Abstract
Many coxsackievirus B isolates bind to human decay-accelerating factor (DAF) as well as to the coxsackievirus and adenovirus receptor (CAR). The first-described DAF-binding isolate, coxsackievirus B3 (CB3)-RD, was obtained during passage of the prototype strain CB3-Nancy on RD cells, which express DAF but very little CAR. CB3-RD binds to human DAF, whereas CB3-Nancy does not. To determine the molecular basis for the specific interaction of CB3-RD with DAF, we produced cDNA clones encoding both CB3-RD and CB3-Nancy and mutated each of the sites at which the RD and Nancy sequences diverged. We found that a single amino acid change, the replacement of a glutamate within VP3 (VP3-234E) with a glutamine residue (Q), conferred upon CB3-Nancy the capacity to bind DAF and to infect RD cells. Readaptation of molecularly cloned CB3-Nancy to RD cells selected for a new virus with the same VP3-234Q residue. In experiments with CB3-H3, another virus isolate that does not bind measurably to DAF, adaptation to RD cells resulted in a DAF-binding isolate with a single amino acid change within VP2 (VP2-138 N to D). Both VP3-234Q and VP2-138D were required for binding of CB3-RD to DAF. In the structure of the CB3-RD-DAF complex determined by cryo-electron microscopy, both VP3-234Q and VP2-138D are located at the contact site between the virus and DAF.
INTRODUCTION
Coxsackieviruses, members of the Enterovirus genus within the Picornavirus family, are common human pathogens (23). Group B coxsackieviruses (CB) are important causes of viral meningitis and myocarditis and have been implicated as a possible trigger for the onset of childhood diabetes.
CB interact with at least two receptors. All isolates so far examined attach to the coxsackievirus and adenovirus receptor (CAR), a 46-kDa transmembrane protein that also serves as a receptor for many adenoviruses (2, 21, 34). In addition, a large subset of CB1, 3, and 5 (including clinical isolates) also binds to decay-accelerating factor (DAF; CD55), a glycosylphosphatidylinositol (GPI)-anchored complement regulatory protein (3, 4, 29). Many other enteroviruses also bind to DAF (1, 16, 26, 30, 36). Because phylogenetically diverse enteroviruses interact with different domains within the DAF molecule, it has been suggested that viruses have evolved independently to bind DAF (26). The evolutionary pressures that gave rise to the DAF-binding phenotype and the role for DAF attachment during infection in vivo are uncertain.
In polarized epithelial cells, CAR is a component of the tight junction (7), a structure at the apical edge of the basolateral membrane that regulates the paracellular flow of ions and macromolecules across intact epithelium (9). No CAR is present on the apical cell membrane, and within the tight junction CAR is inaccessible to the virus; as a result, exposure of the apical surface to CB isolates that do not bind DAF results in little, if any, infection (7). In contrast, DAF is highly expressed on the apical membranes of polarized epithelial cells, and interaction with DAF permits DAF-binding CB to infect polarized epithelial cells (33). DAF functions both to permit virus attachment and to initiate kinase signals (8) essential for subsequent events in virus entry.
The first DAF-binding CB isolate to be identified was CB3-RD, originally obtained by Crowell and colleagues after blind passage of the prototype CB3-Nancy strain on RD rhabdomyosarcoma cells (27). Unlike the Nancy strain, CB3-RD readily infected RD cells and bound to a 60- to 70-kDa cellular protein (13, 24) that was subsequently identified as DAF (4).
Lindberg et al. (20) constructed a series of genomic chimeras between CB3-RD and a CB3-Nancy isolate (not the isolate from Crowell's lab, however) and identified a genome segment that conferred on the Nancy isolate the capacity to infect and kill RD cells efficiently—a phenotype that likely depends on, but is not necessarily synonymous with, DAF binding. Within this segment (which encodes VP2 and a part of VP3), the wild-type and RD isolates differed at two sites: in RD, a serine was substituted for threonine at position 151 within VP2 (VP2-151S) and a valine was substituted for aspartate at VP2 position 108 (VP2-108V). However, other investigators have questioned whether VP2-151S and VP2-108V account for the RD phenotype (31). Furthermore, in the structure of the CB3-DAF complex determined by cryo-electron microscopy (cryo-EM) (10), neither VP2-151S nor VP2-108V was found to be in contact with DAF.
To determine the molecular basis for the specific interaction of CB3-RD with DAF, we produced cDNA clones encoding both CB3-RD and CB3-Nancy from low-passage virus stocks obtained from Richard Crowell. We found that a single amino acid change, the replacement of a glutamate within VP3 (VP3-234E) with a glutamine residue derived from CB3-RD (VP3-234Q), confers upon CB3-Nancy the capacity to bind DAF and to infect RD cells. To investigate further the routes by which CB3 can adapt to bind DAF, we passed CB3-H3—an isolate that does not bind to DAF although it already possesses VP3-234Q—on RD cells and obtained a DAF-binding isolate with a single amino acid change within VP2 (VP2-138 N to D). Both VP3-234Q and VP2-138D are required for virus attachment to DAF. Both residues are within the site of interaction with DAF, as determined by the published cryo-EM structure of the virus-receptor complex (10), as well as in a newly refined structure (J. D. Yoder, J. O. Cifuente, and S. Hafenstein, unpublished data). These results indicate that single amino acid changes permit CB3 to bind DAF and gain tropism for new cell types.
