Abstract
NS5A plays a critical, yet poorly defined, role in hepatitis C virus genome replication. The protein consists of three domains, each of which is able to bind independently to the 3′ untranslated region (UTR) of the viral positive strand genomic RNA. The peptidyl-prolyl isomerase cyclophilin A (CypA) binds to domain II, catalyzing cis-trans isomerization. CypA inhibitors such as cyclosporine (CsA) have been shown to inhibit hepatitis C virus (HCV) replication. We show here that CypA stimulated domain II RNA binding activity, and this stimulation was abrogated by CsA. An isomerase mutant of CypA (H126Q) failed to bind to domain II and did not stimulate RNA binding. Finally, we demonstrate that the RNA binding of two domain II mutants, the D316E and D316E/Y317N mutants, previously shown to exhibit CypA independence for RNA replication, was unaffected by CypA. This study provides an insight into the molecular mechanism of CypA activity during HCV replication and further validates the use of CypA inhibitors in HCV therapy.
TEXT
Hepatitis C virus (HCV) is an important human pathogen that infects approximately 3% of the global population (170 million individuals), causing persistent infection in many of these patients (17). Therapeutic options are currently limited to a combination of pegylated alpha interferon and ribavirin, with limited success and significant side effects. In the search for new therapies, much interest has focused on inhibitors of cyclophilins (Cyps). Cyps are a family of proteins that possess peptidyl-prolyl isomerase activity; the prototypic member of the family, CypA, is the target for an immunosuppressive drug, cyclosporine (CsA), as the CsA-CypA complex binds to calcineurin and inhibits T-cell activation. CypA (and, controversially, CypB and CypC) has been shown to play a role in HCV replication, binding to both NS5A and the polymerase NS5B. CypA may also act via NS2, thereby modulating cleavage of the polyprotein (3). The isomerase activity of CypA has been shown to be essential for HCV replication (1, 15), and specific residues within NS5A are the target for this isomerase activity (4, 7). Accordingly, CsA and other, nonimmunosuppressive CypA inhibitors, such as DEB025 (Alisporivir), NIM811, and SCY635, have been shown to potently inhibit HCV replication both in vitro and also in a clinical context in an interferon-independent manner. A critical mutation within domain II of NS5A, D320E (genotype 1b numbering), is associated with CypA independence and resistance to DEB025 (4).
Despite this body of knowledge regarding the involvement of CypA in viral replication, the molecular mechanisms underpinning the precise role of this cellular factor remain to be determined. Intriguingly, CypB binds to NS5B and modulates its affinity for RNA in a genotype-dependent fashion (10, 19). In this regard, we (6) and others (8, 9) have recently shown that NS5A also binds to RNA, and we therefore set out to address whether CypA might also modulate this activity.
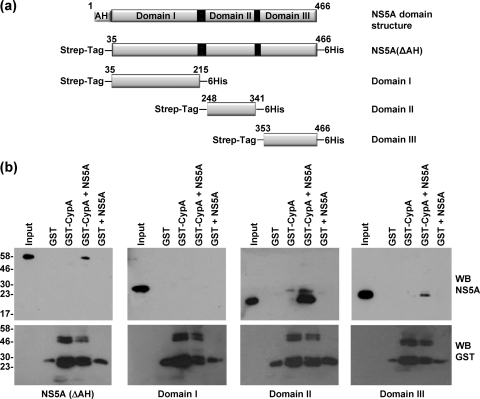
We previously established conditions for the expression and purification of each of the three domains of NS5A, derived from both the genotype 1b J4 isolate and the genotype 2a JFH-1 isolate (6). We further showed that each domain could independently bind to the 3′ untranslated region (UTR) of the virus-positive strand genomic RNA, although the binding of domain III was weak in comparison to that of domains I and II. As CypA has been shown to bind to domain II, we were interested in determining whether CypA might influence the interactions between this domain and RNA. Initially, we focused on the genotype 2a JFH-1 isolate and verified that CypA was able to bind to the individual domains of NS5A. CypA was expressed as a glutathione S-transferase (GST) fusion protein in Escherichia coli and purified as previously described (2). Purified NS5A, lacking the N-terminal amphipathic helix and thus termed NS5A(ΔAH), or each of the individually expressed three domains (18) (Fig. 1 a), were incubated with GST-CypA immobilized on glutathione-agarose beads. Bound NS5A was detected by Western blotting using a sheep polyclonal antiserum (16), and as shown in Fig. 1b, NS5A(ΔAH) and either domain II or III alone bound to GST-CypA, although the binding of domain III was weak. In contrast, domain I did not bind to GST-CypA.
