Abstract
After fusing with the plasma membrane, enveloped poxvirus virions form actin-filled membranous protrusions, called tails, beneath themselves and move toward adjacent uninfected cells. While much is known about the host and viral proteins that mediate formation of actin tails, much less is known about the factors controlling release. We found that the phosphoinositide 5-phosphatase SHIP2 localizes to actin tails. Localization requires phosphotyrosine, Abl and Src family tyrosine kinases, and neural Wiskott-Aldrich syndrome protein (N-WASP) but not the Arp2/Arp3 complex or actin. Cells lacking SHIP2 have normal actin tails but release more virus. Moreover, cells infected with viral strains with mutations in the release inhibitor A34 release more virus but recruit less SHIP2 to tails. Thus, the inhibitory effects of A34 on virus release are mediated by SHIP2. Together, these data suggest that SHIP2 and A34 may act as gatekeepers to regulate dissemination of poxviruses when environmental conditions are conducive.
INTRODUCTION
Orthopoxviruses, including vaccinia virus (VACV), monkeypox virus (MPXV), and variola virus (VARV), are large double-stranded DNA (dsDNA) viruses that cause characteristic umbilicated vesiculopustular skin lesions (pox) (12). VARV is the causative agent of smallpox, and VACV is used for vaccination against smallpox (12). Although smallpox has been eradicated, naturally occurring poxviruses are still of concern to humans. In particular, MPXV is endemic in Africa (57) and has the potential for spread to humans from bushmeat and squirrels (28, 29, 53, 57, 58), and recent outbreaks in the Democratic Republic of Congo have raised the possibility of human-to-human transmission (58). Efforts to understand the capacity for human-to-human transmission among poxviruses have focused on how the virus spreads from cell to cell.
Infection by poxviruses is initiated upon entry of either of two different forms of the virus. The first, called the intracellular mature virus (IMV; also called mature virion [MV]), consists of a viral core surrounded by one or two lipid bilayers derived from an endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC) (21, 59, 61, 71). A second infectious form of the virus, called the extracellular enveloped virus (EEV; also called enveloped virus [EV]) (67), consists of an IMV enveloped in additional membranes derived from the host cell. IMV is released following lysis of host cells (60), whereas the precursors of EEV traffic along microtubules to the cell periphery (15, 20, 56, 75). Upon fusion with the plasma membrane, the doubly enveloped virion stimulates formation of actin-filled membranous protrusions called tails and then disengages from the host cell (67).
Formation of actin tails occurs by a mechanism conserved among VACV, MPXV, and VARV (55). EEV recruits host Abl and Src family tyrosine kinases (39, 40, 54), which phosphorylate viral protein A36 at residues 112 and 132 (40), thereby facilitating recruitment of Nck, Grb2, Wiskott-Aldrich syndrome protein (WASP)-interacting protein (WIP), and neural WASP (N-WASP) (13, 14, 37, 63, 76). Interactions with phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] in the plasma membrane induce conformational changes in N-WASP that allow the protein to bind to and activate Arp2/Arp3 (Arp2/3) complex, a nucleator for actin polymerization (37, 62). The rate of actin-mediated propulsion and actin tail length appear to be a function of the turnover rate and interactions among viral factors and recruited host proteins (8, 76).
While extensive information is available about the viral and host factors that initiate actin polymerization, much less is known about the factors that contribute to virion release. Based on mutation experiments, several viral factors (including F12, F13, A33, A34, B5, and A36) have been implicated in viral release (67) although in many cases, such mutations also affect actin tail formation or specific infectivity, thereby precluding unequivocal determination of the role these proteins play in release. That virus release also depends on cell type (36, 44) indicates that host factors also participate. Reeves et al. separated actin motility from release by demonstrating that redundant Src and Abl family tyrosine kinases mediate tail formation, whereas only Abl family kinases mediate release (54).
Previous work from our lab and from others has implicated phosphatidylinositol-3 (PI3)-kinase activities at several distinct steps of viral maturation though not in formation of actin tails or in release (35, 70, 81). Nevertheless, the observation that host proteins involved with vesicular trafficking, such as Alix, Tsg101 and Eps15, also affect poxviral spread (22) suggests that lipid signaling may also regulate viral dissemination. In this regard, we considered the possibility that other lipid and phosphoinositide (PI) signaling molecules, including lipid phosphatases, might also participate in virion release.
SHIP2 and its related isoform SHIP1 are SH2 domain-containing inositol polyphosphate 5-phosphatases (7, 27, 31, 32, 47). Whereas SHIP1 is expressed in hematopoietic cells, SHIP2 is expressed ubiquitously (18, 65). Both isoforms exhibit PI 5-phosphatase activity with phosphatidylinositol-3,4,5-trisphosphate [PI(3,4,5)P3] and PI(4,5)P2 as substrates (5, 16, 18, 38, 49, 72, 77). SHIP2 was originally identified as a negative regulator of insulin signaling (6), and mutations in SHIP2 have also been linked to metabolic disorders, including type II diabetes (23–25, 33, 66). In addition, SHIP2 has also been implicated in regulating cytoskeletal organization and endocytosis (38, 51, 68, 80). Smith et al. recently demonstrated that SHIP2 localizes to actin protrusions, called pedestals, which form beneath enteropathogenic Escherichia coli (EPEC) cells, and that reduction of SHIP2 levels causes an aberrant pedestal structure (68).
Here, we demonstrate that SHIP2 localizes beneath VACV during actin tail formation in a manner that depends on both tyrosine kinases and N-WASP but not actin. We also show that SHIP2 negatively regulates release of virions and may act as a gatekeeper molecule that limits poxvirus dissemination.
MATERIALS AND METHODS
Cells, viruses, and reagents.
BSC40, 3T3, Abl1−/− Abl2−/−, N-WASPflox/flox, and N-WASP−/− cells (ATCC) were grown in Dulbecco's modified Eagle's medium ([DMEM] Cellgro, MediaTech, Inc., Manassas, VA) supplemented with 10% fetal bovine serum ([FBS] Atlanta Biologicals, Norcross, GA) and 10 IU/ml penicillin and 10 μg/ml streptomycin ([P-S] Cellgro, MediaTech, Inc., Manassas, VA). Src−/− Fyn−/− Yes−/− cells were grown in DMEM (Gibco, Carlsbad, CA) supplemented with 10% FBS and P-S. SHIP2+/+ and SHIP2−/− cells were grown in DMEM (Gibco, Carlsbad, CA) supplemented with 10% FBS, P-S, and 1% of 50 mM β-mercaptoethanol in Dulbecco's phosphate-buffered saline (DPBS). All cells were grown at 37°C in a 5% CO2 incubator. Mouse embryonic fibroblasts were isolated from embryos that were homozygous for a knockout of the SHIP2 (inositol polyphosphate phosphatase-like protein 1 [INPPL1]) gene (6) and were prepared as reported in Zhang et al. (82). Viral strains were grown and propagated as previously described (54). Viral strain WI was provided by Bernard Moss and consisted of the WR strain containing A34 derived from strain IHD-J (3). VACV strain IHD-J expressing luciferase was made by Reeves et al. and consisted of the firefly luciferase gene under the control of a synthetic early/late promoter (55). Titers of all viral strains were determined on BSC40 cells.
