Abstract
By using a rhinosvirus/poliovirus type 1 chimera, PV1(RIPO), with the cognate internal ribosome entry site (IRES) of human rhinovirus type 2 (HRV2), we set out to shed light on the mechanism by which this variant expresses its attenuated phenotype in poliovirus-sensitive, CD155 transgenic (tg) mice and cynomolgus monkeys. Here we report that replication of PV1(RIPO) is restricted not only in human cells of neuronal origin, as was reported previously, but also in cells of murine origin at physiological temperature. This block in replication was enhanced at 39.5°C but, remarkably, it was absent at 33°C. PV1(RIPO) variants that overcame the replication block were derived by serial passage under restrictive conditions in either mouse cells or human neuronal cells. All adapting mutations mapped to the 5′-nontranslated region of PV1(RIPO). Variants selected in mouse cells, but not in human neuronal cells, exhibited increased mouse neurovirulence in vivo. The observed strong mouse-specific defect of PV1(RIPO) at nonpermissive temperature correlated with the translational activity of the HRV2 IRES in this chimeric virus. These unexpected results must be kept in mind when poliovirus variants are tested in CD155 tg mice for their neurovirulent potential, particularly in assays of live attenuated oral poliovirus vaccine lots. Virulence may be masked by adverse species-specific conditions in mouse cells that may not allow accurate prediction of neurovirulence in the human host. Thus, novel poliovirus variants in line for possible development of human vaccines must be tested in nonhuman primates.
INTRODUCTION
Virulence is the most intriguing phenotype of a virus. It is also the most difficult phenotype to understand, because it involves numerous facilities of the virus and the host organism that work together or against each other. These processes can be studied in tissue culture and in animal models, if available. The virulence phenotype of poliovirus (PV), an enterovirus of Picornaviridae, has been studied for over 100 years, yet many of the fundamental steps in infection and disease progression remain unknown.
The host range of poliovirus is restricted to humans and nonhuman primates. This is principally related to the expression of CD155 (pvr), its only cellular receptor. CD155, whose gene expresses four splice variants, is a glycoprotein and cell adhesion protein carrying multiple functions in the host organism (29, 37, 41). The isolation and characterization of the CD155 gene made it possible to construct CD155 transgenic (tg) mice which, on injection with PV, show symptoms of paralysis similar to those of human poliomyelitis (28, 50). The occurrence of classical poliomyelitis in the CD155 tg mice implies that the distribution and function of the receptor in neuronal tissues in the animals are similar to those in humans (28, 50). However, CD155 tg mice cannot be infected orally, the natural route of poliovirus entry in humans, regardless of how the receptor is expressed (different promoters, different splice variants), a phenomenon that is not understood (41). Although Northern blot analyses of human and CD155 tg mouse tissue homogenates have indicated that the transcription of the CD155 gene is widespread (28, 50), the expression of the receptor protein is in most tissues very scarce (2, 54). Exceptions are human enterocytes, where CD155 expression can be clearly demonstrated (22). Nevertheless, in CD155 tg mice lacking the interferon α/β receptor, poliovirus infection is seen in numerous tissues normally spared by viral infection (43). This observation suggests that the apparent restricted tissue tropism seen in infected mice is due predominantly to the innate immune response and not to availability of the CD155 receptor protein.
Picornavirus genomic RNA and mRNA are of identical nucleotide sequences. The virion RNA, however, is linked at its 5′ end to the small protein VPg, which is cleaved off before engagement with ribosomes to yield a 5′-pUp terminus (60). Thus, in contrast to host cellular mRNAs, poliovirus mRNA lacks a 5′-cap structure and controls its translation with an internal ribosomal entry site (IRES) located within the 5′-nontranslated region (5′-NTR) (12, 24, 45).
