Abstract
Some of the earliest studies of retroviral integration targeting reported that sites of gammaretroviral DNA integration were positively correlated with DNase I-hypersensitive sites in chromatin. This led to the suggestion that open chromatin was favorable for integration. More recent deep sequencing experiments confirmed that gammaretroviral integration sites and DNase I cleavage sites are associated in genome-wide surveys. Paradoxically, in vitro studies of integration show that nucleosomal DNA is actually favored over naked DNA, raising the question of whether integration target DNA in chromosomes is wrapped in nucleosomes or nucleosome free. In this study we examined gammaretroviral integration by infecting primary human CD4+ T lymphocytes with a murine leukemia virus (MLV)-based retroviral vector or xenotropic murine leukemia virus-related virus (XMRV), and isolated 32,585 unique integration sites using ligation-mediated PCR and 454 pyrosequencing. CD4+ T lymphocytes were chosen for study because of the particularly dense genome-wide mapping of chromatin features available for comparison. Analysis relative to predicted nucleosome positions showed that gammaretroviruses direct integration into outward-facing major grooves on nucleosome-wrapped DNA, similar to the integration pattern of HIV. Also, a suite of histone modifications correlated with gene activity are positively associated with integration by both MLV and XMRV. Thus, we conclude that favored integration near DNase I-hypersensitive sites does not imply that integration takes place exclusively in nucleosome-free regions.
INTRODUCTION
Integration of a DNA copy of the retroviral RNA genome is an essential step in the viral replication cycle. Integration targeting in cellular chromosomes is not random–early studies reported that gammaretrovirus integration occurred preferentially near DNase I-hypersensitive sites, suggesting integration near promoters in actively transcribed chromatin domains (39, 47, 55). Chromatin structure in such regions has been suggested to be open in some incompletely defined sense, potentially correlating with reduced nucleosome occupancy (for recent work, see references 23, 38, 50, and 52). This led to the proposal that steric hindrance by nucleosome occupancy might prevent retroviral integration.
With the development of methods for studying integration in vitro it became possible to compare integration into nucleosomal and naked DNA templates. Surprisingly, these studies showed that purified integrase preferentially utilizes histone-associated DNA over naked DNA, and the association of DNA with histones increased integration at particular sites which are not favored in naked DNA (41, 42, 45). Integration into nucleosomal DNA in vitro showed a 10-base periodicity, consistent with integration in outward facing DNA major grooves (34, 44, 45). The most kinked regions of nucleosomal DNA were shown to be particularly favored (41, 42). Integration of gammaretroviral DNA into simian virus 40 (SV40) minichromosomes in vivo, in contrast, did not show a periodic pattern (43). This reopened the question of the relationship of gammaretroviral integration and nucleosomal DNA, particularly given the early observations of association with DNase I-hypersensitive sites. In addition, for the early in vitro integration reactions, in many cases only a single viral DNA end was joined to target DNA, leaving open the question of the influence of nucleosome binding on more authentic reactions in which correctly spaced pairs of viral DNA ends become coordinately integrated.
Recent studies using high-throughput sequencing have determined that integration of the prototype gammaretrovirus murine leukemia virus (MLV) preferentially takes place near transcription start sites (8, 9, 14, 17, 33, 56, 61) and regions less than 1 kb from DNase I-hypersensitive sites (5, 28). MLV integration is also favored in areas of high CpG island density, GC content, and several histone methylation marks, all of which are normally associated with the promoters and transcription start sites of active genes. Similar to MLV, integration of XMRV, another gammaretrovirus, favors integration near the same set of features (26, 27). XMRV has been proposed to be a human pathogen (30, 49), but recent data suggest that it may instead be a laboratory contaminant (19, 22, 35, 46). In the studies described below, XMRV serves as a second model gammaretrovirus.
Gammaretroviral integration site selection is distinct from that of HIV. Genome-wide sequencing studies identified active transcription units as the preferred target for HIV integration (33, 51), with the entire length of transcription units favored, not just the transcription start site as with gammaretroviruses. The relationship of HIV integration to predicted nucleosome positions was investigated in a large study of HIV integration (40,000 unique sites), revealing a 10-bp periodicity in sites of high integration frequency that corresponded to outward-facing major grooves on DNA wrapped around a histone octamer (57). A second study characterizing HIV integration in primary human T cells yielded a similar periodic pattern (58). These data establish that the step at which HIV preintegration complexes capture target DNA and commit to integration often takes place on nucleosome-wrapped DNA in chromosomes in vivo. DNA binding proteins in addition to integrase also can bind to histone-associated DNA—for example, binding of NF-κB is known to be almost unaffected by the presence or absence of histone octamers (1).
In this study, we sought to investigate the relationship between nucleosome structure in cellular chromosomes and gammaretrovirus integration in vivo. We studied integration by XMRV or an MLV-based vector in CD4+ T cells. T cells were chosen for study because of the particularly deep genome-wide mapping of epigenetic modification and chromosome-bound proteins available (3, 50, 60). We isolated 32,585 unique integration sites, thereby producing the largest data sets for XMRV and for MLV in CD4+ T cells to date. We analyzed the sequence surrounding each integration site with nucleosome position prediction software (24, 25, 52) in order to identify sites favored for integration on nucleosome-wrapped DNA and further characterized integration frequency near additional forms of genome-wide annotation.
