Abstract
Knotted DNA has potentially devastating effects on cells. By using two site-specific recombination systems, we tied all biologically significant simple DNA knots in Escherichia coli. When topoisomerase IV activity was blocked, either with a drug or in a temperature-sensitive mutant, the knotted recombination intermediates accumulated whether or not gyrase was active. In contrast to its decatenation activity, which is strongly affected by DNA supercoiling, topoisomerase IV unknotted DNA independently of supercoiling. This differential supercoiling effect held true regardless of the relative sizes of the catenanes and knots. Finally, topoisomerase IV unknotted DNA equally well when DNA replication was blocked with hydroxyurea. We conclude that topoisomerase IV, not gyrase, unknots DNA and that it is able to access DNA in the cell freely. With these results, it is now possible to assign completely the topological roles of the topoisomerases in E. coli. It is clear that the topoisomerases in the cell have distinct and nonoverlapping roles. Consequently, our results suggest limitations in assigning a physiological function to a protein based upon sequence similarity or even upon in vitro biochemical activity.
Keywords: DNA supercoiling, gyrase, site-specific recombination, DNA catenane, quinolone, DNA replication
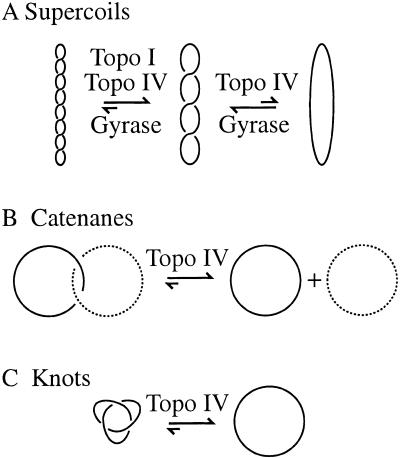
Metabolic processing of the DNA double helix leads to three topological forms that all cells must resolve to survive: linking number (Lk), catenanes, and knots (Fig. 1). The Lk is the number of times that the two strands of the DNA are interwound. In cells, the value of Lk typically is lower than would be most thermodynamically favored (Lk0), and, as a result, the DNA writhes back upon itself, resulting in negative supercoiling (Fig. 1A). Catenanes, two or more DNA molecules interlinked, are important intermediates in DNA replication (Sundin and Varshavsky 1981; Marians 1987; Adams et al. 1992b; Shekhtman et al. 1993; Zechiedrich and Cozzarelli 1995) and recombination (Fig. 1B; for review, see Wasserman and Cozzarelli 1986). A DNA knot is a single DNA molecule entangled with itself (Fig. 1C).
Figure 1.
Topological forms. A single line represents the DNA double helix. No deformation short of severing the DNA backbone can alter the three topological forms of DNA. (A) The torsional stress resulting from the Lk deficit causes the DNA double helix to writhe about itself, termed negative supercoiling. In Escherichia coli cells, gyrase introduces negative supercoils into DNA and is countered by topoisomerase I (topo I) and topoisomerase IV (topo IV), which relax negative supercoils (Menzel and Gellert 1983; Zechiedrich et al. 2000). Topoisomerase I relaxes the DNA partially, and topoisomerase IV relaxes it nearly completely (Zechiedrich et al. 2000). (B) DNA catenanes are two (or more) interlocked DNA rings. Solid and dashed lines distinguish the two rings here. Topoisomerase IV unlinks catenanes generated by replication or recombination in vivo (Adams et al. 1992a; Zechiedrich and Cozzarelli 1995; Zechiedrich et al. 1997). (C) A knot is the entanglement of a single DNA molecule. We show here that topoisomerase IV unknots DNA in vivo.
The effects that negative supercoils or unresolved catenanes have on cellular functioning can be envisioned easily. Negative supercoiling favors any process that requires double helix opening, including DNA replication, transcription, and recombination (Kanaar and Cozzarelli 1992; Kornberg and Baker 1992; Shlyakhtenko et al. 2000). Whereas negative supercoils can have beneficial effects on DNA metabolism when properly regulated, DNA catenanes are mostly, if not exclusively, problematic to the cell if they are left unresolved. For example, catenane links must be entirely eliminated to allow for the partitioning of chromosomes.
Although the effects of knotting on cellular functions have not been evaluated in vivo, the following evidence reveals that knotted DNA may have consequences for cells that are distinct from those effects caused by supercoiling or catenation: (1) In purified systems, gene transcription is significantly impaired on knotted templates (Portugal and Rodriguez-Campos 1996). (2) DNA knots inhibit chromatin assembly in cellular extracts (Rodriguez-Campos 1996). (3) DNA knots may serve as topological barriers between different regions of the chromosome, altering both the structural organization of the genome and whether DNA sites can interact (Staczek and Higgins 1998). (4) An important and potentially lethal cellular consequence of knotted DNA, long familiar to mountain climbers and anglers working with ropes, is that knots weaken lines and lead to breaks (McNally 1993). It has been demonstrated, both by computer simulation (Saitta et al. 1999) and by single molecule studies (Arai et al. 1999), that knots lower the tensile strength of biologically relevant polymers. Therefore, the failure to resolve a knotted genome may lead to double-strand breaks in the DNA, which are potentially lethal events.
Although knots could be problematic, DNA knotting occurs naturally in cells (Shishido et al. 1987; Martin-Parras et al. 1998). Knots result from packaging the long DNA into the small cell and could arise from any process that involves DNA strand breaks, including DNA replication, recombination, repair, and topoisomerization (Liu et al. 1980; Mizuuchi et al. 1980; Pollock and Nash 1983; Spengler et al. 1985; Wasserman and Cozzarelli 1991). Replicated DNA located behind a stalled fork, an event estimated to occur with as many as half of the replication forks in Escherichia coli during aerobic growth conditions (Cox 1998), has been shown to be particularly prone to knotting (Viguera et al. 1996; Sogo et al. 1999). In addition, the transformation of the plectonemic supercoils (Fig. 1A) known to be prevalent in E. coli (Bliska and Cozzarelli 1987) into knots may be thermodynamically favored. Knotted DNA has been calculated to adopt a conformation that is lower in free energy than the plectonemically supercoiled DNA (Vologodskii and Marko 1997; Podtelezhnikov et al. 1999). This process would be particularly detrimental to the cell because once supercoiled DNA is converted to knotted DNA, the benefits of DNA supercoiling are lost (Podtelezhnikov et al. 1999).
In spite of the propensity of cellular DNA to become knotted, and the continuous action of the processes within the cell that can cause knotting, only a low steady-state fraction (<1%) of plasmid DNA is knotted normally (Shishido et al. 1987; Martin-Parras et al. 1998; R.W. Deibler and E.L. Zechiedrich, unpubl.). This low amount suggests that the cell is well equipped to remove knots. Although other possible ways to untie a DNA knot may exist, the most straightforward and least dangerous to the cell is to create a transient break in the DNA backbone, pass DNA through this break (to remove the knot crossing), and reseal the broken DNA. This is the mechanism of a topoisomerase.
Topoisomerases are divided into two types. The type-1 enzymes, topoisomerases I and III in E. coli, alter Lk in steps of one by passing nucleic acids through single-strand breaks in the DNA. The type-2 topoisomerases are gyrase and topoisomerase IV, and they alter Lk in steps of two by passing DNA through a double-strand break. Both of these enzymes are essential (Menzel and Gellert 1983; Kato et al. 1990; Schmid 1990).
Traditionally, gyrase has been considered to be the cellular unknotting enzyme based upon the following observations: (1) Purified gyrase unties several different types of knots in vitro (Liu et al. 1980; Mizuuchi et al. 1980). (2) Quinolone antibiotics, previously thought to target only gyrase, cause an accumulation of knotted plasmids in vivo (Ishii et al. 1991; Adams et al. 1992a). (3) Mutations in gyrase that reduce its supercoiling activity in the cell also lead to an increased accumulation of knots (Shishido et al. 1987). (4) Gyrase seems to be designed to transport a contiguous segment of DNA through itself during the intramolecular supercoiling reaction (Liu and Wang 1978; Kirchhausen et al. 1985; Vologodskii and Cozzarelli 1996). Knots are also an intramolecular substrate.
A role for topoisomerase IV in unknotting DNA in vivo has not been addressed directly. Even studies that demonstrated the role for topoisomerase IV in unlinking catenated replicons could not discern a role for topoisomerase IV in DNA unknotting (Adams et al. 1992b; Khodursky et al. 1995). However, a role for this enzyme may have been overlooked in the aforementioned studies concluding that gyrase unknotted DNA. The quinolones used in these studies would have also inhibited topoisomerase IV, which is now known to be a target of these drugs in vivo (Khodursky et al. 1995). In the strains containing mutations in gyrase, it is possible that additional mutations existed in the genes for topoisomerase IV (Raji et al. 1985). Like gyrase, purified topoisomerase IV is capable of unknotting DNA (Kato et al. 1992). In addition, whereas previous models for type-2 topoisomerase action predicted that knotting and unknotting should be equally likely, topoisomerase IV and its eukaryotic homologs (gyrase was not tested) possess a significant intrinsic preference to unknot rather than knot DNA (Rybenkov et al. 1997a). Additionally, purified topoisomerase III is proficient at knotting and unknotting DNA with single-stranded regions (Du et al. 1995). In conjunction with the RecQ helicase, topoisomerase III has been demonstrated to link intact double-stranded DNA in vitro (Harmon et al. 1999). If topoisomerase III and RecQ can reverse this process in vivo, then topoisomerase III may contribute to unknotting despite being a type-1 enzyme.