Picornavirus IRES elements are highly structured RNA elements that cover an unusually large segment of the viral genome RNA (e.g., 6%, or ∼450 nucleotides [nt]) (36, 61). Although these elements are distinct in structure, they are interchangeable between viruses of different genera and even of different families, yielding novel, viable chimeric viruses, some of which have interesting phenotypes (1, 9, 15, 18, 35). Several studies with IRES-chimeric viruses illustrated the potential contribution of IRES elements toward cell tropism of the virus (9, 15, 25), although tropism is, in fact, governed by a multitude of different determinants. These include (i) an incompatibility of viral proteins with viral proliferation in different hosts. This has been shown for human rhinoviruses that fail to replicate in mouse cells (19, 20, 34, 62). (ii) A restriction of poliovirus proliferation in many tissues of the CD155 tg mice due to a robust host innate immune response (21, 43, 64). (iii) Absence of detectable CD155 expression in specific tissues, as reported, for example, in renal interstitial cells but not in renal glomeruli of CD155 tg mice (22, 51). (iv) Restricted IRES function due to a lack (or insufficient supply) of specific cellular IRES trans-acting factors (ITAFs). An example is the requirement of a neuronal version of the polypyrimidine track binding protein for neurovirulence of the GDVII variant of Theiler's murine encephalomyelitis virus (47).
Gromeier et al. (15) previously constructed a chimeric poliovirus, called PV1(RIPO), in which the cognate poliovirus IRES was exchanged with that of human rhinovirus 2 (Fig. 1 A). This chimera expressed an interesting host range phenotype in that it replicated with wild-type (wt) kinetics in HeLa cells or in cells of glioblastoma multiforme (15, 17) but was highly restricted in human cells of neuronal origin, such as SK-N-MC (15) and HEK-293 cells (4). Remarkably, PV1(RIPO) and PV3(RIPO) (a chimera of wt Leon/37 poliovirus type 3 carrying the human rhinovirus type 2 [HRV2] IRES) are highly attenuated in CD155 tg mice (7, 15); in addition, PV1(RIPO) was also found to be highly attenuated in nonhuman primates after intraspinal injection (7, 16). These phenotypes have led to intense efforts to develop a derivative of PV1(RIPO), called PVSRIPO (a virus in which the IRES of the Sabin 1 vaccine virus has been exchanged with the HRV2 IRES [42]), as an oncolytic agent for the treatment of human glioma (14, 17). Most recently, Gromeier and his colleagues have presented data that explain the favored growth properties of PVSRIPO in glioblastoma multiforme cells (13).
Fig. 1.

Genetic structures of the 5′-NTRs of PV1(M), PN6, and PV1(RIPO). The poliovirus open reading frame (open arrows), AUG initiation codon, the cloverleaf (CL), the IRES, and spacer I (S-I) and spacer II (S-II) are indicated. The linker sequence of 11 nt containing the EcoRI restriction site (in bold) was inserted into S-I of PV1(M) between nt 108 and 109 to construct the PN6 virus. Roman numbers (II, V, and VI) refer to domains of the IRES.
This study was initiated to understand the mechanisms by which PV1(RIPO) is restricted in cells of neuronal origin, hoping to connect this information with the remarkable attenuation phenotype in CD155 tg mice and nonhuman primates.
Earlier experiments showed that at 39.5°C PV1(RIPO) is highly temperature sensitive (ts) in SK-N-MC and in WI-38 cells but not in HeLa cells or HTB-14 cells (a glioblastoma cell line) (6). Here we report that PV1(RIPO) does not replicate at all in mouse cells at 37°C or 39.5°C, whereas replication at 33°C is similar to that of poliovirus type 1 Mahoney strain [PV1(M)]. The strong host-specific ts phenotype has allowed us to map escape mutants at 37°C and study the growth phenotype and neurovirulence in CD155 tg mice. A similar genetic study was carried out to isolate PV1(RIPO) mutants able to replicate in human neuronal cells. We found that all mutations allowing escape from replication restriction mapped to the 5′-NTR. Engineering these 5′-NTR mutations into PV1(RIPO) produced the same phenotypes as that of the escape mutants, an observation suggesting that neither the proteins of PV1(RIPO) nor its 3′-NTR is involved in the escape from growth restriction. Finally, mutants with restored wt replication phenotypes in mouse fibroblasts regained neurovirulence in CD155 tg mice. These data suggest that CD155 tg mice may have limitations in assessing neurovirulence of chimeric viruses such as PV1(RIPO).
MATERIALS AND METHODS
Viruses and cells.