MATERIALS AND METHODS
Preparation of XMRV and MLV-based vector.
To produce XMRV, LNCaP (ATCC CRL-1740) cells maintained in RPMI were seeded (1.6 × 106 cells) in a T25 flask and transfected with pXRMVJ7 (49) and Lipofectamine LTX according to the manufacturers' recommendations. Two days after transfection, medium containing XMRV was collected and diluted 1:1 with RPMI medium. This was used to infect a separate flask of LNCaP cells seeded 1.6 × 106 cells in a T25 flask. The newly infected cells were maintained in culture for 1 month, establishing chronic infection.
In order to produce the MLV-based vector, 293T cells (maintained in Dulbecco modified Eagle medium [DMEM]) were transfected with the MLV vector plasmid pMLV LTR-GFP, the packaging construct pCGP, and pMD.G, which produces the envelope protein VSV-G. Vector containing supernatant was collected after 48 h, filtered, concentrated, and treated with DNase I. PT67 cells (Clontech 631510) were transduced twice with the 293T cell-produced vector to produce a 10A1-pseudotyped, MLV-based vector-producing cell line.
Transduction of primary human CD4+ T cells.
Primary human CD4+T cells were purified by the University of Pennsylvania Immunology Core from mononuclear leukapheresis product using negative selection by adding antibodies specific for HLA-DR, CD21, CD16, CD11b, CD14, and CD8 to peripheral blood lymphocytes (PBLs) and separating the cells with Dynal GAM-coated beads. Purified CD4+ T cells were activated with 5 μg/ml of PHA-L and maintained in RPMI supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml interleukin 2 (IL-2), and 1% penicillin-streptomycin.
CD4+ T cells were infected with XMRV-using spinoculation. Briefly, 5 days postplating, the XMRV-containing medium from chronically infected LNCaP cells was collected and spun to remove cell debris. PHA-L-activated CD4+ T cells were resuspended at 0.5 × 106 cells/ml in XMRV-containing supernatant. The cell suspension was then transferred to a plate and spun at 1,200 × g for 3.5 h at 32°C. Following spinoculation, the cells were spun down and the virus-containing medium was removed and replaced with RPMI containing 100 U/ml IL-2. The cells were maintained for 5 days prior to harvesting of genomic DNA with the DNeasy blood and tissue kit (Qiagen).
CD4+ T cells were transduced with the MLV-based vector using the RetroNectin-bound virus infection method, as described by the manufacturer (Takara). Briefly, vector-containing medium was collected from freshly passaged 10A1 pseudotyped MLV-based vector-producing cells (described earlier) after 48 h of incubation at 37°C. The vector-containing medium was centrifuged to remove cell debris and filtered. The vector was then bound to RetroNectin-coated plates (catalog no. T110A; Takara) by centrifugation at 1,200 × g for 2 h at 32°C. Activated CD4+ T cells at 0.5 × 106 cells/ml were then added to the prepared plates and incubated at 32°C. After 18 h, the transduction procedure was repeated. After both transductions, the cells were maintained for 5 days before harvesting of genomic DNA with the DNeasy blood and tissue kit (Qiagen).
Recovery of integration sites and analysis of integration site distributions.
Recovery of integration sites was performed as previously described (13, 57). Briefly, two recovery methods were used. In one, linkers were ligated to restriction enzyme-digested genomic DNA from infected cells and nested PCR was used to amplify virus-host DNA junctions. In the second, phage Mu transposase was used to install linkers, as described in reference 7. Different batches of samples were separately bar coded with the second pair of PCR primers. PCR-amplified products were purified by binding to beads and sequenced using 454/Roche pyrosequencing (titanium technology). Reads were quality filtered by requiring perfect matches to the long terminal repeat (LTR) linker, bar code, and flanking LTR and mapped to the human genome. All sites were required to align to the human genome within 3 bp of the LTR edge, with the great majority showing no gap. Association to genomic features and histone modifications were performed as described previously (4, 6, 32). Nucleosome prediction was carried out using software available at http://genie.weizmann.ac.il/software/nucleo_prediction.html using 5 kb of human sequence surrounding each integration site. Fourier transform analysis was performed with Statistica (Statsoft).
RESULTS
Isolation and sequencing of integration sites.
To isolate gammaretrovirus integration sites, primary human CD4+ T cells were infected with XMRV or a stock of an MLV-based vector encoding green fluorescent protein (GFP). XMRV infection rates were typically 15% as determined by quantitative PCR (qPCR) (49). MLV-based vector transduction typically yielded 30% GFP-positive cells. Cells were harvested 5 days after infection, and the genomic DNA was purified. Gammaretrovirus integration sites were isolated using ligation-mediated PCR after cleaving genomic DNA with BstYI, NlaIII, MseI, and Tsp509I, followed by linker ligation, or by using Mu transposase in vitro to install linkers in genomic DNA (7). A total of 122,900 sequence reads were obtained, which, after quality filtering and dereplication, yielded 32,585 unique integration sites (Table 1).
Table 1.
