Abstract
Maturation of human cytomegalovirus (HCMV) initiates with nucleocapsids that egress from the nucleus and associate with a juxtanuclear cytoplasmic assembly compartment, where virion envelopment and release are orchestrated. Betaherpesvirus conserved proteins pp150 (encoded by UL32) and pUL96 are critical for HCMV growth in cell culture. pp150 is a capsid-proximal tegument protein that preserves the integrity of nucleocapsids during maturation. pUL96, although expressed as an early protein, acts late during virus maturation, similar to pp150, based on the comparable antigen distribution in UL96, UL32, or UL96/UL32 dual mutant virus-infected cells. pp150 associates with nuclear capsids prior to DNA encapsidation, whereas both pp150 and pUL96 associate with extracellular virus, suggesting that pUL96 is added after pp150. In the absence of pUL96, capsid egress from the nucleus continues; however, unlike wild-type virus infection, pp150 accumulates in the nuclear, as well as in the cytoplasmic, compartment. Ultrastructural evaluation of a UL96 conditional mutant revealed intact nuclear stages but aberrant nucleocapsids accumulating in the cytoplasm comparable to the known phenotype of UL32 mutant virus. In summary, pUL96 preserves the integrity of pp150-associated nucleocapsids during translocation from the nucleus to the cytoplasm.
INTRODUCTION
Human cytomegalovirus (HCMV) tegument proteins are important virus structural components that control a range of events in infection, including transcriptional activation and host cell modulation immediately following delivery to cells, as well as virus assembly, maturation, and egress during the late phase of replication (5, 28). There are only two HCMV tegument components (pp150 and pUL96) (14, 44) that lack any obvious functional or sequence homolog in either alpha- or gammaherpesviruses (27) but are critical for virus replication, based on systematic mutagenesis studies (12, 48). pp150 plays an essential role, stabilizing nucleocapsids during virus maturation (2, 37). pUL96 is known to associate with virus particles in minor quantities (44) and may be a positional homolog of herpes simplex virus 1 (HSV-1) UL14 tegument protein (Andrew Davison, personal communication). Studies of a UL14 mutant virus have yielded an indication of this gene's roles in the translocation of input nucleocapsids into the nucleus (47); hence, characterization of pUL96 would unveil any relationship to this HSV-1 function.
HCMV maturation involves a concerted series of events orchestrated by protein-protein interactions (5, 28). Viral DNA encapsidation and initial tegumentation occur in the nucleus. Nucleocapsids translocate into the cytoplasm, where they mature into virions within the assembly compartment (AC), the site controlling final tegumentation, virus envelopment, and egress from cells (5, 11, 28). pp150, a major virion tegument protein, locates to the nucleus as maturation begins (18, 30), binds to capsids (4), promotes maturation through protein domains required for capsid interaction (2, 4, 37), and remains associated with nucleocapsids that mature into virions (10, 13, 14, 21, 22). pp150 is the only HCMV tegument protein that is known to be absolutely essential for virus maturation. Based on the localization of major capsid protein (MCP) and viral DNA within the AC of UL32 mutant virus-infected cells, this protein was initially thought to control cytoplasmic events in virus maturation (2). Subsequently, however, ultrastructural analysis of UL32 mutant virus-infected cells by transmission electron microscopy (TEM) revealed intact nucleocapsids within the nucleus but abnormal vesicle-like particles in the AC (37), a pattern that indicated disintegration of preformed nucleocapsids in the absence of this critical viral function. A recent cryoelectron microscopy study demonstrated that pp150 is a constituent of the net-like layer of icosahedrally ordered capsid-bound tegument (49), thus supporting the proposed nucleocapsid-stabilizing role of pp150 in virus maturation. Overall, experimental evidence has supported a stepwise process starting with the addition of pp150 to capsids early during tegumentation in the nucleus, in order to promote capsid stability during translocation to the cytoplasm, where maturation steps take place in the AC.
Cytoplasmic steps in HCMV maturation occur mainly in an AC defined by a unique rearrangement of cellular organelles and vesicle transport machinery together with accumulation of viral structural antigens (11, 31, 37). Endosomes are arranged toward the middle of the AC, surrounded by concentric layers of trans-Golgi network (TGN) and cis-Golgi network, with the endoplasmic reticulum outermost (11). During HCMV maturation, the AC controls final envelopment and vesicle trafficking through release of mature virions and dense bodies from the cell (11, 31). These maturation events coincide with virion transport through the AC largely under the control of host vesicle transport machinery. Recent evidence suggests that transport and release of virus particles are also influenced by a viral function, the UL103 gene product (1). HCMV employs TGN- and/or endosome-derived vesicles for envelopment (35, 40), and endosomal trafficking pathways controlled by Rab family members (19, 20, 24) and ESCRT (36) proteins contribute to HCMV maturation. Given that pp150 lacks obvious signals for nuclear import or export, viral and/or cellular proteins may escort pp150 to nuclear sites of encapsidation and/or cytoplasmic sites of virus assembly in the AC. One such interaction of pp150 with Rab6 effector protein Bicaudal D1 influences the localization of pp150 to the AC (20). Thus, cytoplasmic virus maturation is governed by an interplay of viral and host factors.
UL96 mutant viruses are replication deficient (12, 48). Here, we addressed the role of pUL96 in viral replication by constructing and characterizing mutants, as well as by studying the expression kinetics, localization in infected cells, and association of this protein with virus particles. A recently established method of tagging cytomegalovirus open reading frames (ORFs) with the FK506-binding protein destabilization domain (FKBP-DD) (3, 16) provided an opportunity to evaluate conditional UL96 mutant virus. Functional analysis of DD-tagged mutants revealed the benefits of expected virus association of pUL96 to the mutant phenotype. Evidence derived from mutant virus growth and antigen expression properties, combined with ultrastructural studies, implicates pUL96 in preserving integrity of pp150-associated nucleocapsids during translocation into the cytoplasm.
MATERIALS AND METHODS
Sequences and alignments.
Nucleotide sequences from HCMV Merlin (NC_006273) and HCMV TowneBAC (AY315197) were used for comparison and analysis of UL96 sequence conservation. The Merlin and TowneBAC sequences differed by one nucleotide at position 105 (C to T, synonymous codon for leucine 35). The Merlin UL96 amino acid sequence (YP_081543) was aligned with amino acid sequences of pUL96 homologs from rhesus cytomegalovirus (RhCMV; YP_068222), chimpanzee cytomegalovirus (CCMV; AAM00734), human herpesvirus 6 (HHV6; CAA58360), and HHV7 (YP_073808). Both the pairwise and multiple alignments were done in VectorNTI (ver 11; Invitrogen Corporation, Carlsbad, CA) using the ClustalW algorithm.
Cells.
Primary human foreskin-derived fibroblasts (HF) were cultured in Dulbecco's modified Eagle's medium (Invitrogen Corporation, Carlsbad, CA) containing 4.5 g/ml glucose, 10% fetal bovine serum (S1245OH; Atlanta Biologicals, Lawrenceville, GA), 1 mM sodium pyruvate, 2 mM l-glutamine, and 100 U/ml penicillin-streptomycin (Cellgro, Manassas, VA) at 37°C with 5% CO2. HF between passages 5 and 15 were used for transfections and infections. The cell culture medium was changed (for transfections) or additional medium was added (for infections) every other day.
BAC mutagenesis and recombinant viruses.