MATERIALS AND METHODS
Cells.
HeLa cells were maintained in modified Eagle's medium supplemented with l-glutamine, penicillin, streptomycin, nonessential amino acids, and 5% fetal bovine serum; RD cells were maintained in the same medium, but with 10% fetal serum. CHO-CAR (35), CHO-DAF (22), and CHO-pcDNA (35) cells were generated and maintained as described previously.
Infectious viral cDNA clones.
To generate clones of CB3-RD and CB3-Nancy, HeLa cells were infected (1 PFU/cell) for 7 h and total RNA was isolated with Trizol reagent (Invitrogen). Reverse transcription was performed with 5 μg of total RNA, 20 pmol of virus-specific primer incorporating an MluI restriction site (CB3-7381RMlu, 5′-GTACGCGTTTTTTTTTTTTTTTCCGCACCGAATGCGGAGAATTTA-3′), and SuperScript III polymerase (Invitrogen). PCR amplification was then performed with rTth DNA polymerase XL (Applied Biosystems), using primer CB3-7381RMlu and an upstream primer incorporating an SbfI site (5′-GGAATTAACCTGCAGGTTAAAACAGCCTGTGGGTTG-3′. The PCR program was as follows: 1 cycle at 94°C for 1 min, 35 cycles at 94°C for 15 s followed by 8 min at 68°C, and 1 cycle at 72°C for 12 min. PCR products (approximately 7,400 bp) were gel purified and digested with SbfI and MluI (New England BioLabs) and then cloned into plasmid pSport1 downstream of the T7 promoter. Full-length CB3-H3 cDNA was provided by Kirk Knowlton and Sally Huber (18). Specific mutations were introduced by splice overlap extension PCR (11).
Generation of viruses from infectious viral cDNA clones.
For CB3-Nancy, CB3-RD, and their derivatives, viral cDNA was reverse transcribed using T7 polymerase (RiboMAX, Promega), viral RNA quality was assessed by gel electrophoresis, and virus was produced by transfection of RNA into HeLa cells using DMRIE-C (Invitrogen). Three to 5 days after transfection, virus was harvested and purified and titers were determined on HeLa cells as described previously (32). CB3-H3 and H3-VP2-138D were generated by transfection of HeLa cells with DNA, using Lipofectamine 2000 (Invitrogen), and virus was harvested 4 days later.
Radiolabeled virus binding.
CB3-RD and CB3-Nancy were radiolabeled with [35S]methionine and [35S]cysteine as described elsewhere (5). 35S-labeled virus was bound to confluent cell monolayers in 24-well plates for 1 h at room temperature. Unbound virus was removed with three washes with binding buffer, cells were lysed with Solvable detergent (PerkinElmer, Waltham, MA), and cell-bound radioactivity was assessed.
Immunofluorescence staining of virus-infected cells.
For immunofluorescence assays, RD cell monolayers in 8-well Lab-Tek chamber slides (Nalge Nunc International) were infected with viruses (1 PFU/cell); after 72 h, the monolayers were fixed and stained with anti-enterovirus VP1 monoclonal antibody (MAb) (NCL-ENTERO; Leica Microsystems) and fluorescein isothiocyanate (FITC)-labeled secondary antibody as described previously (8).
Adaptation to RD cells.
CB3-Nancy and CB3-H3, derived from molecular clones, were passed five times on HeLa cells to permit variants to accumulate. Virus was then passed four times on RD cells, and the consensus sequence was determined by direct sequencing of reverse transcription (RT)-PCR products. To isolate individual virus variants within the RD cell-adapted population, HeLa cell monolayers in 96-well plates were infected with virus at approximately 0.3 infectious virions per monolayer. Monolayers were observed for the appearance of cytopathic changes, and then viruses from individual wells were expanded and sequenced.
Nucleotide sequence accession numbers.
The sequences of CB3-Nancy and CB3-RD have been deposited in GenBank under accession numbers JN048468 and JN048469, respectively.
RESULTS
CB3-Nancy and CB3-RD differ in 6 amino acids in the P1 region.
To identify the genetic changes that accompanied the adaptation of CB3-Nancy to growth in RD cells, we first determined the nucleotide sequences of the two virus isolates. HeLa cells were infected with CB3-Nancy and CB3-RD, both from low-passage stocks provided by Richard Crowell, and RNA was isolated during the first infection cycle. Viral sequences were reverse transcribed and amplified with PCR primers spanning the entire viral genome. To obtain the consensus sequence for each virus—independently of errors introduced by PCR and sequence variation within the viral quasispecies—PCR products were sequenced directly, without cloning into plasmid vectors.