Integration site data sets used in this study
| Study (reference) | Integration site set | Recovery methoda | Viral vector | Cell type | Total no. of integration sequence reads | No. of unique integration sites | Comments |
|---|---|---|---|---|---|---|---|
| This study | Roth-MLV-CD4T-BstYI/MselI/NlaIII/Tsp509I | Rest. Enz. | MLV | CD4T | 44,609 | 25,753 | Activated human T cells treated with MLV-based retroviral vector |
| This study | Roth-MLV-CD4T-Mu | Mu | MLV | CD4T | 10,119 | 957 | Activated human T cells treated with MLV-based retroviral vector |
| This study | Roth-XMRV-CD4T-BstYI/MseI/NlaIII/Tsp509I | Rest. Enz. | XMRV | CD4T | 61,233 | 5,736 | Activated human T cells treated with XMRV |
| This study | Roth-XMRV-CD4T-Mu | Mu | XMRV | CD4T | 6,939 | 139 | Activated human T cells treated with XMRV |
| Wang et al. (58) | Wang-VA2-June-201-ExVivo-ApoI | Rest. Enz. | HIV | CD4T | 3,544 | 1,163 | Activated human T cells treated with a lentiviral vector ex vivo |
| Wang et al. (58) | Wang-VA2-June-201-ExVivo-Avr | Rest. Enz. | HIV | CD4T | 1,540 | 393 | Activated human T cells treated with a lentiviral vector ex vivo |
| Wang et al. (58) | Wang-VA2-June-202-ExVivo-ApoI | Rest. Enz. | HIV | CD4T | 3,605 | 1,786 | Activated human T cells treated with a lentiviral vector ex vivo |
| Wang et al. (58) | Wang-VA2-June-202-ExVivo-Avr | Rest. Enz. | HIV | CD4T | 1,394 | 627 | Activated human T cells treated with a lentiviral vector ex vivo |
| Wang et al. (58) | Wang-VA2-June-203-ExVivo-ApoI | Rest. Enz. | HIV | CD4T | 4,430 | 2,380 | Activated human T cells treated with a lentiviral vector ex vivo |
| Wang et al. (58) | Wang-VA2-June-203-ExVivo-Avr | Rest. Enz. | HIV | CD4T | 2,700 | 1,277 | Activated human T cells treated with a lentiviral vector ex vivo |
| Wang et al. (57) | Wang-VSVGgfp-Jurkat-454-hetero-Avr | Rest. Enz. | HIV | Jurkat | 71,259 | 19,881 | Jurkat cell cultures infected in vitro |
| Wang et al. (57) | Wang-VSVGgfp-Jurkat-454-hetero-Mse | Rest. Enz. | HIV | Jurkat | 73,237 | 19,356 | Jurkat cell cultures infected in vitro |
Rest. Enz., linker ligation method; Mu, Mu transposase method. See Materials and Methods for details.
As controls, three random positions in the genome were matched with every integration site. Random sites were picked from a set of computationally generated sites equally as distant from the restriction enzyme cleavage site as that used in isolation of the experimental site. In the subsequent statistical analysis, experimental integration sites were compared with matched random controls to minimize the effects of recovery bias.
Integration near transcription start sites and DNase I-hypersensitive sites.
We first confirmed that our MLV and XMRV data sets showed the expected distributions for gammaretroviruses, specifically a preference for integration near transcription start sites and DNase I-hypersensitive sites. Integration less than 2 kb from RefSeq transcription start sites was especially favored for MLV—23.5% of integration events occurred within that window (Fig. 1 A). XMRV also favored integration near transcription start sites (15.6% of sites were less than 2 kb from RefSeq transcription start sites). In contrast, HIV integration was disfavored near transcription start sites. Both gammaretroviruses also favored integration near DNase I cleavage sites (Fig. 1B). Fully 40.9% of MLV integration sites and 36.1% of XMRV integration sites were within 1 kb of a DNase I cleavage site.
Fig. 1.
Gammaretrovirus integration sites are enriched near transcription start sites and DNase I cleavage sites in T cells. (A) Percentage of integration sites found within each interval surrounding transcription start sites. Integration sites near transcription start sites were compiled onto a single start site, and the frequencies were mapped. The x axis shows the distance relative to the transcription start site (at 0). The y axis shows the percentage of integration sites in the indicated window. The color code for each data set is indicated to the right. (B) Percentage of integration sites found within each interval surrounding a DNase I cleavage site. Markings on the graph are as in panel A.
Prediction of nucleosome positions and the relationship to integration frequency.
We investigated the relationship of gammaretroviral integration to nucleosome positions by using nucleosome prediction software to call the locations of nucleosomes in a 5-kb window surrounding each integration site or matched random control. To determine nucleosome locations, we used the DNA-nucleosome interaction model developed by Segal et al. (24, 25, 52). In addition, we reanalyzed the two previously studied HIV integration site data sets from infections of Jurkat cells (57) and T cells (58) in parallel.
We first investigated the frequency of integration at different locations on the nucleosome surface. We extracted those integration sites expected to lie on nucleosome surfaces and quantified the distance from each integration site to the nucleosome dyad axis of symmetry. For the data in Fig. 2, we used the nucleosome prediction method described in reference 52, as was used previously for HIV in references 57 and 58, though several other nucleosome prediction methods yielded similar results (data not shown).