The neurovirulent poliovirus type 1 strain Mahoney [PV1(M)] is the strain used routinely in the laboratory (5). PV1(RIPO) was constructed as described previously (15). The PN6 poliovirus variant carrying an EcoRI restriction site in the 5′-NTR was originally constructed by Trono et al. (58). HRV2 was derived from an infectious cDNA clone kindly provided to us by Tim Skern (University of Vienna, Vienna, Austria). The mouse neuroblastoma cell line Neuro-2A stably expressing CD155 (N2aCD155) (39, 57), mouse L fibroblast cell lines stably expressing CD155 (H20A and L20B) (37, 48), and mouse NIH 3T3 fibroblasts stably expressing CD155 (NIH 3T3CD155 [S. Mueller and E. Wimmer, unpublished data]), all of which are susceptible to wt poliovirus infection, were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 100 μg/ml penicillin-streptomycin and 10% fetal bovine serum. HEK-293 cells, which are human neuroepithelial cells transformed with sheared adenovirus type 5 DNA (42) were a generous gift from M. Gromeier (4). They were maintained in DMEM containing 100 μg/ml penicillin-streptomycin and 10% fetal bovine serum. HeLa (human cervical cancer) cells and human neuroblastoma cell lines SK-N-MC and SK-N-SH were obtained from the American Type Culture Collection (Manassas, VA) and were maintained according to the supplier's specifications.
Serial passages of PV1(RIPO) in SK-N-MC and H20A cells.
The selection of PV1(RIPO) isolates capable of efficient replication in mouse cells and human neuroblastoma cells was carried out according to the following procedure: H20A and SK-N-MC cells were infected at an multiplicity of infection (MOI) of 10 with PV1(RIPO) and incubated at 37°C for 4 days or until the appearance of cytopathic effect (CPE). After 7 blind passages, complete CPE was observed, RNA was extracted from the viral cell lysate, which served as template for reverse transcription-PCR (RT-PCR), and the purified PCR amplicons were used for sequencing reactions.
RNA extraction, RT-PCR, and DNA sequencing.
Viral RNA was extracted from infected cells by using TRIzol solution (Invitrogen) and used as the template for RT-PCR. The Titan one-tube RT-PCR system (Roche, Mannheim, Germany) was used to perform RT-PCRs following the manufacturer's instructions, and the PCR amplicons were purified with the QIAquick gel extraction kit (Qiagen). The sequences of the purified PCR products were determined with oligonucleotide primers and cycle sequencing (ABI Prism Big Dye terminator cycle sequencing ready reaction kit; Applied Biosystems) in an automated sequencer (model 310; Applied Biosystems).
Construction of plasmids.
Several recombinant variants with phenotypes quite distinct from the parent PV1(RIPO) were constructed using different combination of mutations in the 5′-NTR that had been identified in the adapted isolates. For instance, the plasmid for one of the recombinant variants, R-1235 (see below), was constructed as follows and was named R-1235r: cDNA prepared from R-1235 viral RNA was cut with BbrpI and SacI and ligated to a similarly restricted PV1(RIPO) fragment. To construct R-2r, which has a single C134G mutation in the 5′-NTR of PV1(RIPO), site-directed mutagenesis was carried out in a two-step PCR. For the first-step PCR, two PCR fragments, F1 and F2, were amplified by using pT7PV1(RIPO) as template and the primer pair 5′-TTAAAACAGCTCTGGGGTTGTACCCACCCC-3′ and 5′-TCAGTAATCTGGCTGATTACCGCCTATTGGTCTTTGTGAAAAAC-3′ for the F1 fragment and the primer pair 5′-GTCCTGTTTCGAAGCCGCGTTACTAGC-3′ and 5′-AGTTTTTCACAAAGACCAATAGGCGGTAATCAGCCAGATTACTG-3′ for the F2 fragment. The two PCR fragments, F1 and F2 carrying overlapping ends, were then used as templates for the second-step PCR. R-123r, R-12r, R-35r, R-3r, and R-5r were constructed by choosing appropriate restriction endonuclease sites within R-1235r and PV1(RIPO) plasmids and exchanging fragments between these plasmids. Mutations in the final constructs were verified by sequencing using the ABI Prism DNA sequencing kit.
In vitro transcription, transfection, and virus isolation.
HRV2 plasmid was linearized with KpnI. All other plasmids were linearized with DraI. RNAs were synthesized with phage T7 RNA polymerase, and the RNA transcripts were transfected into HeLa R19 cell monolayers by the DEAE-dextran method as described previously (59). The incubation time was 2 to 3 days for full-length viral constructs. Virus titer was determined by a plaque assay, as described before (38).
One-step growth curves at 33°C, 37°C, and 39.5°C.
