Abstract
The microRNA miR-122 and DDX6/Rck/p54, a microRNA effector, have been implicated in hepatitis C virus (HCV) replication. In this study, we demonstrated for the first time that HCV-JFH1 infection disrupted processing (P)-body formation of the microRNA effectors DDX6, Lsm1, Xrn1, PATL1, and Ago2, but not the decapping enzyme DCP2, and dynamically redistributed these microRNA effectors to the HCV production factory around lipid droplets in HuH-7-derived RSc cells. Notably, HCV-JFH1 infection also redistributed the stress granule components GTPase-activating protein (SH3 domain)-binding protein 1 (G3BP1), ataxin-2 (ATX2), and poly(A)-binding protein 1 (PABP1) to the HCV production factory. In this regard, we found that the P-body formation of DDX6 began to be disrupted at 36 h postinfection. Consistently, G3BP1 transiently formed stress granules at 36 h postinfection. We then observed the ringlike formation of DDX6 or G3BP1 and colocalization with HCV core after 48 h postinfection, suggesting that the disruption of P-body formation and the hijacking of P-body and stress granule components occur at a late step of HCV infection. Furthermore, HCV infection could suppress stress granule formation in response to heat shock or treatment with arsenite. Importantly, we demonstrate that the accumulation of HCV RNA was significantly suppressed in DDX6, Lsm1, ATX2, and PABP1 knockdown cells after the inoculation of HCV-JFH1, suggesting that the P-body and the stress granule components are required for the HCV life cycle. Altogether, HCV seems to hijack the P-body and the stress granule components for HCV replication.
INTRODUCTION
Hepatitis C virus (HCV) is the causative agent of chronic hepatitis, which progresses to liver cirrhosis and hepatocellular carcinoma. HCV is an enveloped virus with a positive single-stranded 9.6-kb RNA genome, which encodes a large polyprotein precursor of approximately 3,000 amino acid (aa) residues. This polyprotein is cleaved by a combination of the host and viral proteases into at least 10 proteins in the following order: core, envelope 1 (E1), E2, p7, nonstructural 2 (NS2), NS3, NS4A, NS4B, NS5A, and NS5B (12, 13, 21). The HCV core protein, a nucleocapsid, is targeted to lipid droplets (LDs), and the dimerization of the core protein by a disulfide bond is essential for the production of infectious virus (24). Recently, LDs have been found to be involved in an important cytoplasmic organelle for HCV production (26). Budding is an essential step in the life cycle of enveloped viruses. The endosomal sorting complex required for transport (ESCRT) system has been involved in such enveloped virus budding machineries, including that of HCV (5).
DEAD-box RNA helicases with ATP-dependent RNA-unwinding activities have been implicated in various RNA metabolic processes, including transcription, translation, RNA splicing, RNA transport, and RNA degradation (32). Previously, DDX3 was identified as an HCV core-interacting protein by yeast two-hybrid screening (25, 29, 43). Indeed, DDX3 is required for HCV RNA replication (3, 31). DDX6 (Rck/p54) is also required for HCV replication (16, 33). DDX6 interacts with an initiation factor, eukaryotic initiation factor 4E (eIF-4E), to repress the translational activity of mRNP (38). Furthermore, DDX6 regulates the activity of the decapping enzymes DCP1 and DCP2 and interacts directly with Argonaute-1 (Ago1) and Ago2 in the microRNA (miRNA)-induced silencing complex (miRISC) and is involved in RNA silencing. DDX6 localizes predominantly in the discrete cytoplasmic foci termed the processing (P) body. Thus, the P body seems to be an aggregate of translationally repressed mRNPs associated with the translation repression and mRNA decay machinery.
In addition to the P body, eukaryotic cells contain another type of RNA granule termed the stress granule (SG) (1, 6, 22, 30). SGs are aggregates of untranslating mRNAs in conjunction with a subset of translation initiation factors (eIF4E, eIF3, eIF4A, eIFG, and poly(A)-binding protein [PABP]), the 40S ribosomal subunits, and several RNA-binding proteins, including PABP, T cell intracellular antigen 1 (TIA-1), TIA-1-related protein (TIAR), and GTPase-activating protein (SH3 domain)-binding protein 1 (G3BP1). SGs regulate mRNA translation and decay as well as proteins involved in various aspects of mRNA metabolisms. SGs are cytoplasmic phase-dense structures that occur in eukaryotic cells exposed to various environmental stress, including heat, arsenite, viral infection, oxidative conditions, UV irradiation, and hypoxia. Importantly, several viruses target SGs and stress granule components for viral replication (10, 11, 34, 39). Recent studies suggest that SGs and the P body physically interact and that mRNAs may move between the two compartments (1, 6, 22, 28, 30).
miRNAs are a class of small noncoding RNA molecules ∼21 to 22 nucleotides (nt) in length. miRNAs usually interact with 3′-untranslated regions (UTRs) of target mRNAs, leading to the downregulation of mRNA expression. Notably, the liver-specific and abundant miR-122 interacts with the 5′-UTR of the HCV RNA genome and facilitates HCV replication (15, 17, 19, 20, 31). Ago2 is at least required for the efficient miR-122 regulation of HCV RNA accumulation and translation (40). However, the molecular mechanism(s) for how DDX6 and miR-122 as well as DDX3 positively regulate HCV replication is not fully understood. Therefore, we investigated the potential role of P-body and stress granule components in HCV replication.
MATERIALS AND METHODS
Cell culture.
293FT cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS). HuH-7-derived RSc cured cells, in which cell culture-generated HCV-JFH1 (JFH1 strain of genotype 2a) (37) could infect and effectively replicate, were cultured in DMEM with 10% FBS as described previously (3-5, 23).
Plasmid construction.
To construct pcDNA3-FLAG-DDX6, a DNA fragment encoding DDX6 was amplified from total RNAs derived from RSc cells by reverse transcription (RT)-PCR using KOD-Plus DNA polymerase (Toyobo) and the following pairs of primers: 5′-CGGGATCCAAGATGAGCACGGCCAGAACAGAGAACCCTGTT-3′ (forward) and 5′-CCGCTCGAGTTAAGGTTTCTCATCTTCTACAGGCTCGCT-3′ (reverse). The obtained DNA fragments were subcloned into either BamHI-XhoI site of the pcDNA3-FLAG vector (2), and the nucleotide sequences were determined by BigDye termination cycle sequencing using an ABI Prism 310 genetic analyzer (Applied Biosystems, Foster City, CA).
RNA interference.