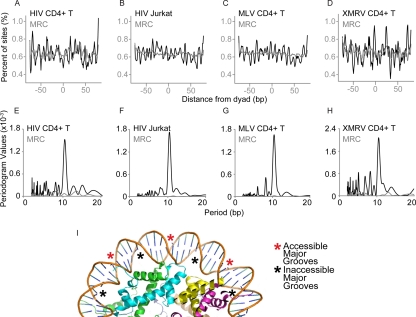
Fig. 2.
Integration frequency relative to predicted nucleosome positions for HIV, MLV, and XMRV. Nucleosome positions were predicted in the 5 kb of sequence surrounding each integration site. The distance of each integration site (black trace) or matched random control (gray trace) from the nucleosome center of symmetry was then determined for each site, and distances for all sites were tabulated. (A to D) Percentage of HIV or gammaretrovirus integration sites present at each position on the nucleosome. The integration site data set tested is marked above each panel. The distances of each integration to the nucleosome center of symmetry were compiled, and the percentage of sites was plotted. Position 0 corresponds to the minor groove at the nucleosome center of symmetry. (E to H) Fourier transform analysis of the data in panel A for each HIV and gammaretrovirus data set. The x axis shows the period of the wave function in panels A to D calculated for each data set (indicated at the top of each panel). The y axis shows the strength of the periodogram value. The results for the integration site data set are shown in black, and the results for the matched random controls are shown in gray (barely visible along the x axis). (I) Accessible and inaccessible DNA major grooves of nucleosome-bound DNA. The diagram shows one gyre of DNA bound to the nucleosome surface, with asterisks indicating accessible (red) or inaccessible (black) major grooves. Coordinate file 1AOI was used to generate the diagram.
Figure 2A to D shows the frequency of retrovirus integration at each position on the nucleosome summarized over all integration sites in each experiment. The graph for the two previously studied HIV data sets (Fig. 2A and B, black line) shows a periodic pattern of high and low values with a minimum at zero, which corresponds to the nucleosome center of symmetry (57, 58). The matched random controls, in contrast, showed little or no variation in frequency relative to positioning on the nucleosome.
For both MLV and XMRV, the positioning of integration sites with respect to the nucleosome dyad also showed a periodic pattern with minima near zero (Fig. 2C and D, black line). The amplitudes and patterns for MLV and XMRV closely resemble those for HIV. No periodic pattern was seen for the gammaretroviral matched random controls (Fig. 2C and D, gray trace on each panel).
Figure 2E to H show Fourier transform analysis of the data in Fig. 2A to D, which summarizes the amplitude and period of the curves. The peak with the highest periodogram value for all four integration site data sets has a value of 10.4 bp, which corresponds to the number of base pairs per turn of the B-DNA double helix (Fig. 2E to H, dark curve). In contrast, the matched random control sites did not show a dominant period (Fig. 2E to H, gray curve barely visible along the x axis). Retroviral DNA integration is known to be favored on outward-facing DNA major grooves in vitro, and previously this periodic pattern was seen for HIV integration sites and interpreted as indicating that the integration is favored at these positions in vivo. We thus infer that gammaretroviral integration also can take place at outward-facing nucleosomes in chromosomes in vivo (diagramed in Fig. 2I).
We also investigated the total fraction of integration sites predicted to be on nucleosomes for each virus. We compared separate versions of the nucleosome prediction software, using several choices for the parameters, and also compared nucleosome occupancy data for T cells generated using chromatin immunoprecipitation with high-throughput sequencing (ChIP-seq) (23, 38, 50, 52). We found that the absolute number of integration sites on nucleosomes was strongly influenced by the software, search parameters, and data sets used. For nucleosome predictions generated as in Fig. 2, about 70% of integration sites were called as nucleosome associated. No consistent differences were seen among the different retroviruses or between experimental integration sites and matched random controls. Comparison to the data on genome-wide mapping of nucleosome positions by ChIP-seq, in contrast, suggested that 50 to 60% of integration sites were nucleosome associated (23, 38, 50, 52). Using these nucleosome positions, a slightly greater proportion of experimental integration sites were called as nucleosomal compared to matched random controls. Thus, the relative frequency of integration on nucleosomes is method dependent and not fully clarified by the available data.
It may seem surprising that periodic patterns of integration on the nucleosome surface are discernible (Fig. 2) despite the divergence of the underlying annotation generated using the different methods for calling nucleosome positions. However, the method of Widom, Segal, and coworkers used here takes account of local sequence features which are well established to influence the energetics of DNA bending. Thus, the predictors seem to be effective at determining the rotational orientation of DNA once nucleosome bound, despite uncertainty in the total percentages on nucleosomes. We return to this issue in the Discussion.
Gammaretroviral integration frequency relative to epigenetic marks and bound proteins.