Here we show that topoisomerase IV unknots DNA in E. coli. Gyrase does not affect unknotting DNA in vivo directly or indirectly through its supercoiling activity. We also show that the rate of topoisomerase IV unknotting is identical in the presence or absence of DNA replication, which demonstrates that topoisomerase IV activity does not require an association with ongoing DNA replication.
Results
Experimental strategy
We generated DNA knots using two different site-specific recombination systems and measured the removal of these recombinant knots following the inhibition of gyrase and/or topoisomerase IV. The site-specific recombination systems allowed us to generate a large number of knots with defined topology and to study the effects of topoisomerases specifically upon the removal, rather than the formation, of knots.
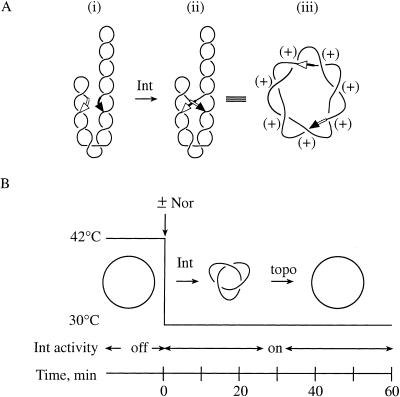
We first used the well-characterized site-specific Int (integrase) recombination system of bacteriophage λ, which the phage employs for integration and excision from the host chromosome. When the Int recombination sites attB and attP are on the same DNA plasmid in an inverted (head-to-head) orientation, site synapsis traps supercoil nodes, which are converted to knot nodes by recombination (Fig. 2A). Int is expressed in E. coli strains harboring a mutant λ lysogen incapable of excision (see Table 1). Recombination, in addition to knotting the plasmid, inverts the DNA between the two sites and destroys the attP and attB sites to form attL and attR to prevent further recombination.
Figure 2.
Int recombination system to knot DNA. (A) Mechanism of Int recombination. Int and the host protein IHF (not shown) bind to the att sites on the negatively supercoiled Int recombination substrate (pJB3.5i or pABCi) with the recombination att sites (arrows) in an inverted orientation to form the synaptic complex (i). Strand exchange, which is shown by the switching of arrowheads (ii), converts the DNA supercoil nodes that were trapped by synapsis into knot nodes. The knotted Int recombination product (iii) is torus, right-handed, and positive. (B) Experimental protocol. Details are described in the text.
Table 1.
E. coli strains
| Strain
|
Genotypea
|
Reference
|
|---|---|---|
| C600 | F-thr-1 leu-6 thi-1 lacYl sup E44 tonA21 | Kato et al. 1988 |
| LZ23 | C600 except gyrAL83zei-723::Tn10 kanr | Khodursky et al. 1995 |
| LZ24 | C600 except zei-723::Tn10 kanr | Khodursky et al. 1995 |
| LZ35 | W3101λ except gyrAL83zei-723::Tn10 kanr | Zechiedrich et al. 1997 |
| LZ36 | W3101λ except zei-723::Tn10 kanr | Zechiedrich et al. 1997 |
| LZ37 | LZ35 except parCK84 | Zechiedrich et al. 1997 |
| LZ38 | LZ36 except parCK84 | Zecheidrich et al. 1997 |
| ParE10 | W3110 except parE10 | Kato et al. 1990 |
| W3101λ | F-λ (P80 red114 xis-l cl857) | Bliska and Cozzarelli 1987 |
| W3110 | F- | Kato et al. 1990 |
| 1643 | C600 except gyrAL83zei-723::Tn10 parCK84, kanr | Khodursky et al. 1995 |
| 1644 | C600 except zei-723::Tn 10parCK84, kanr | Khodursky et al. 1995 |
kan, kanamycin; r, resistant.
To selectively inhibit gyrase or topoisomerase IV, we used an isogenic set of E. coli strains (see Table 1). In these strains, the wild-type alleles of gyrase (gyrA+) and topoisomerase IV (parC+) produce enzymes that are inhibited by norfloxacin, whereas mutant alleles, gyrAr and parCr, produce drug-resistant forms of the enzymes that remain fully active in the presence of the drug.
The experimental protocol is diagrammed in Figure 2B. Cells harboring the plasmid pJB3.5i, or in later experiments pABCi, were grown with shaking to late logarithmic phase (70 Klett units) at 30°C. The cultures were then shifted to 42°C to induce Int expression (Bliska and Cozzarelli 1987). The cultures were shifted back to 30°C to activate the Int protein, which is not functional at 42°C (Bliska and Cozzarelli 1987; Zechiedrich et al. 1997). Upon return to 30°C (Time = 0 min), the quinolone norfloxacin was added to inhibit gyrase and topoisomerase IV, which are the only cellular targets of this drug up to 100-fold greater than the minimal inhibitory concentration (Khodursky et al. 1995). Neither topoisomerase III nor topoisomerase I is inhibited by the norfloxacin concentrations used here (Harmon et al. 1999; Zechiedrich et al. 2000). Plasmid DNA was isolated at various times following the shift back to 30°C and divided into three fractions, which were treated as described below.
Removal of DNA knots generated by Int recombination
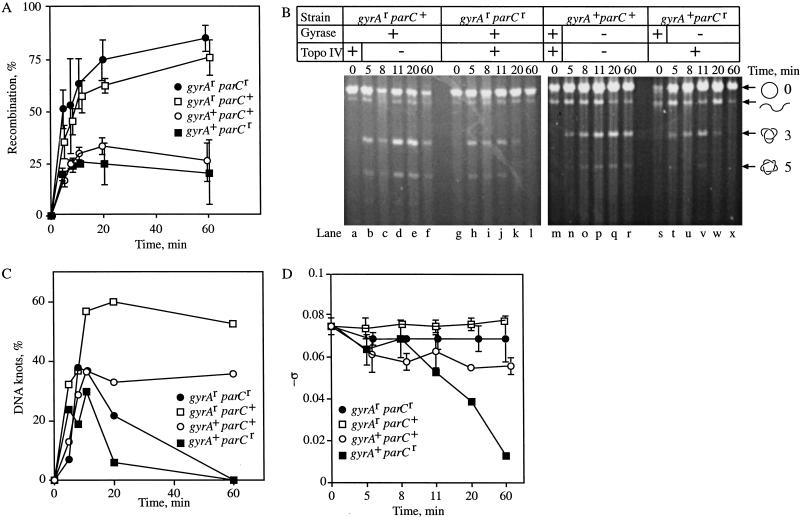
The amount of Int recombination was measured by digesting one fraction of the purified plasmid DNA with BamHI and EcoRI, which generates differently sized products in the parental and inverted orientation. All recombination proceeds through a knotted intermediate (Pollock and Nash 1983). Therefore, the fraction of DNA that underwent recombination equaled the fraction of DNA that was knotted in an experiment. The digestion products were displayed on 1% agarose gels and quantified using scanning densitometry (Nucleovision). There was no detectable recombination before the cells were shifted back to 30°C, indicating that the system was tightly regulated (Fig. 3A, Time = 0 min). Recombination occurred efficiently in all strains following induction of Int (Fig. 3A). In accordance with previous results (Zechiedrich et al. 1997), DNA isolated from cells in which gyrase activity was inhibited by norfloxacin (gyrA+) had less recombination (25%–30% of the total plasmid DNA) than that from cells in which gyrase was functioning (gyrAr, ∼80%). Because DNA supercoiling stimulates Int recombination, the loss of gyrase activity causes less recombination.
Figure 3.
Metabolism of Int recombination knots in vivo. (A) The extent of Int recombination for each strain is shown. Cells were grown as described for Figure 2. Plasmid DNA was isolated and submitted to restriction digestion with EcoRI and BamHI. Samples were analyzed by agarose gel electrophoresis. Shown is the percent of total plasmid DNA recombined with time. No recombination occurred before Int was induced (Time = 0). Error bars are from two experiments that were averaged. (B) Unknotting of Int recombination products. Representative agarose gels of Int recombination in the isogenic strains. In this and subsequent figures, the grid above the gels shows which strain was used and whether gyrase and topoisomerase IV were active (+) or not (−). The positions of nicked circular (○) and linear (wavy line) plasmid, as well as the trefoil (3) and pentafoil (5) knots are shown. (C) Graphic representation of the data. Shown is the percent of the total plasmid DNA that was knotted with time for each strain. Data were quantified by Scanning densitometry with Nucleovision or TechSoftware PhosphorImage analyses using Storm. Each time point was repeated 2–3 times with similar results. (D) A fraction of the purified plasmid DNA used in A and B was subjected to electrophoresis on 1%–1.2% agarose gels that contained (2–20 μg/mL) chloroquine to visualize DNA supercoiling (Zechiedrich et al. 2000). The mean ς was calculated and is shown for each strain over time. Where not shown, the error was smaller than the symbol (except for the gyrA+ parCr strain at 8, 20, and 60 min, each of which was performed once).