The following small interfering RNAs (siRNAs) were used: human ATXN2/ATX2/ataxin-2 (siGENOME SMRT pool M-011772-01-005), human PABP1/PABPC1 (siGENOME SMRT pool M-019598-01-005), human Lsm1 (siGENOME SMRT pool M-005124-01-005), human Xrn1 (siGENOME SMRT pool M-013754-01-005), human G3BP1 (ON-TARGETplus SMRT pool L-012099-00-005), human PATL1 (siGENOME SMRT pool M-015591-00-005), and siGENOME nontargeting siRNA pool 1 (D-001206-13-05) (Dharmacon, Thermo Fisher Scientific, Waltham, MA), as a control. siRNAs (25 nM final concentration) were transiently transfected into RSc cells (3-5, 23) using Oligofectamine (Invitrogen) according to the manufacturer's instructions. Oligonucleotides with the following sense and antisense sequences were used for the cloning of short hairpin RNA (shRNA)-encoding sequences targeted to DDX6 (DDX6i) as well as the control nontargeting shRNA (shCon) in a lentiviral vector: 5′-GATCCCCGGAGGAACTAACTCTGAAGTTCAAGAGACTTCAGAGTTAGTTCCTCCTTTTTGGAAA-3′ (sense) and 5′-AGCTTTTCCAAAAAGGAGGAACTAACTCTGAAGTCTCTTGAACTTCAGAGTTAGTTCCTCCGGG-3′ (antisense) for DDX6i and 5′-GATCCCCGAATCCAGAGGTAATCTACTTCAAGAGAGTAGATTACCTCTGGATTCTTTTTGGAAA-3′ (sense) and 5′-AGCTTTTCCAAAAAGAATCCAGAGGTAATCTACTCTCTTGAAGTAGATTACCTCTGGATTCGGG-3′ (antisense) for shCon. The oligonucleotides described above were annealed and subcloned into the BglII-HindIII site, downstream from an RNA polymerase III promoter of pSUPER (8), to generate pSUPER-DDX6i and pSUPER-shCon, respectively. To construct pLV-DDX6i and pLV-shCon, the BamHI-SalI fragments of the corresponding pSUPER plasmids were subcloned into the BamHI-SalI site of pRDI292, an HIV-1-derived self-inactivating lentiviral vector containing a puromycin resistance marker allowing for the selection of transduced cells (7). pLV-DDX3i, described previously (3), was used.
Lentiviral vector production.
The vesicular stomatitis virus G protein (VSV-G)-pseudotyped HIV-1-based vector system was described previously (27, 44). The lentiviral vector particles were produced by the transient transfection of the second-generation packaging construct pCMV-ΔR8.91 (27, 44), the VSV-G-envelope-expressing plasmid pMDG2, as well as pRDI292 into 293FT cells with FuGene6 reagent (Roche Diagnostics, Mannheim, Germany).
HCV infection experiments.
The supernatants were collected from cell culture-generated HCV-JFH1 (37)-infected RSc cells (3-5, 23) at 5 days postinfection and stored at −80°C after filtering through a 0.45-μm filter (Kurabo, Osaka, Japan) until use. For infection experiments with HCV-JFH1, RSc cells (1 × 105 cells/well) were plated onto 6-well plates and cultured for 24 h. We then infected the cells at a multiplicity of infection (MOI) of 1 or 4. The culture supernatants were collected at 24 h postinfection, and the levels of the core protein were determined by an enzyme-linked immunosorbent assay (ELISA) (Mitsubishi Kagaku Bio-Clinical Laboratories, Tokyo, Japan). Total RNA was also isolated from the infected cellular lysates by using an RNeasy minikit (Qiagen, Hilden, Germany) for analysis of intracellular HCV RNA. The infectivity of HCV-JFH1 in the culture supernatants was determined by a focus-forming assay at 48 h postinfection. HCV-JFH1-infected cells were detected by using anti-HCV core (CP-9 and CP-11 mixture).
Quantitative RT-PCR analysis.
The quantitative RT-PCR analysis of HCV RNA was performed by real-time LightCycler PCR (Roche) as described previously (3-5, 14, 23). We used the following forward and reverse primer sets for the real-time LightCycler PCR: 5′-ATGAGTCATGTGGCAGTGGA-3′ (forward) and 5′-GCTGGCTGTACTTCCTCCAC-3′ (reverse) for DDX3, 5′-ATGAGCACGGCCAGAACAGA-3′ (forward) and 5′-TTGCTGTGTCTGTGTGCCCC-3′ (reverse) for DDX6, 5′-TGACGGGGTCACCCACACTG-3′ (forward) and 5′-AAGCTGTAGCCGCGCTCGGT-3′ (reverse) for β-actin, and 5′-AGAGCCATAGTGGTCTGCGG-3′ (forward) and 5′-CTTTCGCAACCCAACGCTAC-3′ (reverse) for HCV-JFH1.
Preparation of anti-PATL1 antibody.
The anti-PATL1 antiserum was raised in rabbits using the glutathione S-transferase (GST)-fused PATL1 Ct (C-terminal region of PATL1, aa 450 to 770) as an antigen, and immunoglobulins were affinity purified by using the maltose-binding protein (MBP)-fused PATL1 Ct that was immobilized on an N-hydroxysuccinimide (NHS) column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden).
Preparation of LDs.
Lipid droplets (LDs) were prepared as described previously (26). Cells were pelleted by centrifugation at 1,500 rpm. The pellet was resuspended in hypotonic buffer (50 mM HEPES [pH 7.4], 1 mM EDTA, 2 mM MgCl2) supplemented with a protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan) and was incubated for 10 min at 4°C. The suspension was homogenized with 30 strokes of a glass Dounce homogenizer using a tight-fitting pestle (Wheaton, Millville, NJ). A 1/10 volume of 10× isotonic buffer {0.2 M HEPES (pH 7.4), 1.2 M potassium acetate (KoAc), 40 mM magnesium acetate [Mg(oAc)2], and 50 mM dithiothreitol (DTT)} was added to the homogenate. The nuclei were removed by centrifugation at 2,000 rpm for 10 min at 4°C. The supernatant was collected and centrifuged at 16,000 × g for 10 min at 4°C. The supernatant was mixed with an equal volume of 1.04 M sucrose in isotonic buffer (50 mM HEPES, 100 mM KCl, 2 mM MgCl2, and protease inhibitor cocktail). The solution was set in a 13.2-ml Polyallomer centrifuge tube (Beckman Coulter, Brea, CA). One milliliter of isotonic buffer was loaded onto the sucrose mixture. The tube was centrifuged at 100,000 × g in an SW41Ti rotor (Beckman Coulter) for 1 h at 4°C. After the centrifugation, the LD fraction on the top of the gradient solution was recovered in phosphate-buffered saline (PBS). The collected LD fraction was used for Western blot analysis.
Western blot analysis.