Bacterial artificial chromosome (BAC) recombineering protocols (38) were followed to mutate regions of UL96 as reported earlier for other CMV proteins (37, 43), with some modifications. Briefly, Escherichia coli DH10B carrying TowneBAC and pSIM6 was made electroporation competent by growth to an optical density at 600 nm of 0.4 to 0.6 and two washes with ice-cold nanopure water. A PCR amplicon carrying 50 bp of homology on either flank of the desired deletion region from TowneBAC in addition to the complete kanamycin resistance (Kanr)-Bacillus subtilis levansucrase (SacB) cassette was introduced into competent cells by electroporation. Kanr colonies were selected on LB-kanamycin plates and screened for sucrose sensitivity. Sucrose-sensitive colonies were further screened by PCR for the proper orientation and location of the Kanr-SacB cassette. Colonies positive by PCR screening were further evaluated following overnight growth, BAC DNA preparation, and digestion with several restriction enzymes to confirm the expected restriction fragment length polymorphism. From the selected BAC clone (ΔUL96BAC, pON5017), additional clones were derived following the introduction of a PCR fragment containing wild-type (WT) UL96 (rescued BAC, pON5018), an epitope tag (Myc-UL96-BAC, pON5019), or stop mutants (S1, pON5020; S2, pON5021; S3, pON5022; S4, pON5023) and screening for the loss of kanamycin resistance, as well as sucrose sensitivity. UL96S1-BAC was further modified to replace UL32 with a Kanr-SacB cassette to generate Δ32-96SBAC (pON5024). pON5017 was also engineered to insert UL96 with an FKBP-DD tag (3) at the C terminus (UL96DDBAC, pON5025). The UL96 region from these viruses was sequenced to confirm the presence of the desired mutation, as well as the absence of any undesired modifications. At 9 days posttransfection (dpt), supernatants from HF cultures transfected with replication-competent BACs were harvested for recovery of recombinant viruses. Proteins fused to the DD are expected to be functionally restored in the presence of a ligand (Shield-1) (3). Viruses recovered from UL96DDBAC transfections/infections of cells cultured in the presence of 1 μM Shield-1 were termed UL96DDBAC(+) because these conditions provided the potential for either pUL96 incorporation into the virion or pUL96-mediated changes in virion quality, whereas those recovered from transfections/infections in the absence of Shield-1 were termed UL96DDBAC(−). Thus, the requirement to construct a UL96-expressing cell line to grow UL96 mutant virus was circumvented. However, the DD fusion approach was challenging due to the unavailability of good commercial antibodies, especially against the C-terminal FKBP-DD fusion protein; the cost of Shield-1, which makes growing large quantities of virus unreasonably expensive; and the need to optimize the FKBP-DD fusion location and Shield-1 concentration for each individual protein. Single-step (multiplicity of infection [MOI] of 3.0) or multiple-step (MOI of 0.01) growth curves were obtained by infecting confluent HF in triplicate and harvesting cells plus supernatants at designated time points postinfection. These samples were mixed with an equal volume of triple-autoclaved milk and sonicated (Sonicator 3000; Misonix Incorporated, Farmingdale, NY) before freezing or direct titration. The samples were diluted 10- to 100,000-fold for titration by fluorescent-focus assays on confluent HF, where enhanced green fluorescent protein-positive (eGFP+) foci (focus-forming units or FFU) were counted at 10 days postinfection (dpi) by epifluorescence microscopy.
Virus and capsid purification.
Extracellular CMV particles and nuclear B capsids were purified using established protocols for herpesviruses (13, 22, 39), with some modifications. Briefly, HF monolayers were infected with TowneBAC or Myc-UL96-BAC virus (MOI of 5) and harvested at 4 to 5 dpi. Medium was clarified of any cellular debris by low-speed centrifugation (2,000 × g, 10 min), and virus particles were pelleted (∼20,000 × g, 1 h). These extracellular virus (ECV) particles were purified on a 30% sucrose cushion (SW 32Ti rotor, Beckman L-90 ultracentrifuge, 24,000 rpm, 1 h). For nuclear B capsids, cell pellets were washed in 1× phosphate-buffered saline (PBS) and incubated in hypotonic buffer (20 mM Tris-HCl, pH 7.5) for 20 min to swell before the addition of Triton X-100 (1.5% final concentration) to lyse the cells for 30 min. Nuclei were spun at 2,000 × g for 10 min, resuspended in TNE (500 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, pH 8.0), sonicated (Sonicator 3000; Misonix Incorporated, Farmingdale, NY) for 10 s, and spun at 14,000 rpm for 30 s in a microcentrifuge. The above step was repeated once, and combined supernatants were loaded onto a 20 to 50% discontinuous sucrose-in-TNE gradient before centrifugation (SW-41 rotor, Beckman L-80 ultracentrifuge, 24,000 rpm, 1 h). Capsids were observed as visible-light-scattering bands with A capsids forming a thin upper band around the 30% gradient and B capsids forming a thick lower band around the 35% gradient; the C-capsid band was faint. These B capsids were harvested by puncturing the side of the centrifuge tube with a 23-gauge needle and washed once in TNE before concentration by centrifugation (SW-41 rotor, Beckman L-80 ultracentrifuge, 24,000 rpm, 1 h). The identity and purity of harvested capsids were confirmed by electron microscopy. The capsids or ECV particles were resuspended in 2% SDS-containing lysis buffer and run on a 4 to 20% gradient polyacrylamide gel before transfer to a polyvinylidene difluoride membrane and analysis by immunoblotting.
Drug inhibition and release experiments.
For determination of pUL96 kinetics, confluent HF monolayers were pretreated with cycloheximide (CH; 50 μg/ml; purchased from Acros Organics, Geel, Belgium) for 1 h and then infected with Myc-UL96-BAC virus (MOI of 3.0) in medium containing CH (50 μg/ml) for 1 h, washed, and thereafter incubated for 8 h in the presence of CH (50 μg/ml). After this, HF were washed four times with medium and incubated in drug-free medium. Phosphonoformate (PFA; 300 μg/ml; Sigma) or 2-bromo-5,6-dichloro-1-(β-d-ribofuranosyl)benzimidizole (BDCRB; 20 μM; a gift from John Drach) was added at 1 h postinfection (hpi) and maintained for 48 hpi. In some experiments, labeled BDCRB block release, BDCRB added at 1 hpi was maintained until 96 hpi, when cultures were washed four times with medium and then further incubated in drug-free medium. Triplicate samples were used for all of the drug block release studies. Samples of infected cells and cell-free medium were harvested at designated time points and stored at −80°C before titration by fluorescent-focus assays on HF.
Antibodies, immunofluorescence assays, and immunoblot analyses.
Mouse monoclonal antibodies to pp28 (CH19), gB (2F12), pp65 (UL83, CH12), and pUL44 (ICP-36, CH13) were purchased from Virusys Corporation, Sykesville, MD, and MAB810 (IE1, IE2) was purchased from Millipore, Billerica, MA. Monoclonal antibodies against pp150 (36-14) (31), gMgN (14-16A) (6), and MCP (28-4) (9) were kindly provided by William Britt, University of Alabama, Birmingham. Monoclonal anti-c-myc antibody (9E10; Santa Cruz Biotechnology Inc., Santa Cruz, CA) or chicken anti-myc antibody (Gallus Immunotech Inc., Fergus, Ontario, Canada) was used to detect c-myc-tagged proteins. Fluorescent-label-tagged secondary antibodies (Alexa-594 anti-mouse IgG [H+L], Alexa-488 anti-mouse IgG [H+L], and Alexa-488 anti-chicken IgG [H+L]) were purchased from Molecular Probes-Invitrogen Corporation (Carlsbad, CA) and used in immunofluorescence assays (IFA) described below. Hoechst 33258 (AnaSpec Corporation, San Jose, CA) staining identified the nuclei in IFA. Coomassie brilliant blue (0.025%; Thermo Fisher Scientific, Rockford, IL), dissolved in a solution containing 40% methanol, 10% acetic acid, and 50% nanopure water, was used for staining of SDS-polyacrylamide gels. For immunoblot analyses, peroxidase-labeled horse anti-mouse IgG (Vector Laboratories, Burlingame, CA) was used as the secondary antibody. Anti-β-actin antibody (AC-74; Sigma-Aldrich, St. Louis, MO) was used as a control for sample loading in immunoblot assays. Blots were detected using ECL Western blotting detection reagents (GE Healthcare, Buckinghamshire, United Kingdom).
Microscopy.