CB3-Nancy and CB3-RD were found to differ at 13 nucleotide (nt) positions (Table 1). Two nucleotide differences were found within the 5′ untranslated region (UTR), and one within the 3′ UTR. Within the protein-coding region, 10 nucleotide differences were observed. These resulted in six predicted changes at the amino acid level, all within the P1 region that encodes the capsid proteins VP1 to VP4 (Fig. 1). To generate infectious cDNA clones encoding CB3-Nancy and CB3-RD, we amplified full-length cDNA copies of the two genomes and inserted them into a plasmid vector downstream of a T7 promoter. When full-length RNAs were transcribed in vitro and transfected into HeLa cells, viruses that maintained the phenotypes of the original virus stocks were produced: cloned CB3-RD readily infected RD cells, as determined by a fluorescence focus assay, and radiolabeled CB3-RD bound to human DAF expressed on CHO-DAF cells. In contrast, cloned CB3-Nancy infected HeLa cells, but not RD cells, and showed no measurable binding to human DAF (Fig. 2 and 3).
Table 1.
Comparison of CB3-Nancy and CB3-RD nucleotide and amino acid sequences
| Gene | Position (nt) | CB3-Nancya |
CB3-RDa |
||
|---|---|---|---|---|---|
| nt | aa | nt | aa change | ||
| 5′ UTR | 125 | G | - | A | - |
| 474 | C | - | C(T) | - | |
| 511 | T | - | T(A) | - | |
| 529 | T(C) | - | T | - | |
| 578 | G | - | A | - | |
| 586 | T(C) | - | T | - | |
| VP2 | 987 | C | A | T | V |
| 1380 | C | A | T | V | |
| VP3 | 1861 | G | K | A | - |
| 2011 | C | Y | C(T) | - | |
| 2430 | C | T | T | I | |
| 2432 | T | S | A | T | |
| 2438 | G | E | C | Q | |
| VP1 | 2726 | T | L | A | I |
| 2A | 3496 | C | S | A | - |
| 2C | 4044 | A | N | A(G) | -(S) |
| 3C | 5485 | C | A | C(T) | - |
| 5893 | C | H | T | - | |
| 3D | 6478 | G | Q | G(A) | - |
| 7163 | T | L | C | - | |
| 3′ UTR | 7334 | C | - | T | - |
Parentheses indicate residues in the cDNA clone that differ from the consensus sequence determined for each virus; hyphens indicate no change. aa, amino acid.
Fig. 1.
Amino acid sequence differences between CB3-Nancy and CB3-RD. Regions where the sequences of CB3-Nancy and CB3-RD diverge are shown. Conserved amino acids are marked with hyphens. Sequence differences are in bold.
Fig. 2.
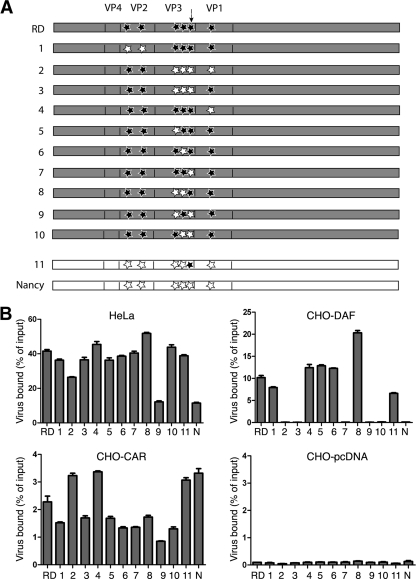
Mapping of amino acid residues required for CB3-RD interaction with DAF. (A) Mutant constructs were made by overlap extension PCR from full-length cDNA clones encoding CB3-RD (in gray) or CB3-Nancy (in white). Mutations are indicated with stars: black stars represent amino acid residues from CB3-RD, and open stars represent amino acid residues from CB3-Nancy. An arrow indicates VP3-234. (B) CHO cells stably transfected with DAF (CHO-hDAF) and CAR (CHO-hCAR) were incubated with radiolabeled viruses at room temperature for 1 h to measure binding to specific receptors. Data are presented as percentages of input virus bound to cells ± the standard deviation (SD) for triplicate samples. Mutants of CB3-RD in which VP3-234Q was replaced by 234E (constructs 2, 3, 7, 9, and 10) lost the capacity to bind DAF. A mutant of CB3-Nancy in which VP3-234E was replaced by 234Q (construct 11) acquired the capacity to bind DAF. N, Nancy.
Fig. 3.