One-step growth experiments in different human and mouse cell lines were carried out as follows. Cell monolayers in 35-mm plastic culture dishes were washed with DMEM and inoculated at an MOI of 10 with the virus to be tested. The dishes were rocked for 30 min at room temperature, and the cells were thoroughly washed to remove unbound virus and placed at 33°C, 37°C, or 39.5°C. At 0, 2, 4, 6, 8, 12, 24, and 48 h postinfection (p.i.), the dishes were subjected to three consecutive freeze-thaw cycles, and the viral titers of the supernatants were determined by plaque assay on HeLa cells, as described before (38).
Poliovirus luciferase replicons and luciferase assays.
We previously described a wt replicon, PV1(M)-luc, in which the PV capsid coding sequence was replaced by that of the firefly luciferase gene (32). In addition we constructed two new chimeric replicons that, in analogy to their full-length infectious counterparts, carry either the wt HRV2 IRES [PV1(RIPO)-luc] or a mouse-adapted HRV2 IRES (R-1235r-luc). In vitro-transcribed replicon RNA was transfected into monolayers (35-mm-diameter dishes) of HeLa, SK-N-MC, and L20B cells by a modified DEAE-dextran transfection method (40) and incubated at 33°C, 37°C, or 39.5°C in DMEM with 2% fetal calf serum. At different time points posttransfection, the growth medium was removed from the dishes and the cells were washed gently with 2 ml of phosphate-buffered saline (PBS). The cells were lysed in passive lysis buffer (Promega), and the firefly luciferase activity was measured by methods described previously (63) with a firefly luciferase substrate kit (Promega). The rate of viral translation was assayed by incubating transfected cells in the presence of 2 mM guanidine hydrochloride (GnHCl), a potent inhibitor of PV replication. RNA replication of a construct, on the other hand, can be assessed by considering the ratio of luficerase signals obtained in the absence and presence of GnHCl.
Antibodies.
Anti-2C monoclonal antibodies (MAbs) (46) and antiactin mouse MAb (Calbiochem) were used as primary antibodies for Western blot analysis.
Immunoblot analysis.
For immunoblot analysis, cell lysates of HeLa R19, SK-N-MC, and L20B cells collected 9 h postinfection were separated by SDS-PAGE (12% acrylamide). Following electrotransfer to polyvinylidene difluoride membranes, the membranes were blocked with 5% skim milk in 0.1% Tween 20–PBS for 0.5 h. Membranes were probed with anti-2C monoclonal antibodies at a 1:200 dilution overnight at 4°C. The membranes were washed three times and incubated with horseradish peroxidase-conjugated anti-mouse immunoglobin G for 2 h. After three washes, proteins were visualized with an enhanced chemiluminescence reagent kit (Amersham International Plc.) according to the manufacturer's recommended procedure. The same blot was reused, and the same steps were repeated to detect actin. For this purpose, antiactin (Ab-1) mouse MAb (JLA20; Calbiochem) was used as primary antibody and goat anti-mouse IgM conjugated to horseradish peroxidase (Calbiochem) was used as secondary antibody.
Neurovirulence assays in mice.
Groups of four CD155 tg mice (28) were inoculated intracerebrally with virus amounts ranging from 102 to 108 PFU (30 μl/mouse) as previously described (41). Mice were examined daily for 21 days after inoculation for paralysis and/or death. The virus titer that induced paralysis or death in 50% of the mice (PLD50) was calculated by the method of Reed and Muench (49). All experiments involving mice were conducted in compliance with institutional IACUC regulations and federal guidelines.
RESULTS
Sequence alignments of spacer I in the 5′-NTR of human enteroviruses and human rhinoviruses reveal a highly conserved region.
Throughout this study we used the original PV1(RIPO) (15) rather than PVSRIPO virus to avoid potential complications of data interpretation caused by the presence of a PV1 Sabin capsid, which confers a ts phenotype to the virus (27). The construction of PV1(RIPO) (15) made use of an EcoRI restriction site linker that was originally inserted at nt 108 into the parental poliovirus by Trono and colleagues and reported not to affect the resulting poliovirus variant (PN6) (58). The unstructured RNA sequence between the cloverleaf and the IRES element, where the insertion was made, is now referred to as spacer I (Fig. 1; see also data posted at http://ms.cc.sunysb.edu/∼smueller/Jahan/FigureS1.html) (56, 61). This region has recently been shown to harbor important determinants of virus replication (5, 8, 57). We therefore reexamined the sequence context of the resulting spacer I in the PV1(RIPO) chimera. Upon sequence alignment of entero- and rhinovirus genomes, we observed that spacer I harbors a highly conserved sequence corresponding to nucleotide positions 101 to 112 of the PV genome, with the core sequence GTAACTTAGAAG (http://ms.cc.sunysb.edu/∼smueller/Jahan/FigureS1.html). The EcoRI linker insertion disrupts this conserved sequence and increases the length of spacer I, e.g., the distance between the cloverleaf and IRES, in both PN6 and PV1(RIPO). This posed the question of whether this EcoRI linker insertion contributes to the viral phenotype in different cell types or to neurovirulence. Therefore, we included variant PN6 in many of our following analyses.