Samples were prepared using established protocols for IFA and confocal fluorescence microscopy. Briefly, cells (HF) were grown on coverslip inserts in 24-well tissue culture dishes. After transfections/infections, at the endpoint, cells were fixed in 3.7% formaldehyde for 10 min and incubated in 50 mM NH4Cl in 1× PBS for 10 min to reduce autofluorescence. This was followed by washing in 1× PBS, incubation in 0.5% Triton X-100 for 20 min to permeabilize the cells, and finally washing and incubation with primary and secondary antibodies at a 1:1,000 dilution in 0.1% bovine serum albumin in 1× PBS. Coverslips were retrieved from the wells, mounted on glass slides with a drop of mounting medium (Gel/Mount; Biomeda, Foster City, CA), and dried overnight before imaging. Images were acquired on a Carl Zeiss LSM 510 META confocal fluorescence microscope or a Zeiss Axio Imager A1 epifluorescence microscope using a 40× or 100× objective. Samples for TEM were prepared by infecting HF with UL96DDBAC(−) (MOI of 0.01), UL96DDBAC(+) (MOI of 3.0), or TowneBAC (MOI of 3.0) virus. At the endpoint, cells were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) for 2 h at room temperature. Cells were then washed with the same buffer and postfixed with buffered 1.0% osmium tetroxide at room temperature for 1 h. Following several washes with 0.1 M cacodylate buffer, cells were dehydrated with ethanol, infiltrated, and embedded in Eponate 12 resin (Ted Pella Inc., Redding, CA). Cell culture plates were cracked with a hammer to release the resin after it had solidified, and ultrathin sections (60 to 70 nm) of monolayer cells were cut and counterstained using uranyl acetate and lead citrate. Examination of ultrathin sections was carried out on a Hitachi H-7500 TEM operated at 75 kV, and images were captured using a Gatan BioScan (Pleasanton, CA) charge-coupled device camera. The images were acquired and analyzed with the Digital Micrograph (Pleasanton, CA) software.
RESULTS
Role of pUL96 in virus replication.
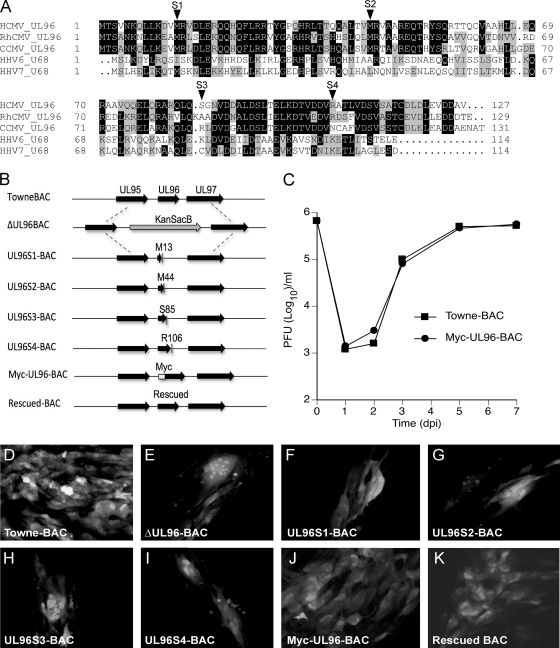
In order to assess the presence of conserved regions, alignment of amino acid sequences of pUL96 homologs from primate betaherpesviruses was performed (Fig. 1A). While there are no obvious conserved domains across all of these betaherpesviruses, the alignment suggests significant sequence similarity among these small 114- to 131-amino-acid (aa) proteins with pairwise identities of 18.9% (HCMV versus HHV7), 19.7% (HCMV versus HHV6), 57.4% (HCMV versus RhCMV), and 68.7% (HCMV versus CCMV). Earlier studies reported UL96 as critical for virus replication based on deletion or transposon insertion mutation of the ORF (12, 48). Given that UL96 is located in a transcriptionally complex region of the viral genome (45) where deletions or transposon insertions may interfere with neighboring or overlapping genes, we constructed UL96 mutants that would have little chance of impacting the expression or function of other genes. To accomplish this goal, a UL96 deletion in TowneBAC (ΔUL96BAC) was constructed and repaired with WT UL96 or fragments containing individual translation stop mutations at defined locations (Fig. 1B), namely, amino acids M13, M44, S85, and R106 (S1, S2, S3, and S4, respectively). These mutations were specifically designed (i) to disrupt the two N-terminal methionines (M13, M44) in order to assess any role of these potential translation start sites and (ii) to truncate the C-terminal region of the protein to various lengths. These mutations would not be predicted to interfere with flanking ORFs in the viral genome. ΔUL96BAC transfections resulted in two distinct phenotypes: small syncytial (multinucleated) foci (∼70%) (Fig. 1E) and mononucleate infected cells (∼30%) (data not shown). All of the stop mutant BACs (S1 to S4) showed plaque phenotypes (Fig. 1F to I) indistinguishable from that of ΔUL96BAC (Fig. 1E) and clearly distinct from that of the considerably larger and nonsyncytial TowneBAC plaques (Fig. 1D), suggesting that all of the evaluated UL96 mutants exhibit defects. The similarity of the phenotypes of all of the stop mutants suggested the presence of a single translation start site and indicated that the C-terminal region of pUL96 is required for function. TowneBAC was also engineered to tag pUL96 with a c-myc epitope at the N terminus (Myc-UL96-BAC), a modification that apparently did not alter protein function based on plaque size and phenotype (Fig. 1J). The Myc-UL96-BAC and parental TowneBAC viruses exhibited similar viral replication kinetics (Fig. 1C). Finally, ΔUL96BAC was genetically rescued to express UL96 similar to TowneBAC and this rescued BAC showed a plaque phenotype similar to that of TowneBAC (Fig. 1K). In combination, these results confirm that pUL96 is critical for virus replication.
Fig. 1.
pUL96 is a betaherpesvirus conserved protein critical for HCMV growth in cell culture. (A) Amino acid sequence alignment of pUL96 homologs from HCMV (YP_081543), RhCMV (YP_068222), CCMV (AAM00734), HHV6 (CAA58360), and HHV7 (YP_073808). Identical amino acids (black) and similar amino acids (gray) are highlighted. (B) Schematic of the UL95-UL97 region in TowneBAC (top line) with other bacmid constructs shown below. ΔUL96BAC is depicted with a KanSacB insert replacing UL96, UL96S1-BAC through UL96S4-BAC are depicted with the positions of amino acids (M13, M44, S85, and R106) that were replaced with novel termination codons to prematurely terminate UL96, Myc-UL96-BAC is depicted with the position of an amino-terminal epitope tag on UL96, and rescued BAC, constructed from the ΔUL96BAC, is depicted with repaired UL96. (C) Single-step growth curves of TowneBAC and Myc-UL96-BAC viruses. HF were infected (MOI of 3.0), total viruses (cells plus medium) were harvested at the indicated times postinfection, and titers were determined on HF monolayers. The viral titers represent the averages of triplicate experiments, and standard errors are within symbols. The data point at 0 dpi indicates the input virus dose. (D to K) Typical eGFP+ plaque phenotypes of the indicated BAC constructs at 10 dpt in HF. Original magnification, ×400.
pUL96, an early protein, associates with virus particles in the cytoplasm of infected cells.
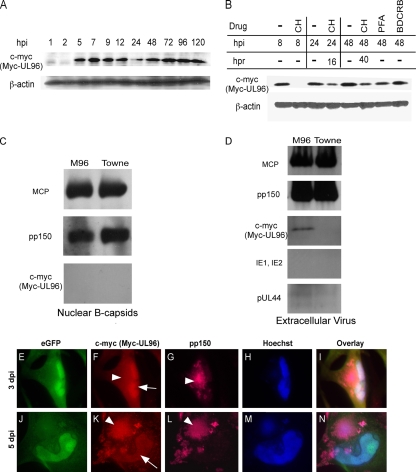
A comprehensive transcriptional analysis of the HCMV UL93-to-UL99 region revealed that UL96 is an early gene (45). We employed Myc-UL96-BAC and immunoblot analyses to assess the time course of pUL96 expression. Unusual for a putative virus structural protein, Myc-UL96 (∼14 kDa) was detected as early as 5 hpi and reached a steady state by 7 hpi (Fig. 2A), indicating early kinetics. Detection of this tagged protein of expected size, as predicted from the amino acid sequence, indicated the presence of a bona fide start codon at the location of epitope tag insertion. This immunoblot signal was not derived from input virion protein, given that Myc-UL96 was absent in the 1- and 2-hpi samples and expression of Myc-UL96 was blocked when cells were pretreated (1 h), infected (1 h), and maintained (8 h) in CH (50 μg/ml) (Fig. 2B). The expression was partially restored after washing out of the CH with normal medium at 8 hpi and incubation of cells without drug for an additional 16 h (to 24 hpi) or 40 h (to 48 hpi) (Fig. 2B). Levels of Myc-UL96 were only slightly reduced in the presence of the viral DNA replication inhibitor PFA (300 μg/ml) through 48 hpi (Fig. 2B). The encapsidation inhibitor BDCRB has been reported to have a minimal impact on viral DNA replication or protein accumulation (42), and treatment did not affect Myc-UL96 levels (Fig. 2B). These characteristics of Myc-UL96 expression indicate that pUL96 is a delayed early (β) gene, consistent with transcript analysis (45). Further, the expression pattern of pUL96, including the plateau of accumulated levels well in advance of DNA replication, suggested that this protein could function at any time, starting at the early phase of viral replication.