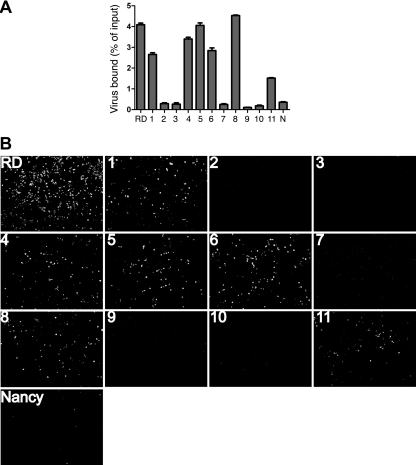
Mapping of amino acid residues required for interaction with RD cells. (A) Percentages of bound radiolabeled virus, determined as for preceding figures. (B) Virus infection. RD cell monolayers were incubated with viruses (10 PFU/cell) at 37°C for 72 h and then stained with anti-enterovirus VP1 MAb to measure virus infection. All DAF-binding virus strains infected RD cells, whereas non-DAF-binding strains did not.
The sequences of cloned CB3-Nancy and CB3-RD showed eight additional nucleotide differences that had not been observed in the consensus sequences (Table 1, in parentheses); these may have resulted from PCR errors, or they may reflect sequence diversity among the many individual viral genomes that arise during replication. These nucleotide differences had no effect on the predicted protein sequence, with one exception: in the CB3-RD cDNA clone, a guanine residue at position 4044 resulted in a single amino acid change within the nonstructural protein 2C, which is unlikely to affect virus interaction with DAF.
VP3-234Q is essential for CB3-RD attachment to DAF.
The consensus sequences revealed six amino acid differences between the capsid proteins of CB3-RD and CB3-Nancy. To determine which of these differences accounts for the different avidities of the two viruses for DAF, we introduced mutations into the CB3-RD molecular clone to replace each of the divergent RD amino acids with the corresponding Nancy residues (Fig. 2A, constructs 1 to 10). Mutant viruses were generated by transfection of HeLa cells with viral RNAs transcribed in vitro, and each virus was radiolabeled by growth in [35S]cysteine and [35S]methionine. All of the mutant viruses bound detectably to HeLa and CHO-CAR cells (Fig. 2B); less than 0.1% of input virus bound to control CHO-pcDNA cells.
Mutation en bloc of the three divergent residues within VP3 (231I, 232T, and 234Q; construct 3) ablated virus binding to DAF expressed on CHO-DAF cells, but mutation of the divergent residues within VP2 (construct 1) or VP1 (construct 4) had no effect. Mutation of VP3-234Q ablated virus attachment to DAF (construct 7); in contrast, mutation of the other divergent VP3 residues, either singly or in combination (constructs 5, 6, and 8), had no effect. These results suggested that VP3-234Q is essential for CB3-RD attachment to DAF. Introduction of this single residue into CB3-Nancy (construct 11) permitted attachment to DAF, thus confirming its importance for DAF interaction.
VP3-234Q is essential for CB3-RD infection of RD cells.
The tropism of CB3-RD for RD cells has been related to its avidity for DAF, although the relationship between DAF binding and RD cell infection has been questioned by some investigators (31). Each of the radiolabeled mutant viruses that had bound to CHO-DAF cells also bound to RD cells (Fig. 3A); the lower levels of binding to RD cells most likely reflect the low level of expression of DAF on these cells (12). When RD cells were exposed to mutant viruses, infection by each of the DAF-binding viruses was detectable by immunofluorescence staining for VP1 (Fig. 3B); infection was not detected with any of the viruses that did not bind DAF. Infection of RD cells by CB3-Nancy with VP3-234Q (construct 11) indicates that this residue, which is essential for attachment to DAF, is also a critical determinant of tropism for RD cells.
Passing cloned CB3-Nancy on RD cells reselects for VP3-234Q.
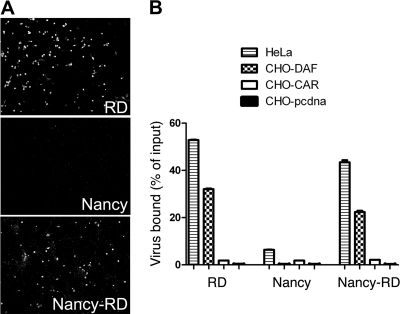
CB3-RD was originally selected by passing CB3-Nancy on RD cells. To determine whether a mutation to VP3-234Q was the sole path by which CB3-Nancy could acquire avidity for DAF and tropism for RD cells, we passed CB3-Nancy, derived from the molecular clone, on RD cells. After four passages, we obtained a new virus stock (which we called CB3-Nancy-RD) that readily infected RD cells (Fig. 4 A) and which also bound to DAF (Fig. 4B). Direct sequencing of RT-PCR products spanning the capsid region revealed a single substitution (G to C) at nucleotide 2438, resulting in the same mutation of VP3-234 from E to Q that we had observed for the original CB3-RD isolate.