The growth restriction of PV1(RIPO) in human neuronal is not absolute.
Previously PV1(RIPO) was described to be replication deficient in the neuroblastoma cell line SK-N-MC (15), a phenotype that is much enhanced at 39.5°C (6). To assess the generality of this behavior in neuronal cells, we extended the ts analysis to two additional neuronal cells lines, HEK-293 and SK-N-SH. The growth phenotypes of PV1(RIPO) and PN6 in HeLa R19 was first analyzed at 33°C, 37°C, and 39.5°C and found to be similar to that of PV1(M) (Fig. 2 A). When tested in three different human cell lines of neuronal origin (Fig. 2B to D), PN6 replicated with similar growth kinetics as PV1(M) at all three temperatures (closed triangles), whereas the growth of PV1(RIPO) in SK-N-MC cells was increasingly inhibited with increasing temperature (Fig. 2B), confirming previous results (4, 6, 15). In contrast to SK-N-MC cells, the proliferation of PV1(RIPO) in HEK-293 cells was virtually unaffected by temperature, as it was equally inhibited at all three temperatures tested (Fig. 2C). In contrast to both SK-N-MC cells and HEK-293 cells, the proliferation of PV1(RIPO) in SK-N-SH cells was completely restored at 33°C and showed only a slight ts phenotype (Fig. 3 D). Our data suggest that the neuron-specific phenotype of PV1(RIPO) is neither absolute nor general, but rather is quite variable between different human neuronal cells. Furthermore, since the EcoRI linker has little if any effect on replication of PN6 in cells of human origin at any temperature, we conclude that the HRV2 IRES alone rather than the EcoRI linker is responsible for the observed phenotype of PV1(RIPO) in human neuronal cells.
Fig. 2.
One-step growth curves of PV1(RIPO) in different human cell lines. Graphs indicate growth of PV1(M), PN6, and PV1(RIPO) in human nonneuronal cell line HeLa R19 (A) and in human neuronal cell lines SK-N-MC (B), HEK-293 (C), and SK-N-SH (D). Cells were infected at an MOI of 10 and incubated at 33°C, 37°C, and 39.5°C. The virus titers were determined by plaque assay on monolayers of HeLa R19 cells, as described in Materials and Methods.
Fig. 3.
One-step growth curves of PV1(RIPO) in different mouse cell lines. Graphs show growth of PV1(M), PN6, and PV1(RIPO) in mouse nonneuronal cell lines L20B (A) and NIH 3T3CD155 (B) and mouse neuronal cell line N2aCD155 (C). Cells were infected at an MOI of 10 and incubated at 33°C, 37°C, and 39.5°C. The virus titers were determined by plaque assay on monolayers of HeLa R19 cells, as described in Materials and Methods.
The ts phenotype of HRV2 is exacerbated in neuronal cells.
Due to its life cycle, which is generally confined to the cooler upper airway epithelium, HRV2 is naturally temperature sensitive when grown at elevated temperatures (10). It is not known whether the ts phenotype is caused by the HRV2 IRES or by other determinants, as experiments to define the genetics of ts of HRV2 have not been reported. However, since PV1(RIPO) analysis revealed a marked ts phenotype in neuronal cells, as a consequence of its HRV2 IRES, the question arose as to whether the parental HRV2 virus displays a similar behavior. One-step growth experiments with HRV2 in HeLa and SK-N-MC cells confirmed the general ts phenotype at 39.5°C (see data posted at http://ms.cc.sunysb.edu/∼smueller/Jahan/FigureS2.html). Still, HRV2 grew surprisingly well at 37°C in HeLa cells, with near-wt kinetics, but hardly at all in SK-N-MC cells. While the HRV2 phenotype is much more severe, it points to a similar trend as that observed with PV1(RIPO), in that the phenotype is exacerbated in neuronal cells. It is possible, therefore, that the natural ts phenotype of HRV2 results from the combination of several ts determinants mapping to the IRES as well as to HRV2-specific proteins.