Microscopy.
Various cell lines used for microscopy were plated onto poly-d-lysine (PDL)-collagen-coated glass coverslips and infected with ∼106 PFU of WR or virus expressing an F13-green fluorescent protein (F13-GFP) fusion for 16 h. After 16 h, cells were fixed as previously described (54) and processed for microscopy. To detect DNA, cells were stained with 17 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI). Actin was visualized using phalloidin-564 (Molecular Probes/Invitrogen, Carlsbad, CA). The following antibodies for microscopy were used: anti-SHIP1 (Cell Signaling Technology, Beverly, MA), anti-SHIP2 (anti-INPPL1; Novus Biologicals, Littleton, CO), anti-lamellipodium (LPD) (anti-RAPH; Sigma, St. Louis, MO), and anti-SHC (BD Transduction Labs) were used at a 1:50 dilution; anti-Xpress (Invitrogen, Carlsbad, CA) was used at 1:200; and anti-phosphotyrosine (PY) (4G10; Millipore, Billerica, MA) and anti-Myc (9E10; Millipore, Billerica, MA) were used at 1:250. Anti-B5 was a gift from Jay Hooper, USAMRIID, and was isolated from rabbits vaccinated with a B5 DNA vaccine. Anti-B5 was added to cells after permeabilization and used at a 1:5,000 dilution. Tyrosine kinase inhibitors, imatinib mesylate (Gleevec) or dasatinib (Sprycel), were added to cells at 10 μM for 30 min after a 15.5-h infection. For transfections, cells were transfected with various SHIP2 constructs (49, 82) or Myc-SHIP1 using FuGene 6 (Roche, Indianapolis, IN) for 48 h. We did not observe a difference in the numbers of actin tails formed on infected transfected cells.
Comet assays.
Experiments with vaccinia virus were conducted under biosafety level 2 (BSL-2) conditions. One hundred or 10 PFU of vaccinia virus strain WR, IHD-J, vRB12 (WR ΔF13), or WI was diluted in 500 μl of 2% FBS-DMEM and added to monolayers of naïve BSC40, SHIP2+/+, or SHIP2−/− cells in six-well dishes. Virus was allowed to adsorb to and enter the cells for 1 h at 37°C in 5% CO2. Unbound virus was then removed by washing monolayers with 1 ml of PBS. Medium was then replaced with 10% FBS-DMEM. Two days after infection, monolayers were fixed and stained with crystal violet solution (0.1% crystal violet and 20% ethanol).
EEV and CAV measurements.
To quantify the amount of EEV and cell-associated virus (CAV) produced, SHIP2+/+ and SHIP2−/− cells were grown in six-well dishes in triplicate wells. Cell numbers were quantified, and cells were infected at a multiplicity of infection (MOI) of 5 or 0.01 with strain IHD-J. Virus was diluted in 500 μl of 2% FBS in DMEM and allowed to adsorb to cells for 1 h. Unbound virus was then removed by washing cells three times with PBS and then adding 1.5 ml of 10% FBS in DMEM. To quantify EEV, supernatant was removed 24 or 48 h later and spun at 400 × g for 10 min to remove cells. IMV was neutralized with 1:1,000 10F5 (anti-L1) antibody (a gift from Jay Hooper, USAMRIID) for 1 h at 37°C; the supernatant was then diluted and added to naïve BSC40 cell monolayers, and plaques were enumerated after 2 days. To quantify CAV, monolayers were scraped into 1 ml of 2% FBS in DMEM, and virus was released through three freeze-thaw cycles. CAV was diluted and added to naïve BSC40 monolayers.
RNA interference (RNAi).
To knock down SHIP2 protein, BSC40 cells in duplicate six-well plates were transfected with 50 nM RNA complementary to SHIP2 (INPPL1 On-Targetplus small interfering RNA [siRNA] human sequences 1, 2, 3, and 4; Dharmacon) using RNAiMAX Lipofectamine reagent (Invitrogen, Carlsbad, CA). After 3 days cells were infected with 100 PFU of VACV strain IHD-J for 48 h. After this time plaques were fixed and stained with crystal violet. To confirm protein knockdown, BSC40 cells in triplicate six-well plates were transfected with 50 nM RNA complementary to SHIP2 for 3 days, and protein knockdown was confirmed by Western blotting with anti-SHIP2 (anti- INPPL1 at 1:500; Novus Biologicals, Littleton, CO) and anti-glyceraldehyde-3-phosphate dehydrogenase ([GAPDH] 1:1,000; Sigma-Aldrich, St. Louis, MO).
Luciferase assay.
To quantify viral entry, SHIP2+/+ and SHIP2−/− cells were grown in six-well dishes in triplicate wells. Cell numbers were quantified, and cells were infected at a multiplicity of infection of 5 with strain IHD-J expressing luciferase (55). Virus was diluted in 500 μl of cold 2% FBS in DMEM and allowed to adsorb to cells for 1 h at 4°C. Unbound virus was then removed by washing cells three times with cold PBS and adding 1.5 ml of prewarmed 10% FBS in DMEM. After a 2-h infection cells were lysed, and protein concentration was determined using a Bio-Rad DC (detergent-compatible) Protein Assay kit (Bio-Rad, Hercules, CA). Luciferase expression was measured using 20 μg of protein with a Bright-Glo luciferase assay system (Promega, Madison, WI) and a Synergy HT BioTek plate reader.
Specific infectivity.
Specific infectivity was calculated based on a method using the optical density at 260 nm (OD260) as described by Parrish and Moss (42). Briefly, SHIP2+/+ and SHIP2−/− cells were grown in three 150-cm2 tissue culture dishes and infected with ∼108 PFU/ml IHD-J for 3 days. To liberate virus, flasks were subjected to three freeze-thaw cycles. Medium and cell debris were purified from medium by spinning samples at 10,000 rpm in a JA-10 rotor for 1.25 h. Cell debris and virus were resuspended in 2 ml of 2% FBS in DMEM. To remove cell debris, samples were spun at 400 × g for 10 min. Virus was further purified through a 36% sucrose cushion, followed by two, 25 to 40% sucrose gradients. Viral particles were quantified by the OD260, where 1 OD260 unit is equivalent to 1.2 × 1010 particles. To calculate the number of infectious particles, purified virus was diluted in 2% FBS in DMEM and added to naïve monolayers, and the plaques were enumerated 2 days later. Specific infectivity was calculated as 136.3 ± 46.5 particles/PFU for SHIP2+/+ cells and 80.0 ± 18.0 particles/PFU for SHIP2−/− cells.
Western analysis.