Fig. 2.
Myc-pUL96 expression, association with virus particles, and localization in infected cells. (A) Immunoblot time course (hpi) of Myc-UL96 expression following Myc-UL96-BAC virus infection of HF, using total cell lysates and detection with antibodies to c-myc or β-actin (loading control). (B) Immunoblot analyses similar to that in panel A but depicting the impact of CH, PFA, or BDCRB treatment on Myc-UL96 expression. Shown are the accumulated levels of Myc-UL96 or β-actin from Myc-UL96-BAC virus-infected cells that were either cultured with the indicated drugs until harvest at the indicated times (hpi) or cultured with the indicated drugs for 8 h, washed, and then cultured in normal medium until harvest at the times indicated as hours postrelease (hpr). (C and D) Detection of the indicated antigens (MCP, pp150, c-myc [pUL96], IE1, IE2, and pUL44) in immunoblot assays of sucrose gradient-purified nuclear B capsids (C) or ECV particles (D) from Myc-UL96-BAC (M96) or TowneBAC (Towne) infections. (E to N) IFA detection of Myc-UL96 and pp150 in HF infected with Myc-UL96-BAC virus and fixed for processing at 3 (E to I) or 5 (J to N) dpi. Nuclear (arrows) and cytoplasmic (arrowheads) viral antigens are indicated. Original magnification, ×1,000.
To study whether pUL96 was associated with virus particles, nuclear B capsids or ECV particles from TowneBAC or Myc-UL96-BAC infections were purified, denatured, and run on an SDS-polyacrylamide gel. Based on Coomassie brilliant blue staining, the quality and composition of nuclear B capsids from TowneBAC or Myc-UL96-BAC infections appeared similar, as was the case for ECV particles from these infections (data not shown). Again, samples were run on SDS-polyacrylamide gel, transferred to membrane, and immunoblotted to detect pp150 or c-myc (Myc-UL96). Both nuclear B capsids and ECV particles incorporated pp150, but only ECV particles incorporated pUL96 (Fig. 2C and D). pp150 was detected at levels similar to those of MCP (intrinsic control) in these experiments. The ECV particle preparation was free of intracellular contaminants based on the absence of nonstructural immediate-early 1 (IE1), IE2, and pUL44 antigens (Fig. 2D). Expression and localization of pUL96 were investigated using IFA on Myc-UL96-BAC virus-infected HF. Based on the detection of the c-myc tag and consistent with the predicted properties of a small-sized protein (25), Myc-UL96 remained diffuse throughout the nucleus and cytoplasm of infected cells from 8 through 72 hpi (Fig. 2F and I and data not shown). At later times (day 5), significant accumulation of Myc-UL96 occurred in the AC, as well as in the nucleus (Fig. 2K and N). At this time, Myc-UL96 accumulation in the AC (Fig. 2K) coincided with the pp150 localization pattern (Fig. 2L and N). In summary, pUL96 is expressed from early through late times postinfection and is present in the nucleus, as well as the cytoplasm, of infected cells. Cytoplasmic localization of pUL96 shifts from diffuse at earlier times to concentrated in the AC, where pp150 and maturing virion-associated nucleocapsids accumulate as infection proceeds to the late phase. This, along with the evidence that pUL96 associates with ECV particles but not nuclear B capsids, is most consistent with this protein being acquired as nucleocapsids translocate from the nucleus to the cytoplasm of infected cells.
Phenotypic characterization of UL96 mutant and UL96/UL32 dual mutant viruses.
Given the similarities in pp150 and pUL96 localization and the known importance of pp150 during cytoplasmic virus maturation, we investigated whether these two proteins impact the same stage of virus maturation. TowneBAC was engineered to carry a stop mutation at the beginning (M13) of the UL96 ORF together with a deletion of the UL32 ORF (Δ32-96SBAC). This BAC did not produce any plaques or infectious virus when transfected into HF, similar to the phenotype of a UL32 deletion mutant (2, 32). Cells transfected with TowneBAC, ΔUL96BAC, or Δ32-96SBAC at 10 dpt were compared for localization of viral structural proteins MCP, pp65, pp28, pp150, gB, and gMgN with the following results, which are summarized in Table 1. (i) In parental TowneBAC-transfected cells, the expected patterns were observed with tegument proteins (pp28, pp65, pp150), as well as with glycoproteins (gB, gMgN) and MCP. Specifically, pp28, pp150, gB, and gMgN localized to the AC (Fig. 3A to A2, C to C2, D to D2, and E to E2), pp65 showed a predominantly cytoplasmic pattern with little nuclear staining (Fig. 3B to B2), and MCP was detected only in the nucleus (Fig. 3F to F2). (ii) In ΔUL96BAC, a majority of transfected cells showed the expected syncytial phenotype (Fig. 3G to L2). In these cells, pp28 localized to the cytoplasm in a central AC-like region (Fig. 3G to G2), consistent with its function late in virus maturation. pp65, a constituent of virions, as well as dense bodies (21, 44), showed predominantly nuclear staining with less-intense cytoplasmic staining (Fig. 3H to H2), a pattern distinct from that obtained with TowneBAC transfections. pp65 was not detected in nuclei of cells adjacent to those infected with ΔUL96BAC (Fig. 3H to H2), in contrast to TowneBAC (Fig. 3B to B2) or ΔUL32BAC infections (37). Defects in the cytoplasmic accumulation and intercellular translocation of pp65 during UL96 mutation suggest that pUL96 may alter dense-body maturation. pp150 was detected in the nuclei, as well as in the AC (Fig. 3I to I2), of mutant virus-infected cells. The detection of pp150 in the nuclei of UL96 mutant virus-infected cells in a pattern that was distinct from that of parental virus-infected cells raised the possibility that pUL96 may promote pp150 association with maturing nucleocapsids, an event that seems to be critical for capsid stabilization based on the studies using UL32 mutants (37). gB (Fig. 3L to L2) and gMgN (Fig. 3K to K2) were localized in the cytoplasm in an AC-like region, suggesting that the stages of virus maturation where these proteins function are intact. MCP (Fig. 3J to J2) was localized predominantly in the nucleus, although antigen was also detected in the cytoplasm, suggesting instability of nucleocapsids in the cytoplasm, as observed during ΔUL32BAC virus infections (37). (iii) Transfection of Δ32-96SBAC resulted in single eGFP+ cells that contained one to three nuclei at 10 dpt (Fig. 3M to R2), similar to transfection with UL32 mutant viruses (37) and in contrast to the more syncytial cells seen during ΔUL96BAC transfections, suggesting that disruption of pp150 suppresses the syncytium formation observed with the UL96 mutant. Δ32-96SBAC-transfected cells expressed a range of viral antigens. pp28 localized in the cytoplasm in an AC-like location (Fig. 3M to M2), consistent with its function late in virus maturation. pp65 localized predominantly to primary transfected cell nuclei with little staining in the cytoplasm (Fig. 3N to N2), similar to ΔUL96BAC but distinct from TowneBAC. As a control, pp150 failed to be detected in double mutant-infected cells (Fig. 3O to O2). gB and gMgN were localized in the cytoplasm in an AC-like location (Fig. 3P to P2 and Q to Q2), suggesting that the virus maturation functions of these proteins remained intact. MCP was detected in the nucleus, as well as in the cytoplasm (Fig. 3R to R2), a pattern reminiscent of events within ΔUL32BAC-transfected cells (37), indicating that the abnormal MCP localization pattern previously characterized for pp150-deficient viruses (2, 37) was preserved in the double mutant. In summary, viral antigen patterns in cells infected with the double mutant were similar to those of either single mutant, suggesting a replication block at the same point in late stages of infection.
Table 1.
