 for raw sPMCA score; P = 0.17 for relative PrPCWD burden [determined by a post hoc test]). What is also apparent in these analysis is that as the obex score increased, there was generally less individual variation in peripheral PrPCWD accumulation (as represented by a more tight clustering of individuals with an obex score of 6 than of individuals with an obex score of 3 or 4) (Fig. 3a and c). The peripheral distribution and intensity of PrPCWD, conversely, were not significantly related to the source of the inoculum (F4,17 = 1.67 and P = 0.072 for the raw sPMCA score; F4,17 = 1.42 and P = 0.185 for the relative PrPCWD burden [determined by PERMANOVA]) (Fig. 3b and d).
DISCUSSION
Among the transmissible spongiform encephalopathies (TSEs), chronic wasting disease is unique in its high level of transmissibility and, thus, prevalence in free-ranging populations. Here we report the presence of substantial prion-converting activity in excretory tissues proximate to the saliva, urine, and feces shed by CWD-infected deer. While these results do not speak to infectivity, our previous research has shown that conventional test-negative, sPMCA-positive tissues and bodily fluids may indeed harbor infectivity (17, 18). The use of sPMCA as a surrogate for a bioassay to detect PrPCWD in excretory tissues may help to explain the efficient horizontal transmission of CWD but also raises questions regarding the source and mechanism of prion shedding from glandular and mucosal tissues.
Historical cross-sectional studies of cervid herds where CWD is endemic have largely failed to demonstrate, by conventional immunohistochemistry or Western blotting, protease-resistant prions in organs involved in the production and excretion of saliva, urine, or feces (2, 14, 49). These assays rely on proteolytic, heat, and/or formic acid treatments, practices which may preclude the identification of recently described protease-sensitive forms of infectious prion proteins, denoted sPrPRES (9, 22, 39, 40, 52). Anecdotally, however, protease-resistant PrP has been identified in ectopic lymphoid aggregates in the kidneys of CWD-exposed deer by IHC, although no conclusions could be drawn regarding any relationship of this phenomenon with prionuria (19). Protease-resistant PrP has also been demonstrated in the lingual epithelium of hamsters inoculated with the hyper (HY) strain of transmissible mink encephalopathy (10) and in the salivary glands of sheep naturally exposed to scrapie (55). Alternatively, bioassays have demonstrated infectious prions in the salivary glands of scrapie-infected goats (16), the tongue and nasal epithelium of bovine spongiform encephalopathy (BSE)-infected cattle (3), and the urine of animals with concurrent nephritis (18, 42). As sPMCA was also shown to be capable of amplifying both protease-resistant and -sensitive forms of PrPSc (36), the absence of protease-resistant forms in these tissues would not preclude positive amplification, since other evidence exists for protease-sensitive infectious prions (9, 22, 36). Taken together, the above-described observations raise the following interesting questions regarding the mechanisms involved in prionsialia and prionuria. (i) From where do the infectious prions in bodily fluids arise? (ii) Are the infectious prions in excreta present as traditional, protease-resistant forms at very low levels or as a more elusive protease-sensitive species? (iii) Are infectious prions transmitted in cell-free or cell-associated forms?
Excreta—urine, saliva, and feces—are made up of components from the organs and tissues responsible for their production, including aqueous, cellular, and proteinaceous constituents. We hypothesized that these organs may be involved in prion production and/or excretion, and we found a range of CWD prion amplifications from such tissues. Variation in CWD prion-amplifying activities in tissues significantly correlated with the apparent PrPCWD burden within the obex of the individual animal, while a trending relationship was observed between peripheral distribution and intensity and the source of inoculum and route of exposure (e.g., whole blood i.v. and urine and feces p.o., etc.). Perhaps with more animals per group or longer incubation times, this overall trend between inoculum and peripheral distribution may have become statistically significant.
These two findings point to an increased risk of bodily fluid infectivity with disease progression and, potentially, source of exposure. The tissue distribution variation with the route of inoculation has been described previously for both viral and bacterial infections (6, 29, 37, 45), so this finding is perhaps not surprising. Likewise, in the case of neurotropic viruses, virus levels in peripheral tissue often positively correlate with the central nervous system burden (8). Evaluation of the prion distribution in tissues of naturally infected cervids may reveal patterns corresponding with those described in this study, i.e., widespread peripheral accumulation of prions that vary according to the central nervous system burden and, potentially, the exposure source.