BSC40 or CRL2019 cells were grown in 10-cm tissue culture dishes. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (Cell Signaling, Beverly, MA), and protein concentration was determined using a Bio-Rad DC Protein Assay kit (Bio-Rad, Hercules, CA). One hundred micrograms of protein was separated by SDS-PAGE; proteins were then transferred to nitrocellulose membranes. Those were blocked with 3% milk in Tris-buffered saline containing 0.05% Tween 20 (TBST) for 1 h. Membranes were probed with anti-SHIP1 (1:1,000), anti-SHIP2 (1:500), or anti-tubulin (1:5,000; Abcam, Cambridge, MA) in blocking solution for an additional hour. Bands were detected using anti-mouse or anti-rabbit antibodies conjugated to horseradish peroxidase ([HRP] GE Healthcare, United Kingdom), and blots were developed using Pierce ECL Western Blotting Substrate (Thermo Scientific, Waltham, MA).
RESULTS
The inositol phosphatase SHIP2 localizes to VACV actin tails.
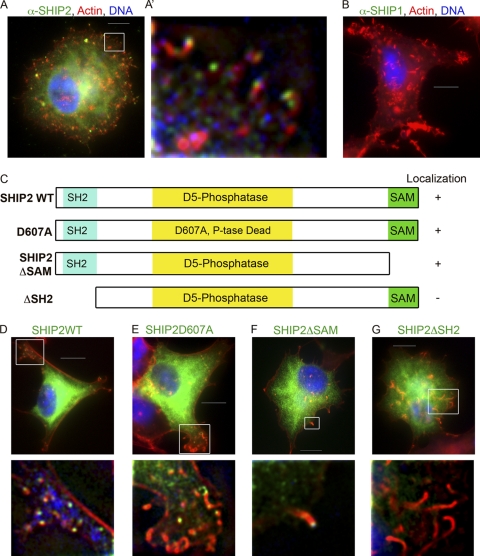
To explore a role for PI signaling in actin tail formation and virion release, we first assessed whether PI kinases or phosphatases localized beneath virions. We could not detect localization of type I PI3-kinases with several antisera (data not shown). However, an endogenous protein recognized by an antibody to the phosphoinositide phosphatase SHIP2 was enriched relative to the cytoplasm at the tips of actin tails and directly apposed to the virion (Fig. 1 A and A′). Localization of SHIP2 was apparent in the majority of actin tails in BSC40 cells. The localization appeared specific as antiserum that detects SHIP1 but not SHIP2 did not recognize epitopes in actin tails (Fig. 1B). Western analysis confirmed that SHIP1 was not expressed at detectable levels in BSC40 cells, in contrast to SHIP2 (see Fig. S1A and B in the supplemental material). However, exogenously expressed SHIP1 did localize to tails in BSC40 cells (see Fig. S1C).
Fig. 1.
SHIP2 is recruited to VACV actin tails via its SH2 domain. (A) Images of BSC40 cells infected with VACV strain WR and stained with antibodies recognizing endogenous SHIP2. Panel A′ shows a magnification of the boxed region in panel A. (B) Images of BSC40 cells infected with VACV strain WR and stained with antibodies recognizing endogenous SHIP1. See also Fig. S1 in the supplemental material. (C) Domain organization of SHIP2 and SHIP2 mutants. (D to G) Images of BSC40 cells expressing Xpress-SHIP2-WT, or SHIP2 mutants Xpress-SHIP2-D607A, Xpress-SHIP2-ΔSAM, or Xpress-SHIP2-ΔSH2. Note that deletion of the SH2 domain prevented localization to actin tails. Images in the lower panels are magnifications of the respective boxed regions. Scale bar, 10 μm. P-tase, phosphatase; α, anti.
To identify the domains of SHIP2 necessary for recruitment to actin tails, we transiently transfected vectors expressing wild type (WT) SHIP2 (SHIP2-WT) or SHIP2 variants containing point mutations or deletions of particular domains into BSC40 cells (Fig. 1C). Overexpressed SHIP2-WT (Fig. 1D) localized to the tops of tails, as did a phosphatase dead mutant (SHIP-D607A) (Fig. 1E), and a sterile alpha motif (SAM) domain deletion mutant (SHIP2-ΔSAM) (Fig. 1F). The localization of SHIP2-ΔSAM to actin tails appeared consistent with the localization of exogenous SHIP1, which lacks a SAM domain (see Fig. S1C in the supplemental material). In contrast, SHIP2 lacking the SH2 domain (SHIP2-ΔSH2) did not localize to actin tails (Fig. 1G). Together, these data suggest that the SH2 domain is required for localization of SHIP2 to actin tails; however, neither phosphatase activity nor the SAM domain appeared required for localization.
Localization of SHIP2 requires Abl and Src family tyrosine kinases and N-WASP but not actin.
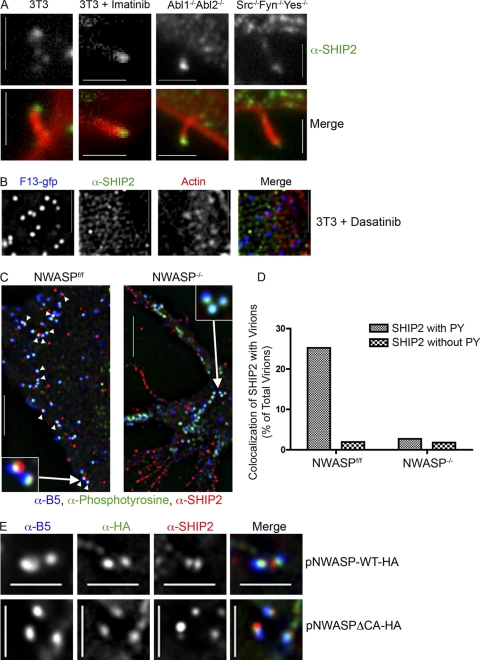
Observations with SHIP2-ΔSH2 indicated that tyrosine phosphorylation might be required for localization of SHIP2 to actin tails. We next assessed whether localization of SHIP2 was evident in cell lines lacking particular tyrosine kinases or in cells treated with tyrosine kinase inhibitors. SHIP2 remained localized to actin tails formed on Abl1−/− Abl2−/− cells, Src−/− Fyn−/− Yes−/− cells, and on cells treated with 10 μM imatinib mesylate (STI-571), an inhibitor of Abl family kinases (Fig. 2 A) (64). However, in cells treated with dasatinib (BMS-354825), an inhibitor of both Abl and Src family kinases and actin tails (55), no localization of SHIP2 was evident apposed to virions (Fig. 2B).
Fig. 2.
Localization of SHIP2 requires Abl and Src family tyrosine kinases and N-WASP but not actin. (A) Images of endogenous SHIP2 in actin tails formed by WR on 3T3 cells, 3T3 cells treated with 10 μM imatinib mesylate (inhibits Abl1 and Abl2), Abl1−/− Abl2−/− cells, or Src−/− Fyn−/− Yes−/− cells. Scale bar, 2 μm. (B) Endogenous SHIP2 does not appear localized with F13-GFP virions following dasatinib treatment, which inhibits both Src and Abl family kinases and blocks actin tails. Scale bar, 2 μm. (C) Images of colocalization of SHIP2 with vaccinia virus protein B5 and PY in N-WASPflox/flox or N-WASP−/− cells. Arrows indicate colocalization of all three proteins, and insets are magnifications of the indicated WR virions. Scale bar, 5 μm. See also Fig. S2 in the supplemental material. (D) Quantification of colocalization of B5 fluorescence, SHIP2, and PY in images such as those in panel C. Data are expressed as a percentage of total virions on which SHIP2 was colocalized with B5 with or without PY staining in N-WASPflox/flox cells (n = 1,244 virions) and N-WASP−/− cells (n = 1,133 virions). (E) Localization of SHIP2 to WR virions does not require actin. BSC40 cells were transfected with pNWASP-WT-HA (where HA is the hemagglutinin A epitope tag) or pNWASP-ΔCA-HA and infected with VACV. Colocalization of SHIP2 with B5 was evident in cells transfected with either N-WASP-WT-HA or N-WASP-ΔCA-HA. Scale bar, 2 μm.