Fig. 4.
Adaptation of cloned CB3-Nancy to growth in RD cells selects for DAF binding. (A) Infection of RD cells by the readapted virus, Nancy-RD. (B) The readapted virus binds to DAF.
To determine whether less common mutations—not evident from the CB3-Nancy-RD consensus sequence—also permit adaptation of CB3-Nancy to RD cells, we isolated clonal virus populations by limiting dilution from the CB3-Nancy-RD stock and directly sequenced the region surrounding nt 2438. VP3-234Q was seen in 45 of the 47 cloned viruses examined; the 2 remaining clones did not bind to DAF and did not infect RD cells. Thus, mutation to VP3-234Q is the dominant—if not the sole—route by which CB3-Nancy acquires RD cell tropism and avidity for DAF.
VP2-138D is also important for CB3 interaction with DAF.
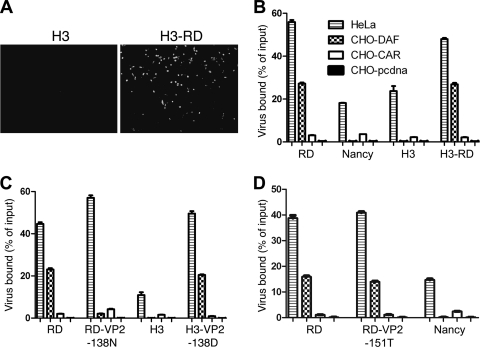
We had previously observed (unpublished data) that CB3-H3, a cardiovirulent virus isolate originally obtained from the heart of an infected mouse (14), did not bind measurably to DAF. The nucleotide sequence of CB3-H3 (18) reveals that this virus possesses the VP3-234Q residue important for CB3-RD attachment with DAF, consistent with the idea that other capsid residues are also essential for the interaction. We generated CB3-H3 from a molecular clone and passed it in RD cells to obtain a variant (CB3-H3-RD) that infected RD cells (Fig. 5 A) and bound to DAF (Fig. 5B). Direct sequencing of the CB3-H3-RD capsid region revealed that the adapted virus had undergone a single amino acid change within VP2 (nt 1361, A to G; VP2-138 N to D).
Fig. 5.
VP2-138D is also important for virus interaction with DAF. (A) Adaptation of CB3-H3 to growth in RD cells. RD cells were exposed to CH3-H3 or the adapted variant CB3-H3-RD and then stained at 72 h with anti-VP1 antibody to detect infection. (B) RD-adapted CB3-H3 binds to DAF. CHO cells expressing DAF or CAR, control CHO-pcDNA cells, and HeLa cells were incubated with radiolabeled viruses, and virus binding was measured as described in Materials and Methods. (C) In H3-VP2-138D, VP2-138D was introduced into cloned H3; this virus gained the capacity to bind DAF. In RD-VP2-138N, VP2-138D in the RD clone was replaced with VP2-138N; this virus lost the capacity to bind DAF. (D) Replacement of VP2-151S with T does not affect binding of CB3-RD to DAF.
Introduction of VP2-138D into cloned CB3-H3 (Fig. 5C) resulted in a virus with the capacity to bind DAF, confirming the importance of this residue for DAF interaction. Like CB3-H3-RD, both CB3-Nancy and CB3-RD have a D residue at VP2-138. We replaced this residue in the RD clone with the VP2-138N observed in CB3-H3. The resulting virus showed a dramatic loss in its avidity for DAF (Fig. 5C), indicating that VP2-138D, like VP3-234Q, is important for the interaction of CB3 with DAF.
Mutation of VP2-151S does not affect virus attachment to DAF.
As discussed above, it had previously been reported that sequence differences at VP2-108 and VP2-151 accounted for the differences in RD cell tropism observed for CB3-Nancy and CB3-RD (20). In particular, replacement of VP2-151T with S was believed to be important for RD cell tropism; substitution of V for D at VP2-108, which is not exposed on the virus surface, was thought less likely to be involved. We introduced into the CB3-RD backbone mutations replacing VP2-151S with T and VP2-108V with D, both together and singly. CB3-RD with VP2-151T bound to DAF as efficiently as did virus with VP2-151S (Fig. 5D), indicating that VP2-151S is not required for virus interaction with DAF. No virus with VP2-108D was obtained, and we suspect that the capsid structure does not tolerate aspartate at this position.
VP3-234Q and VP2-138D are in close contact with DAF in the virus-receptor complex.