Summary of IFA localization patterns and relative nuclear versus cytoplasmic fluorescence intensities of viral structural antigen staininga
| Antigen | TowneBAC |
ΔUL96BAC |
*ΔUL32BAC |
Δ32-96SBAC |
||||
|---|---|---|---|---|---|---|---|---|
| Nuc | Cyt | Nuc | Cyt | Nuc | Cyt | Nuc | Cyt | |
| pp28 | − | +++ | − | +++ | − | +++ | − | +++ |
| pp65 | + | +++ | +++ | + | + | +++ | +++ | ++ |
| pp150 | − | +++ | ++ | +++ | − | − | − | − |
| gB | − | +++ | − | +++ | − | +++ | − | +++ |
| gMgN | − | +++ | − | +++ | − | +++ | − | +++ |
| MCP | +++ | − | +++ | + | +++ | +++ | +++ | +++ |
Symbols for TowneBAC-, ΔUL96BAC-, ΔUL32BAC-, and Δ32-96SBAC-infected cells: −, not detected; +, weak; ++, medium; +++, bright. Representative IFA images for TowneBAC, ΔUL96BAC, and Δ32-96SBAC infections are shown in Fig. 3, and *ΔUL32BAC results are based on earlier reports (2, 37). Nuc, nuclear; Cyt, cytoplasmic.
Fig. 3.
Localization of the indicated viral antigens in TowneBAC (A to F2), ΔUL96BAC (G to L2), and Δ32-96SBAC (M to R2) bacmid transfections determined by IFA at 10 dpt. Shown are the groups of three panels obtained from the same field that include single-color images of the viral antigens (red, left panels), DNA detected by Hoechst 33258 (blue, middle panels), or a composite (overlay, right) that also includes virally encoded eGFP fluorescence (green). Nuclear (arrows) and cytoplasmic (arrowheads) viral antigens are indicated. Original magnification, ×1,000.
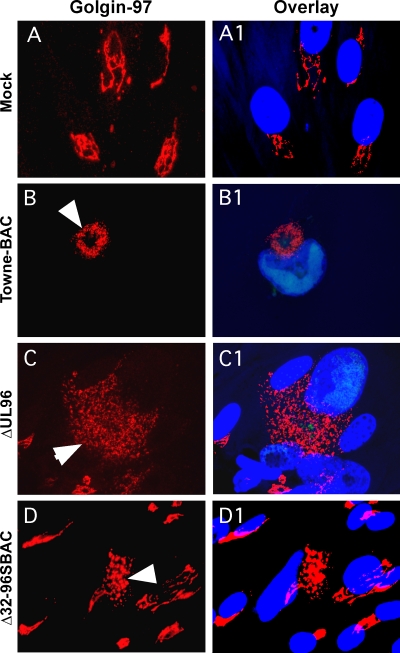
To investigate the organization of the AC following the transfection of ΔUL96BAC or Δ32-96SBAC in comparison to that of the parental virus, we evaluated the TGN marker golgin-97. The TGN was localized as perinuclear stacks of membranes in mock-transfected cells (Fig. 4A and A1) but took the expected ring form (Fig. 4B and B1) in parental virus-infected cells (11, 36). Based on this marker, TGN was fragmented and enlarged, possibly as a result of syncytium formation, but was still positioned in an AC-like location during ΔUL96BAC infection (Fig. 4C and C1). In Δ32-96SBAC-transfected cells (Fig. 4D and D1), the TGN appeared as a compact mass in the vicinity of the nucleus, reminiscent of the UL32 mutant virus (37). These abnormal TGN patterns indicate that the normal AC architecture is disorganized in ΔUL96BAC and that double mutant virus-infected cells show the same phenotype as ΔUL32BAC-infected cells (37). The similarity of compromise in AC structure between double mutant and ΔUL32BAC virus infections is consistent with pUL96 and pp150 impacting the same pathway during virus maturation.
Fig. 4.
Localization patterns of golgin-97 (red) in HF mock transfected (A and A1) or transfected with TowneBAC (B and B1), ΔUL96BAC (C and C1), and Δ32-96SBAC (D and D1) bacmids and fixed at 10 dpt for IFA. Infected cells (arrowheads) were identified by the fluorescence of virally encoded eGFP (not shown). The overlay images (A1, B1, C1, and D1) also include Hoechst 33258 staining, which identifies the nucleus (blue). Original magnification, ×1,000.
pUL96 is important for virus maturation.
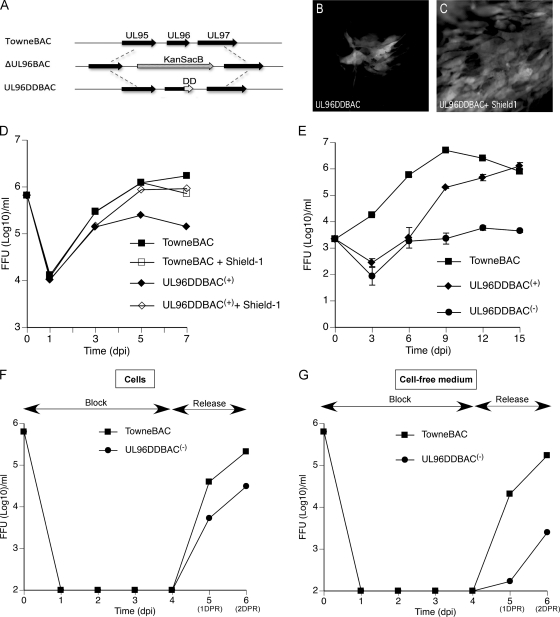
To create a conditional UL96 mutant, an FKBP-DD (3) was fused at the carboxyl terminus (UL96DDBAC) of the protein in the TowneBAC genome, using ΔUL96BAC as an intermediate in BAC recombineering (Fig. 5 A). The addition of this DD was predicted to result in the degradation of the fusion protein (3, 16), resulting in a mutant phenotype. In fact, UL96DDBAC transfections resulted in small syncytial foci (Fig. 5B) similar to those obtained with any of the other UL96 mutants, including ΔUL96BAC, suggesting that the DD had the predicted impact on pUL96 function. Importantly, the phenotype was restored to the parental virus plaque size when 1 μM Shield-1, which was predicted to induce proper protein folding, was present in the medium (Fig. 5C). Thus, the DD-regulated protein functioned in a Shield-1-dependent fashion, as expected (3). This allowed conditional expression, as well the opportunity to produce high-titer UL96DDBAC virus stocks and, subsequently, the evaluation of replication properties under conditions where pUL96 would be expected to be functional or nonfunctional.
Fig. 5.
Characterization of a conditional UL96 mutant virus. (A) Schematic of the genome structure of UL96DDBAC (bottom), constructed from ΔUL96BAC (middle), indicating the location of the FKBP-DD relative to the native locus (top). (B and C) eGFP+ plaque phenotype of UL96DDBAC upon transfection into HF at 10 dpt in the absence (B) or presence (C) of 1 μM Shield-1 in the medium. (D) Single-step growth curves of TowneBAC and UL96DDBAC(+) viruses recovered from cells cultured in the presence or absence of 1 μM Shield-1. HF were infected (MOI of 3.0), total viruses (cells plus medium) were harvested at the indicated times postinfection, and titers were determined on HF monolayers without Shield-1. eGFP+ FFU were counted at 10 dpi. The viral titers represent the averages of triplicate experiments, and standard errors are within the symbols. The data point at 0 dpi indicates the input virus dose. (E) Multiple-step growth curves of TowneBAC, UL96DDBAC(+), and UL96DDBAC(−) viruses. HF were infected (MOI of 0.01), total viruses (cells plus medium) were harvested at the indicated times postinfection, and titers were determined and plotted as described above. (F and G) BDCRB block release experiments were performed as described in Materials and Methods. The titers of total cell-associated (F) and cell-free medium-associated (G) viruses were determined and plotted as described above.