Fig. 1.
GST-CypA interacts with domains II and III, but not domain I, of NS5A. (a) Schematic diagram of the functional domains of NS5A and the design of the constructs used in the article (JFH-1 genotype 2a NS5A protein numbering). The NS5A constructs all lacked the N-terminal amphipathic helix (AH) and contained an N-terminal Strep tag and a C-terminal hexahistidine tag (6). The full-length construct is referred to as NS5A(ΔAH). (b) Purified GST, or GST-CypA (40 μg), was immobilized on 20 μl glutathione-agarose beads and incubated with 5 μg purified NS5A(ΔAH) or individual domains overnight at 4°C in binding buffer (40 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 10 mm dithiothreitol [DTT], 50 μg/ml bovine serum albumin [BSA], 10 μg/ml yeast tRNA [Ambion]) (6). Beads were washed three times, eluted in SDS-PAGE sample buffer, and analyzed by Western blotting (WB) with a sheep polyclonal antibody to NS5A (16) or a mouse anti-GST antibody (Serotec).
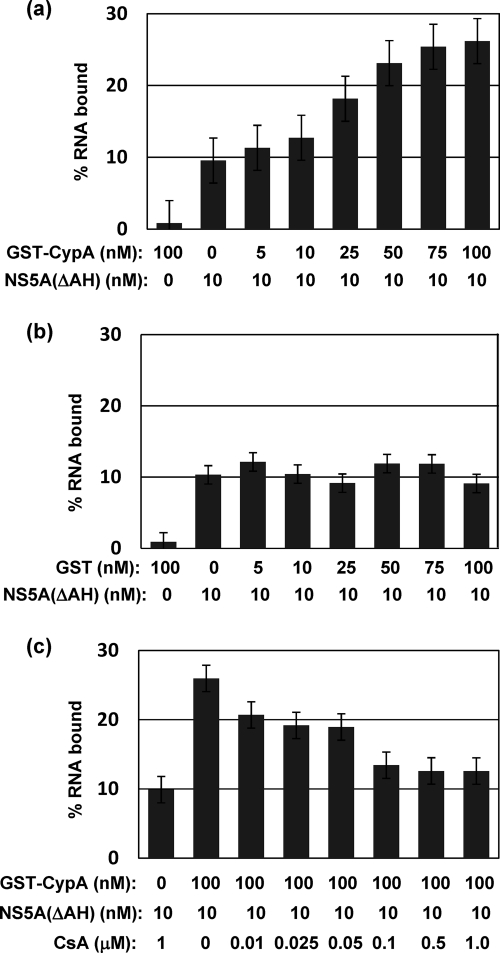
We then investigated the effects of GST-CypA on the ability of NS5A(ΔAH) to bind the 3′ UTR RNA. A 32P-labeled RNA of 236 bases in length, corresponding to the 3′ UTR of the JFH-1 isolate, was produced by in vitro transcription and binding to NS5A(ΔAH) assessed using a filter binding assay as previously described (6, 8). Constant amounts of both NS5A(ΔAH) (10 nM) and 32P-labeled 3′ UTR RNA (1 nM) were incubated with increasing concentrations of GST-CypA (0 to 100 nM) or GST alone as a negative control. We had previously shown that under these reaction conditions, a concentration of 10 nM NS5A(ΔAH) was not saturating (6). As shown in Fig. 2 a, the addition of GST-CypA resulted in a dose-dependent increase in the fraction of RNA bound, up to a maximum 2.5-fold increase at 100 nM GST-CypA. Addition of GST alone had no effect, even at the maximal concentration used (Fig. 2b). In the absence of NS5A(ΔAH), neither GST-CypA nor GST at the maximal concentration used bound to RNA above the background level (left hand bar).
Fig. 2.
CypA stimulates RNA binding by NS5A in a CsA-sensitive fashion. NS5A(ΔAH) (10 nM) was preincubated with increasing concentrations of GST-CypA (a) or GST (b) at 4°C in binding buffer. 32P-labeled 3′ UTR RNA (1 nM) was added, and binding reactions proceeded for 30 min in binding buffer prior to washing and filter binding as described previously (6). The left hand bar shows the background RNA binding of either GST-CypA or GST in the absence of NS5A(ΔAH). (c) NS5A(ΔAH) (10 nM), 32P-labeled 3′ UTR RNA (1 nM), and GST-CypA (100 nM) were incubated with increasing concentrations of CsA (0 to 1 μM). The left hand bar shows the background RNA binding of NS5A(ΔAH) in the presence of CsA (1 μM) and the absence of GST-CypA.