A second portion of the plasmid was nicked with DNase I to analyze DNA unknotting. The electrophoretic mobility of several types of knots has been studied extensively (Wasserman and Cozzarelli 1991; Stasiak et al. 1996; Vologodskii et al. 1998). The knots in our experiments were identified by comparison to catenane and knot ladders generated by Int (Spengler et al. 1985) and T4 type-2 topoisomerase (Wasserman and Cozzarelli 1991). An ethidium-bromide-stained agarose gel is shown in Figure 3B. The amount of knotting was quantified and is shown graphically in Figure 3C. When both gyrase and topoisomerase IV were functioning (gyrAr parCr), all DNA knots were removed by 60 min, even in the presence of norfloxacin (Fig. 3B, lanes g–l; Fig. 3C, line with filled circles). In contrast, DNA knots persisted when both gyrase and topoisomerase IV were inhibited (gyrA+ parC+) with norfloxacin (Fig. 3B, lanes m–r; Fig. 3C, line with open circles). Because there was no significant metabolism of knots in the absence of both gyrase and topoisomerase IV, we conclude that no significant additional unknotting activity is present.
In cells where topoisomerase IV was functioning, but gyrase was inhibited with norfloxacin (gyrA+ parCr), knots were removed by 60 min (Fig. 3B, lanes s–x; Fig. 3C, line with filled boxes). The knot removal by topoisomerase IV, alone, was similar to the results when both enzymes were functioning. We found no unknotting when topoisomerase IV was inhibited and gyrase remained fully functional in the gyrAr parC+ strain (Fig. 3B, lanes a–f; Fig. 3C, line with open boxes). Even after 60 min, nearly all the recombined DNA remained knotted. This result shows topoisomerase IV unties right-handed, torus knots.
To confirm that norfloxacin had affected specifically gyrase or topoisomerase IV, we used the third portion of purified plasmid DNA to measure DNA supercoiling. As shown in Figure 1A, DNA supercoiling is determined by gyrase countering topoisomerases I and IV (Zechiedrich et al. 2000). When gyrase is inhibited, topoisomerases I and IV relax DNA. When topoisomerase IV is inhibited, the DNA attains a more negatively supercoiled steady-state level (Fig. 1A). The plasmid DNA from the experiment was subjected to electrophoresis in agarose gels containing chloroquine, which allows separation of the supercoil topoisomers. Shown is −ς with time after the addition of norfloxacin. We found that the topoisomerases were inhibited as expected (Fig. 3D). When both enzymes remained active with drug, in the gyrAr parCr strain, DNA supercoiling remained near normal levels (Fig. 3D, line with filled circles). When only topoisomerase IV was inhibited (gyrAr parC+), the DNA became slightly more negatively supercoiled (Fig. 3D, line with open boxes). In the gyrA+ parC+ strain, the DNA relaxed to a ς of (−) 0.057 through the action of topoisomerase I alone (Fig. 3D, line with open circles). When topoisomerase IV remained functional and gyrase was inhibited (gyrA+ parCr), the DNA became almost completely relaxed (Fig. 3D, line with filled boxes). These supercoiling measurements have an additional importance that is described below.
Removal of DNA knots generated by Hin recombination
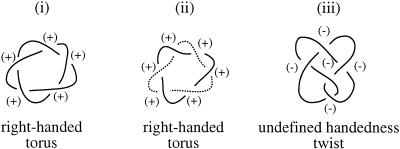
Int-mediated DNA knots and the catenated intermediates of DNA replication are toroidal, right-handed, and positive in structure. Therefore, it could be possible that our Int knots were removed by topoisomerase IV because of their similarity to catenanes generated by DNA replication (Fig. 4, cf. i and ii), and not because topoisomerase IV normally unknots all DNA in the cell. Furthermore, in strains containing a mutant gyrase, knots were found to accumulate that were negative and belonged to the twist family (Shishido et al. 1987), a family defined by characteristic twist nodes (see Fig. 4A,iii). This finding suggests that gyrase may have a role in untying knots with a negative twist topology. To determine how negative twist knots are resolved, we utilized an in vivo E. coli DNA knotting system based upon the Hin invertase site-specific recombination system from Salmonella typhimurium.
Figure 4.
Topology of knots and catenanes. As in Figures 1 and 2, the DNA double helix is shown as a single line. Int recombination produces toroidal, right-handed, positive knots (i). DNA replication produces catenated intermediates that are also toroidal, right-handed, and positive (ii) (Adams et al. 1992b). The two rings are distinguished by solid and dashed lines. Hin recombination produces negative twist knots that contain elements of both right- and left-handedness and, therefore, have undefined handedness (iii).
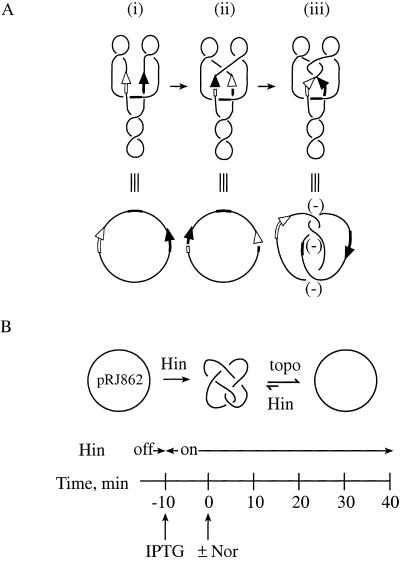
The mechanism that Hin uses to knot DNA is shown in Figure 5A. Hin and the protein Fis bind to the hix sites and the enhancer, respectively, of a negatively supercoiled plasmid substrate with the geometry shown (Heichman and Johnson 1990). Hin cleaves the hix sites, leaving a 2-nucleotide overhang (Fig. 5A), and the DNA strands rotate 180° in a right-handed direction. If the DNA has wild-type hix sites, the overhangs created by cleavage are identical, and religation can occur after a single 180° strand rotation. However, in the mutant recombination substrate used here, Hin cleavage produces overhangs that are not complementary (Heichman et al. 1991). A 360° strand rotation, which will return the hix sites to the original parental orientation, must take place so that religation can occur, which results in a negative trefoil knot (Fig. 5A,iii; Heichman et al. 1991). If additional 360° strand rotations take place, twist knots with five, seven, or more nodes result (Heichman et al. 1991). Hin can also tie additional knot nodes into plasmids that are already knotted to form what is referred to as composite knots. The products of Hin recombination, including composite knots, have a negative twist topology. The majority of knots isolated from cells have this topology (Shishido et al. 1987; Heichman et al. 1991; Ishii et al. 1991; R.W. Deibler and E.L. Zechiedrich, unpubl.).
Figure 5.
Hin recombination system to knot DNA. (A) Hin recombination. Hin and Fis (not shown) bind to the hix sites, denoted by arrows, and the enhancer (thickened line), respectively, on negatively supercoiled DNA to form the synaptic complex shown (i). Hin makes a double-stranded DNA staggered break in each of the hix sites to leave two nucleotide overhangs (Johnson and Simon 1985; Johnson et al. 1986; Heichman and Johnson 1990). With a wild-type substrate, the DNA overhangs are complementary and DNA religation would occur after strand exchange (ii). In the Hin substrate used here, pRJ826 (Heichman et al. 1991), an additional right-handed rotation restores the DNA to its original configuration and religation can take place (Heichman et al. 1991) (iii). The hix sites are not altered by recombination and serve as substrates for further recombination. (B) Experimental protocol. Details are described in the text.
We cotransformed an isogenic set of E. coli strains derived from the C600 parental background (see Table 1) with the Hin recombination substrate pRJ862, and pKH66, which contains the hin gene under control of the IPTG-inducible tac promoter. The C600 strains contained the various combinations of wild-type and drug-resistant gyrase and topoisomerase IV alleles as used above (see Table 1). The assay is shown in Figure 5B. Cells were grown with shaking at 37°C to mid-logarithmic phase (30 Klett units). IPTG was added to a final concentration of 1 mM. Following a 10-min incubation to allow Hin expression, norfloxacin was added (Time = 0 min) to inhibit the type-2 topoisomerases. Because this system does not lead to the destruction of the recombination sites as recombination by Int does, Hin continuously recombines the DNA to form new knots. Consequently, the cellular topoisomerases constantly remove these knots as they are formed, to reach a steady-state level of knotting of ∼5% of the total plasmid population in all strains (Fig. 6B, Time = 0).