To determine whether localization of SHIP2 requires other components of VACV actin tails, we next assessed SHIP2 localization in fibroblast cell lines derived from N-WASPflox/flox or N-WASP−/− mice and infected with VACV. Cell lines deficient in N-WASP fail to form tails (69); therefore, we measured the colocalization of SHIP2 with phosphotyrosine and anti-B5, components of cell-associated virions. In N-WASPflox/flox cells, 25% of virions colocalized with both SHIP2 and phosphotyrosine (Fig. 2C and D; see also Fig. S2 in the supplemental material). In contrast, only 1.9% of virions lacking a phosphotyrosine signal colocalized with SHIP2. In N-WASP−/− cells, the percentage of virions colocalizing with SHIP2 was similar whether phosphotyrosine was present or not (1.8 to 2.7%) and similar to that observed in N-WASPflox/flox cells for virions lacking phosphotyrosine. These data suggest that N-WASP and phosphotyrosine together are required for localization of SHIP2. To rule out the possibility that localization of SHIP2 required the Arp2/3 complex or actin, we assessed localization of SHIP2 in BSC40 cells expressing N-WASP with a deletion of the cofilin and acidic domains (N-WASP-ΔCA), which fails to recruit the Arp2/3 complex and thereby precludes formation of actin tails (26). As shown in Fig. 2E, SHIP2, N-WASP-ΔCA, and the virion (detected with anti-B5 monoclonal antibody [MAb]) colocalized. Collectively, these data suggest that activity of Abl or Src family kinases, perhaps acting redundantly (54), is required for localization of SHIP2 and that localization depends on N-WASP but not on the Arp2/3 complex or actin. The interaction between SHIP2 and N-WASP appears to be indirect as we were unable to detect a direct association of SHIP2 with N-WASP in immunoprecipitation experiments (data not shown).
SHIP2 does not regulate formation of actin tails.
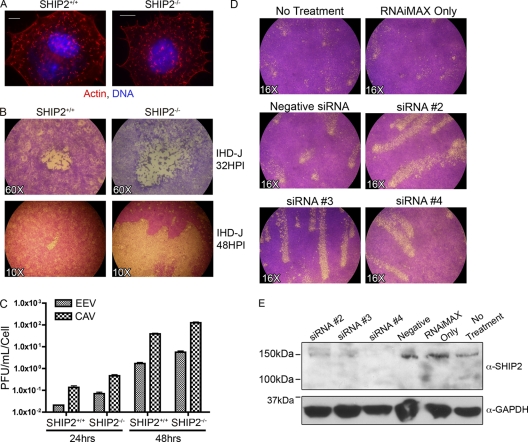
Localization of SHIP2 beneath virions on actin tails raised the possibility that the protein might regulate either actin tails or virion release or both. To test these possibilities, we first assessed actin tails in embryonic fibroblasts derived from SHIP2+/+ and SHIP2−/− mice. As shown in Fig. 3 A, the numbers and sizes of actin tails appeared similar in the two cell types, as did the velocities of virions on tails (data not shown). We also did not observe a difference in the numbers or localization of B5-positive virions outside these cells (data not shown). We confirmed that SHIP1 was not expressed in either SHIP2+/+ or SHIP2−/− cells, SHIP2 was not expressed in the SHIP2−/− cells, and SHIP2 was evident only on the tops of actin tails found in SHIP2+/+ but not in SHIP2−/− cells (see Fig. S3 in the supplemental material). Previous work by Smith et al. identified two interacting partners of SHIP2, SHC and LPD, which localize to EPEC pedestals (68), and Krause et al. identified LPD as localizing to VACV actin tails (30). We confirmed that LPD as well as SHC localized to the tips of VACV tails (See Fig. S4A and B). However, both SHC and LPD were recruited to tails in both SHIP2+/+ cells and SHIP2−/− cells. Together, these data suggest that SHIP2 is required for neither the formation of actin tails nor the recruitment of LPD or SHC.
Fig. 3.
SHIP2 regulates virion dissemination independent of actin tail formation. (A) VACV strain WR forms actin tails on SHIP2+/+ cells and SHIP2−/− cells. Cells were stained for actin (red) and DNA (blue). Scale bar, 10 μm. See also Fig. S3 and S4 in the supplemental material. (B) VACV strain IHD-J forms larger comet tails in SHIP2−/− cells than in SHIP2+/+ cells. Cells were infected for 32 h (upper panels; magnification, ×60) or 48 h (lower panels; magnification, ×10) and stained with crystal violet to visualize plaques. (C) Quantification of EEV and CAV of strain IHD-J from SHIP2+/+ and SHIP2−/− cells at 24 and 48 h postinfection. (D) SHIP2 knockdown with siRNA enhances virion release relative to control cells. BSC40 cells were left untreated or were treated with Lipofectamine reagent (RNAiMAX), negative siRNA, or either of three siRNAs that target SHIP2 (2, 3, or 4) for 3 days; cells were then infected with VACV strain IHD-J for 2 days and stained and visualized at a magnification of ×16. (E) The upper panel shows Western analysis with SHIP2 antiserum of lysates of BSC40 cells left untreated or treated as described for panel D. The lower panel shows Western analysis of lysates with GAPDH antiserum to confirm equivalent loading of samples.
SHIP2 regulates virion dissemination by inhibiting release of EEV.
We next investigated the effects of SHIP2 on plaque formation. To do this, SHIP2+/+ or SHIP2−/− cells were infected with VACV strain IHD-J, which releases large numbers of EEV particles (3). At 32 h postinfection, plaques formed by IHD-J are similar in size to those seen with other strains (e.g., WR) (Fig. 3B, upper panel). However, unlike WR plaques, IHD-J plaques are associated with an archipelago of smaller plaques, termed “comets,” which are evident 48 h after infection and which are indicative of enhanced EEV release in this strain (1, 19). Characteristic plaques formed by IHD-J were evident on SHIP2+/+ and SHIP2−/− cells by 32 h (Fig. 3B, upper panel) though plaques were slightly larger on SHIP2−/− cells. Comets visualized at 48 h postinfection were significantly larger in SHIP2−/− cells than those in SHIP2+/+ cells, often merging with adjacent comets and extending across the plate (Fig. 3B, lower panel). We next measured the amount of EEV released by SHIP2+/+ or SHIP2−/− cells into the supernatant. In accordance with the plaque assays, ∼3-fold more EEV and CAV grew in SHIP2−/− cells than in SHIP2+/+ cells at a low MOI (0.01) (Fig. 3C), consistent with increased viral spread in the monolayer. We did not observe a difference in the ratio of EEV to CAV between the two cell types, and viral replication was similar at an MOI of 5, suggesting that SHIP2 does not affect viral replication (data not shown). To corroborate data from SHIP2−/− cells, we next knocked down SHIP2 in BSC40 cells. BSC40 cells treated with either of three siRNAs specific to SHIP2 (sequences 2, 3, and 4) exhibited larger comets than those seen with negative siRNA, the transfection reagent (RNAiMAX), or untreated cells (Fig. 3D). Knockdown of SHIP2 was confirmed by Western analysis (Fig. 3E).