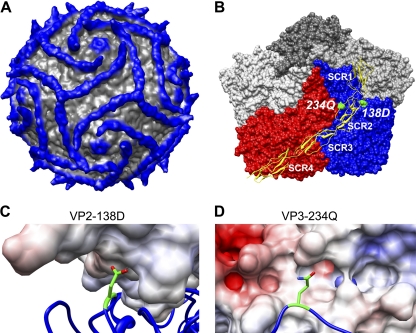
The three-dimensional reconstruction of CB3-RD complexed with the extracellular domain of DAF, previously obtained by cryo-EM (10), has now been refined to 9-Å resolution (Yoder, Cifuente, and Hafenstein, unpublished). As shown in the new structure, the predominant interactions between virus and receptor occur between the “puff region,” formed by residues in VP2, and the second short consensus repeat (SCR2) domain of DAF; the junction between SCRs 2 and 3 is also in close proximity to the virus surface, and several residues within capsid proteins VP1 and VP3 make additional contacts with DAF. Both VP3-234Q and VP2-138D make direct contacts with DAF, on opposite sides of the SCR2 domain (Fig. 6). VP3-234Q is likely to interact with DAF residue Thr70, and VP2-138D with DAF residue Val89. The two interactions effectively anchor SCR2 to the virus surface.
Fig. 6.
Critical capsid residues are in contact with DAF. (A) Cryo-EM reconstruction of DAF bound to CVB3-RD at 9-Å resolution displayed at 1 sigma. Density further than 160 Å from the center of the virus is shown in blue. (B) Surface representation of CVB3-RD protomers surrounding one 5-fold icosahedral axis of the virus with DAF shown in yellow ribbon. The DAF molecule attaches at the 3-fold vertex by way of the C-terminal His tag; SCRs 4 and 3 stretch across the virus surface of the red protomer (in standard orientation), crossing the canyon, and SCR2 interacts with the puff region of the blue (neighboring) protomer. SCR1 makes no contacts, rising above the virus surface. Residues VP2-138D and VP3-234Q are highlighted in green. (C) Close-up view of residue VP2-138D (in green, stick rendering) with DAF colored according to electrostatic potential. The light blue coloring of DAF indicates an overall slightly positive charge in the region nearest to VP2-138D. (D) Close-up view of residue VP3-234Q (in green, stick rendering with oxygen and nitrogen colored red and blue, respectively) with DAF surface rendered and colored according to electrostatic potential; the side chain configuration shown is derived from the crystal structure of CB3-RD and may differ in virus-DAF complex. The light-red coloring of DAF indicates a negative charge in the region nearest to VP2-234Q.
DISCUSSION
Although all group B coxsackieviruses use CAR as a receptor for attachment and infection, isolates differ in their capacities to bind DAF. The results presented here demonstrate that single amino acid changes allow nonbinding CB3 isolates to interact with DAF and thus broaden their tropism. Mutation of VP3-234E to Q permitted CB3-Nancy to bind DAF and infect RD cells. With CB3-H3, in which the identity of VP3 residue 234 was already Q, an additional mutation of VP2-138N to D had a similar effect. Mutation of either residue in CB3-RD resulted in a loss of avidity for DAF, indicating that both are important for virus interaction with this receptor. Consistent with these results, both residues are located at the site of contact between the virus and DAF, as determined by the cryo-EM structure.
The observation that VP3-234Q and VP2-138D interact directly with SCR2 is consistent with earlier data indicating that SCR2—but not SCRs 1, 3, and 4—is indispensable for virus attachment (4). Although we identified two critical capsid residues, the new structure of the CB3-DAF complex shows at least 15 capsid residues in close proximity to SCR2, some of which are also likely to contribute to the avidity of the virus-receptor interaction. These residues, which are largely conserved among the virus isolates we examined here, may provide a surface that accommodates DAF in a way that is either stabilized by VP3-234Q and VP2-138D or destabilized by VP3-234E and VP3-138N. Neither of the amino acid changes we identified is likely to perturb the overall conformation of the virus surface, but it is interesting that both involve charge changes. Upon examining the electrostatic surface of DAF, we found that VP2-138D points into a positively charged pocket (Fig. 6 C); replacement of the negatively charged aspartate residue with asparagine may destabilize this interaction. The positively charged amide moiety of VP3-234Q is in contact with a negatively charged patch on the SCR2 surface; replacement of the glutamine with a negatively charged glutamate may also have a destabilizing effect (Fig. 6D).
Other investigators have recently suggested that, consistent with our conclusions, VP3-234Q is important for CB3-RD interaction with DAF (although this was not tested by mutagenesis) (6). However, our results do not support the conclusion of Lindberg and colleagues (20) that VP2-151S and/or VP2-108V accounts for the tropism of CB3-RD for RD cells, and they do not suggest that these residues are important for interaction with DAF. These residues are present in both the CB3-Nancy and CB3-RD isolates we studied and thus cannot be responsible for the differences in tropism we observed. We found that mutation of VP2-151S had no effect on virus interaction with DAF and were unable to generate virus with VP2-108D. It is interesting that the viruses studied by Lindberg et al. (one with tropism for RD cells and one without) both possessed the VP2-138D residue we found to be important for DAF interaction and that—consistent with our observations—the RD-tropic virus had undergone a replacement of VP3-234E with Q, in addition to the changes at VP2-108 and VP2-151 (20).