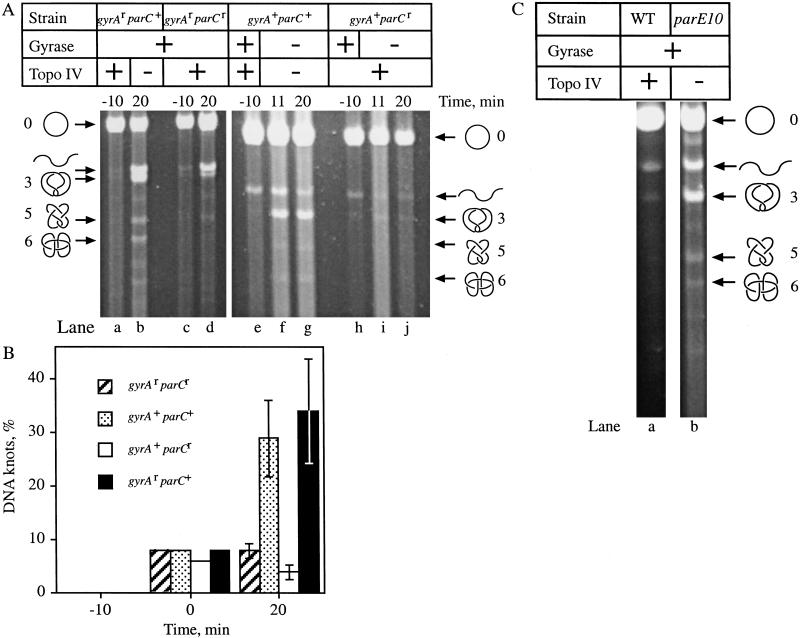
Figure 6.
Metabolism of Hin recombination knots in vivo. (A) Unknotting of Hin recombination products. Hin recombination was carried out and plasmid DNA was isolated, nicked with DNase I, and subjected to electrophoresis through 1% agarose gels. Ethidium bromide-stained agarose gels are shown. The positions of nicked circular (○), linear (wavy line), and knotted (3,5,6) DNA are indicated. (B) Graphic representation of the data from two experiments. The data are presented as the percent of total plasmid DNA knotted for three different time points, after the addition of IPTG (−10 min), after the addition of norfloxacin (Nor) (0 min), and 20 min following the addition of Nor (20 min). (C) Gel showing knotting in parE10 strain. Cells containing the Hin recombination expression vector, pKH66, and the substrate, pRJ862, were grown as described for Figure 5. However, before the addition of IPTG, wild-type (WT) (lane a) and parE10 mutant (lane b) cells were shifted to 42°C to inactivate the mutant topoisomerase IV. The positions of nicked circular (○), linear (wavy line), and knotted (3,5,6) DNA are shown. This experiment was repeated with similar results.
At various times following the induction of Hin expression, plasmid DNA was isolated, nicked, and visualized by high-resolution agarose gel electrophoresis (Sundin and Varshavsky 1981). As with the knots generated by Int, when both gyrase and topoisomerase IV were drug resistant (gyrAr parCr), DNA knots formed by Hin recombination in vivo were untied in the presence of norfloxacin (Fig. 6A, lanes c,d; Fig. 6B, hatched bar). Knotting remained near the 5% level as had been observed in the absence of drug. When both gyrase and topoisomerase IV were inhibited by norfloxacin (gyrA+ parC+), knots accumulated (Fig. 6A, lanes e–g). Roughly 30% of the total plasmid DNA was knotted 20 min after the addition of norfloxacin in the gyrA+ parC+ strain (Fig. 6B, stippled bar).
Topoisomerase IV alone (gyrA+ parCr) unknotted DNA to a similar level as when both gyrase and topoisomerase IV were active (Fig. 6A, cf. lanes j and d; Fig. 6B, cf. white to hatched bar at 20 min). However, in the absence of topoisomerase IV, a functional gyrase (gyrAr parC+) could not untie the Hin-mediated knots (Fig. 6A, lanes a,b). In fact, the highest levels of DNA knotting were seen in this strain; ∼35% of the total plasmid DNA was knotted (Fig. 6B, black bar). This finding most likely results from gyrase-mediated supercoiling stimulating Hin recombination, because Hin recombination is known to be greatly affected by DNA supercoiling. More recombination, in turn, leads to more knots being tied. We conclude that topoisomerase IV, not gyrase, resolves the knotted negative twist products of Hin recombination.
DNA unknotting in conditionally lethal topoisomerase IV mutants
In the experiments described above, the antibiotic norfloxacin was used to inhibit topoisomerase IV. Indeed, the previous experiments examining the in vivo role of topoisomerase IV in removing DNA catenanes produced by site-specific recombination also relied solely upon norfloxacin-based inhibition of topoisomerase IV because these systems are thermosensitive (Zechiedrich et al. 1997). Although we thought it unlikely, it was possible that our results were affected by the presence of norfloxacin. Therefore, we repeated the Hin recombination experiments in a strain containing a conditionally lethal mutation in the parE gene of topoisomerase IV, parE10. This mutant strain fails to segregate its chromosomes, accumulates catenated intermediates of DNA replication, and is nonviable at 42°C (Kato et al. 1990; Adams et al. 1992b; Zechiedrich and Cozzarelli 1995). However, the parE10 mutant has wild-type topoisomerase IV activity at the permissive temperature (30°C) as measured by in vivo decatenation (Zechiedrich and Cozzarelli 1995) and relaxation assays (Zechiedrich et al. 2000).
Because Hin has been shown to be thermolabile in vitro (Johnson and Simon 1985), we measured Hin-mediated knotting in the gyrAr parC+ strain at 42°C. The experiment was carried out as for Figure 6A. The results at 42°C were indistinguishable from those obtained at 37°C (not shown). This shows that Hin is fully functional at 42°C in vivo, and could be used in conjunction with the temperature-sensitive topoisomerase IV mutant.
DNA knotting was compared for the parE10 mutant and its wild-type parental strain at 42°C. Cultures were grown at 30°C with shaking to mid-logarithmic phase (35 Klett units). IPTG was added to induce Hin expression (Time = −10). After 10 min, cells were shifted to 42°C to inactivate topoisomerase IV. Plasmid DNA was isolated 40 min later. The accumulation of catenated replication intermediates (Adams et al. 1992b; Zechiedrich and Cozzarelli 1995) confirmed that topoisomerase IV had been inactivated (data not shown). At 30°C, Hin tied knots and topoisomerase IV untied them to a steady-state level in either strain of ∼5% (data not shown). However, Hin-mediated DNA knots accumulated in the parE10 mutant (Fig. 6C, lane b) and not in the wild-type strain at 42°C (Fig. 6C, lane a). Therefore, knotted recombination products accumulate in the absence of topoisomerase IV regardless of the means used to inactivate the enzyme.
Effect of negative supercoiling on DNA unknotting
Previous studies showed a marked effect of DNA supercoiling on decatenation by topoisomerase IV both in vivo (Zechiedrich et al. 1997) and with purified components (Ullsperger and Cozzarelli 1996). In addition, as explained above, gyrase mutants with a reduced supercoiling activity have higher levels of knotting than wild-type cells (Shishido et al. 1987). A possible explanation for these findings is that gyrase, which has a pervasive effect on DNA metabolism, contributes to unknotting in vivo by supercoiling DNA. This might be expected because negative supercoiling increases the free energy of a knot (Shaw and Wang 1993). In addition, negative supercoiling may group the knot nodes together (Marko 1999) or facilitate the localization of a preferred topoisomerase IV cleavage site at a knot node. Therefore, we examined the effect of negative supercoiling on topoisomerase IV unknotting in vivo.
In our experiments, we observed no turnover of knots in parC+ strains treated with norfloxacin (see Fig. 3C). However, through its supercoiling activity, gyrase stimulated decatenation in these same strains under identical conditions (Zechiedrich et al. 1997). Therefore, gyrase-mediated supercoiling did not stimulate unknotting and did stimulate decatenating by topoisomerase IV treated with norfloxacin. In the parCr strains, the rate and extent of unknotting was the same whether gyrase was blocked by drug (gyrA+) or not (gyrA+) (Fig. 3C, compare filled squares to filled circles). These findings are similar to those found in vitro, where the rate of unknotting of Int products by purified topoisomerase IV without norfloxacin was also shown to be unaffected by negative supercoiling (Ullsperger and Cozzarelli 1996).