The stock of mutant virus grown in the presence of Shield-1 was termed UL96DDBAC(+) because these conditions should promote stability and the association of pUL96DD with viral particles. In contrast, virus produced in the absence of Shield-1 was termed UL96DDBAC(−). Compared to the parental virus in the absence of Shield-1, UL96DDBAC(+) virus exhibited a 10-fold or greater defect in growth at 5 and 7 dpi but not at earlier times (day 3; Fig. 5D) in a single-step growth analysis. In the presence of 1 μM Shield-1, viruses recovered from UL96DDBAC(+) infections replicated nearly as well as parental TowneBAC (Fig. 5D), suggesting that UL96 function was normalized (3). Importantly, Shield-1 did not have a deleterious effect on the replication of the parental virus (Fig. 5D). Combined, these data indicate that pUL96 is important to sustain viral replication. Multiple-step growth curves (Fig. 5E) revealed that the UL96DDBAC(−) virus was defective and replicated more slowly than both the UL96DDBAC(+) and parental viruses, manifesting an up-to-10,000-fold reduced level of viral progeny (Fig. 5E). The UL96DDBAC(+) virus showed an intermediate phenotype, with an up-to-1,000-fold defect (Fig. 5E), but produced sufficient UL96DDBAC(−) progeny that could be utilized for further experiments. Overall, these experiments revealed conditions where the expected stabilization of pUL96DD benefited virus replication.
To distinguish the role of pUL96 during virus maturation and release as opposed to early events in replication, HF were infected with UL96DDBAC(−) or the parental virus at an MOI of 3.0 and incubated for 4 days in the presence of the encapsidation inhibitor BDCRB (20 μM) to synchronize the infection. The BDCRB block was then released, and cells were incubated further for 2 days. Analysis of cell-associated (Fig. 5F) and released virus infectivity (Fig. 5G) revealed a lag in UL96DDBAC(−) virus replication compared to that obtained with TowneBAC. In summary, these results demonstrate that pUL96 controls a virus maturation step involving efficient accumulation of cell-associated virus preceding release.
Localization of viral antigens in conditional pUL96 mutant virus-infected cells.
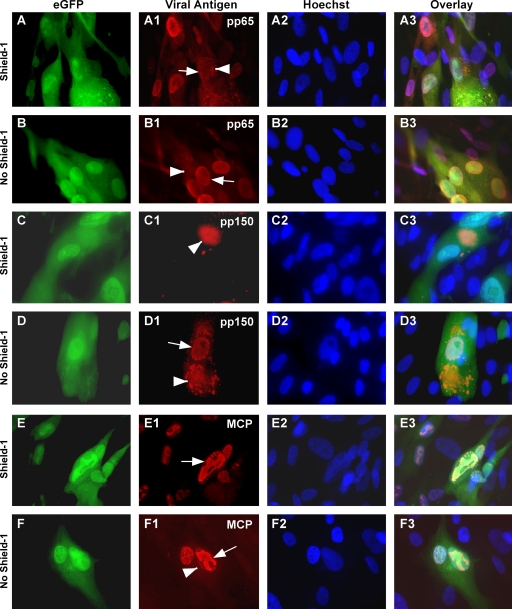
To study the stage of virus replication affected by UL96, localization of selected virus antigens was determined in UL96DDBAC(−)-infected cells. In the presence of Shield-1, when pUL96 should be stable, pp65 localized to the cytoplasm, as well as to the nucleus (Fig. 6A to A3) at 5 dpi, a phenotype comparable to, but distinct from, that obtained in TowneBAC infections, probably due to the expected absence of delivered pUL96 in UL96DDBAC(−) infections. In the absence of Shield-1, when pUL96 should be unstable, the localization appeared predominantly nuclear with little diffused pp65 in the cytoplasm, similar to that obtained with other UL96 mutants (Fig. 6B to B3). Thus, pp65 localization indicated a defect in its nuclear-to-cytoplasmic translocation during infection with the UL96DDBAC(−) virus. pp150 localized exclusively to the cytoplasm in the presence of Shield-1 (Fig. 6C to C3) but, like other UL96 mutants, was detected in the nucleus as well as in the cytoplasm in its absence (Fig. 6D to D3), indicating defects in translocation. MCP localized exclusively to the nucleus in the presence (Fig. 6E to E3), but not in the absence of Shield-1, where minor levels of MCP were also detected in the cytoplasm (Fig. 6F to F3), suggesting capsid instability. In summary, the phenotype of UL96DDBAC(−) virus resembled that of other UL96 mutants under conditions where pUL96 should be unstable but resembled that of TowneBAC when pUL96 should be stable.
Fig. 6.
Localization of pp65 (A1, A3, B1, and B3), pp150 (C1, C3, D1, and D3), or MCP (E1, E3, F1, and F3) in UL96DDBAC(−) virus infections in the presence or absence of 1 μM Shield-1 at 5 dpi determined by IFA. Shown are the groups of four horizontal panels obtained from the same field, including single-color images of the virally encoded eGFP (green, panels A, B, C, D, E, and F), viral antigens (red, panels A1, B1, C1, D1, E1, and F1), DNA detected by Hoechst 33258 (blue, panels A2, B2, C2, D2, E2, and F2), and a composite (overlay, panels A3, B3, C3, D3, E3, and F3). Nuclear (arrows) and cytoplasmic (arrowheads) viral antigens are indicated. Original magnification, ×1,000.
pUL96 is critical for the stability of nucleocapsids in the cytoplasm.
To further investigate UL96 mutant virus maturation defects, as well as to examine whether these defects are reversed when pUL96DD should be stable and thus incorporated in virions, electron microscopy of the following samples was performed: (i) HF infected with UL96DDBAC(−) virus at an MOI of 0.01 in the absence of Shield-1 (This infection should lack both virion pUL96 and de novo synthesis of pUL96, conditions considered most similar to a UL96 deletion virus.), (ii) HF infected with UL96DDBAC(+) virus at an MOI of 3.0 in the absence of Shield-1 (This infection should deliver virion pUL96 but not support a contribution of newly synthesized pUL96. TEM of these samples would assess any advantages of the expected delivery of virion pUL96 to virus maturation.), (iii) HF infected with UL96DDBAC(+) virus at an MOI of 3.0 in the presence of 1 μM Shield-1 (This infection should have virion pUL96, as well as newly synthesized pUL96; therefore, this condition should be similar in phenotype to that seen with TowneBAC virus.), (iv) HF infected with TowneBAC virus at an MOI of 3.0 in the absence of Shield-1 (as a positive control), and (v) HF infected with TowneBAC virus at an MOI of 3.0 in the presence of 1 μM Shield-1 (to control for any nonspecific impact of Shield-1 on the infected cell phenotype).
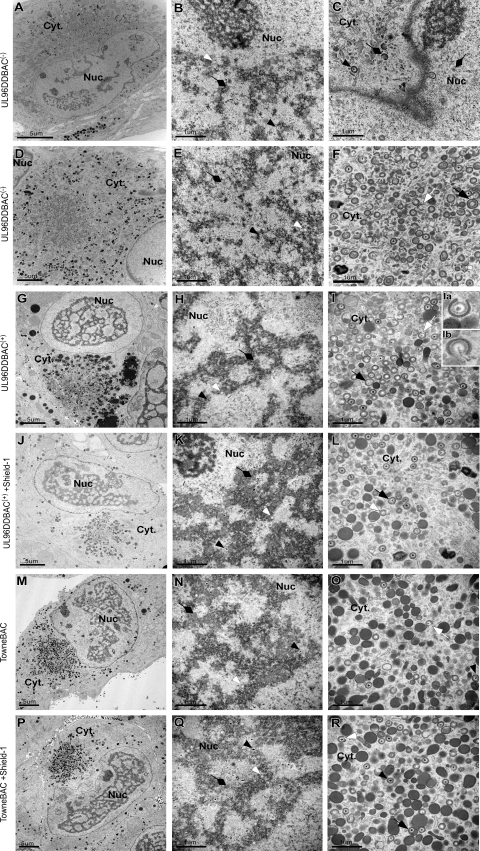
As expected, UL96DDBAC(−) virus (MOI of 0.01) infections resulted in multinucleate and mononucleate infected cells, both having a rudimentary AC visible at 4 dpi upon examination by TEM (Fig. 7A and D), as suggested earlier by IFA. All three types of capsid (A, lacking a scaffold or DNA core; B, containing a scaffold; C, containing a DNA core) were observed in the nucleus (Fig. 7B and E). The primary envelopment process at the inner nuclear membrane appeared intact (Fig. 7C). The AC contained numerous virus particles, although more than 95% exhibited irregular morphologies such as large size (>10%) and partial envelopment (>20%) (Fig. 7F). Most of these particles lacked an obvious electron-dense DNA core (Fig. 7F). Dense bodies were not detected in the cytoplasm of these cells (Fig. 7C and F), consistent with the IFA for pp65. Thus, nuclear stages of virus replication remained intact, while virus maturation in the cytoplasm was severely compromised in UL96DDBAC(−) virus infections, a phenotype comparable to that obtained with ΔUL32BAC (37). In the absence of Shield-1, UL96DDBAC(+) virus-infected cells (MOI = 3.0) remained mononucleate (Fig. 7G) and the nuclei contained all of the capsid types indistinguishable from those seen in Shield-1-treated mutant virus or parental virus infections (Fig. 7H, K, N, and Q). However, many (>20%) virus particles in the AC-like region appeared partially enveloped (Fig. 7I, inset Ia) and some of these (2 to 5%) appeared to be losing viral DNA (Fig. 7I, inset Ib), characteristics we interpreted as consistent with virus particle instability. Dense bodies were detected in the cytoplasm in these infections. Thus, the defects observed during UL96DDBAC(−) infection were only partially reversed when UL96DDBAC(+) was used, although these data showed that expected delivered pUL96 or residual activity of the pUL96DD protein in UL96DDBAC(+) contributed to maturation efficiency. The phenotypes of UL96DDBAC(+) plus Shield-1, TowneBAC, and TowneBAC plus Shield-1 infections were indistinguishable, with similar numbers and morphologies when nuclear or cytoplasmic virus particles were compared (Fig. 7J to R). In summary, these results show that pUL96 provides a critical function supporting the accumulation of mature nucleocapsids in the cytoplasm. The genetic evidence suggests that pUL96 confers stability on capsids as they translocate from the nucleus to the cytoplasm.