CsA is a cyclic nonribosomal peptide that binds to CypA; the resulting complex inhibits calcineurin-mediated T-cell activation and is thus immunosuppressive. CsA and other CypA inhibitors also represent a potent new class of anti-HCV agents (5). To test whether CsA was able to block the CypA-mediated enhancement of NS5A RNA binding, filter binding assays were performed in the presence of a constant amount of NS5A(ΔAH) (10 nM), 32P-labeled 3′ UTR RNA (1 nM), and GST-CypA (100 nM), together with an increasing concentration of CsA (0 to 1 μM). As shown in Fig. 2c, CsA was indeed able to inhibit the CypA-mediated enhancement of NS5A 3′ UTR RNA binding, although in the absence of CypA, CsA had no effect on the ability of NS5A to bind the 3′ UTR RNA. We conclude from these data that CypA binds to NS5A and enhances binding to the 3′ UTR RNA and that this enhancement is dependent on the peptidyl-prolyl isomerase activity of CypA.
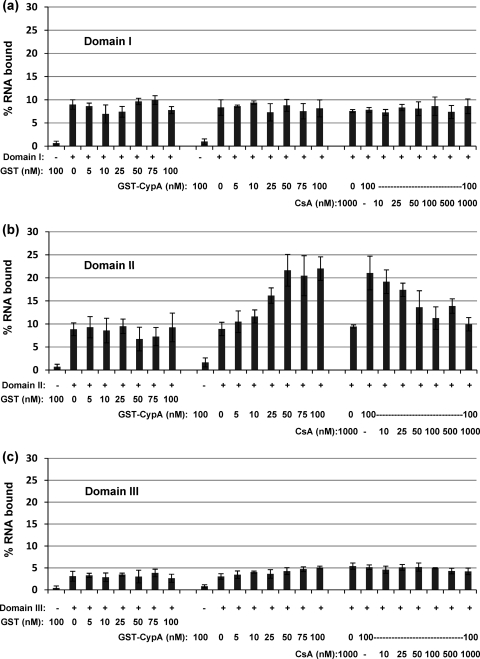
We had previously shown that each of the three domains of NS5A was able to bind independently to the 3′ UTR RNA, so we therefore asked the question of whether the RNA binding activities of each of the three domains were also modulated by CypA. We again performed filter binding assays using constant amounts of both NS5A (10 nM) and 32P-labeled 3′ UTR RNA (1 nM) together with increasing concentrations of either GST-CypA (0 to 100 nM) or GST alone as a negative control. Remarkably, only the RNA binding of domain II was significantly affected by CypA (Fig. 3 b); again, at the maximal concentration of GST-CypA (100 nM), there was a 2.5-fold increase in the fraction of RNA bound. Furthermore, the addition of CsA resulted in a dose-dependent inhibition of the CypA-mediated enhancement. Neither GST-CypA nor CsA had any effect on the RNA binding of domain I or III (Fig. 3a and c), despite the fact that domain III was able to bind to CypA, albeit inefficiently compared to domain II (Fig. 1b). These data show that the effect of CypA on the RNA binding of NS5A can be completely attributed to its influence on domain II.
Fig. 3.
CypA stimulates RNA binding by NS5A domain II in a CsA-sensitive fashion. Purified NS5A domain I (a), domain II (b), or domain III (c) was preincubated with the indicated concentrations of GST, GST-CypA, and CsA for 30 min at 4°C in binding buffer as described above. 32P-labeled 3′ UTR RNA (1 nM) was added, and binding reactions proceeded for 30 min at 4°C in binding buffer. −, no NS5A protein; +, 10 nM NS5A protein.