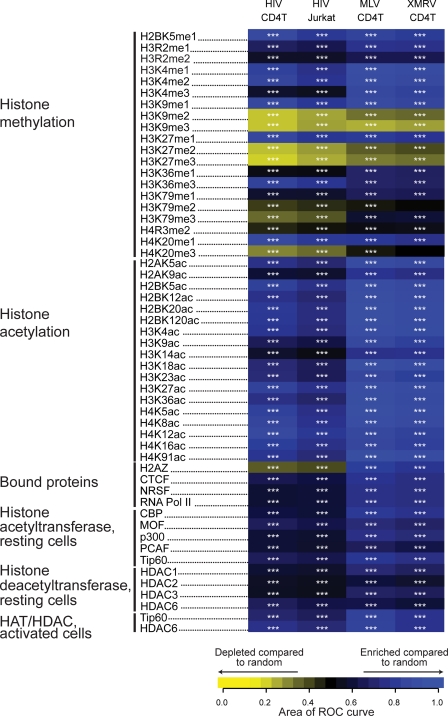
To explore further potential associations between integration targeting and nucleosome structure, we compared integration site density with genome-wide annotation of positions of histone posttranslational modification and bound cellular proteins in CD4+ T cells (3, 50, 60). Figure 3 summarizes the data as a heat map indicating associations between genomic annotation and integration frequency. For each tile on the heat map, the relative frequency of integration near a genomic feature is compared to the frequency in matched random controls using the ROC area method (4), and trends are indicated by the color code. The statistical significance of comparisons to matched random controls is reported by the asterisks on each tile.
Fig. 3.
Integration frequency relative to sites of histone posttranslational modification and chromatin-bound proteins in T cells. Integration site data sets are indicated in the columns; epigenetic marks and bound proteins (quantified using ChIP-seq data) are indicated in the rows. ChIP-seq data for T cells was from references 3, 50, and 60. The frequency of integration sites relative to the matched random controls was quantified using the ROC area method (4). Each tile in the heat map summarizes the trends in the integration site data set versus matched random controls for each form of annotation indicated at the left side of the panel. An ROC area of 0.5 indicates no distinction between integration site data sets and the matched random controls for the ChIP-seq-annotated feature. Values greater than 0.5 indicate a positive association; values less than 0.5 indicate a negative association. ROC areas were calculated using a 10-kb window surrounding integration sites. Enrichment of a particular feature relative to the integration site data set is indicated in blue, and depletion of a feature is indicated in yellow. ***, P < 0.001. For methods used in the statistical tests see reference 4.
Histone methylation.
Integration sites for both HIV and gammaretroviruses are enriched in regions containing histone methylation marks associated with active genes and promoter regions, as anticipated from previous literature (17, 21, 26, 57, 58). For example, H2BK5me1, H3K4me1, H3K4me2, H3K9me1, H3K27me1, and H4K20me1 modifications, all known to be associated with actively transcribed genes and their promoter regions, were all highly enriched near HIV and gammaretrovirus integration sites compared to results for matched random controls. In contrast, histone methylation marks associated with silent genes and promoters were depleted in areas surrounding retrovirus integration sites. In some cases, these histone modifications mediate effects on chromatin structure. Di- and trimethylations of H3K9 are known to recruit HP1, a protein that mediates gene silencing, a state unfavorable for retrovirus integration (2). H3K27me2 and H3K27me3, additional marks of inactive chromatin, were also depleted near both HIV and gammaretrovirus integration sites.
Histone acetylation.
Unlike histone methylation, histone acetylation is mainly associated with actively transcribed genes. When 18 histone acetylation marks were examined, enrichment was found within a 10-kb window around both HIV and gammaretrovirus integration sites. Although the majority of acetylation is associated with active genes, there is some positional preference. For example, H2AK9ac, H3K9ac, H3K18ac, and H3K36ac are more frequently found in the region surrounding the transcription start site. Accordingly, these modifications are slightly less enriched surrounding HIV integration sites, which are found primarily within transcription units.
Bound proteins.
We next analyzed the association of gammaretrovirus and HIV integration sites with binding sites for host cell proteins. The modified histone H2A.Z is found associated with active gene promoters and is rarely present within transcription units. As expected, the 10-kb region surrounding HIV integration sites was depleted of the binding of H2A.Z, while the 10 kb surrounding gammaretrovirus integration sites was enriched, reflecting the known integration preferences of gammaretroviruses and indicating that H2A.Z-containing promoters are among those favored for integration. The zinc finger protein CCCTC-binding factor CTCF is known to be involved in myriad functions, including gene activation, repression, and insulation (40). There is also evidence for its involvement in intra- and interchromosomal contacts. CTCF binding is correlated with gene density, and it most often binds in intergenic regions. CTCF binding is enriched near HIV integration sites and is highly enriched near gammaretrovirus integration sites, attributable to the tendency of gammaretroviruses to integrate near gene boundaries. REST, also known as neuron restrictive silencing factor (NRSF), regulates gene expression by recruiting chromatin-modifying proteins (37). The region surrounding HIV integration was slightly enriched in REST binding, as was the region surrounding gammaretrovirus integration sites. Bound RNA polymerase II (Pol II) was found to be enriched near both HIV and gammaretrovirus integration sites. Bound histone acetyltransferases (HATs) and histone deacetyltransferases (HDACs) were enriched near both HIV and gammaretroviruses. HATs and HDACs work together to specify the acetylation state of chromatin. Recent genome-wide studies show that both HATs and HDACs bind to transcriptionally active genes and that the binding is mediated by phosphorylated RNA Pol II (60). Thus, the localization of HATs and HDACs at active transcription units explains their enrichment surrounding retrovirus integration sites.