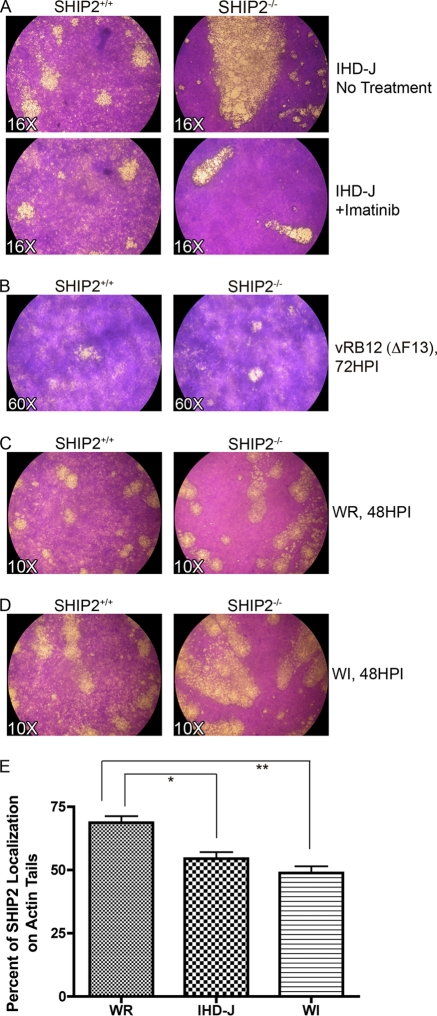
To confirm that the large comets formed on the SHIP2−/− cells were specifically due to increased release of EEV, we carried out two additional experiments. Reeves et al. found that release of EEV required activity of Abl family tyrosine kinases (54). In accordance with the idea that more EEV is released from SHIP2−/− cells than from SHIP2+/+ cells, we found that the Abl family tyrosine kinase inhibitor imatinib mesylate blocked comets in SHIP2+/+ cells and reduced the size and extent of comets in SHIP2−/− cells (Fig. 4 A). Second, we infected both cells types with WR vRB12, a virus lacking F13L (2), a gene required to form EEV and comets. As shown in Fig. 4B, vRB12 formed similarly sized plaques on SHIP2+/+ and SHIP2−/− cells but did not form comets on either cell type. We could find no evidence to support the possibility that differences in infectivity or cellular migration rates could account for the apparent increase in the size of comets in SHIP2−/− cells. Furthermore, luciferase under the control of an early/late promoter was similarly expressed in both cell types at 2 h postinfection, indicating that increased viral entry did not account for enhanced comet size (see Fig. S5 in the supplemental material). In addition, specific infectivity levels of VACV produced in SHIP2−/− and SHIP2+/+ cells were similar, as were the numbers of plaques formed on both cell types, and no differences in rates of movement of SHIP2+/+ and SHIP2−/− cells were evident (data not shown). Collectively, these data suggest that SHIP2 inhibits release of EEV.
Fig. 4.
A34 and SHIP2 negatively regulate virion dissemination. (A) Release of virions from SHIP2+/+ and SHIP2−/− cells is sensitive to imatinib mesylate. Cells were infected with VACV IHD-J for 48 h postinfection and stained with crystal violet. Plaques were visualized by microscopy at a magnification of ×16. Imatinib mesylate (10 μM) was added to cells at 1 h postinfection. (B) Comets formed on SHIP2+/+ and SHIP2−/− cells are specifically due to EEV. Cells were infected for 72 h with WR vRB12, a virus lacking the F13L gene and unable to form EEV. Although plaques formed on the two cell types were of similar sizes, no comets were evident. Plaques were visualized at a magnification of ×60. (C) Plaques formed by VACV strain WR on SHIP2+/+ and SHIP2−/− cells 48 h postinfection (magnification, ×10). (D) Plaques formed by VACV strain WI on SHIP2+/+ and SHIP2−/− cells at 48 h postinfection (magnification, ×10). (E) Quantification of SHIP2 recruitment to actin tails in cells infected with VACV strains WR, IHD-J, or WI. WR (n = 2597 tails, 45 cells) recruits more SHIP2 to tails than IHD-J (n = 2,046 tails, 58 cells) (P = 0.0003) or WI (n = 2,763 tails, 32 cells) (P = 0.0001). HPI, hours postinfection.
VACV protein A34 mediates the effects of SHIP2 on inhibition of EEV release.
The observation that deletion of A34 in VACV enhances release of virus (34) suggests that A34, a component of EEV, acts as an inhibitor of release. In accordance with this idea, the VACV strain IHD-J, which contains a mutation in A34, releases more EEV and forms larger comets than strain WR, and a WR strain containing A34 derived from IHD-J (called WI) forms larger comets than WR (3). The large comets evident in SHIP2−/− cells compared to those in SHIP2+/+ cells led us to hypothesize that A34 may act via SHIP2. To test this possibility, we assessed the effects of WR and WI comets formed on SHIP2−/− and SHIP2+/+ cells. As shown in Fig. 4C, WR was unable to form comets on SHIP2+/+ cells but did form comets on SHIP2−/− cells (Fig. 4C). WI produced small comets on SHIP2+/+ cells but large comets on SHIP2−/− cells (Fig. 4D), reminiscent of those seen with IHD-J in these cells (Fig. 3B). We hypothesized that the capacity of IHD-J and WI to form larger comets than WR was due to differences in the recruitment of SHIP2 to actin tails in these strains. As shown in Fig. 4E, an inverse correlation exists between EEV release and the efficacy of SHIP2 recruitment to actin tails. Thus, whereas strain WR recruited SHIP2 to 69% of actin tails, strains IHD-J and WI recruited SHIP2 to 54% and 49% of tails, respectively, a statistically significant difference compared to WR (P < 0.0003 and P < 0.0001, respectively) (Fig. 4E). Collectively, these data suggest the following:(i) that recruitment of SHIP2, via its SH2 domain, requires N-WASP and the Abl and Src family tyrosine kinases but that these proteins alone are not sufficient; (ii) that viral protein A34 recruits SHIP2 to actin tails; and (iii) that SHIP2 at least in part mediates inhibition of release by A34.