Cells were lysed in a buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 4 mM EDTA, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate (SDS), 1 mM DTT, and 1 mM phenylmethylsulfonyl fluoride. Supernatants from these lysates were subjected to SDS-polyacrylamide gel electrophoresis, followed by immunoblot analysis using anti-DDX3 (catalog no. 54257 [NT] and 5428 [IN]; Anaspec, San Jose, CA), anti-DDX6 (A300-460A; Bethyl Laboratories, Montgomery, TX), anti-adipose differentiation-related protein (ADFP; GTX110204; GeneTex, San Antonio, TX), anti-calnexin (NT; Stressgen, Ann Arbor, MI), anti-HCV core (CP-9 and CP-11; Institute of Immunology, Tokyo, Japan), anti-β-actin antibody (A5441; Sigma, St. Louis, MO), anti-ATX2/SCA2 antibody (A302-033A; Bethyl), anti-PABP (sc-32318 [10E10]; Santa Cruz Biotechnology, Santa Cruz, CA), anti-PABP (ab21060; Abcam, Cambridge, United Kingdom), anti-G3BP1 (611126; BD Transduction Laboratories, San Jose, CA), anti-LSM1 (LS-C97364, Life Span Biosciences, Seattle, WA), anti-HSP70 (610607; BD), anti-XRN1 (A300-443A; Bethyl), or anti-PATL1 antibody.
Immunofluorescence and confocal microscopic analysis.
Cells were fixed in 3.6% formaldehyde in PBS, permeabilized in 0.1% NP-40 in PBS at room temperature, and incubated with anti-DDX3 antibody (54257 [NT] and 5428 [IN]; Anaspec), anti-DDX3X (LS-C64576; Life Span), anti-DDX6 (A300-460A; Bethyl), anti-HCV core (CP-9 and CP-11), anti-ATX2/SCA2 antibody (A302-033A; Bethyl), anti-ataxin-2 (611378; BD), anti-PABP (ab21060; Abcam), anti-G3BP1 (A302-033A; Bethyl), anti-LSM1 (LS-C97364; Life Span), anti-XRN1 (A300-443A; Bethyl), anti-Dcp2 (A302-597A; Bethyl), anti-human Ago2 (011-22033; Wako, Osaka, Japan), or anti-PATL1 antibody at a 1:300 dilution in PBS containing 3% bovine serum albumin (BSA) for 30 min at 37°C. The cells were then stained with fluorescein isothiocyanate (FITC)-conjugated anti-rabbit antibody (Jackson ImmunoResearch, West Grove, PA) at a 1:300 dilution in PBS containing BSA for 30 min at 37°C. Lipid droplets and nuclei were stained with borondipyrromethene (BODIPY) 493/503 (Molecular Probes, Invitrogen) and DAPI (4′,6-diamidino-2-phenylindole), respectively, for 15 min at room temperature. Following extensive washing in PBS, the cells were mounted onto slides using a mounting medium of 90% glycerin-10% PBS with 0.01% p-phenylenediamine added to reduce fading. Samples were viewed under a confocal laser scanning microscope (LSM510; Zeiss, Jena, Germany).
Statistical analysis.
A statistical comparison of the infectivities of HCV in the culture supernatants between the knockdown cells and the control cells was performed by using the Student t test. P values of less than 0.05 were considered statistically significant. All error bars indicate standard deviations.
RESULTS
HCV infection hijacks the P-body components.
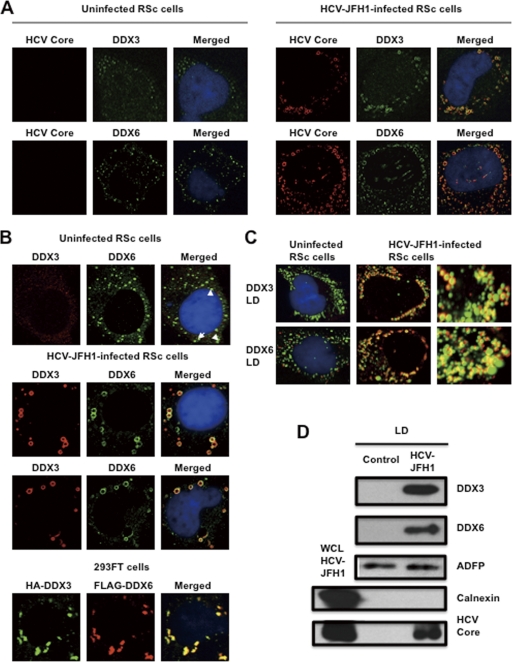
To investigate the potential role of P-body components in the HCV life cycle, we first examined the alteration of the subcellular localization of DDX3 or DDX6 by HCV-JFH1 infection using confocal laser scanning microscopy as previously described (2), since both DDX3 and DDX6 were identified previously as P-body components (6). For this, we used HuH-7-derived RSc cells, in which cell culture-generated HCV-JFH1 (JFH1 strain of genotype 2a) (37) can infect and effectively replicate (3, 4, 23). HCV-JFH1-infected RSc cells at 60 h postinfection were stained with anti-HCV core antibody, anti-DDX3, and/or anti-DDX6. Lipid droplets (LDs) and nuclei were stained with BODIPY 493/503 and DAPI (4′,6-diamidino-2-phenylindole), respectively. Samples were viewed under a confocal laser scanning microscope. Although we observed that endogenous DDX3 localized in faint cytoplasmic foci in uninfected RSc cells, DDX3 relocalized, formed ringlike structures, and colocalized with the HCV core protein in response to HCV-JFH1 infection (Fig. 1 A). On the other hand, endogenous DDX6 was localized in the evident cytoplasmic foci termed P bodies in the uninfected cells (Fig. 1A). DDX6 also relocalized, formed ringlike structures, and colocalized with the core protein in response to HCV-JFH1 infection (Fig. 1A). Although we failed to observe that most of the P bodies of DDX6 perfectly colocalized with DDX3 in uninfected RSc cells (Fig. 1B), we observed a few P bodies of DDX6 colocalized with DDX3 in the uninfected cells (Fig. 1B, arrowheads). Intriguingly, we found that endogenous DDX3 colocalized with endogenous DDX6 in HCV-JFH1-infected cells (Fig. 1B). To further confirm this finding, pHA-DDX3 (41) and pcDNA3-FLAG-DDX6 were cotransfected into 293FT cells. Consequently, we observed that hemagglutinin (HA)-DDX3 colocalized with FLAG-DDX6 in 293FT cells coexpressing HA-DDX3 and FLAG-DDX6 (Fig. 1B), suggesting cross talk of DDX3 with DDX6. Recently, LDs have been found to be involved in an important cytoplasmic organelle for HCV production (26). Indeed, both DDX3 and DDX6 were recruited around LDs in response to HCV infection, while these proteins did not colocalize with LDs in uninfected naïve RSc cells (Fig. 1C). Furthermore, both DDX3 and DDX6 accumulated in the LD fraction of the HCV-JFH1-infected RSc cells; however, we could not detect both proteins in the LD fraction from uninfected control cells (Fig. 1D), suggesting that DDX3 and DDX6 are recruited around LDs in response to HCV infection.