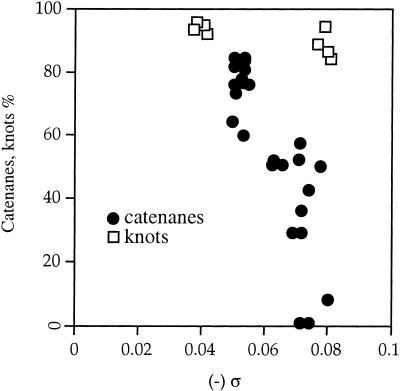
An important parameter that may distinguish the catenanes from the previous experiments from the knot substrates used here is their different lengths. The two rings of the catenane were 2.9 kb and 0.6 kb. The earlier in vitro experiments were carried out with 7.2-kb Int knots only (Ullsperger and Cozzarelli 1996). To determine whether size had an effect on the DNA supercoiling dependence of topoisomerase IV-mediated decatenation, we constructed new 7.8-kb Int substrates—pABCi, which forms knots, and pABCd, which forms catenated rings of 3.2 kb and 4.6 kb. Each ring formed from pABCd is approximately the size of the knot formed in the Int substrate plasmid pJB3.5i used above. We repeated the experiments described for Figure 3B with pABCi to test whether the removal of larger knots was also unaffected by supercoiling. As before, topoisomerase IV untied this 7.8-kb knot, and gyrase had no effect in vivo (data not shown). We plotted the amount of knotted pABCi as a function of negative supercoiling (Fig. 7). The rate and extent of topoisomerase IV unknotting was the same in all cases, including when the DNA was relaxed, in the gyrA+ parCr strain, or hypernegatively supercoiled, in the gyrAr parC+ strain. Supercoiling did not appreciably enhance unknotting of pABCi. However, negative supercoiling strongly stimulated the topoisomerase IV-mediated resolution of the pABCd catenanes (data not shown). Therefore the size of the rings does not alter the effect of supercoiling on decatenation. These findings demonstrate that over a twofold range of DNA length, decatenation by topoisomerase IV is strongly stimulated and unknotting is unaffected by negative supercoiling.
Figure 7.
Effect of DNA supercoiling (ς) on unknotting. The amount of plasmid DNA that had recombined and was knotted in the parC+-containing strains was measured. The supercoiling values were measured as described in Figure 3D. The correlation between knotting (Y axis) and ς (X axis) is shown for plasmid pABCi (open boxes). The correlation between accumulated catenanes and ς for plasmid pJB3.5d (filled circles) is shown for comparison (Zechiedrich et al. 1997).
Topoisomerase IV unknotting is independent of DNA replication
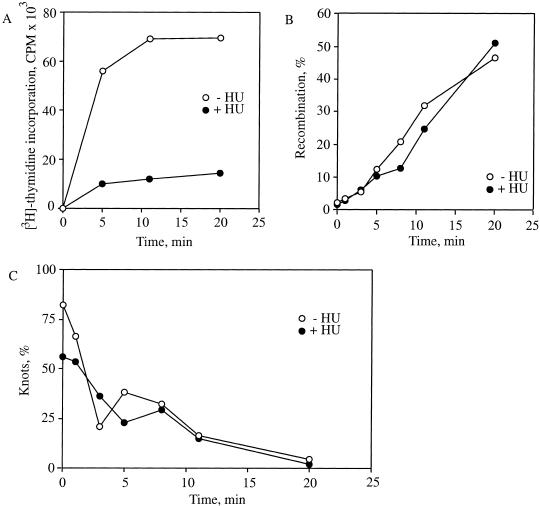
The physiological roles of the topoisomerases have been frequently studied in the context of DNA replication. Both topoisomerase IV and gyrase are involved in the synthesis and segregation of DNA (for review, see Levine et al. 1998). The overexpression of the γ subunit of the replicative DNA polymerase III holoenzyme suppressed the conditional lethality of the parE10 mutant used above in our studies (Levine and Marians 1998). This finding suggested an association between the DNA replication machinery and topoisomerase IV. The knots formed by Int do not require DNA replication. Additionally, in contrast to decatenation by topoisomerase IV, which is very rapid, unknotting is slow enough to be measurable. Therefore, we were able to test whether topoisomerase IV-mediated unknotting was obligatorily associated with DNA replication. We tied knots in pJB3.5i with Int as described in Figure 2. We then inhibited DNA synthesis with 50 mM hydroxyurea (Adams et al. 1992b) and monitored topoisomerase IV unknotting in the absence and presence of replication. The inhibition of DNA replication did not affect the recombination by λ Int, which reached 60% by 60 min in both the presence and absence of hydroxyurea (Fig. 8A). Hydroxyurea blocked DNA replication by ∼90% as measured by 3H-thymidine incorporation (Fig. 8B). The kinetics of unknotting by topoisomerase IV were identical in the presence or absence of hydroxyurea (Fig. 8C). Therefore, topoisomerase IV unknotting activity is neither dependent upon nor obligatorily associated with DNA replication.
Figure 8.
Effect of DNA replication on unknotting. (A) Inhibition of DNA replication by hydroxyurea. The LZ36 strain was grown with shaking to mid-logarithmic growth phase (70 Klett units). DNA replication rates were measured by monitoring [methyl-3H]-thymidine incorporation in the absence (open circles) or presence (filled circles) of 50 mM hydroxyurea. This experiment was performed twice with the same results. (B) Effect of hydroxyurea on rates of Int recombination. Int recombination was measured as described for Figure 2 in the absence (open circles) and presence (filled circles) of 50 mM hydroxyurea. This experiment was performed twice with the same results. (C) Effect of hydroxyurea on knot resolution by topoisomerase IV. Experiments were performed as described in Figure 2 except samples were incubated with 50 mM hydroxyurea. Plotted is the percentage of the total recombined plasmid that was knotted in the absence (open circles) or presence (filled circles) of 50 mM hydroxyurea with time. This experiment was carried out twice with the same results.
Discussion
We have determined that topoisomerase IV is the sole unknotting enzyme for E. coli. We generated knotted plasmid DNA in vivo using two different site-specific recombination systems, λ Int and S. typhimurium Hin. These two systems produce knots with different topological parameters and together represent most, if not all, of the DNA knots that form within cells (Shishido et al. 1987; R.W. Deibler and E.L. Zechiedrich, unpubl.). We have shown that although topoisomerase IV is both the decatenating and unknotting enzyme of the cell, it carries out these two processes differently. Although supercoiling promotes decatenation by topoisomerase IV, regardless of the size of the DNA rings in the catenanes, the resolution of knots is unaffected by negative supercoiling. Finally, we have shown that topoisomerase IV unknotting is independent of DNA replication.
Our finding that topoisomerase IV unknots DNA in vivo suggests new interpretations of the data that implicated gyrase in unknotting. Often these studies used antibiotics to inhibit gyrase (Ishii et al. 1991; Adams et al. 1992b). However, at the concentrations of the drugs used, topoisomerase IV would also have been inhibited (Khodursky et al. 1995; Zechiedrich et al. 1997). Therefore, knots produced by the normal metabolism of DNA may have persisted in the cell because there was insufficient topoisomerase IV function to remove the knots. In support of this, we show here that even highly negatively supercoiled DNA and active gyrase cannot assist in knot removal when topoisomerase IV is inhibited. An alternative explanation is that in the presence of quinolones, gyrase not only becomes inhibited in terms of its supercoiling activity, but also becomes more prone to knot DNA. Indeed, others have noted that there is only a subtle distinction as to which strand is passed and which is cleaved that gyrase must recognize if it is to supercoil rather than to knot DNA (Vologodskii and Cozzarelli 1996), and either drugs or mutation may affect this distinction. Because we used site-specific recombination to generate knotted DNA, obtained the same results using two different means to inhibit topoisomerase IV, and monitored the resolution rather than the formation of knots, we are confident that the possibility of a defective gyrase knotting DNA does not affect the interpretations of our data.
Gyrase introduces supercoils, but does not unknot or decatenate in the cell
As part of its enzymatic mechanism, gyrase wraps ∼135 bp of DNA about itself (Liu and Wang 1978), whereas topoisomerase IV binds only 34 bp (Peng and Marians 1995). The deletion of the wrapping domain of gyrase results in a protein that functions similarly to topoisomerase IV both biochemically and genetically in that it decatenates well in vitro and can partially suppress a topoisomerase IV temperature-sensitive mutant (Kampranis and Maxwell 1996). Therefore, DNA wrapping makes gyrase uniquely suited for the introduction of supercoils (Liu and Wang 1978; Kirchhausen et al. 1985). However, it is not clear why this wrapping, which facilitates the transport of a contiguous DNA strand, is specific to supercoiling rather than to unknotting. The preferred substrate for gyrase is thought to be a left-handed positive supercoil (for review, see Kornberg and Baker 1992). Gyrase has been proposed to act by inverting the left-handed positive DNA crossing to a right-handed negative node. Importantly, we found that gyrase did not unknot either the right-handed trefoils produced by Int or the left-handed trefoils generated by Hin. If gyrase simply inverted left-handed nodes to form right-handed nodes, then gyrase should have at least unknotted the trefoil knots produced by Hin. Instead, left-handed trefoils were the most prevalent types of knot observed when topoisomerase IV was inhibited and gyrase functioned (Fig. 3B, lanes a–f; Fig. 6A, lane b). If gyrase inverted all of the positive crossings, then the trefoils produced by Int should have been resolved, and they were not. These results show that neither handedness alone nor sign alone can explain the inability of gyrase to unknot DNA.