DISCUSSION
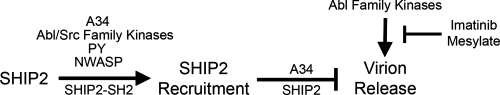
Here, we explore the role of the phosphoinositide 5-phosphatase SHIP2 in release of VACV from infected cells. We found that SHIP2 localizes to actin tails in an SH2-dependent manner, suggesting that phosphorylated tyrosines on cellular or viral proteins within the tail mediate recruitment. Host Src and Abl family kinases localize to and redundantly form tails (14, 54), and we find that these kinases are also required for recruitment of SHIP2 (Fig. 2). Abl but not Src family kinases also appear to play a role in enhancing release of EEV (54). In this regard, comets formed on SHIP2+/+ and SHIP2−/− cells are blocked by imatinib mesylate, a specific inhibitor of Abl family kinases (54). Thus, Abl family kinases play dual but antagonistic roles in EEV release, by both facilitating recruitment of a release inhibitor (SHIP2) and promoting virion release (Fig. 5).
Fig. 5.
Overview of SHIP2 recruitment and regulation of virion release.
Src and Abl family tyrosine kinases phosphorylate the viral protein A36 at two sites, suggesting that phosphorylated A36 may directly recruit SHIP2 (40). However, this seems unlikely because phospho-A36 recruits N-WASP via Nck and WIP (14, 37), and SHIP2 recruitment also depends on N-WASP. We cannot rule out the possibility that factors distal to N-WASP may recruit SHIP2. Whereas Grb2 is one such candidate (63), the Arp2/3 complex and actin do not appear to be involved as SHIP2 does not affect actin tails and as N-WASP-ΔCA, which does not recruit the Arp2/3 complex, still recruits SHIP2. In this regard, Smith et al. found that SHIP2 appears to regulate host lipids and actin polymerization in EPEC pedestals (68), which resemble actin tails formed by vaccinia virus (17). However, our data do not recapitulate the EPEC phenotype, suggesting that VACV utilizes SHIP2 in a manner distinct from EPEC.
Still unresolved is whether SHIP2 inhibits virion release via its catalytic activity or, alternatively, serves as a scaffold to recruit effectors. Two known SHIP2 binding partners, SHC and LPD, are recruited to VACV actin tails, though by a mechanism that appears to be independent of SHIP2 (see Fig. S3 in the supplemental material). Nevertheless, other SHIP2 effectors have been described, including epidermal growth factor receptor (EGFR), filamin, p130Cas, Cbl, vinexin, Arap3, APS, JIP-1, and intersectin (11, 38, 41, 43, 48, 50, 52, 73, 78, 79).
Another possibility is that SHIP2 phosphatase activity is required to inhibit release of EEV. In this regard, we have attempted to localize PH domains that specifically recognize various phosphatidylinositol phosphates (PIPs) on actin tails. Although some PH domains do appear to localize, point mutants that abolish binding to lipid moieties in vitro also localize, suggesting that recruitment is nonspecific (S. McNulty and D. Kalman, unpublished data). Furthermore, we have been unable to detect Akt-PH, Akt, or Akt-(P)S473 at the tops of tails (McNulty and Kalman, unpublished). Notably, we cannot rule out the possibility that expression of the PH domain alone does not compete effectively with intact proteins that utilize multiple binding sites, a phenomenon we observed previously with localization of proline-rich regions and SH2 domains from tyrosine kinases in EPEC pedestals (4).
Do viral proteins participate with SHIP2 to regulate release of EEV? The observation that deletion of A34 or point mutations within A34 (K151E) enhance release of EEV (3) raises the possibility that the normal function of A34 is to suppress EEV release. Our data suggest that A34 mediates recruitment of SHIP2 and that SHIP2 at least in part mediates inhibition of release by A34 (Fig. 4). A34 exists in a complex with viral proteins A33, A36, B5, and F13 (45, 46); therefore, it remains possible that SHIP2 tethers a complex of host and viral proteins that regulate viral release. Other intrinsic host defense molecules have been recently described, including tetherin and viperin, which are induced in an interferon-dependent manner. Tetherin is antagonized by the HIV protein Vpu, and viperin is antagonized by a cellular protein, farnesyl diphosphate synthase (FPPS), which facilitates virion release (9, 74). In contrast, VACV regulation of release through SHIP2 appears distinct. Rather than antagonizing release, A34 cooperates with SHIP2 to inhibit release. A34 is a transmembrane glycoprotein with homology to C-type lectins (10) and may detect the presence of leukocytes, cell adhesion molecules, or endocytic receptors. In so doing, A34 may play a gatekeeper role so as to limit release except when environmental conditions are conducive to dissemination.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R56A105896101A2 and R01A107246201A2 to D.K. and by a grant from the Fonds de la Recherche Scientifique Médicale to C.E.
We thank Jay Hooper (USAMRIID) for sharing antibodies and Bernard Moss (NIH) for sharing the WI virus. We also thank Jack Taunton (University of California, San Francisco) for sharing the N-WASPflox/flox and N-WASP−/− cells. We also thank Sam Speck, Eric Hunter, Victor Faundez, Aron Lukacher, Ed Mocarski, and Alyson Swimm (Emory University) for insightful comments and discussion.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Blasco R., Moss B. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J. Virol. 65:5910–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blasco R., Moss B. 1992. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J. Virol. 66:4170–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blasco R., Sisler J. R., Moss B. 1993. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J. Virol. 67:3319–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bommarius B., et al. 2007. Enteropathogenic Escherichia coli Tir is an SH2/3 ligand that recruits and activates tyrosine kinases required for pedestal formation. Mol. Microbiol. 63:1748–1768 [DOI] [PubMed] [Google Scholar]
- 5. Chi Y., et al. 2004. Comparative mechanistic and substrate specificity study of inositol polyphosphate 5-phosphatase Schizosaccharomyces pombe synaptojanin and SHIP2. J. Biol. Chem. 279:44987–44995 [DOI] [PubMed] [Google Scholar]
- 6. Clement S., et al. 2001. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature 409:92–97 [DOI] [PubMed] [Google Scholar]
- 7. Damen J. E., et al. 1996. The 145-kDa protein induced to associate with Shc by multiple cytokines is an inositol tetraphosphate and phosphatidylinositol 3,4,5-triphosphate 5-phosphatase. Proc. Natl. Acad. Sci. U. S. A. 93:1689–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dodding M. P., Way M. 2009. Nck- and N-WASP-dependent actin-based motility is conserved in divergent vertebrate poxviruses. Cell Host Microbe 6:536–550 [DOI] [PubMed] [Google Scholar]