Fig. 1.
Dynamic recruitment of DDX3 and DDX6 around lipid droplets (LDs) in response to HCV-JFH1 infection. (A) HCV-JFH1 disrupts the P-body formation of DDX6. Cells were fixed at 60 h postinfection and were then examined by confocal laser scanning microscopy. Cells were stained with anti-HCV core (CP-9 and CP-11 mixture) and either anti-DDX3 (54257 and 54258 mixture) or anti-DDX6 (A300-460A) antibody and then visualized with FITC (DDX3 or DDX6) or Cy3 (core). Images were visualized by using confocal laser scanning microscopy. The two-color overlay images are also exhibited (merged). Colocalization is shown in yellow. (B) HCV-JFH1 recruits DDX3 or DDX6 around LDs. Cells were stained with either anti-DDX3 or anti-DDX6 antibody and were then visualized with Cy3 (red). Lipid droplets and nuclei were stained with BODIPY 493/503 (green) and DAPI (blue), respectively. A high-magnification image is also shown. (C) Colocalization of DDX3 with DDX6. HCV-JFH1-infected RSc cells at 60 h postinfection were stained with anti-DDX3X (LS-C64576) and anti-DDX6 (A300-460A) antibodies. 293FT cells cotransfected with 100 ng of pcDNA3-FLAG-DDX6 and 100 ng of pHA-DDX3 (41) were stained with anti-FLAG-Cy3 and anti-HA-FITC antibodies (Sigma). (D) Association of DDX3 and DDX6 with LDs in response to HCV-JFH1 infection. The LD fraction and whole-cell lysates (WCL) were collected from uninfected RSc cells (control) or HCV-JFH1-infected RSc cells at 5 days postinfection. The results of Western blot analyses of DDX3, DDX6, and the HCV core protein as well as the LD marker ADFP and the endoplasmic reticulum (ER) marker calnexin in the LD fraction are shown.
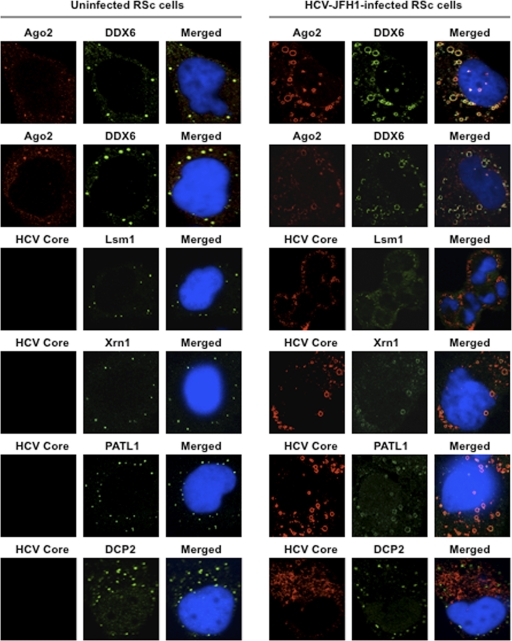
These results suggest that HCV-JFH1 infection disrupts P-body formation. Therefore, we further examined whether or not HCV-JFH1 disrupts the P-body formations of other microRNA effectors, including Ago2; the Sm-like protein Lsm1, which is a subunit of heptameric-ring Lsm1-7, involved in decapping; the 5′-to-3′ exonuclease Xrn1; the decapping activator PATL1; and the decapping enzyme DCP2 (6, 21, 30). As expected, HCV-JFH1 disrupted the P-body formations of Ago2, Lsm1, and Xrn1 as well as PATL1 (Fig. 2). Lsm1, Xrn1, or PATL1 relocalized, formed ringlike structures, and colocalized with the HCV core protein in response to HCV-JFH1 infection, whereas they were localized predominantly in P bodies in uninfected RSc cells (Fig. 2). In fact, we observed that DDX6 colocalized with Ago2, a P-body marker (Fig. 2). In contrast, HCV-JFH1 failed to disrupt the P-body formation of DCP2 (Fig. 2). Thus, these results suggest that HCV disrupts P-body formation through the hijacking of P-body components.
Fig. 2.
HCV disrupts the P-body formation of microRNA effectors. Uninfected RSc cells and HCV-JFH1-infected RSc cells at 72 h postinfection were stained with anti-human AGO2 (011-22033) and anti-DDX6 (A300-460A) antibodies. The cells were also stained with anti-HCV core and anti-Lsm1 (LS-C97364), anti-Xrn1 (A300-443A), anti-PATL1, or anti-DCP2 (A302-597A) antibodies and were examined by confocal laser scanning microscopy.
HCV hijacks stress granule components.