Fig. 7.
TEM of UL96DDBAC(−), UL96DDBAC(+), or TowneBAC virus-infected HF. Cells were infected with UL96DDBAC(−) virus (MOI of 0.01) and fixed for TEM at 4 dpi (A to F). Representative images of mononucleate cells (A to C) and multinucleate cells (D to F) are shown. Cells infected with UL96DDBAC(+) (G to L) or TowneBAC (M to R) virus at an MOI of 3.0 in the absence (G to I, M to O) or presence (J to L, P to R) of 1 μM Shield-1 and fixed for TEM at 5 dpi are shown. Particles appearing to undergo partial envelopment (Ia) or loss of genomic DNA (Ib) are magnified and shown in the insets. Nuc, nucleus; Cyt, cytoplasm; white arrowheads, A capsids; black arrowheads, C capsids; diamondheads, B capsids; white arrows, partially enveloped capsids; black arrows, morphologically abnormal capsids; bars, 5 μm (A, D, G, J, M, and P) and 1 μm (B, C, E, F, H, I, K, L, N, O, Q, and R).
DISCUSSION
This study demonstrated that pUL96, a betaherpesvirus-specific protein, (i) is critical for viral replication and has a functional C-terminal domain, (ii) is expressed as an early protein that accumulates along with pp150 in the AC, (iii) is very important for the accumulation of virions and dense bodies in the AC, (iv) plays a critical role late in virus replication during virus maturation, (v) acts at a stage in virus maturation similar to that of pp150, and (vi) like pp150, stabilizes virus particles during their translocation from the nucleus to the cytoplasm. pUL96 is present in ECV particles but not in nuclear B capsids; however, pp150 can be detected in both of these particle types. Thus, pUL96 is most likely to be added to virus particles later in virus maturation than pp150. Importantly, UL96, UL32, and UL96/UL32 dual mutant viruses all exhibit similar replication defects based on viral and cellular antigen localization, indicating that these two gene products act in a concerted fashion within the maturation pathway.
Virus maturation involves specific steps controlled by viral proteins. In this study, we show that preformed capsids in the nucleus acquire pp150 before DNA encapsidation, as suggested by earlier immunofluorescence localization studies (30). After this initial tegumentation, nucleocapsids egress from the nucleus following disruption of the nuclear lamina mediated by the CMV UL50 and UL53 proteins (7, 8, 26, 29). Several tegument proteins are added in the cytoplasm prior to the envelopment and cellular egress of virus particles. pp150 is an essential capsid-proximal tegument protein that stabilizes nucleocapsids during nucleus-to-cytoplasm translocation. pUL96 appears to play a very important role in augmenting this stabilization process. Like many of the maturation functions that have been characterized, UL96 mutants are not completely replication defective. For example, virus maturation is influenced by the viral protein kinase pUL97, a component of viral tegument that influences nuclear egress (23), organization of the AC, and tegumentation of virus particles (17), but UL97 null mutants replicate, albeit poorly. Virus envelopment is regulated by pp28, pUL35, and pUL71 in a similar augmenting rather than essential manner (32–34, 46). Finally, the exocytosis efficiency of enveloped virions and dense bodies is influenced by one other tegument protein, pUL103 (1), possibly acting together with cellular ESCRT machinery (36). Thus, pUL96, although not essential, is required for efficient virus replication.
We have shown that (i) pp150 is added to nuclear capsids prior to the packaging of viral DNA, although its primary role is at the stage when nucleocapsids translocate to the cytoplasm (2, 37) and (ii) pUL96 and pp150 have interdependent functions in virus maturation. Based on the evidence that UL96 mutants show a defect at the cytoplasmic stage of virus maturation, this protein must be active as nucleocapsids translocate to the cytoplasm to prevent their disintegration. We found that pUL96 is made starting early during infection, which sparked investigation of pUL96 function during the early phases of virus replication. Similar numbers of primary infected HF expressed IE1 and IE2 antigens at early times postinfection when infected with an equivalent MOI of TowneBAC or UL96DDBAC(−) virus (data not shown), suggesting that virus entry was intact. Also, an early replication defect was ruled out because mutant virus-infected cells progressed to late stages of infection based on viral antigen localization and TEM. pUL96 is sufficiently small (14 kDa) to diffuse freely (25) between the nucleus and the cytoplasm and therefore could function in either compartment. This protein accumulates within cytoplasmic AC along with maturing virus and is most likely to be added during nucleocapsid translocation or in the AC because it is associated with virus particles that are released from cells. Given the well-established properties of pp150 binding tightly to nucleocapsids (10, 15, 41, 49) and localization to nuclear replication compartments in close proximity to nucleocapsids during early infection (30), an accumulation of pp150 in the nucleus during UL96 mutant virus infection suggested the involvement of pUL96 in the translocation of pp150-bound nucleocapsids out of the nucleus.
To study the role of pUL96 in virus maturation, we devised a conditional UL96 knockdown construct by using an FKBP-DD fusion strategy. As is the case for other HCMV maturation functions, a UL96-expressing cell line has proven to be technically challenging. TEM studies of UL96DDBAC(−)- and UL96DDBAC(+)-infected cells revealed that nucleocapsids translocated from the nucleus to the cytoplasm, although they exhibited a compromised morphology and altered size (Fig. 7C, F, and I). The preponderance of aberrant virus particles suggested a high particle-to-PFU ratio in UL96 mutants. The irregular morphology of pUL96 mutant capsids was less severe than that reported for the pp150 mutant (37), and the maturation block in the UL96/UL32 dual mu- tant virus was similar to that of the UL32 mutant, based on antigen localization. This, along with the evidence that pp150 associates with nuclear capsids before DNA encapsidation, indicated that the essential UL32 function is upstream of the UL96 function. Given the low levels of pUL96 detected on semipure virus particles by mass spectrometry (44), particularly in relation to the abundant levels of pp150 found (21, 44) (Fig. 2D), we believe that this protein is likely to confer capsid stability by influencing pp150 incorporation or orientation on capsids. It is unlikely that these two proteins form a stable complex due to the very low levels of pUL96 associated with virus particles. Additional biophysical and biochemical investigations are under way to address this issue more directly.
Based on this study, pUL96 functions during virus maturation and not during early phases of infection. In contrast, HSV-1 UL14, a potential homolog, is important for efficient nuclear targeting of incoming capsids (47). Thus, UL14 and UL96 do not appear to act at similar replication stages. Lack of dense bodies in the cytoplasm, compromised cell-to-cell spread of pp65, and a predominant nuclear accumulation of pp65 during UL96 mutant infections indicate that this protein also influences the maturation and release of dense bodies. Thus, pUL96 contributes to the stability of nucleocapsids together with pp150 to ensure efficient translocation from the nucleus to the cytoplasmic AC, while also influencing the maturation of capsidless dense bodies.