- 9. Douglas J. L., et al. 2010. The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog. 6:e1000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duncan S. A., Smith G. L. 1992. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J. Virol. 66:1610–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dyson J. M., et al. 2001. The SH2-containing inositol polyphosphate 5-phosphatase, SHIP-2, binds filamin and regulates submembraneous actin. J. Cell Biol. 155:1065–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fenner F. 1996. Poxviruses, p. 2673–2702 In Knipe D. M., et al. (ed.), Fields virology, 3rd ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 13. Frischknecht F., et al. 1999. Tyrosine phosphorylation is required for actin-based motility of vaccinia but not Listeria or Shigella. Curr. Biol. 9:89–92 [DOI] [PubMed] [Google Scholar]
- 14. Frischknecht F., et al. 1999. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 401:926–929 [DOI] [PubMed] [Google Scholar]
- 15. Geada M. M., Galindo I., Lorenzo M. M., Perdiguero B., Blasco R. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 82:2747–2760 [DOI] [PubMed] [Google Scholar]
- 16. Giuriato S., et al. 2002. SHIP2 overexpression strongly reduces the proliferation rate of K562 erythroleukemia cell line. Biochem. Biophys. Res. Commun. 296:106–110 [DOI] [PubMed] [Google Scholar]
- 17. Gouin E., Welch M. D., Cossart P. 2005. Actin-based motility of intracellular pathogens. Curr. Opin. Microbiol. 8:35–45 [DOI] [PubMed] [Google Scholar]
- 18. Habib T., Hejna J. A., Moses R. E., Decker S. J. 1998. Growth factors and insulin stimulate tyrosine phosphorylation of the 51C/SHIP2 protein. J. Biol. Chem. 273:18605–18609 [DOI] [PubMed] [Google Scholar]
- 19. Herrera E., Lorenzo M. M., Blasco R., Isaacs S. N. 1998. Functional analysis of vaccinia virus B5R protein: essential role in virus envelopment is independent of a large portion of the extracellular domain. J. Virol. 72:294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hollinshead M., et al. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hollinshead M., Vanderplasschen A., Smith G. L., Vaux D. J. 1999. Vaccinia virus intracellular mature virions contain only one lipid membrane. J. Virol. 73:1503–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Honeychurch K. M., Yang G., Jordan R., Hruby D. E. 2007. The vaccinia virus F13L YPPL motif is required for efficient release of extracellular enveloped virus. J. Virol. 81:7310–7315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ishida S., et al. 2006. Association of SH-2 containing inositol 5′-phosphatase 2 gene polymorphisms and hyperglycemia. Pancreas 33:63–67 [DOI] [PubMed] [Google Scholar]
- 24. Kagawa S., et al. 2005. Impact of SRC homology 2-containing inositol 5′-phosphatase 2 gene polymorphisms detected in a Japanese population on insulin signaling. J. Clin. Endocrinol. Metab. 90:2911–2919 [DOI] [PubMed] [Google Scholar]
- 25. Kaisaki P. J., et al. 2004. Polymorphisms in type II SH2 domain-containing inositol 5-phosphatase (INPPL1, SHIP2) are associated with physiological abnormalities of the metabolic syndrome. Diabetes 53:1900–1904 [DOI] [PubMed] [Google Scholar]
- 26. Kalman D., et al. 1999. Enteropathogenic E. coli acts through WASP and Arp2/3 complex to form actin pedestals. Nat. Cell Biol. 1:389–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kavanaugh W. M., et al. 1996. Multiple forms of an inositol polyphosphate 5-phosphatase form signaling complexes with Shc and Grb2. Curr. Biol. 6:438–445 [DOI] [PubMed] [Google Scholar]
- 28. Khodakevich L., Jezek Z., Messinger D. 1988. Monkeypox virus: ecology and public health significance. Bull. World Health Organ. 66:747–752 [PMC free article] [PubMed] [Google Scholar]
- 29. Khodakevich L., Szczeniowski M., Manbu ma D., Jezek Z., Marennikova S., Nakano J., Messinger D. 1987. The role of squirrels in sustaining monkeypox virus transmission. Trop. Geogr. Med. 39:115–122 [PubMed] [Google Scholar]
- 30. Krause M., et al. 2004. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev. Cell 7:571–583 [DOI] [PubMed] [Google Scholar]
- 31. Lioubin M. N., et al. 1996. p150Ship, a signal transduction molecule with inositol polyphosphate-5-phosphatase activity. Genes Dev. 10:1084–1095 [DOI] [PubMed] [Google Scholar]
- 32. Liu Y., Bankaitis V. A. 2010. Phosphoinositide phosphatases in cell biology and disease. Prog. Lipid Res. 49:201–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marion E., et al. 2002. The gene INPPL1, encoding the lipid phosphatase SHIP2, is a candidate for type 2 diabetes in rat and man. Diabetes 51:2012–2017 [DOI] [PubMed] [Google Scholar]
- 34. McIntosh A. A., Smith G. L. 1996. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J. Virol. 70:272–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McNulty S., et al. 2010. Multiple phosphatidylinositol 3-kinases regulate vaccinia virus morphogenesis. PLoS One 5:e10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meiser A., Boulanger D., Sutter G., Krijnse Locker J. 2003. Comparison of virus production in chicken embryo fibroblasts infected with the WR, IHD-J, and MVA strains of vaccinia virus: IHD-J is most efficient in trans-Golgi network wrapping and extracellular enveloped virus release. J. Gen. Virol. 84:1383–1392 [DOI] [PubMed] [Google Scholar]
- 37. Moreau V., et al. 2000. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat. Cell Biol. 2:441–448 [DOI] [PubMed] [Google Scholar]
- 38. Nakatsu F., et al. 2010. The inositol 5-phosphatase SHIP2 regulates endocytic clathrin-coated pit dynamics. J. Cell Biol. 190:307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Newsome T. P., Scaplehorn N., Way M. 2004. SRC mediates a switch from microtubule- to actin-based motility of vaccinia virus. Science 306:124–129 [DOI] [PubMed] [Google Scholar]
- 40. Newsome T. P., Weisswange I., Frischknecht F., Way M. 2006. Abl collaborates with Src family kinases to stimulate actin-based motility of vaccinia virus. Cell Microbiol. 8:233–241 [DOI] [PubMed] [Google Scholar]
- 41. Onnockx S., et al. 2008. The association between the SH2-containing inositol polyphosphate 5-phosphatase 2 (SHIP2) and the adaptor protein APS has an impact on biochemical properties of both partners. J. Cell. Physiol. 214:260–272 [DOI] [PubMed] [Google Scholar]
- 42. Parrish S., Moss B. 2006. Characterization of a vaccinia virus mutant with a deletion of the D10R gene encoding a putative negative regulator of gene expression. J. Virol. 80:553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Paternotte N., et al. 2005. SHIP2 interaction with the cytoskeletal protein vinexin. FEBS J. 272:6052–6066 [DOI] [PubMed] [Google Scholar]
- 44. Payne L. G. 1979. Identification of the vaccinia hemagglutinin polypeptide from a cell system yielding large amounts of extracellular enveloped virus. J. Virol. 31:147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Perdiguero B., Blasco R. 2006. Interaction between vaccinia virus extracellular virus envelope A33 and B5 glycoproteins. J. Virol. 80:8763–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Perdiguero B., Lorenzo M. M., Blasco R. 2008. Vaccinia virus A34 glycoprotein determines the protein composition of the extracellular virus envelope. J. Virol. 82:2150–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pesesse X., Deleu S., De Smedt F., Drayer L., Erneux C. 1997. Identification of a second SH2-domain-containing protein closely related to the phosphatidylinositol polyphosphate 5-phosphatase SHIP. Biochem. Biophys. Res. Commun. 239:697–700 [DOI] [PubMed] [Google Scholar]