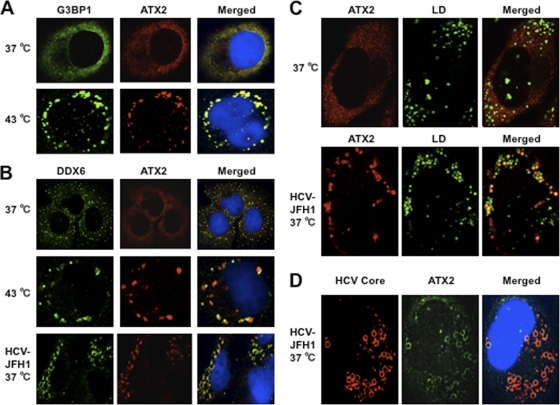
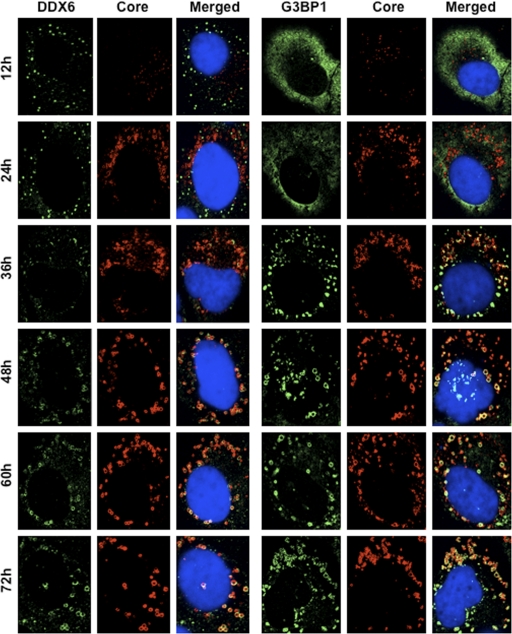
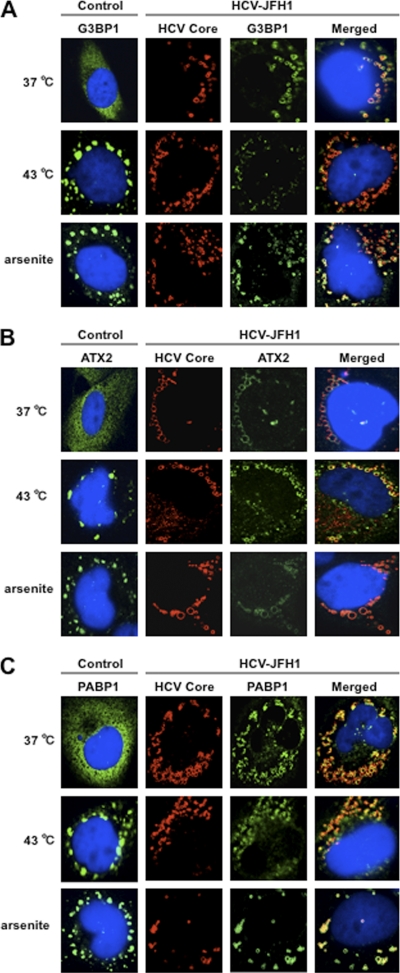
Since Nonhoff et al. recently reported that DDX6 interacted with ataxin-2 (ATX2) (28), we examined the potential cross talk among DDX6, ATX2, and HCV. Although ATX2 and G3BP1, a well-known stress granule component (36), were dispersed in the cytoplasm at 37°C, both proteins formed discrete aggregates termed stress granules and colocalized with each other in response to heat shock at 43°C for 45 min, indicating that ATX2 is also stress granule component (Fig. 3 A). We did not observe prominent colocalization between DDX6 and ATX2 at 37°C (Fig. 3B). In contrast, we found that DDX6 was recruited, juxtaposed, and partially colocalized with stress granules of ATX2 in response to heat shock at 43°C for 45 min in the uninfected RSc cells (Fig. 3B). Notably, ATX2 was recruited, formed the ring-like structures, and partially colocalized with DDX6 in response to HCV-JFH1 infection even at 37°C (Fig. 3B). Furthermore, we noticed that ATX2 was recruited around LDs in HCV-JFH1-infected cells at 72 h postinfection, while ATX2 did not colocalize with LDs in uninfected cells (Fig. 3C), suggesting the colocalization of ATX2 with the HCV core protein in infected cells. Indeed, ATX2 colocalized with the HCV core protein in HCV-JFH1-infected RSc cells at 37°C (Fig. 3D). Moreover, HCV-JFH1 infection induced the colocalization of the core protein with other stress granule components, G3BP1 or PABP1 as well as ATX2 (Fig. 4 and 5). To further confirm our findings, we examined the time course of the redistribution of DDX6 and G3BP1 after inoculation with HCV-JFH1. Consequently, we still detected the P-body formation of DDX6 and dispersed G3BP1 in the cytoplasm, and we did not observe a colocalization between the HCV core protein and DDX6 at 12 and 24 h postinfection (Fig. 4). In contrast, we found that the P-body formation of DDX6 began to be disrupted at 36 h postinfection (Fig. 4). Consistently, G3BP1 formed stress granules at 36 h postinfection (Fig. 4). We then noticed a ringlike formation of DDX6 or G3BP1 and colocalization with the HCV core protein after 48 h postinfection (Fig. 4), suggesting that the disruption of P-body formation and the hijacking of P-body and stress granule components occur in a late step of HCV infection.
Fig. 3.
Dynamic redistribution of ataxin-2 (ATX2) around LDs in response to HCV-JFH1 infection. (A) ATX2 is a stress granule component. RSc cells were incubated at 37°C or 43°C for 45 min. Cells were stained with anti-G3BP1 (A302-033A) and anti-ATX2 (A93520) antibodies and were examined by confocal laser scanning microscopy. (B) Dynamic redistribution of DDX6 and ATX2 in response to heat shock or HCV infection. RSc cells after heat shock at 43°C for 45 min or 72 h after inoculation with HCV-JFH1 were stained with anti-DDX6 and anti-ATX2 (A93520) antibodies. (C) HCV relocalizes ataxin-2 to LDs. HCV-JFH1-infected RSc cells at 72 h postinfection were stained with anti-ATX2 (A93520) antibody and BODIPY 493/503. (D) ATX2 colocalizes with the HCV core protein. HCV-JFH1-infected RSc cells at 72 h postinfection were stained with anti-ATX2/SCA2 (A301-118A) and anti-HCV core antibodies.
Fig. 4.
Dynamic redistribution of DDX6 and G3BP1 in response to HCV-JFH1 infection. RSc cells at the indicated times (hours) after inoculation with HCV-JFH1 were stained with anti-HCV core and either anti-DDX6 (A300-460A) or anti-G3BP1 (A302-033A) antibodies.
Fig. 5.
HCV suppresses stress granule formation in response to heat shock or treatment with arsenite. Naïve RSc cells or HCV-JFH1-infected RSc cells at 72 h postinfection were incubated at 37°C or 43°C for 45 min. Cells were also treated with 0.5 mM arsenite for 30 min. Cells were stained with anti-HCV core and anti-G3BP1 (A), anti-ATX2 (B), or anti-PABP1 (ab21060) (C) antibodies and were examined by confocal laser scanning microscopy.
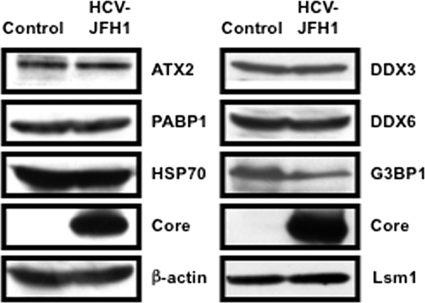
We then examined whether or not HCV-JFH1 infection could affect the stress granule formation of G3BP1, ATX2, or PABP1 in response to heat shock or treatment with arsenite. These stress granule components dispersed in the cytoplasm at 37°C, whereas these proteins formed stress granules in response to heat shock at 43°C for 45 min or treatment with 0.5 mM arsenite for 30 min (Fig. 5). In contrast, stress granules were not formed in HCV-JFH1-infected cells at 72 h postinfection in response to heat shock at 43°C for 45 min (Fig. 5), suggesting that HCV-JFH1 infection suppresses stress granule formation in response to heat shock or treatment with arsenite. Intriguingly, G3BP1, ATX2, or PABP1 still colocalized with the HCV core protein even under the above-described stress conditions (Fig. 5). Furthermore, Western blot analysis of cell lysates of uninfected or HCV-JFH1-infected cells at 72 h postinfection showed similar protein expression levels of ATX2, PABP1, HSP70, DDX3, DDX6, and Lsm1 but not G3BP1 (Fig. 6), suggesting that HCV-JFH1 infection does not affect host mRNA translation.