ACKNOWLEDGMENTS
We are thankful to Hong Yi at the Robert P. Apkarian Integrated Electron Microscopy Core at Emory University for help with electron microscopy. Confocal microscopy was performed at the Cell Imaging and Microscopy Core at Emory Winship Cancer Institute. We acknowledge Louise McCormick, Jenny Ahlqvist, and Wade Gibson for critical reading of the manuscript and Mocarski lab members for useful discussions during this project. Tom Wandless, Stanford University, generously provided Shield-1 reagent and destabilization domain constructs. BDCRB was kindly provided by John Drach, University of Michigan. William J. Kaiser and Linda J. Roback helped with the cell culture in the Mocarski lab.
Public Health Service grant RO1 AI20211 and the Georgia Cancer Coalition funded this research.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Ahlqvist J., Mocarski E. 2011. Cytomegalovirus UL103 controls virion and dense body egress. J. Virol. 85:5125–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. AuCoin D. P., Smith G. B., Meiering C. D., Mocarski E. S. 2006. Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J. Virol. 80:8199–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Banaszynski L. A., Chen L. C., Maynard-Smith L. A., Ooi A. G., Wandless T. J. 2006. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126:995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baxter M. K., Gibson W. 2001. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J. Virol. 75:6865–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Britt W. 2007. CMV maturation and egress, p. 311–323In Arvin A. M., Mocarski E. S., Moore P., Whitley R., Yamanishi K., Campadelli-Fiume G., Roizman B. (ed.), Human herpesviruses: biology, therapy and immunoprophylaxis. Cambridge Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 6. Britt W. J., Auger D. 1985. Identification of a 65 000 dalton virion envelope protein of human cytomegalovirus. Virus Res. 4:31–36 [DOI] [PubMed] [Google Scholar]
- 7. Buser C., Walther P., Mertens T., Michel D. 2007. Cytomegalovirus primary envelopment occurs at large infoldings of the inner nuclear membrane. J. Virol. 81:3042–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Camozzi D., et al. 2008. Remodelling of the nuclear lamina during human cytomegalovirus infection: role of the viral proteins pUL50 and pUL53. J. Gen. Virol. 89:731–740 [DOI] [PubMed] [Google Scholar]
- 9. Chee M., Rudolph S. A., Plachter B., Barrell B., Jahn G. 1989. Identification of the major capsid protein gene of human cytomegalovirus. J. Virol. 63:1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen D. H., Jiang H., Lee M., Liu F., Zhou Z. H. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10–16 [DOI] [PubMed] [Google Scholar]
- 11. Das S., Vasanji A., Pellett P. E. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol. 81:11861–11869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dunn W., et al. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223–14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gibson W. 1983. Protein counterparts of human and simian cytomegaloviruses. Virology 128:391–406 [DOI] [PubMed] [Google Scholar]
- 14. Gibson W. 1981. Structural and nonstructural proteins of strain Colburn cytomegalovirus. Virology 111:516–537 [DOI] [PubMed] [Google Scholar]
- 15. Gibson W. 1996. Structure and assembly of the virion. Intervirology 39:389–400 [DOI] [PubMed] [Google Scholar]
- 16. Glass M., Busche A., Wagner K., Messerle M., Borst E. M. 2009. Conditional and reversible disruption of essential herpesvirus proteins. Nat. Methods 6:577–579 [DOI] [PubMed] [Google Scholar]
- 17. Goldberg M. D., et al. 2011. Human cytomegalovirus UL97 kinase and non-kinase functions mediate viral cytoplasmic secondary envelopment. J. Virol. 85:3375–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hensel G., Meyer H., Gartner S., Brand G., Kern H. F. 1995. Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32). J. Gen. Virol. 76(Pt. 7):1591–1601 [DOI] [PubMed] [Google Scholar]
- 19. Homman-Loudiyi M., Hultenby K., Britt W., Soderberg-Naucler C. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-Golgi network 46, and mannosidase II. J. Virol. 77:3191–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Indran S. V., Ballestas M. E., Britt W. J. 2010. Bicaudal D1-dependent trafficking of human cytomegalovirus tegument protein pp150 in virus-infected cells. J. Virol. 84:3162–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Irmiere A., Gibson W. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118–133 [DOI] [PubMed] [Google Scholar]
- 22. Irmiere A., Gibson W. 1985. Isolation of human cytomegalovirus intranuclear capsids, characterization of their protein constituents, and demonstration that the B-capsid assembly protein is also abundant in noninfectious enveloped particles. J. Virol. 56:277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krosky P. M., Baek M. C., Coen D. M. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77:905–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krzyzaniak M. A., Mach M., Britt W. J. 2009. HCMV-encoded glycoprotein M (UL100) interacts with Rab11 effector protein FIP4. Traffic 10:1439–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Macara I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Milbradt J., Webel R., Auerochs S., Sticht H., Marschall M. 2010. Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus. J. Biol. Chem. 285:13979–13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mocarski E. S. 2007. Betaherpesvirus genes and their functions, p. 202–228In Arvin A. M., Mocarski E. S., Moore P., Whitley R., Yamanishi K., Campadelli-Fiume G., Roizman B. (ed.), Human herpesviruses: biology, therapy and immunoprophylaxis. Cambridge Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 28. Mocarski E. S., Jr., Shenk T., Pass R. F. 2006. Cytomegaloviruses, p. 2701–2772In Knipe D. M., Howley P. M. (ed.), Fields virology, 5th edition Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 29. Muranyi W., Haas J., Wagner M., Krohne G., Koszinowski U. H. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854–857 [DOI] [PubMed] [Google Scholar]
- 30. Sampaio K. L., Cavignac Y., Stierhof Y. D., Sinzger C. 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J. Virol. 79:2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sanchez V., Greis K. D., Sztul E., Britt W. J. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schauflinger M., et al. 2011. The tegument protein UL71 of human cytomegalovirus is involved in late envelopment and affects multivesicular bodies. J. Virol. 85:3821–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schierling K., Buser C., Mertens T., Winkler M. 2005. Human cytomegalovirus tegument protein ppUL35 is important for viral replication and particle formation. J. Virol. 79:3084–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seo J. Y., Britt W. J. 2007. Cytoplasmic envelopment of human cytomegalovirus requires the postlocalization function of tegument protein pp28 within the assembly compartment. J. Virol. 81:6536–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Severi B., Landini M. P., Govoni E. 1988. Human cytomegalovirus morphogenesis: an ultrastructural study of the late cytoplasmic phases. Arch. Virol. 98:51–64 [DOI] [PubMed] [Google Scholar]
- 36. Tandon R., AuCoin D. P., Mocarski E. S. 2009. Human cytomegalovirus exploits ESCRT machinery in the process of virion maturation. J. Virol. 83:10797–10807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tandon R., Mocarski E. S. 2008. Control of cytoplasmic maturation events by cytomegalovirus tegument protein pp150. J. Virol. 82:9433–9444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thomason L. C., et al. 2007. Recombineering: genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. Chapter 1:Unit 1.16 [DOI] [PubMed] [Google Scholar]
- 39. Thomsen D. R., Newcomb W. W., Brown J. C., Homa F. L. 1995. Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J. Virol. 69:3690–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tooze J., Hollinshead M., Reis B., Radsak K., Kern H. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163–178 [PubMed] [Google Scholar]
- 41. Trus B. L., Gibson W., Cheng N., Steven A. C. 1999. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J. Virol. 73:2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Underwood M. R., et al. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Upton J. W., Kaiser W. J., Mocarski E. S. 2010. Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7:302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Varnum S. M., et al. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wing B. A., Huang E. S. 1995. Analysis and mapping of a family of 3′-coterminal transcripts containing coding sequences for human cytomegalovirus open reading frames UL93 through UL99. J. Virol. 69:1521–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Womack A., Shenk T. 30 November 2010, posting date Human cytomegalovirus tegument protein pUL71 is required for efficient virion egress. MBio 1(5):e00282–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamauchi Y., et al. 2008. The UL14 tegument protein of herpes simplex virus type 1 is required for efficient nuclear transport of the alpha transinducing factor VP16 and viral capsids. J. Virol. 82:1094–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu D., Silva M. C., Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396–12401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu X., et al. 2011. Biochemical and structural characterization of the capsid-bound tegument proteins of human cytomegalovirus. J. Struct. Biol. 174:451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]