The detection of amplifiable prions in peripheral excretory tissues call into question (i) whether peripheral prion amplification occurs in situ or, alternatively, whether excretory tissues merely serve a more passive role in prion excretion and (ii) the kinetics of prion excretion, i.e., how the levels of prion excretion and infectivity in bodily fluids change over the course of the disease. It is generally accepted that TSE pathogenesis follows a pattern of centripetal spread from the periphery to the central nervous system (either solely via the peripheral nervous system or after amplification in the lymphoreticular system), followed by central amplification and centrifugal spread back to the periphery (4, 5, 11, 15, 53, 54). We found that some peripheral tissue levels rivaled those in the obex of the same animal, perhaps suggesting concurrent early dissemination to the central nervous system and peripheral organs. A further understanding of the kinetics of centripetal and centrifugal prion dissemination could emanate from a sequential sPMCA evaluation of the peripheral prion distribution in exposed animals.
In summary, the present study demonstrates for the first time prion-amplifying activity in organs and tissues associated with prion shedding. The ultimate source and mechanism of release into bodily fluids remain unknown. The elevated and consistent activity found in salivary gland and urinary bladder may suggest an active role in prion excretion. These findings minimally warrant additional, more detailed, and longitudinal studies of the nature and kinetics of excreted prions.
ACKNOWLEDGMENTS
We sincerely thank all of those who have made important contributions to the manuscript, including David Osborne and Sally Dahmes, without whom the primary experiments with cervids could not have been performed, and Michael Miller, who provided initial samples of CWD+ brain and bodily fluids. Candace Mathiason was integral in assisting with the design and implementation of blinded sPMCA of tissues. Other notable contributors who provided assistance with assay development and interpretation include Timothy Kurt and Amy Nalls. For long-term care and sample collection from source deer and transgenic mice, we sincerely thank Sheila Hays, Kelly Anderson, and Jeanette Hayes-Klug. Without each of their contributions and unique skills, this work could not have been completed.
Footnotes
Published ahead of print on 27 April 2011.
REFERENCES
- 1. Angers R. C., et al. 2009. Chronic wasting disease prions in elk antler velvet. Emerg. Infect. Dis. 15:696–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balachandran A., et al. 2010. Experimental oral transmission of chronic wasting disease to red deer (Cervus elaphus elaphus): early detection and late stage distribution of protease-resistant prion protein. Can. Vet. J. 51:169–178 [PMC free article] [PubMed] [Google Scholar]
- 3. Balkema-Buschmann A., et al. 2011. BSE infectivity in the absence of detectable PrP(Sc) accumulation in the tongue and nasal mucosa of terminally diseased cattle. J. Gen. Virol. 92:467–476 [DOI] [PubMed] [Google Scholar]
- 4. Bartz J. C., Dejoia C., Tucker T., Kincaid A. E., Bessen R. A. 2005. Extraneural prion neuroinvasion without lymphoreticular system infection. J. Virol. 79:11858–11863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beekes M., McBride P. A. 2007. The spread of prions through the body in naturally acquired transmissible spongiform encephalopathies. FEBS J. 274:588–605 [DOI] [PubMed] [Google Scholar]
- 6. Bravata D. M., et al. 2007. Inhalational, gastrointestinal, and cutaneous anthrax in children. A systematic review of cases: 1900 to 2005. Arch. Pediatr. Adolesc. Med. 161:896–905 [DOI] [PubMed] [Google Scholar]
- 7. Browning S. R., et al. 2004. Transmission of prions from mule deer and elk with chronic wasting disease to transgenic mice expressing cervid PrP. J. Virol. 78:13345–13350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a. Castilla J., et al. 2005. Detection of prions in blood. Nat. Med. 11:982–985 [DOI] [PubMed] [Google Scholar]
- 8. Charlton K. M., Casey G. A., Campbell J. B. 1983. Experimental rabies in skunks: mechanisms of infection of the salivary glands. Can. J. Comp. Med. 47:363–369 [PMC free article] [PubMed] [Google Scholar]
- 9. Colby D. W., et al. 2010. Protease-sensitive synthetic prions. PLoS Pathog. 6:e1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeJoia C., Moreaux B., O'Connell K., Bessen R. A. 2006. Prion infection of oral and nasal mucosa. J. Virol. 80:4546–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dormont D. 2002. Prion diseases: pathogenesis and public health concerns. FEBS Lett. 529:17–21 [DOI] [PubMed] [Google Scholar]