Fig. 6.
Host protein expression levels in response to HCV-JFH1 infection. The results of the Western blot analyses of cellular lysates with anti-ATX2/SCA2 antibody (A301-118A), anti-PABP1 (ab21060), anti-HSP70 (610607), anti-HCV core, anti-β-actin, anti-DDX3 (54257 [NT] and 5428 [IN] mixture), anti-DDX6 (A300-460A), anti-G3BP1 (611126), or anti-LSM1 (LS-C97364) antibody in HCV-JFH1-infected RSc cells at 72 h postinfection as well as in naïve RSc cells are shown.
P-body and stress granule components are required for HCV replication.
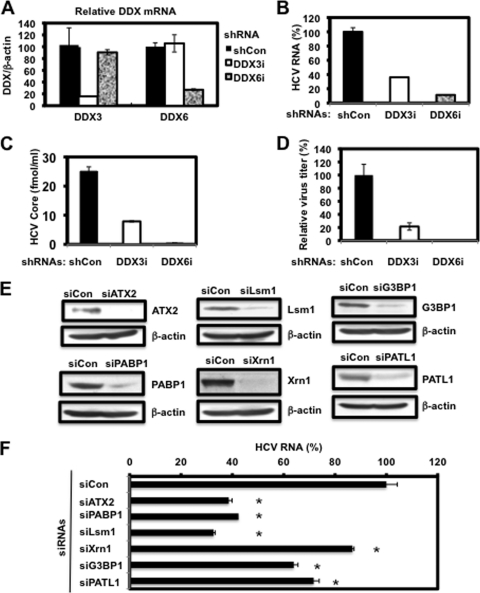
Finally, we investigated the potential role of P-body and stress granule components in the HCV life cycle. We first used lentiviral vector-mediated RNA interference to stably knock down DDX6 as well as DDX3 in RSc cells. We used puromycin-resistant pooled cells 10 days after lentiviral transduction in all experiments. Real-time LightCycler RT-PCR analysis of DDX3 or DDX6 demonstrated a very effective knockdown of DDX3 or DDX6 in RSc cells transduced with lentiviral vectors expressing the corresponding shRNAs (Fig. 7 A). Importantly, shRNAs did not affect cell viabilities (data not shown). We next examined the levels of HCV core and the infectivity of HCV in the culture supernatants as well as the level of intracellular HCV RNA in these knockdown cells 24 h after HCV-JFH1 infection at an MOI of 4. The results showed that the accumulation of HCV RNA was significantly suppressed in DDX3 or DDX6 knockdown cells (Fig. 7B). In this context, the release of the HCV core protein and the infectivity of HCV in the culture supernatants were also significantly suppressed in these knockdown cells (Fig. 7C and D). This finding suggested that DDX6 is required for HCV replication, like DDX3. To further examine the potential role of other P-body and stress granule components in HCV replication, we used RSc cells transiently transfected with a pool of siRNAs specific for ATX2, PABP1, Lsm1, Xrn1, G3BP1, and PATL1 as well as a pool of control siRNAs (siCon) following HCV-JFH1 infection. In spite of the very effective knockdown of each component (Fig. 7E), the siRNAs used in these experiments did not affect cell viabilities (data not shown). Consequently, the accumulation of HCV RNA was significantly suppressed in ATX2, PABP1, or Lsm1 knockdown cells (Fig. 7F), indicting that ATX2, PABP1, and Lsm1 are required for HCV replication. In contrast, the level of HCV RNA was not affected in Xrn1 knockdown cells (Fig. 7F), suggesting that Xrn1 is unrelated to HCV replication. Furthermore, we observed a moderate effect of siG3BP1 and siPATL1 on HCV RNA replication (Fig. 7F). Altogether, HCV seems to hijack the P-body and stress granule components around LDs for HCV replication.
Fig. 7.
Requirement of P-body and stress granule components for HCV replication. (A) Inhibition of DDX3 or DDX6 mRNA expression by the shRNA-producing lentiviral vector. Real-time LightCycler RT-PCR for DDX3 or DDX6 was also performed for β-actin mRNA in RSc cells expressing shRNA targeted to DDX3 (DDX3i) or DDX6 (DDX6i) or the control nontargeting shRNA (shCon) in triplicate. Each mRNA level was calculated relative to the level in RSc cells transduced with the control nontargeting lentiviral vector (shCon), which was assigned as 100%. Error bars in this panel and other panels indicate standard deviations. (B) Levels of intracellular genome-length HCV-JFH1 RNA in the cells at 24 h postinfection at an MOI of 4 were monitored by real-time LightCycler RT-PCR. Results from three independent experiments are shown. Each HCV RNA level was calculated relative to the level in RSc cells transduced with a control lentiviral vector (shCon), which was assigned as 100%. (C) The levels of HCV core in the culture supernatants from the stable knockdown RSc cells 24 h after inoculation of HCV-JFH1 at an MOI of 4 were determined by ELISA. Experiments were done in triplicate, and columns represent the mean core protein levels. (D) The infectivity of HCV in the culture supernatants from stable-knockdown RSc cells 24 h after inoculation of HCV-JFH1 at an MOI of 4 was determined by a focus-forming assay at 24 h postinfection. Experiments were done in triplicate, and each virus titer was calculated relative to the level in RSc cells transduced with a control lentiviral vector (shCon), which was assigned as 100%. (E) Inhibition of ATX2, PABP1, Lsm1, Xrn1, G3BP1, or PATL1 protein expression by 72 h after transient transfection of RSc cells with a pool of control nontargeting siRNA (siCon) or a pool of siRNAs specific for ATX2, PABP1, Lsm1, Xrn1, G3BP1, or PATL1 (25 nM), respectively. The results of Western blot analyses of cellular lysates with anti-ATX2, anti-PABP1, anti-Lsm1, anti-Xrn1, anti-G3BP1, anti-PATL1, or anti-β-actin antibody are shown. (F) Levels of intracellular genome-length HCV-JFH1 RNA in the cells at 48 h postinfection at an MOI of 1 were monitored by real-time LightCycler RT-PCR. RSc cells were transiently transfected with a pool of control siRNA (siCon) or a pool of siRNAs specific for ATX2, PABP1, Lsm1, Xrn1, G3BP1, and PATL1 (25 nM). At 48 h after transfection, the cells were inoculated with HCV-JFH1 at an MOI of 1 and incubated for 2 h. The culture medium was then changed and incubated for 22 h. Experiments were done in triplicate, and each HCV RNA level was calculated relative to the level in RSc cells transfected with a control siRNA (siCon), which was assigned as 100%. Asterisks indicate significant differences compared to the control treatment (*, P < 0.01).