Gammaretroviral integration frequency relative to genomic landmarks.
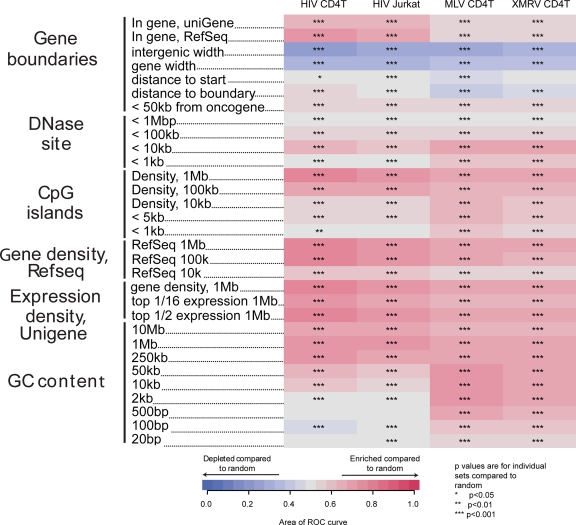
We also tested associations of HIV and gammaretrovirus integration sites with additional genomic landmarks (Fig. 4). Gammaretroviruses favored integration near transcription start sites and in regions enriched in DNase I sites, CpG islands, gene density, highly expressed genes, and high GC content (Fig. 4, right two columns), as reported previously. HIV integration is more frequent within transcription units and near associated features, including gene density, CpG island density, and highly expressed genes (33, 51). Both gammaretroviruses and HIV show favored integration in broad regions (>2 kb) of high GC content. However, in shorter regions (<2 kb) HIV shows less strong favoring of GC-rich regions or even a preference for AT, while gammaretroviruses favor high GC regions of all window sizes.
Fig. 4.
Integration frequency relative to genomic features. Associations between integration site data sets and genomic features are shown using the ROC areas as in Fig. 3. Enrichment of a feature near integration sites relative to matched random controls is depicted in red, and depletion near integration sites is indicated in blue. Asterisks indicate statistical differences compared to matched random controls: *, 0.05 > P > 0.01; **, 0.01 > P > 0.001; ***, P < 0.001. For more information on constructing and interpreting genomic feature heat maps, see reference 3 and supplementary text 1, “Guide to Interpreting Genomic Heat Maps Summarizing Integration Site Distributions,” attached to reference 36.
DISCUSSION
We report here the largest integration site data sets yet generated for MLV and XMRV and use them to investigate the extent to which chromosomal target DNA in vivo is associated with nucleosomes. We used nucleosome prediction methods to call positions of nucleosomes in the regions surrounding integration sites. Integration sites from MLV, XMRV, and HIV were aligned relative to the nucleosome dyad axis, and integration frequencies were compared along the nucleosome-wrapped DNA chain. This showed a periodic increased frequency of integration at specific positions on the nucleosome (Fig. 2), with peaks separated by approximately 10.4 bp, the number of bases per turn in the B-DNA helix. A minimum was found at the nucleosome dyad axis, indicating disfavored integration at the DNA minor groove at the dyad, consistent with the known placement of the initial points of viral DNA joining at phosphates on either side of a target DNA major groove (41, 45). Maxima in the integration intensity corresponded to alternate major grooves. No such pattern was seen for matched random controls. We infer that gammaretrovirus integration in vivo is favored at outward facing major grooves of DNA wrapped around nucleosomes, as with HIV (57, 58). Thus, gammaretrovirus integration can take place in nucleosomal DNA despite favoring targets quite close to DNase I-hypersensitive sites.
The recently published structure of the prototype foamy virus (PFV) intasome bound to model target DNA showed the target DNA to be bent away from the integration complex, roughly as expected for nucleosome-wrapped DNA (31). Thus, data for all the retroviruses studied—PFV, HIV, MLV, and XMRV—suggests that capture of integration targets often takes place when DNA is wrapped on nucleosomes.
It is perhaps surprising that we were able to detect the periodic pattern of integration frequency relative to nucleosome annotation given that the total nucleosome occupancy at integration sites differed depending on the annotation method used. However, the nature of the prediction method, which incorporates well-studied characteristics of DNA bending, appears to be favorable for detecting the periodic pattern. In DNA bends, AA, TT, and TA dinucleotide steps tend to expand the major groove, while GC steps tend to contract the major groove, so that bending is achieved by a 5-bp half-turn spacing of A/T-rich and G/C-rich sequences (48). Given these sequence-directed characteristics, the conformation of a DNA chain on the nucleosome surface can be inferred relatively well, allowing, for example, the design of DNAs that bind to the core histone octamer particularly tightly (42, 59). Absolute occupancy is dependent on additional factors which are not encoded in the DNA sequence, such as concentrations of the reactants and interactions of nucleosomes with other proteins, and is therefore more difficult to calculate. Thus, the use of computationally generated nucleosome positions for identifying periodic integration intensity is effective even when calling the absolute nucleosome occupancy is more difficult.