In addition to CB3-RD and CB3-H3-RD, a number of other CB3 isolates—CB3-PD and PD1 (28), CB3-HA (25), and CB3-Nancy/New (31)—have been shown to bind DAF. Although an isolate initially cloned by Kandolf and colleagues (15, 17) was reported to lack avidity for DAF (25), we (unpublished observations) and others (31) have found that this virus binds DAF quite well. Sequences of all the DAF-binding isolates show conservation of VP2-138D. VP3-234Q is conserved among all the isolates but Nancy/New, which has another uncharged residue, leucine, at this position.
The observation that single amino acid changes permit significant shifts in receptor tropism may have significance for understanding the pathogenesis of viral infection in vivo. As CB3 initiates infection in the gastrointestinal (GI) tract, enters the bloodstream, and spreads to target organs, it is likely to encounter diverse cell types that vary greatly in expression and localization of CAR, DAF, and other potential receptor molecules. DAF-binding variants within a virus population may enjoy a selective growth advantage at some sites, such as the intestinal lumen, where DAF is readily accessible but CAR is not; we previously found that passage of CB3-Nancy on polarized intestinal epithelial cells selected for DAF-binding variants (33). At other sites, DAF-binding viruses may be at a disadvantage. Because of error-prone RNA replication, picornaviruses exist as quasispecies, with a broad genomic diversity; experiments with poliovirus have demonstrated that virus populations experience multiple evolutionary bottlenecks during infection and spread through the host (19). Given the ease with which CB3 undergoes changes in DAF tropism during in vitro culture, it would not be surprising if similar changes occur as viruses encounter anatomic barriers in vivo.
ACKNOWLEDGMENTS
We thank Sally Huber and Kirk Knowlton for the CB3-H3 cDNA clone, Karin Klingel and Reinhard Kandolf for pCVB3-T7, and the CHOP Nucleic Acids Core for DNA sequencing.
This work was supported by grants from the NIH (AI52281 to J.M.B. and AI07927 to S.H.) and by the Plotkin Endowed Chair in Infectious Diseases at the Children's Hospital of Philadelphia.
Footnotes
Published ahead of print on 11 May 2011.
REFERENCES
- 1. Bergelson J. M., et al. 1994. Decay-accelerating factor (CD55), a glycosylphosphatidylinositol-anchored complement regulatory protein, is a receptor for several echoviruses. Proc. Natl. Acad. Sci. U. S. A. 91:6245–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bergelson J. M., et al. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320–1323 [DOI] [PubMed] [Google Scholar]
- 3. Bergelson J. M., et al. 1997. Clinical coxsackievirus B isolates differ from laboratory strains in their interaction with two cell surface receptors. J. Infect. Dis. 175:697–700 [DOI] [PubMed] [Google Scholar]
- 4. Bergelson J. M., et al. 1995. Coxsackievirus B3 adapted to growth in RD cells binds to decay-accelerating factor (CD55). J. Virol. 69:1903–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bergelson J. M., et al. 1993. Infection by echoviruses 1 and 8 depends on the α2 subunit of human VLA-2. J. Virol. 67:6847–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carson S. D., Chapman N. M., Hafenstein S., Tracy S. 2011. Variations of coxsackievirus B3 capsid primary structure, ligands, and stability are selected for in a coxsackievirus and adenovirus receptor-limited environment. J. Virol. 85:3306–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cohen C. J., et al. 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. U. S. A. 98:15191–15196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coyne C. B., Bergelson J. M. 2006. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124:119–131 [DOI] [PubMed] [Google Scholar]
- 9. Fanning A. S., Mitic L. L., Anderson J. M. 1999. Transmembrane proteins in the tight junction barrier. J. Am. Soc. Nephrol. 10:1337–1345 [DOI] [PubMed] [Google Scholar]
- 10. Hafenstein S., et al. 2007. Interaction of decay-accelerating factor with coxsackievirus B3. J. Virol. 81:12927–12935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 12. Hsu K.-H. L., Lonberg-Holm K., Alstein B., Crowell R. L. 1988. A monoclonal antibody specific for the cellular receptor for the group B coxsackieviruses. J. Virol. 62:1647–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hsu K.-H. L., Paglini S., Alstein B., Crowell R. L. 1990. Identification of a second cellular receptor for a coxsackievirus B3 variant, CB3-RD, p. 271–277 In Brinton M., Heinz F. (ed.), New aspects of positive-strand RNA viruses. American Society for Microbiology, Washington, DC [Google Scholar]