DISCUSSION
So far, the P body and stress granules have been implicated in mRNA translation, RNA silencing, and RNA degradation as well as viral infection (1, 6, 22, 30). Host factors within the P body and stress granules can enhance or limit viral infection, and some viral RNAs and proteins accumulate in the P body and/or stress granules. Indeed, the microRNA effectors DDX6, GW182, Lsm1, and Xrn1 negatively regulate HIV-1 gene expression by preventing the association of viral mRNA with polysomes (9). In contrast, miRNA effectors such as DDX6, Lsm1, PatL1, and Ago2 positively regulate HCV replication (Fig. 7B and F) (16, 31, 33). We have also found that DDX3 and DDX6 are required for HCV RNA replication (3) (Fig. 7B) and that DDX3 colocalized with DDX6 in HCV-JFH1-infected RSc cells (Fig. 1B), suggesting that DDX3 comodulates the DDX6 function in HCV RNA replication. In this regard, the liver-specific miR-122 interacts with the 5′-UTR of the HCV RNA genome and positively regulates HCV replication (15, 17, 19, 20, 31). Since miRNAs usually interact with DDX6 and Ago2 in miRISC and are involved in RNA silencing, DDX6 and Ago2 may be required for miR-122-dependent HCV replication. Indeed, quite recently, a study showed that Ago2 is required for miR-122-dependent HCV RNA replication and translation (40). However, little is known regarding how miR-122 and DDX6 positively regulate HCV replication. Accordingly, we have shown that these miRNA effectors, including DDX6, Lsm1, Xrn1, and Ago2, accumulated around LDs and the HCV production factory and colocalized with the HCV core protein in response to HCV infection (Fig. 1 and 2). However, the decapping enzyme DCP2 did not accumulate and colocalize with the core protein (Fig. 2). Consistent with this finding, Scheller et al. reported previously that the depletion of DCP2 by siRNA did not affect HCV production (33). Since HCV harbors the internal ribosome entry site (IRES) structure in the 5′-UTR of the HCV genome instead of a cap structure, unlike HIV-1, DCP2 may not be recruited on the HCV genome and utilized for HCV replication. Otherwise, DCP2 may determine whether or not DDX6 and miRNAs positively or negatively regulate target mRNA.
Furthermore, we have demonstrated that HCV infection hijacks the P-body and stress granule components around LDs (Fig. 1, 2, 4, and 5). We have found that the P-body formation of DDX6 began to be disrupted at 36 h postinfection (Fig. 4). Consistently, G3BP1 formed stress granules at 36 h postinfection. We then observed the ringlike formation of DDX6 or G3BP1 and colocalization with the HCV core protein after 48 h postinfection, suggesting that the disruption of P-body formation and the hijacking of P-body and stress granule components occur at a late step of HCV infection. Furthermore, HCV infection could suppress stress granule formation in response to heat shock or treatment with arsenite (Fig. 5). In this regard, West Nile virus and dengue virus, of the family Flaviviridae, interfere with stress granule formation and P-body assembly through interactions with T cell intracellular antigen 1 (TIA-1)/TIAR (11). Moreover, PABP1 and G3BP1, stress granule components, are known to be common viral targets for the inhibition of host mRNA translation (34, 39). In fact, HIV-1 and poliovirus proteases cleave PABP1 and/or G3BP1 and suppress stress granule formation during viral infection (34, 39). On the other hand, HCV infection transiently induced stress granules at 36 h postinfection (Fig. 4) and did not cleave PABP1 (Fig. 6); however, HCV could suppress stress granule formation in response to heat shock or treatment with arsenite through hijacking their components around LDs, the HCV production factory (Fig. 5). Consistently, Jones et al. showed that HCV transiently induces stress granules of enhanced green fluorescent protein (EGFP)-G3BP at 36 h after infection with the cell culture-generated HCV (HCVcc) reporter virus Jc1FLAG2 (p7-nsGluc2A); however, those authors did not show the recruitment of EGFP-G3BP to LDs (18). Although we do not know the exact reason for this apparent discrepancy, several possible explanations can be offered. First, those authors examined the localization of EGFP-G3BP within 48 h postinfection, and we observed it at later times (Fig. 4). Second, they used only EGFP-tagged G3BP instead of endogenous G3BP1. Third, they used a Jc1FLAG2 (p7-nsGluc2A) clone, and an HCV-JFH1 clone could markedly induce the recruitment of the core protein to LDs compared to that of Jc1. Also, Jangra et al. failed to observe the recruitment of DDX6 to LDs at 2 days after infection with HJ3-5 virus (16). Accordingly, we also observed that most of the DDX6 still formed intact P bodies at earlier times (12 h or 24 h postinfection). Importantly, we observed the recruitment of DDX6 to LDs 48 h later (Fig. 4). Furthermore, those authors did not show the ringlike structure formation of the HJ3-5 core protein around LDs, unlike the JFH1 core protein that we used in this study. The interaction of the HCV core protein with DDX6 may explain the recruitment of P-body components to LDs. However, we do not yet know whether the P-body function(s) can be performed on LDs. At least, HCV infection did not affect the translation of several host mRNAs with 5′ caps and 3′ poly(A) tails despite the disruption of P-body formation at 72 h postinfection (Fig. 6), suggesting that HCV does not affect P-body function and that HCV recruits functional P bodies to LDs.
We need to address the potential role of stress granule components, such as PABP1, in HCV replication/translation, since the HCV genome does not harbor the 3′ poly(A) tail. Intriguingly, we have found that the accumulation of HCV RNA was significantly suppressed in PABP1 knockdown RSc cells (Fig. 7F). In this regard, Tingting et al. demonstrated previously that G3BP1 and PABP1 as well as DDX1 were identified as the HCV 3′-UTR RNA-binding proteins by proteomic analysis and that G3BP1 was required for HCV RNA replication (35). Yi et al. also reported that G3BP1 was associated with HCV NS5B and that G3BP1 was required for HCV RNA replication (42). We observed a moderate effect of siG3BP1 on HCV RNA replication (Fig. 7F). In contrast, the accumulation of HCV RNA was significantly suppressed in ATX2 and Lsm1 knockdown cells as well as in PABP1 knockdown cells (Fig. 7F), suggesting that ATX2, Lsm1, and PABP1 are required for HCV replication.