What proportion of gammaretrovirus integration sites are on nucleosomes? The periodic pattern indicates that target capture for some integration events involves nucleosome-bound DNA, but even the lowest “trough” positions in the periodic pattern show detectable integration frequency. The finding of the trough sites may indicate that some of the integration events take place on DNA that is not associated with nucleosomes. However, it is also possible that an error in the nucleosome calls or positioning of DNA on histones falsely suggests that integration takes place in trough positions. These uncertainties prevent definitive assessment of the proportion of integration events on nucleosomes, and we conclude only that we have evidence that some gammaretroviral target capture events involve nucleosome-associated DNA.
One model for favored MLV integration at transcription start sites would hold that integration is disfavored due to wrapping in nucleosomes, so that promoters are favored due to a lack of bound nucleosomes. Potentially consistent with this, detailed mapping of nucleosome positions indicates that the ∼50 to 100 bp at the transcription start site is depleted for nucleosomes (for a review, see reference 23). However, the data presented here argue against the restricted access model because at least some of the gammaretroviral integration takes place on nucleosome-wrapped DNA, as shown by the periodic pattern, and the region for favored gammaretrovirus integration extends ∼2 kb in each direction (Fig. 1), a region that is wider than the nucleosome-free region.
If absence of nucleosomes does not account for favored MLV integration at transcription start sites, then how do MLV preintegration complexes recognize favored sites? For HIV, favored integration in active transcription units is a result of tethering by the LEDGF/p75 transcriptional mediator protein. HIV integrase binds to a C-terminal domain of LEDGF/p75 (10, 11, 16, 29). The N-terminal domains of LEDGF/p75 direct binding to active transcription units (15). Depleting cells for LEDGF/p75 reduces the favoring of integration in transcription units (12, 32, 53), and swapping the LEDGF/p75 N-terminal domains for other chromatin binding domains retargets HIV DNA integration to new locations (18, 20, 54). One of the goals of this study was to probe the deep annotation of T cells for strongly correlated features, potentially providing clues to the nature of a possible tethering protein for gammaretroviruses. However, analysis to date has not disclosed any single strong candidate that predicts gammaretroviral integration patterns. Rather, a collection of histone marks and bound proteins associated with active transcription units all are correlated, so that no single candidate for a tethering factor stands out. Going forward, as more genome-wide annotation accumulates for T cells it will be possible to use the data sets reported here to assess correlations with integration intensity and newly mapped chromatin-bound proteins, allowing additional candidate factors to be investigated.
ACKNOWLEDGMENTS
We are grateful to members of the Bushman laboratory for help and suggestions.
This work was supported by NIH grants AI52845 and AI082020, the University of Pennsylvania Center for AIDS Research, and the Penn Genome Frontiers Institute with a grant with the Pennsylvania Department of Health. S.L.R. was supported by NIH training grant T32 AI-007632.
The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Footnotes
Published ahead of print on 11 May 2011.
REFERENCES
- 1. Angelov D., et al. 2004. The histone octamer is invisible when NF-kappa B binds to the nucleosome. J. Biol. Chem. 279:42374–42382 [DOI] [PubMed] [Google Scholar]
- 2. Bannister A. J., et al. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120–124 [DOI] [PubMed] [Google Scholar]
- 3. Barski A., et al. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837 [DOI] [PubMed] [Google Scholar]
- 4. Berry C., Hannenhalli S., Leipzig J., Bushman F. D. 2006. Selection of target sites for mobile DNA integration in the human genome. PLoS Comp. Biol. 2:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Biasco L. A., et al. 2011. Integration profile of retroviral vector in gene therapy treated patients is cell-specific according to gene expression and chromatin conformation of target cell. EMBO Mol. Med. 3:89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brady T., et al. 2009. Integration target site selection by a resurrected human endogenous retrovirus. Genes Dev. 23:633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brady T., et al. 16 March 2011, posting date. A method to sequence and quantify DNA integration for monitoring outcome in gene therapy. Nucleic Acids Res. doi: 10.1093/nar/gkr140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cattoglio C., et al. 2007. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood 110:1770–1778 [DOI] [PubMed] [Google Scholar]
- 9. Cattoglio C., et al. 2010. High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood 116:5507–5517 [DOI] [PubMed] [Google Scholar]
- 10. Cherepanov P., Ambrosio A. L., Rahman S., Ellenberger T., Engelman A. 2005. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl. Acad. Sci. U. S. A. 102:17308–17313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cherepanov P., et al. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372–381 [DOI] [PubMed] [Google Scholar]
- 12. Ciuffi A., et al. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11:1287–1289 [DOI] [PubMed] [Google Scholar]
- 13. Ciuffi A., et al. 2009. Methods for integration site distribution analyses in animal cell genomes. Methods 47:261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deichmann A., et al. 2007. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Invest. 117:2225–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Rijck J., Bartholomeeusen K., Ceulemans H., Debyser Z., Gijsbers R. 2010. High-resolution profiling of the LEDGF/p75 chromatin interaction in the ENCODE region. Nucleic Acids Res. 38:6135–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emiliani S., et al. 2005. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 280:25517–25523 [DOI] [PubMed] [Google Scholar]