There are at least three possible explanations for why gyrase activity is limited to the introduction of negative supercoils: (1) Gyrase can somehow assess simultaneously both the topological sign and the handedness of a node and will perform DNA strand passage only at a left-handed, positive node. Because at least the simple twist and torus knots cannot be left-handed and positive, as plectonemic supercoils are, knots are not an efficient substrate for gyrase. It was suggested previously that gyrase can sense or regulate the DNA segment being transported (Vologodskii and Cozzarelli 1996), but the physical mechanism gyrase might employ for this is unclear. (2) Gyrase can more easily wrap a supercoil node about itself than a knot node. (3) The conformation of a supercoil and a knot are distinct in terms of a local property, such as the angle between the segments forming the node, the flexibility of the nodes, or the physical tightness of the node. Which of these models is correct remains to be seen.
Topoisomerase IV is sensitive to global DNA topology
As stated above, type-2 topoisomerases modify DNA topology by binding to a crossover formed between two DNA duplexes (Zechiedrich and Osheroff 1990), cleaving one of the duplexes, passing the other through this transiently open gate, and resealing the break in the backbone (for review, see Wang 1996). It would therefore appear that a locally acting type-2 topoisomerase would be unaffected by the global topology of a DNA molecule. However, the results presented here, as well as discoveries by others, contradict this straightforward model. A type-2 topoisomerase will remove a catenane before it removes a supercoil node, even when supercoil nodes are in vast excess (Roca and Wang 1994; Ullsperger and Cozzarelli 1996). In addition, these enzymes will hydrolyze ATP more efficiently in the presence of a catenane node than a supercoil node (Anderson et al. 1998). The type-2 topoisomerases that have been tested preferentially unknot rather than knot DNA, which also indicates that these enzymes can recognize the global topology of DNA (Rybenkov et al. 1997a). Because supercoiling stimulates DNA decatenating, but not unknotting by topoisomerase IV, there must be some difference between knots and catenanes. This result is in contrast to reactions carried out by purified gyrase in vitro, where both decatenating and unknotting of DNA by gyrase are stimulated by negative supercoiling (Liu et al. 1980; Mizuuchi et al. 1980), and reveals that this effect may not be simply the result of yet uncharacterized conformational differences between knots and catenanes. Recently, it has been suggested that topoisomerase IV preferentially relaxes positive over negative supercoils because of the opposite handedness of these substrates (Crisona et al. 2000). However, we find that the distinction made by topoisomerase IV between knots and catenanes occurs with substrates of the same handedness and may represent an additional level of substrate recognition by topoisomerase IV. Other models, even those that suggested that a type-2 topoisomerase might track along the DNA, cannot explain this effect (Rybenkov et al. 1997a; Yan et al. 1999).
Topoisomerase IV does not require DNA replication for activity
Our results demonstrate that topoisomerase IV does not require active DNA replication to unknot DNA. This is important because the resolution of DNA knots, as well as the maintenance of homeostatic DNA supercoiling, should be necessary at times other than when DNA is being synthesized. As suggested previously, an interaction with the replication machinery may be important for topoisomerase IV to unlink replicating DNA (Levine and Marians 1998). However, our results demonstrate that at least knotted DNA is freely accessible to topoisomerase IV.
Physiological roles of the topoisomerases
It is now possible to assign completely the topological roles of the topoisomerases in E. coli (Fig. 1). DNA supercoiling (Fig. 1A) is regulated by topoisomerase I, gyrase, and topoisomerase IV: Gyrase introduces negative supercoiling, and this action is countered by topoisomerase I and topoisomerase IV, which relax negative supercoils (Menzel and Gellert 1983; Zechiedrich et al. 2000). Decatenation of linked DNA molecules (Fig. 1B) is carried out by topoisomerase IV regardless of the mechanisms by which the catenanes arose (Zechiedrich and Cozzarelli 1995; Zechiedrich et al. 1997). Gyrase contributes to decatenation by supercoiling the catenane to make a better substrate for topoisomerase IV (Zechiedrich et al. 1997). Gyrase may be additionally important in decatenation because DNA supercoiling has been shown to reduce the equilibrium probability that two DNAs become catenated (Rybenkov et al. 1997b). DNA unknotting (Fig. 1C) is carried out exclusively by topoisomerase IV. Topoisomerase III does not appear to play a role in regulating these topological forms of DNA, and its biological significance remains unknown.
It is clear, therefore, that topoisomerases in the cell have distinct and nonoverlapping roles. Although gyrase and topoisomerase IV can both unknot and decatenate DNA in vitro, only topoisomerase IV performs these activities within the cell. Consequently, our results suggest limitations in assigning to a protein a physiological function based upon sequence similarity (ParC and ParE are 36% and 40% homologous to GyrA and GyrB, respectively) or even upon in vitro biochemical activity.
Topoisomerases do not carry out in the cell all the reactions that they have been shown to carry out in the test tube (Table 2). What prevents enzyme promiscuity in the cell? It seems that each topoisomerase is designed inherently to carry out specific functions. It is also possible that unknown cellular factors play a role in limiting these enzymes to their specific activities. Topoisomerase IV removes crossings from DNA, whether they result from supercoils, catenanes, or knots, but it does not remove all of these crossings blindly. Clearly, removing all catenane links is required for chromosome segregation and removing knot links is important for DNA metabolism. However, the removal of all DNA supercoils, a difficult task in the presence of gyrase, would be detrimental to the cell. The differences in the removal of these topological forms probably reflect the differences in how vital the removal of that problem is to cell survival.
Table 2.
Topoisomerases in E. coli
| Topoisomerase
|
Type
|
Genes
|
In vitro activities
|
In vivo role
|
|---|---|---|---|---|
| I | 1 | topA | Relax (−) supercoils | Relax (−) supercoils |
| Decatenate/catenate | ||||
| Unknot/knot | ||||
| Gyrase | 2 | gyrA, gyrB | Introduce (−) supercoils | Introduce (−) supercoils |
| Decatenate/catenate | ||||
| Unknot/knot | ||||
| III | 1 | topB | Relax (−) supercoils | ? |
| Decatenate/catenate | ||||
| Unknot/knot | ||||
| IV | 2 | parC, parE | Relax (−) supercoils | Relax (−) supercoils |
| Relax (+) supercoils | Relax (+) supercoilsa | |||
| Decatenate/catenate | Decatenate | |||
| Unknot/knot | Unknot |
Materials and methods
Reagents
Restriction enzymes and proteinase K were obtained from Boehringer Mannheim. Supercoiled DNA ladder, 1-kb ladder, and calf thymus topoisomerase I were obtained from GIBCO BRL. T4 DNA ligase was obtained from New England Biolabs. [α-32P]dCTP, [methyl-3H]-thymidine, and the Multiprime DNA-labeling kit were purchased from Amersham Pharmacia. Norfloxacin, chloroquine, RNase A, and DNase I were obtained from Sigma. Nylon Zeta-Probe Blotting Membranes were from Bio-Rad. Purified type-2 topoisomerase from bacteriophage T4 was a gift from K.N. Kreuzer, Duke University. All other chemicals and reagents were analytical grade or better.
Bacterial strains and plasmids
The bacterial strains used are listed in Table 1 and have been described previously (Kato et al. 1990; Khodursky et al. 1995; Zechiedrich and Cozzarelli 1995; Zechiedrich et al. 1997). We used the serine-to-leucine mutation at amino acid position 83 of gyrase and the serine-to-lysine mutation at amino acid position 84 of topoisomerase IV because these mutations provide the highest levels of drug resistance both in vitro and in vivo (Yoshida et al. 1988; Khodursky et al. 1995; Zechiedrich et al. 1997). Int knotting plasmid pJB3.5i (Hildebrandt and Cozzarelli 1995), Hin knotting substrate pRJ862 (Heichman et al. 1991), and Hin expression vector pKH66 (Johnson and Simon 1985) have been described previously. The 7.8-kb Int catenane substrate, pABCi, was constructed by ligating the cat gene (kindly provided by T. Palzkill, Baylor College of Medicine), which encodes chloramphenicol acetyl transferase into the unique HindIII site of pAB3 (Dröge and Cozzarelli 1989) and pAB4 (Wasserman et al. 1988). The 7.8-kb Int catenating substrate pABCd was made by cloning the same cat cassette into HindIII-digested pAB4.
Int recombination
The Int recombination strategy was described previously (Bliska and Cozzarelli 1987; Zechiedrich et al. 1997) and is shown schematically in Figure 2B. The λ lysogenic strains containing an Int recombination plasmid were grown while shaking at 30°C to a cell density of 70 Klett units (83 Klett units = A600 of 1.0 for C600 strains) in LB medium containing the appropriate antibiotic: ampicillin for cells containing pJB3.5i and chloramphenicol for cells with pABCi or pABCd. At this density, cultures were shifted to 42°C for 10 min, which inactivates the thermosensitive cI857 repressor and induces expression of Int. Because the Int recombinase is not active at 42°C the cultures were returned to 30°C, where recombination can be initiated at Time = 0. Norfloxacin was added to the cultures at the time of shift down to a final concentration of 60 μM. Within ∼20 min, recombination is complete. When not inhibited, a topoisomerase (topo) unties the knotted recombination intermediates.