Taking these results together, this study has demonstrated for the first time that HCV hijacks P-body and stress granule components around LDs. This hijacking may regulate HCV RNA replication and translation. Indeed, we have found that the accumulation of genome-length HCV-O (genotype 1b) (14) RNA was markedly suppressed in DDX6 knockdown O cells (data not shown). More importantly, these P-body and stress granule components may be involved in the maintenance of the HCV RNA genome without 5′ cap and 3′ poly(A) tail structures in the cytoplasm for long periods, since the hijacking of P-body and stress granule components by HCV occurred at later times.
ACKNOWLEDGMENTS
We thank D. Trono for the lentiviral vector system, T. Wakita for HCV-JFH1, and K. T. Jeang for pHA-DDX3. We also thank T. Nakamura and K. Takeshita for their technical assistance.
This work was supported by a grant-in-aid for scientific research (C) from the Japan Society for the Promotion of Science (JSPS); by a grant-in-aid for research on hepatitis from the Ministry of Health, Labor, and Welfare of Japan; and by the Viral Hepatitis Research Foundation of Japan. M.K. was supported by a research fellowship from the JSPS for young scientists.
Footnotes
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Anderson P., Kedersha N. 2007. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150 [DOI] [PubMed] [Google Scholar]
- 2. Ariumi Y., et al. 2003. Distinct nuclear body components, PML and SMRT, regulate the trans-acting function of HTLV-1 Tax oncoprotein. Oncogene 22:1611–1619 [DOI] [PubMed] [Google Scholar]
- 3. Ariumi Y., et al. 2007. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J. Virol. 81:13922–13926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ariumi Y., et al. 2008. The DNA damage sensors ataxia-telangiectasia mutated kinase and checkpoint kinase 2 are required for hepatitis C virus RNA replication. J. Virol. 82:9639–9646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ariumi Y., et al. 2011. The ESCRT system is required for hepatitis C virus production. PLoS One 6:e14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beckham C. J., Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bridge A. J., Pebernard S., Ducraux A., Nicoulaz A. L., Iggo R. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34:263–264 [DOI] [PubMed] [Google Scholar]
- 8. Brummelkamp T. R., Bernard R., Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553 [DOI] [PubMed] [Google Scholar]
- 9. Chable-Bessia C., et al. 2009. Suppression of HIV-1 replication by microRNA effectors. Retrovirology 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cristea I. M., et al. 2010. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J. Virol. 84:6720–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Emara M. M., Brinton M. A. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U. S. A. 104:9041–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hijikata M., Kato N., Ootsuyama Y., Nakagawa M., Shimotohno K. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. U. S. A. 88:5547–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hijikata M., et al. 1993. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 90:10773–10777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ikeda M., et al. 2005. Efficient replication of a full-length hepatitis C virus genome, strain O, in cell culture, and development of a luciferase reporter system. Biochem. Biophys. Res. Commun. 329:1350–1359 [DOI] [PubMed] [Google Scholar]
- 15. Jangra R. K., Yi M., Lemon S. M. 2010. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J. Virol. 84:6615–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jangra R. K., Yi M., Lemon S. M. 2010. DDX6 (Rck/p54) is required for efficient hepatitis C virus replication but not IRES-directed translation. J. Virol. 84:6810–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ji H., et al. 2008. MicroRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 27:3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones C. T., et al. 2010. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat. Biotechnol. 28:167–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]
- 20. Jopling C. L., Schütz S., Sarnow P. 2008. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kato N., et al. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. U. S. A. 87:9524–9528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kedersha N., Anderson P. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 431:61–81 [DOI] [PubMed] [Google Scholar]
- 23. Kuroki M., et al. 2009. Arsenic trioxide inhibits hepatitis C virus RNA replication through modulation of the glutathione redox system and oxidative stress. J. Virol. 83:2338–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kushima Y., Wakita T., Hijikata M. 2010. A disulfide-bonded dimer of the core protein of hepatitis C virus is important for virus-like particle production. J. Virol. 84:9118–9127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mamiya N., Worman H. J. 1999. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J. Biol. Chem. 274:15751–15756 [DOI] [PubMed] [Google Scholar]
- 26. Miyanari Y., et al. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097 [DOI] [PubMed] [Google Scholar]
- 27. Naldini L., et al. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267 [DOI] [PubMed] [Google Scholar]
- 28. Nonhoff U., et al. 2007. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell 18:1385–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Owsianka A. M., Patel A. H. 1999. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257:330–340 [DOI] [PubMed] [Google Scholar]
- 30. Parker R., Sheth U. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25:635–646 [DOI] [PubMed] [Google Scholar]
- 31. Randall G., et al. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104:12884–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rocak S., Linder P. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 5:232–241 [DOI] [PubMed] [Google Scholar]
- 33. Scheller N., et al. 2009. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc. Natl. Acad. Sci. U. S. A. 106:13517–13522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith R. W., Gray N. K. 2010. Poly(A)-binding protein (PABP): a common viral target. Biochem. J. 426:1–11 [DOI] [PubMed] [Google Scholar]
- 35. Tingting P., Caiyun F., Zhigang Y., Pengyuan Y., Zhenghong Y. 2006. Subproteomic analysis of the cellular proteins associated with the 3′ untranslated region of the hepatitis C virus genome in human liver cells. Biochem. Biophys. Res. Commun. 347:683–691 [DOI] [PubMed] [Google Scholar]
- 36. Tourrière H., et al. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37. Wakita T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weston A., Sommerville J. 2006. Xp54 and related (DDX6-like) RNA helicase: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 34:3082–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. White J. P., Cardenas A. M., Marissen W. E., Lloyd R. E. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305 [DOI] [PubMed] [Google Scholar]
- 40. Wilson J. A., Zhang C., Huys A., Richardson C. D. 2011. Human Ago2 is required for efficient miR-122 regulation of HCV RNA accumulation and translation. J. Virol. 85:2342–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yedavalli V. S., Neuveut C., Chi Y. H., Kleiman L., Jeang K. T. 2004. Requirement of DDX3 DAED box RNA helicase for HIV-1 Rev-RRE export function. Cell 119:381–392 [DOI] [PubMed] [Google Scholar]
- 42. Yi Z., et al. 2006. Subproteomic study of hepatitis C virus replicon reveals Ras-GTPase-activating protein binding protein 1 as potential HCV RC component. Biophys. Biochem. Res. Commun. 350:174–178 [DOI] [PubMed] [Google Scholar]
- 43. You L. R., et al. 1999. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J. Virol. 73:2841–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zufferey R., Nagy D., Mandel R. J., Naldini L., Trono D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871–875 [DOI] [PubMed] [Google Scholar]