- 17. Felice B., et al. 2009. Transcription factor binding sites are genetic determinants of retroviral integration in the human genome. PLoS One 4:e4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferris A. L., et al. 2010. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc. Natl. Acad. Sci. U. S. A. 107:3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garson J. A., Kellam P., Towers G. J. 2011. Analysis of XMRV integration sites from human prostate cancer tissues suggests PCR contamination rather than genuine human infection. Retrovirology 8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gijsbers R., et al. 2010. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol. Ther. 18:552–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hacein-Bey-Abina S., et al. 2008. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 118:3132–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hue S., et al. 2010. Disease-associated XMRV sequences are consistent with laboratory contamination. Retrovirology 7:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang C., Pugh B. F. 2009. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10:161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaplan N., et al. 2010. Nucleosome sequence preferences influence in vivo nucleosome organization. Nat. Struct. Mol. Biol. 17:918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaplan N., et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458:362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim S., et al. 2008. Integration site preference of xenotropic murine leukemia virus-related virus, a new human retrovirus associated with prostate cancer. J. Virol. 82:9964–9977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim S., et al. 2010. Fidelity of target site duplication and sequence preference during integration of xenotropic murine leukemia virus-related virus. PLoS One 5:e10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lewinski M. K., et al. 2006. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Llano M., et al. 2006. An essential role for LEDGF/p75 in HIV integration. Science 314:461–464 [DOI] [PubMed] [Google Scholar]
- 30. Lombardi V. C., et al. 2009. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science 326:585–589 [DOI] [PubMed] [Google Scholar]
- 31. Maertens G. N., Hare S., Cherepanov P. 2010. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468:326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marshall H., et al. 2007. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One 2:e1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mitchell R. S., et al. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Muller H. P., Varmus H. E. 1994. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 13:4704–4714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oakes B., et al. 2010. Contamination of human DNA samples with mouse DNA can lead to false detection of XMRV-like sequences. Retrovirology 7:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ocwieja K. E., et al. 2011. HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog. 7:e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ooi L., Wood I. C. 2007. Chromatin crosstalk in development and disease: lessons from REST. Nat. Rev. Genet. 8:544–554 [DOI] [PubMed] [Google Scholar]
- 38. Osipov S. A., Preobrazhenskaya O. V., Karpov V. L. 2010. Chromatin structure and transcription regulation in Saccharomyces cerevisiae. Mol. Biol. 44:856–869 [PubMed] [Google Scholar]
- 39. Panet A., Cedar H. 1977. Selective degradation of integrated murine leukemia proviral DNA by deoxyribonucleases. Cell 11:933–940 [DOI] [PubMed] [Google Scholar]
- 40. Phillips J. E., Corces V. G. 2009. CTCF: master weaver of the genome. Cell 137:1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pruss D., Bushman F. D., Wolffe A. P. 1994. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. U. S. A. 91:5913–5917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pruss D., Reeves R., Bushman F. D., Wolffe A. P. 1994. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J. Biol. Chem. 269:25031–25041 [PubMed] [Google Scholar]
- 43. Pryciak P., Muller H. P., Varmus H. E. 1992. Simian virus 40 minichromosomes as targets for retroviral integration in vivo. Proc. Natl. Acad. Sci. U. S. A. 89:9237–9241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pryciak P. M., Sil A., Varmus H. E. 1992. Retroviral integration into minichromosomes in vitro. EMBO J. 11:291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pryciak P. M., Varmus H. E. 1992. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell 69:769–780 [DOI] [PubMed] [Google Scholar]
- 46. Robinson M. J., et al. 2010. Mouse DNA contamination in human tissue tested for XMRV. Retrovirology 7:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rohdewohld H., Weiher H., Reik W., Jaenisch R., Breindl M. 1987. Retrovirus integration and chromatin structure: Moloney murine leukemia proviral integration sites map near DNase I-hypersensitive sites. J. Virol. 61:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Satchwell S. C., Drew H. R., Travers A. A. 1986. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 191:659–675 [DOI] [PubMed] [Google Scholar]
- 49. Schlaberg R., Choe D. J., Brown K. R., Thaker H. M., Singh I. R. 2009. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc. Natl. Acad. Sci. U. S. A. 106:16351–16356 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50. Schones D. E., et al. 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132:887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schroder A. R., et al. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521–529 [DOI] [PubMed] [Google Scholar]
- 52. Segal E., et al. 2006. A genomic code for nucleosome positioning. Nature 442:772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shun M. C., et al. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 21:1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Silvers R. M., et al. 2010. Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Hum. Gene Ther. 21:337–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vijaya S., Steffan D. L., Robinson H. L. 1986. Acceptor sites for retroviral integrations map near DNase I-hypersensitive sites in chromatin. J. Virol. 60:683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang G. P., et al. 2010. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood 115:4356–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang G. P., Ciuffi A., Leipzig J., Berry C. C., Bushman F. D. 2007. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 17:1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang G. P., et al. 2009. Analysis of lentiviral vector integration in HIV plus study subjects receiving autologous infusions of gene modified CD4+T cells. Mol. Ther. 17:844–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang X., Bryant G. O., Floer M., Spagna D., Ptashne M. 2011. An effect of DNA sequence on nucleosome occupancy and removal. Nat. Struct. Mol. Biol. 18:507–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang Z., et al. 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138:1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu X., Li Y., Crise B., Burgess S. M. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749–1751 [DOI] [PubMed] [Google Scholar]