Hin recombination
Strains containing the Hin expression vector pKH66 and Hin substrate pRJ862 were grown while shaking at 37°C. Because Hin recombination is dependent on the Fis protein, it is very sensitive to the growth phase of the bacteria (Johnson and Simon 1985). Fis protein levels have been shown to decrease as bacterial cultures approach stationary phase (Ball et al. 1992). Therefore, in our experiments, cultures were grown to mid-logarithmic phase (30–35 Klett units). IPTG was added to a final concentration of 1 mM to induce Hin expression from pKH66. Cultures were allowed to shake at 37°C for an additional 10 min before the addition of norfloxacin to a final concentration of 60 μM (Time = 0). Aliquots (∼13 mL) of cells were then either plunged into liquid nitrogen or immediately lysed, and the plasmid DNA was isolated. Aliquots were taken before the addition of IPTG, following the 10-min induction of Hin expression, and at various time points after the addition of norfloxacin. Experiments using temperature-sensitive topoisomerase IV mutants were carried out in the same manner except that cells were grown at 30°C, and instead of norfloxacin addition, a shift to 42°C blocked topoisomerase IV (Time = 0). The cells remained at 42°C with shaking for the duration of the experiment (20–40 min).
DNA analyses
The alkaline lysis procedure was used to isolate plasmid DNA (Sambrook et al. 1989). The isolated plasmid DNA was treated with RNase A (20–100 μg/mL final concentration) and divided into three fractions. One fraction of plasmid DNA was singly nicked with DNase I in the presence of ethidium bromide and analyzed by high resolution agarose (0.8%–1%) gels (Sundin and Varshavsky 1981). The DNA was transferred to a Zeta-Probe nylon membrane and probed with radiolabeled linear pJB3.5i, pABCi, or pRJ862. The probe was made using the Multiprime DNA Labeling System from Amersham. DNA knot ladders were generated with T4 topoisomerase (Wasserman and Cozzarelli 1991).
The second fraction of plasmid DNA was analyzed on TAE agarose (1.0%–1.2%) gels containing chloroquine (2–20 μg/mL). The supercoiling levels were determined by the band counting method (Keller 1975). ς was calculated according to the formula ς = ΔLk/Lk0, with Lk0 = 10.45 bp/turn. The third DNA plasmid fraction isolated from the Int experiments was digested with EcoRI and BamHI (pJB3.5i). Similarly, SalI digestion was used to measure pABCi recombination. Quantification of bands on agarose gels was carried out using either the Storm PhosphorImaging system or Nucleovision densitometry software from Nucleotech.
DNA replication inhibition
Cultures were grown with shaking at 30°C to late-logarithmic phase (70 Klett units). To block replication, 50 mM hydroxyurea was added (Adams et al. 1992b). After 10 min, [methyl-3H]-thymidine (1 μL/mL of cells), with specific activity of 70–86 Ci/mmole (1 mCi/mL) was added. At 1, 3, 5, 8, 11, and 20 min following the addition of 3H-thymidine, 0.75-mL aliquots of cells were removed from the culture and added to 0.75 mL stop solution (75% ethanol; 21 mM sodium acetate at pH 5.3; 2 mM EDTA; 2% phenol) at room temperature. 3H-Thymidine incorporation was measured according to Sambrook et al. (1989). Briefly, aliquots were passed through Whatman GF/C filters using a Millipore manifold vacuum apparatus. The filters were subsequently washed, dried, and submitted to scintillation counting.
Acknowledgments
We thank R.C. Johnson and S.K. Merickel for the Hin expression vector and substrate, for technical assistance, and for sharing unpublished results. We are grateful to H. Ikeda, J.-I. Kato, S. Nakamura, and H. Suzuki for strains and to K.N. Kreuzer for purified T4 topoisomerase. We thank T. Palzkill for reagents; I. Darcy, A. Landy, and J.F. Marko for insightful discussions; and P.J. Hastings, R.C. Johnson, R.R. Sinden, M.E. Sowa, A.V. Vologodskii, and members of the laboratory for critically reading the manuscript. E.L. Zechiedrich is a New Investigator in the Toxicological Sciences for the Burroughs Wellcome Fund. This work was supported also by a Curtis Hankamer Research Award and by a Fellowship (to R.W. Deibler) from the Program in Mathematics and Molecular Biology at Florida State University, with funding from the National Science Foundation under Grant DMS-9406348.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL elz@bcm.tmc.edu; FAX (713) 798-7375.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.872301.
References
- Adams DE, Bliska JB, Cozzarelli NR. Cre–lox recombination in E. coli cells: Mechanistic differences from the in vitro reaction. J Mol Biol. 1992a;226:661–673. doi: 10.1016/0022-2836(92)90623-r. [DOI] [PubMed] [Google Scholar]
- Adams DE, Shekhtman EM, Zechiedrich EL, Schmid MB, Cozzarelli NR. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992b;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- Anderson VE, Gootz TD, Osheroff N. Topoisomerase IV catalysis and the mechanism of quinolone action. J Biol Chem. 1998;273:17879–17885. doi: 10.1074/jbc.273.28.17879. [DOI] [PubMed] [Google Scholar]
- Arai Y, Yasuda R, Akashi K, Harada Y, Miyata H, Kinosita KJ, Itoh H. Tying a molecular knot with optical tweezers. Nature. 1999;399:446–448. doi: 10.1038/20894. [DOI] [PubMed] [Google Scholar]
- Ball CA, Osuna R, Ferguson KC, Johnson RC. Dramatic changes in FIS levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska JB, Cozzarelli NR. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987;194:205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Cox MM. A broadening view of recombinational DNA repair in bacteria. Genes Cells. 1998;3:65–78. doi: 10.1046/j.1365-2443.1998.00175.x. [DOI] [PubMed] [Google Scholar]
- Crisona NJ, Strick TR, Bensimon D, Croquette V, Cozzarelli NR. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes & Dev. 2000;14:2881–2892. doi: 10.1101/gad.838900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröge P, Cozzarelli NR. Recombination of knotted substrates by Tn3 resolvase. Proc Natl Acad Sci. 1989;86:6062–6066. doi: 10.1073/pnas.86.16.6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SM, Wang HY, Tse-Dinh YC, Seeman NC. Topological transformations of synthetic DNA knots. Biochemistry. 1995;34:673–682. doi: 10.1021/bi00002a035. [DOI] [PubMed] [Google Scholar]
- Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: A conserved mechanism for control of DNA recombination. Mol Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- Heichman KA, Johnson RC. The Hin invertasome: Protein-mediated joining of distant recombination sites at the enhancer. Science. 1990;249:511–517. doi: 10.1126/science.2166334. [DOI] [PubMed] [Google Scholar]
- Heichman KA, Moskowitz IPG, Johnson RC. Configuration of DNA strands and mechanism of strand exchange in the Hin invertasome as revealed by analysis of recombinant knots. Genes & Dev. 1991;5:1622–1634. doi: 10.1101/gad.5.9.1622. [DOI] [PubMed] [Google Scholar]
- Hildebrandt ER, Cozzarelli NR. Comparison of recombination in vitro and in E. coli cells: Measure of the effective concentration of DNA in vivo. Cell. 1995;81:331–340. doi: 10.1016/0092-8674(95)90386-0. [DOI] [PubMed] [Google Scholar]
- Ishii S, Murakami T, Shishido K. Gyrase inhibitors increase the content of knotted DNA species of plasmid pBR322 in Escherichia coli. J Bacteriol. 1991;173:5551–5553. doi: 10.1128/jb.173.17.5551-5553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RC, Simon MI. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985;41:781–791. doi: 10.1016/s0092-8674(85)80059-3. [DOI] [PubMed] [Google Scholar]
- Johnson RC, Bruist MF, Simon MI. Host protein requirements for in vitro site-specific DNA inversion. Cell. 1986;46:531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- Kampranis SC, Maxwell A. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc Natl Acad Sci. 1996;93:14416–14421. doi: 10.1073/pnas.93.25.14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, Cozzarelli N R. Roles of supercoiled DNA structure in DNA transactions. Curr Opin Struct Biol. 1992;2:369–379. [Google Scholar]
- Kato J-I, Nishimura Y, Imamura R, Niki H, Hiraga S, Susuki H. New topoisomerase essential for chromosome segregation in Escherichia coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- Kato J-I, Suzuki H, Ikeda H. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J Biol Chem. 1992;267:25676–25684. [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci. 1975;72:4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodursky AB, Zechiedrich EL, Cozzarelli NR. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodursky AB, Peter BJ, Schmid MB, DeRisi J, Botstein D, Brown PO, Cozzarelli NR. Analysis of topoisomerase function in bacterial replication fork movement: Use of DNA microarrays. Proc Natl Acad Sci. 2000;97:9419–9424. doi: 10.1073/pnas.97.17.9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T, Wang JC, Harrison SC. DNA gyrase and its complexes with DNA: Direct observation by electron microscopy. Cell. 1985;41:933–943. doi: 10.1016/s0092-8674(85)80074-x. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Baker TA. DNA replication. 2nd ed. New York: W.H. Freeman; 1992. [Google Scholar]