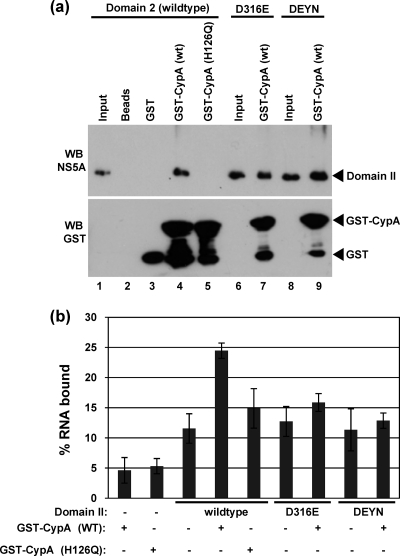
To confirm the dependence on the peptidyl-prolyl isomerase activity of CypA, we utilized a mutant (H126Q) which lacks this activity. This mutant had previously been shown to be unable to bind to full-length NS5A of genotype 1b (2), as expected; therefore, we showed that domain II of JFH- 1 NS5A was also unable to bind to GST-CypA(H126Q) (Fig. 4 a, lane 5). Consistent with this observation, GST-CypA(H126Q) was unable to stimulate the RNA binding activity of domain II (Fig. 4b). As a further control, we generated two mutant forms of domain II, the D316E and D316E/Y317N (DEYN) mutants, containing amino acid substitutions associated with CypA independence and CsA resistance of virus RNA replication. Note that residue D316 of genotype 2a (JFH-1) NS5A corresponds to D320 in genotype 1b, due to a 4-amino-acid deletion toward the N-terminal end of domain II in JFH-1 (12). Both of these mutations retained the ability to bind the 3′ UTR RNA at a level similar to that observed for wild-type domain II; however, the RNA binding ability of both mutants was nonresponsive to CypA. However, this was not due to the inability of these mutants to bind CypA, as both bound as well as the wild type (Fig. 4a), consistent with previously published data (2, 20).
Fig. 4.
CypA stimulation of domain II RNA binding requires isomerase activity. (a) GST-CypA binding assays were performed as described in the legend to Fig. 1. Mutant forms of NS5A domain II were generated by QuikChange mutagenesis (primer sequences are available upon request). (b) RNA binding assays were performed as described in the legend to Fig. 2. Where indicated (+), GST-CypA (wild type [WT] or H126Q) were included at 100 nM.
Our data are consistent with the notion that CypA is able to stimulate the binding of NS5A to the 3′ UTR RNA. This stimulation requires both a stable interaction between CypA and domain II of NS5A and the CypA-catalyzed cis/trans isomerization of peptidyl-prolyl bonds within domain II. The interaction of CypA with NS5A in the absence of isomerase activity is not sufficient to mediate the stimulation of NS5A RNA binding. However, it has previously been noted that CsA disrupts the NS5A-CypA interaction (4), so the observation that CsA abolishes the CypA enhancement of NS5A RNA binding may be due to this disruption rather than inhibition of isomerase activity.
The role of CypA in HCV RNA replication remains undefined, although various mechanisms have been proposed. These include recruitment of NS5B into the replicase (15) and stimulation of polyprotein proteolytic cleavage (11). What is clear is that the isomerase activity of CypA is required (1, 15). Furthermore, recent biochemical studies have demonstrated that CypA catalyzes cis/trans isomerization of peptidyl-prolyl bonds within domain II of NS5A, in particular the peptide bond preceding residue P319 (in genotype 1b) or P315 (in genotype 2a JFH-1) (4, 7), although other prolines within domain II may also be substrates for cis/trans isomerization (7). Domain II has been shown to be natively unstructured (13, 14), and there is good evidence that CypA-catalyzed cis/trans isomerization could result in a conformational change (4, 20). This conformational change, to a more extended form at the CypA substrate binding site, is also induced by the D316E mutation, explaining the CypA independence of the D316E replicon. Our data show that domain II containing the D316E mutation binds as well as the wild type to both CypA and RNA; however, CypA does not stimulate RNA binding (Fig. 4). These data suggest that the D316E mutation blocks some additional event required for stimulation of RNA binding by CypA, for example, cis/trans isomerization at residues other than P315 within domain II. Structural data for the domain II-RNA complex with or without CypA would be needed to address this question, although it is notable that, using a mass spectrometric approach, the C. Cameron laboratory recently identified two peptide sequences within domain II of genotype 1b NS5A that interacted with poly(U) RNA (9), residues 263 to 277 and 295 to 311. The latter peptide contains two conserved proline residues (although overall conservation with JFH-1 is poor within this sequence) and a basic cluster; it is thus conceivable that the conformation of this putative RNA binding motif could be modulated by cis/trans isomerization.
In conclusion, our data provide the first biochemical evidence for a functional role for CypA-NS5A interaction in the process of HCV RNA replication that can be recapitulated in vitro. In this regard, we propose that the observed role of CypA in the enhancement of HCV RNA replication may be dependent, at least in part, on its stimulation of the RNA binding ability of NS5A. One note of caution should be considered: if this were the case, the D316E/Y317N mutant would be expected to bind the RNA more efficiently than the wild type, and our data do not support this conclusion (Fig. 4b). However, as yet, the RNA binding motifs in NS5A remain undefined, and it may be that the D316E/Y317N mutation also directly impacts the RNA binding of domain II. Experiments for addressing these questions are under way in our laboratory.