- 14. Huber S. A., Haisch C., Lodge P. A. 1990. Functional diversity in vascular endothelial cells: role in coxsackievirus tropism. J. Virol. 64:4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kandolf R., Hofschneider P. H. 1985. Molecular cloning of the genome of a cardiotropic coxsackie B3 virus: full-length reverse-transcribed recombinant cDNA generates infectious virus in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 82:4818–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karnauchow T. M., et al. 1996. The HeLa cell receptor for enterovirus 70 is decay-accelerating factor (CD55). J. Virol. 70:5143–5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klump W. M., Bergmann I., Muller B. C., Ameis D., Kandolf R. 1990. Complete nucleotide sequence of infectious coxsackievirus B3 cDNA: two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J. Virol. 64:1573–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Knowlton K. U., Jeon E. S., Berkley N., Wessely R., Huber S. 1996. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J. Virol. 70:7811–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuss S. K., Etheredge C. A., Pfeiffer J. K. 2008. Multiple host barriers restrict poliovirus trafficking in mice. PLoS Pathog. 4:e1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lindberg A. M., Crowell R. L., Zell R., Kandolf R., Pettersson U. 1992. Mapping of the RD phenotype of the Nancy strain of coxsackievirus B3. Virus Res. 24:187–196 [DOI] [PubMed] [Google Scholar]
- 21. Martino T. A., et al. 2000. The coxsackie-adenovirus receptor (CAR) is used by reference strains and clinical isolates representing all six serotypes of coxsackievirus group B and by swine vesicular disease virus. Virology 271:99–108 [DOI] [PubMed] [Google Scholar]
- 22. Milstone A. M., et al. 2005. Interaction with coxsackievirus and adenovirus receptor, but not with decay-accelerating factor (DAF), induces A-particle formation in a DAF-binding coxsackievirus B3 isolate. J. Virol. 79:655–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Modlin J. F. 2010. Coxsackieviruses, echoviruses, newer enteroviruses, and parechoviruses. In Mandel G. L., Bennet J. E., Dolin R. (ed.), Principles and practice of infectious diseases, 7 ed. Churchill Livingstone/Elsevier, New York, NY [Google Scholar]
- 24. Mohanty J. G., Crowell R. L. 1993. Attempts to purify a second cellular receptor for a coxsackievirus B3 variant, CB3-RD from HeLa cells. Virus Res. 29:305–320 [DOI] [PubMed] [Google Scholar]
- 25. Pasch A., Kupper J. H., Wolde A., Kandolf R., Selinka H. C. 1999. Comparative analysis of virus-host cell interactions of haemagglutinating and non-haemagglutinating strains of coxsackievirus B3. J. Gen. Virol. 80:3153–3158 [DOI] [PubMed] [Google Scholar]
- 26. Powell R. M., Ward T., Goodfellow I., Almond J. W., Evans D. J. 1999. Mapping the binding domains on decay accelerating factor (DAF) for haemagglutinating enteroviruses: implications for the evolution of a DAF-binding phenotype. J. Gen. Virol. 80:3145–3152 [DOI] [PubMed] [Google Scholar]
- 27. Reagan K. J., Goldberg B., Crowell R. L. 1984. Altered receptor specificity of coxsackievirus B3 after growth in rhabdomyosarcoma cells. J. Virol. 49:635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schmidtke M., et al. 2000. Attachment of coxsackievirus B3 variants to various cell lines: mapping of phenotypic differences to capsid protein VP1. Virology 275:77–88 [DOI] [PubMed] [Google Scholar]
- 29. Shafren D. R., et al. 1995. Coxsackieviruses B1, B3, and B5 use decay accelerating factor as a receptor for cell attachment. J. Virol. 69:3873–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shafren D. R., Dorahy D. J., Ingham R. A., Burns G. F., Barry R. D. 1997. Coxsackievirus A21 binds to decay-accelerating factor but requires intercellular adhesion molecule 1 for cell entry. J. Virol. 71:4736–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shafren D. R., Williams D. T., Barry R. D. 1997. A decay-accelerating factor-binding strain of coxsackievirus B3 requires the coxsackievirus-adenovirus receptor protein to mediate lytic infection of rhabdomyosarcoma cells. J. Virol. 71:9844–9848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shepley M. P., Sherry B., Weiner H. L. 1988. Monoclonal antibody identification of a 100-kDa membrane protein in HeLa cells and human spinal cord involved in poliovirus attachment. Proc. Natl. Acad. Sci. U. S. A. 85:7743–7747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shieh J. T. C., Bergelson J. M. 2002. Interaction with decay-accelerating factor facilitates coxsackievirus B infection of polarized epithelial cells. J. Virol. 76:9474–9480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tomko R. P., Xu R., Philipson L. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. U. S. A. 94:3352–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang X., Bergelson J. M. 1999. Coxsackievirus and adenovirus receptor cytoplasmic and transmembrane domains are not essential for coxsackievirus and adenovirus infection. J. Virol. 73:2259–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ward T., et al. 1994. Decay-accelerating factor CD55 is identified as the receptor for echovirus 7 using CELICS, a rapid immuno-focal cloning method. EMBO J. 13:5070–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]