- Levine C, Marians KJ. Identification of dnaX as a high-copy suppressor of the conditional lethal and partition phenotypes of the parE10 allele. J Bacteriol. 1998;180:1129–1134. doi: 10.1128/jb.180.5.1232-1240.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine C, Hiasa H, Marians KJ. DNA gyrase and topoisomerase IV: Biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim Biophys Acta. 1998;1400:29–43. doi: 10.1016/s0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- Liu LF, Wang JC. DNA–DNA gyrase complex: The wrapping of the DNA duplex outside the enzyme. Cell. 1978;15:979–984. doi: 10.1016/0092-8674(78)90281-7. [DOI] [PubMed] [Google Scholar]
- Liu LF, Liu C-C, Alberts BM. Type II DNA topoisomerases: Enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980;19:697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- Marians KJ. DNA gyrase-catalyzed decatenation of multiply linked DNA dimers. J Biol Chem. 1987;262:10362–10368. [PubMed] [Google Scholar]
- Marko JF. Coupling of intra- and intermolecular linkage complexity of two DNAs. Phys Rev E. 1999;59:900–912. [Google Scholar]
- Martin-Parras L, Lucas I, Martinez-Robles ML, Hernandez P, Krimer DB, Hyrien O, Schvartzman JB. Topological complexity of different populations of pBR322 as visualized by two-dimensional agarose gel electrophoresis. Nucleic Acids Res. 1998;26:3424–3432. doi: 10.1093/nar/26.14.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally T. The Complete Book of Fly Fishing. 1993. Ragged Mountain Press, Camden, ME. [Google Scholar]
- Menzel R, Gellert M. Regulation of the genes for E. coli DNA gyrase: Homeostatic control of DNA supercoiling. Cell. 1983;34:105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K, Fisher LM, O'Dea MH, Gellert M. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc Natl Acad Sci. 1980;77:1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Marians K. The interaction of Escherichia coli topoisomerase IV with DNA. J Biol Chem. 1995;270:25286–25290. doi: 10.1074/jbc.270.42.25286. [DOI] [PubMed] [Google Scholar]
- Podtelezhnikov AA, Cozzarelli NR, Vologodskii AV. Equilibrium distributions of topological states in circular DNA: Interplay of supercoiling and knotting. Proc Natl Acad Sci. 1999;96:12974–12979. doi: 10.1073/pnas.96.23.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock TJ, Nash HA. Knotting of DNA caused by a genetic rearrangement. Evidence for a nucleosome-like structure in site-specific recombination of bacteriophage λ. J Mol Biol. 1983;170:1–18. doi: 10.1016/s0022-2836(83)80224-1. [DOI] [PubMed] [Google Scholar]
- Portugal J, Rodriguez-Campos A. T7 RNA polymerase cannot transcribe through a highly knotted DNA template. Nucleic Acids Res. 1996;24:4890–4894. doi: 10.1093/nar/24.24.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raji A, Zabel DJ, Laufer CS, Depew RE. Genetic analysis of mutations that compensate for loss of Escherichia coli DNA topoisomerase I. J Bacteriol. 1985;162:1173–1179. doi: 10.1128/jb.162.3.1173-1179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca J, Wang JC. DNA transport by a type II DNA topoisomerase—Evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Campos A. DNA knotting abolishes in vitro chromatin assembly. J Biol Chem. 1996;271:14150–14155. doi: 10.1074/jbc.271.24.14150. [DOI] [PubMed] [Google Scholar]
- Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997a;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- Rybenkov VV, Vologodskii AV, Cozzarelli NR. The effect of ionic conditions on the conformations of supercoiled DNA. II. Equilibrium catenation. J Mol Biol. 1997b;267:312–323. doi: 10.1006/jmbi.1996.0877. [DOI] [PubMed] [Google Scholar]
- Saitta AM, Soper PD, Wasserman E, Klein ML. Influence of a knot on the strength of a polymer strand. Nature. 1999;399:46–48. doi: 10.1038/19935. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmid MB. A locus affecting nucleoid segregation in Salmonella typhimurium. J Bacteriol. 1990;172:5416–5424. doi: 10.1128/jb.172.9.5416-5424.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SY, Wang JC. Knotting of a DNA chain during ring closure. Science. 1993;260:533–536. doi: 10.1126/science.8475384. [DOI] [PubMed] [Google Scholar]
- Shekhtman EM, Wasserman SA, Solomon MJ, Cozzarelli NR. Stereostructure of replicative DNA catenanes from eukaryotic cells. New J Chem. 1993;17:757–763. [Google Scholar]
- Shishido K, Komiyama N, Ikawa S. Increased production of a knotted form of plasmid pBR322 DNA in Escherichia coli DNA topoisomerase mutants. J Mol Biol. 1987;195:215–218. doi: 10.1016/0022-2836(87)90338-x. [DOI] [PubMed] [Google Scholar]
- Shlyakhtenko LS, Hsieh P, Grigoriev M, Potaman VN, Sinden RR, Lyubchenko YL. A cruciform structural transition provides a molecular switch for chromosome structure and dynamics. J Mol Biol. 2000;296:1169–1173. doi: 10.1006/jmbi.2000.3542. [DOI] [PubMed] [Google Scholar]
- Sogo JM, Stasiak A, Martinez-Robles ML, Krimer DB, Hernandez P, Schvartzman JB. Formation of knots in partially replicated DNA molecules. J Mol Biol. 1999;286:637–643. doi: 10.1006/jmbi.1998.2510. [DOI] [PubMed] [Google Scholar]
- Spengler SJ, Stasiak A, Cozzarelli NR. The stereostructure of knots and catenanes produced by phage l integrative recombination: Implications for mechanism and DNA structure. Cell. 1985;42:325–334. doi: 10.1016/s0092-8674(85)80128-8. [DOI] [PubMed] [Google Scholar]
- Staczek P, Higgins NP. Gyrase and Topo IV modulate chromosome domain size in vivo. Mol Microbiol. 1998;29:1435–1448. doi: 10.1046/j.1365-2958.1998.01025.x. [DOI] [PubMed] [Google Scholar]
- Stasiak A, Katritch V, Bednar J, Michoud D, Dubochet J. Electrophoretic mobility of DNA knots. Nature. 1996;384:122. doi: 10.1038/384122a0. [DOI] [PubMed] [Google Scholar]
- Sundin O, Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: Dissection of final stages of SV40 DNA replication. Cell. 1981;25:659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- Ullsperger C, Cozzarelli NR. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J Biol Chem. 1996;271:31549–31555. doi: 10.1074/jbc.271.49.31549. [DOI] [PubMed] [Google Scholar]
- Viguera E, Hernandez P, Krimer DB, Boistov AS, Lurz R, Alonso JC, Schvartzman JB. The ColE1 unidirectional origin acts as a polar replication fork pausing site. J Biol Chem. 1996;271:22414–22421. doi: 10.1074/jbc.271.37.22414. [DOI] [PubMed] [Google Scholar]
- Vologodskii AV, Cozzarelli N R. Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys J. 1996;70:2548–2556. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vologodskii AV, Marko JF. Extension of torsionally stressed DNA by external force. Biophys J. 1997;73:123–132. doi: 10.1016/S0006-3495(97)78053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vologodskii AV, Crisona NJ, Laurie B, Pieranski P, Katritch V, Dubochet J, Stasiak A. Sedimentation and electrophoretic migration of DNA knots and catenanes. J Mol Biol. 1998;278:1–3. doi: 10.1006/jmbi.1998.1696. [DOI] [PubMed] [Google Scholar]
- Wang JC. DNA Topoisomerases. Ann Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- Wasserman SA, Cozzarelli NR. Biochemical topology: Applications to DNA recombination and replication. Science. 1986;232:951–960. doi: 10.1126/science.3010458. [DOI] [PubMed] [Google Scholar]
- ————— Supercoiled DNA-directed knotting by T4 topoisomerase. J Biol Chem. 1991;266:20567–20573. [PubMed] [Google Scholar]
- Wasserman SA, White JH, Cozzarelli NR. The helical repeat of double-stranded DNA varies as a function of catenation and supercoiling. Nature. 1988;334:448–450. doi: 10.1038/334448a0. [DOI] [PubMed] [Google Scholar]
- Yan J, Magnasco MO, Marko JF. Kinetic proofreading mechanism for disentanglement of DNA by topoisomerases. Nature. 1999;401:932–935. doi: 10.1038/44872. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kojima T, Yamagishi J-I, Nakamura S. Quinolone-resistant mutations of the gyrA gene of Escherichia coli. Mol Gen Genet. 1988;211:1–7. doi: 10.1007/BF00338386. [DOI] [PubMed] [Google Scholar]
- Zechiedrich EL, Cozzarelli NR. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes & Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- Zechiedrich EL, Osheroff N. Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J. 1990;9:4555–4562. doi: 10.1002/j.1460-2075.1990.tb07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechiedrich EL, Khodursky AB, Cozzarelli NR. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes & Dev. 1997;11:2580–2592. doi: 10.1101/gad.11.19.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DMJ, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J Biol Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]