- 12. Eklund C. M., Kennedy R. C., Hadlow W. J. 1967. Pathogenesis of scrapie virus infection in the mouse. J. Infect. Dis. 117:15–22 [DOI] [PubMed] [Google Scholar]
- 13. Foster J. D., Parnham D. W., Hunter N., Bruce M. 2001. Distribution of the prion protein in sheep terminally affected with BSE following experimental oral transmission. J. Gen. Virol. 82:2319–2326 [DOI] [PubMed] [Google Scholar]
- 14. Fox K. A., Jewell J. E., Williams E. S., Miller M. W. 2006. Patterns of PrPCWD accumulation during the course of chronic wasting disease infection in orally inoculated mule deer (Odocoileus hemionus). J. Gen. Virol. 87:3451–3461 [DOI] [PubMed] [Google Scholar]
- 15. Glatzel M., Aguzzi A. 2000. Peripheral pathogenesis of prion diseases. Microbes Infect. 2:613–619 [DOI] [PubMed] [Google Scholar]
- 16. Hadlow W. J., et al. 1974. Course of experimental scrapie virus infection in the goat. J. Infect. Dis. 129:559–567 [DOI] [PubMed] [Google Scholar]
- 17. Haley N., Mathiason C., Zabel M. D., Telling G. C., Hoover E. 2009. Detection of sub-clinical CWD infection in conventional test-negative deer long after oral exposure to urine and feces from CWD+ deer. PLoS One 4:e7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haley N. J., Seelig D. M., Zabel M. D., Telling G. C., Hoover E. A. 2009. Detection of CWD prions in urine and saliva of deer by transgenic mouse bioassay. PLoS One 4:e4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hamir A. N., Kunkle R. A., Miller J. M., Hall S. M. 2006. Abnormal prion protein in ectopic lymphoid tissue in a kidney of an asymptomatic white-tailed deer experimentally inoculated with the agent of chronic wasting disease. Vet. Pathol. 43:367–369 [DOI] [PubMed] [Google Scholar]
- 20. International Society for Infectious Diseases 24 January 2010, posting date Chronic wasting disease, cervid—USA (02) (Virginia). Archive number 20100124.0261. International Society for Infectious Diseases, Brookline, MA: http://www.promedmail.org [Google Scholar]
- 21. International Society for Infectious Diseases 3 March 2010, posting date Chronic wasting disease, cervid—USA (03) (Missouri). First report. Archive number 20100303.0697. International Society for Infectious Diseases, Brookline, MA: www.promedmail.org [Google Scholar]
- 22. Jansen C., et al. 2010. The first case of protease-sensitive prionopathy (PSPr) in the Netherlands: a patient with an unusual GSS-like clinical phenotype. J. Neurol. Neurosurg. Psychiatry 81:1052–1055 [DOI] [PubMed] [Google Scholar]
- 23. Keane D. P., et al. 2008. Chronic wasting disease in a Wisconsin white-tailed deer farm. J. Vet. Diagn. Invest. 20:698–703 [DOI] [PubMed] [Google Scholar]
- 24. Kitamoto T., Mohri S., Tateishi J. 1989. Organ distribution of proteinase-resistant prion protein in humans and mice with Creutzfeldt-Jakob disease. J. Gen. Virol. 70(Pt. 12):3371–3379 [DOI] [PubMed] [Google Scholar]
- 25. Kurt T. D., et al. 2007. Efficient in vitro amplification of chronic wasting disease PrPRES. J. Virol. 81:9605–9608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kurt T. D., Telling G. C., Zabel M. D., Hoover E. A. 2009. Trans-species amplification of PrP(CWD) and correlation with rigid loop 170N. Virology 387:235–243 [DOI] [PubMed] [Google Scholar]
- 27. Landis J. R., Koch G. G. 1977. The measurement of observer agreement for categorical data. Biometrics 33:159–174 [PubMed] [Google Scholar]
- 28. Ligios C., et al. 2007. Intraepithelial and interstitial deposition of pathological prion protein in kidneys of scrapie-affected sheep. PLoS One 2:e859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a. Lowry R. 2011. VassarStats: website for statistical computation. http://faculty.vassar.edu/lowry/VassarStats.html
- 29. Maisner A., Neufeld J., Weingartl H. 2009. Organ- and endotheliotropism of Nipah virus infections in vivo and in vitro. Thromb. Haemost. 102:1014–1023 [DOI] [PubMed] [Google Scholar]
- 30. Mathiason C. K., et al. 2010. B cells and platelets harbor prion infectivity in the blood of deer infected with chronic wasting disease. J. Virol. 84:5097–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mathiason C. K., et al. 2009. Infectious prions in pre-clinical deer and transmission of chronic wasting disease solely by environmental exposure. PLoS One 4:e5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mathiason C. K., et al. 2006. Infectious prions in the saliva and blood of deer with chronic wasting disease. Science 314:133–136 [DOI] [PubMed] [Google Scholar]