- 48. Pesesse X., et al. 2001. The Src homology 2 domain containing inositol 5-phosphatase SHIP2 is recruited to the epidermal growth factor (EGF) receptor and dephosphorylates phosphatidylinositol 3,4,5-trisphosphate in EGF-stimulated COS-7 cells. J. Biol. Chem. 276:28348–28355 [DOI] [PubMed] [Google Scholar]
- 49. Pesesse X., et al. 1998. The SH2 domain containing inositol 5-phosphatase SHIP2 displays phosphatidylinositol 3,4,5-trisphosphate and inositol 1,3,4,5-tetrakisphosphate 5-phosphatase activity. FEBS Lett. 437:301–303 [DOI] [PubMed] [Google Scholar]
- 50. Prasad N., Topping R. S., Decker S. J. 2001. SH2-containing inositol 5′-phosphatase SHIP2 associates with the p130Cas adapter protein and regulates cellular adhesion and spreading. Mol. Cell. Biol. 21:1416–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Prasad N. K., Decker S. J. 2005. SH2-containing 5′-inositol phosphatase, SHIP2, regulates cytoskeleton organization and ligand-dependent down-regulation of the epidermal growth factor receptor. J. Biol. Chem. 280:13129–13136 [DOI] [PubMed] [Google Scholar]
- 52. Raaijmakers J. H., et al. 2007. The PI3K effector Arap3 interacts with the PI(3,4,5)P3 phosphatase SHIP2 in a SAM domain-dependent manner. Cell Signal. 19:1249–1257 [DOI] [PubMed] [Google Scholar]
- 53. Reed K. D., et al. 2004. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 350:342–350 [DOI] [PubMed] [Google Scholar]
- 54. Reeves P. M., et al. 2005. Disabling poxvirus pathogenesis by inhibition of Abl-family tyrosine kinases. Nat. Med. 11:731–739 [DOI] [PubMed] [Google Scholar]
- 55. Reeves P. M., et al. 2011. Variola and monkeypox utilize conserved mechanisms of virion motility and release that depend on Abl and Src family tyrosine kinases. J. Virol. 85:21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rietdorf J., et al. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992–1000 [DOI] [PubMed] [Google Scholar]
- 57. Rimoin A. W., et al. 2007. Endemic human monkeypox, Democratic Republic of Congo, 2001-2004. Emerg. Infect. Dis. 13:934–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rimoin A. W., et al. 2010. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc. Natl. Acad. Sci. U. S. A. 107:16262–16267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Risco C., et al. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Roberts K. L., Smith G. L. 2008. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 16:472–479 [DOI] [PubMed] [Google Scholar]
- 61. Rodriguez J. R., Risco C., Carrascosa J. L., Esteban M., Rodriguez D. 1997. Characterization of early stages in vaccinia virus membrane biogenesis: implications of the 21-kilodalton protein and a newly identified 15-kilodalton envelope protein. J. Virol. 71:1821–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rohatgi R., Ho H. Y., Kirschner M. W. 2000. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 150:1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Scaplehorn N., et al. 2002. Grb2 and Nck act cooperatively to promote actin-based motility of vaccinia virus. Curr. Biol. 12:740–745 [DOI] [PubMed] [Google Scholar]
- 64. Schindler T., et al. 2000. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science 289:1938–1942 [DOI] [PubMed] [Google Scholar]
- 65. Schurmans S., et al. 1999. The mouse SHIP2 (Inppl1) gene: complementary DNA, genomic structure, promoter analysis, and gene expression in the embryo and adult mouse. Genomics 62:260–271 [DOI] [PubMed] [Google Scholar]
- 66. Sleeman M. W., et al. 2005. Absence of the lipid phosphatase SHIP2 confers resistance to dietary obesity. Nat. Med. 11:199–205 [DOI] [PubMed] [Google Scholar]
- 67. Smith G. L., Vanderplasschen A., Law M. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 83:2915–2931 [DOI] [PubMed] [Google Scholar]
- 68. Smith K., Humphreys D., Hume P. J., Koronakis V. 2010. Enteropathogenic Escherichia coli recruits the cellular inositol phosphatase SHIP2 to regulate actin-pedestal formation. Cell Host Microbe 7:13–24 [DOI] [PubMed] [Google Scholar]
- 69. Snapper S. B., et al. 2001. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat. Cell Biol. 3:897–904 [DOI] [PubMed] [Google Scholar]
- 70. Soares J. A., et al. 2009. Activation of the PI3K/Akt pathway early during vaccinia and cowpox virus infections is required for both host survival and viral replication. J. Virol. 83:6883–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sodeik B., et al. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Taylor V., et al. 2000. 5′ phospholipid phosphatase SHIP-2 causes protein kinase B inactivation and cell cycle arrest in glioblastoma cells. Mol. Cell. Biol. 20:6860–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vandenbroere I., Paternotte N., Dumont J. E., Erneux C., Pirson I. 2003. The c-Cbl-associated protein and c-Cbl are two new partners of the SH2-containing inositol polyphosphate 5-phosphatase SHIP2. Biochem. Biophys. Res. Commun. 300:494–500 [DOI] [PubMed] [Google Scholar]
- 74. Wang X., Hinson E. R., Cresswell P. 2007. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2:96–105 [DOI] [PubMed] [Google Scholar]
- 75. Ward B. M., Moss B. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 75:11651–11663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weisswange I., Newsome T. P., Schleich S., Way M. 2009. The rate of N-WASP exchange limits the extent of ARP2/3-complex-dependent actin-based motility. Nature 458:87–91 [DOI] [PubMed] [Google Scholar]
- 77. Wisniewski D., et al. 1999. A novel SH2-containing phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase (SHIP2) is constitutively tyrosine phosphorylated and associated with src homologous and collagen gene (SHC) in chronic myelogenous leukemia progenitor cells. Blood 93:2707–2720 [PubMed] [Google Scholar]
- 78. Xie J., et al. 2008. The docking properties of SHIP2 influence both JIP1 tyrosine phosphorylation and JNK activity. Cell Signal. 20:1432–1441 [DOI] [PubMed] [Google Scholar]
- 79. Xie J., Vandenbroere I., Pirson I. 2008. SHIP2 associates with intersectin and recruits it to the plasma membrane in response to EGF. FEBS Lett. 582:3011–3017 [DOI] [PubMed] [Google Scholar]
- 80. Yu J., et al. 2010. MicroRNA-205 promotes keratinocyte migration via the lipid phosphatase SHIP2. FASEB J. 24:3950–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zaborowska I., Walsh D. 2009. PI3K signaling regulates rapamycin-insensitive translation initiation complex formation in vaccinia virus-infected cells. J. Virol. 83:3988–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang J., et al. 2007. SHIP2 controls PtdIns(3,4,5)P(3) levels and PKB activity in response to oxidative stress. Cell Signal. 19:2194–2200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.