Acknowledgments
We are grateful to Takaji Wakita for the HCV genotype 2a (JFH-1) molecular clone.
This work was supported by the Wellcome Trust. T.L.F. was a postgraduate student within the Wellcome Trust 4-year Ph.D. program, entitled “The Molecular Basis of Biological Mechanisms,” awarded to the Astbury Centre for Structural Molecular Biology. Work in the N.J.S. laboratory is supported by Yorkshire Cancer Research.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Chatterji U., et al. 2009. The isomerase active site of cyclophilin A is critical for hepatitis C virus replication. J. Biol. Chem. 284:16998–17005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chatterji U., et al. 2010. HCV resistance to cyclosporin A does not correlate with a resistance of the NS5A-cyclophilin A interaction to cyclophilin inhibitors. J. Hepatol. 53:50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ciesek S., et al. 2009. Cyclosporine A inhibits hepatitis C virus nonstructural protein 2 through cyclophilin A. Hepatology 50:1638–1645 [DOI] [PubMed] [Google Scholar]
- 4. Coelmont L., et al. 2010. DEB025 (Alisporivir) inhibits hepatitis C virus replication by preventing a cyclophilin A induced cis-trans isomerisation in domain II of NS5A. PLoS One 5:e13687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fischer G., Gallay P., Hopkins S. 2010. Cyclophilin inhibitors for the treatment of HCV infection. Curr. Opin. Invest. Drugs 11:911–918 [PubMed] [Google Scholar]
- 6. Foster T. L., Belyaeva T., Stonehouse N. J., Pearson A. R., Harris M. 2010. All three domains of the hepatitis C virus nonstructural NS5A protein contribute to RNA binding. J. Virol. 84:9267–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hanoulle X., et al. 2009. Hepatitis C Virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. J. Biol. Chem. 284:13589–13601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang L., et al. 2005. Hepatitis C virus non-structural protein 5A (NS5A) is a RNA-binding protein. J. Biol. Chem. 280:36417–36428 [DOI] [PubMed] [Google Scholar]
- 9. Hwang J., et al. 2010. Hepatitis C virus nonstructural protein 5A: biochemical characterization of a novel structural class of RNA-binding proteins. J. Virol. 84:12480–12491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ishii N., et al. 2006. Diverse effects of cyclosporine on hepatitis C virus strain replication. J. Virol. 80:4510–4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaul A., et al. 2009. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 5:e1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuiken C., Yusim K., Boykin L., Richardson R. 2005. The Los Alamos hepatitis C sequence database. Bioinformatics 21:379–384 [DOI] [PubMed] [Google Scholar]
- 13. Liang Y., Kang C. B., Yoon H. S. 2006. Molecular and structural characterization of the domain 2 of hepatitis C virus non-structural protein 5A. Mol. Cells 22:13–20 [PubMed] [Google Scholar]
- 14. Liang Y., Ye H., Kang C. B., Yoon H. S. 2007. Domain 2 of nonstructural protein 5A (NS5A) of hepatitis C virus is natively unfolded. Biochemistry 46:11550–11558 [DOI] [PubMed] [Google Scholar]
- 15. Liu Z., Yang F., Robotham J. M., Tang H. 2009. Critical role of cyclophilin A and its prolyl-peptidyl isomerase activity in the structure and function of the hepatitis C virus replication complex. J. Virol. 83:6554–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Macdonald A., et al. 2003. The hepatitis C virus NS5A protein inhibits activating protein-1 (AP1) function by perturbing Ras-ERK pathway signalling. J. Biol. Chem. 278:17775–17784 [DOI] [PubMed] [Google Scholar]
- 17. Shepard C. W., Finelli L., Alter M. J. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558–567 [DOI] [PubMed] [Google Scholar]
- 18. Tellinghuisen T. L., Marcotrigiano J., Gorbalenya A. E., Rice C. M. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576–48587 [DOI] [PubMed] [Google Scholar]
- 19. Watashi K., et al. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19:111–122 [DOI] [PubMed] [Google Scholar]
- 20. Yang F., et al. 2010. A major determinant of cyclophilin dependence and cyclosporine susceptibility of hepatitis C virus identified by a genetic approach. PLoS Pathog. 6:e1001118. [DOI] [PMC free article] [PubMed] [Google Scholar]