- 33. Miller M. W., Williams E. S., Hobbs N. T., Wolfe L. L. 2004. Environmental sources of prion transmission in mule deer. Emerg. Infect. Dis. 10:1003–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mulcahy E. R., Bartz J. C., Kincaid A. E., Bessen R. A. 2004. Prion infection of skeletal muscle cells and papillae in the tongue. J. Virol. 78:6792–6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a. Murayama Y., et al. 2007. Urinary excretion and blood level of prions in scrapie-infected hamsters. J. Gen. Virol. 88:2890–2898 [DOI] [PubMed] [Google Scholar]
- 35. Notari S., et al. 2010. Multiorgan detection and characterization of protease-resistant prion protein in a case of variant CJD examined in the United States. PLoS One 5:e8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pastrana M. A., et al. 2006. Isolation and characterization of a proteinase K-sensitive PrPSc fraction. Biochemistry 45:15710–15717 [DOI] [PubMed] [Google Scholar]
- 37. Prentice M. B., Rahalison L. 2007. Plague. Lancet 369:1196–1207 [DOI] [PubMed] [Google Scholar]
- 38. Saborio G. P., Permanne B., Soto C. 2001. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411:810–813 [DOI] [PubMed] [Google Scholar]
- 39. Safar J., et al. 1998. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 4:1157–1165 [DOI] [PubMed] [Google Scholar]
- 40. Safar J. G., et al. 2005. Diagnosis of human prion disease. Proc. Natl. Acad. Sci. U. S. A. 102:3501–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sakaguchi S., et al. 1993. Kinetics of infectivity are dissociated from PrP accumulation in salivary glands of Creutzfeldt-Jakob disease agent-inoculated mice. J. Gen. Virol. 74(Pt. 10):2117–2123 [DOI] [PubMed] [Google Scholar]
- 42. Seeger H., et al. 2005. Coincident scrapie infection and nephritis lead to urinary prion excretion. Science 310:324–326 [DOI] [PubMed] [Google Scholar]
- 43. Seelig D. M., Mason G. L., Telling G. C., Hoover E. A. 2010. Pathogenesis of chronic wasting disease in cervidized transgenic mice. Am. J. Pathol. 176:2785–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sigurdson C. J. 2008. A prion disease of cervids: chronic wasting disease. Vet. Res. 39:41. [DOI] [PubMed] [Google Scholar]
- 45. Singh G. K., Singh N. P., Garg S. K. 1987. Studies on pathogenesis of fowl pox: virological study. Acta Virol. 31:417–423 [PubMed] [Google Scholar]
- 46. Siso S., et al. 2006. Prion protein in kidneys of scrapie-infected sheep. Vet. Rec. 159:327–328 [DOI] [PubMed] [Google Scholar]
- 47. Siso S., et al. 2008. Occurrence and cellular localization of PrPd in kidneys of scrapie-affected sheep in the absence of inflammation. J. Pathol. 215:126–134 [DOI] [PubMed] [Google Scholar]
- 48. Soto C., Saborio G. P., Anderes L. 2002. Cyclic amplification of protein misfolding: application to prion-related disorders and beyond. Trends Neurosci. 25:390–394 [DOI] [PubMed] [Google Scholar]
- 49. Spraker T. R., et al. 1997. Spongiform encephalopathy in free-ranging mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus) and Rocky Mountain elk (Cervus elaphus nelsoni) in northcentral Colorado. J. Wildl. Dis. 33:1–6 [DOI] [PubMed] [Google Scholar]
- 50. Tamguney G., et al. 2009. Asymptomatic deer excrete infectious prions in faeces. Nature 461:529–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tattum M. H., Jones S., Pal S., Collinge J., Jackson G. S. 2010. Discrimination between prion-infected and normal blood samples by protein misfolding cyclic amplification. Transfusion 50:996–1002 [DOI] [PubMed] [Google Scholar]
- 52. Thackray A. M., Hopkins L., Bujdoso R. 2007. Proteinase K-sensitive disease-associated ovine prion protein revealed by conformation-dependent immunoassay. Biochem. J. 401:475–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Unterberger U., Voigtlander T., Budka H. 2005. Pathogenesis of prion diseases. Acta Neuropathol. 109:32–48 [DOI] [PubMed] [Google Scholar]
- 54. van Keulen L. J., Bossers A., van Zijderveld F. 2008. TSE pathogenesis in cattle and sheep. Vet. Res. 39:24. [DOI] [PubMed] [Google Scholar]
- 55. Vascellari M., et al. 2007. PrPSc in salivary glands of scrapie-affected sheep. J. Virol. 81:4872–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Williams E. S., Young S. 1980. Chronic wasting disease of captive mule deer: a spongiform encephalopathy. J. Wildl. Dis. 16:89–98 [DOI] [PubMed] [Google Scholar]
- 57. Williams E. S., Young S. 1982. Spongiform encephalopathy of Rocky Mountain elk. J. Wildl. Dis. 18:465–471 [DOI] [PubMed] [Google Scholar]