PV1(RIPO) has a mouse cell-specific propagation defect at 37°C that is rescued at 33°C.
Since the restriction of PV1(RIPO) in human neuronal cells is less general than previously thought, especially at physiological temperatures, we wondered whether the strong attenuation of this virus in CD155 tg mice could have additional mouse-specific determinants. We therefore tested the growth phenotype of PV1(RIPO) in several CD155-expressing mouse cell lines, namely, L20B (37, 48) and NIH 3T3CD155, both fibroblast cell lines, and N2aCD155, a mouse neuroblastoma cell line (39). The cells were infected with PV1(M), PN6, or PV1(RIPO) at 33°C, 37°C, and 39.5°C (Fig. 3). Whereas PV1(M) and PN6 replicated well in all cell lines at 33°C and 37°C, PN6 expressed a ts phenotype at 39.5°C in NIH 3T3CD155α cells when infected at an MOI of 10 (Fig. 3). When infections were carried out at a low MOI (0.01), a condition under which weak phenotypes are generally enhanced, PN6 was ts in all mouse cells (see data posted at http://ms.cc.sunysb.edu/∼smueller/Jahan/FigureS3.html), an observation suggesting that the EcoRI linker under the specific conditions of the experiment does contribute the observed change of host tropism. Surprisingly, PV1(RIPO) did not replicate in any mouse cell line at either 37°C or 39.5°C (Fig. 3). In stark contrast, all three cell types supported replication at 33°C, at near-wt levels. At 33°C, therefore, even cells of neuronal origin (N2aCD155 cells) were an adequate substrate for PV1(RIPO) replication (Fig. 3C). These results indicate that PV1(RIPO) has a mouse cell-specific propagation defect at or above physiological temperature, which may be the primary determinant of its attenuation in CD155 tg mice.
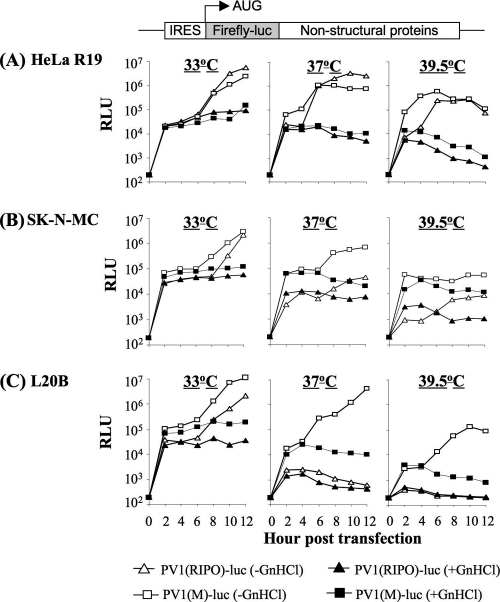
PV1(RIPO) is defective in IRES-mediated translational initiation at the restricted temperature.
After CD155-mediated internalization, the released PV genome RNA serves as mRNA for the translation of a single polyprotein, followed by proteolytic processing and by genome replication (44, 60). In order to determine at which stage of the life cycle the observed attenuation of PV1(RIPO) is manifested, we used two luciferase replicons, PV1(M)-Luc and PV1(RIPO)-Luc, that contain the respective IRES elements in the same context as in their parent viruses, PV1(M) and PV1(RIPO). The capacities for IRES-mediated translation and RNA replication were assessed in HeLa R19 cells, SK-N-MC cells, and mouse L20B cells by transfection with in vitro-transcribed replicon RNA at 33°C, 37°C, and 39.5°C. To differentiate between the luciferase signals due to translation from the input viral RNA and from newly replicated RNA, one portion of the transfected cells received 2 mM GnHCl (Fig. 4), a potent inhibitor of PV RNA replication without any toxic effect on cellular processes or viral translation (3, 23, 33). Translation and replication mediated by the HRV2 IRES in case of the PV1(RIPO)-luc replicon were similar to those of the PV1(M)-luc replicon in HeLa R19 cells, although the kinetics of PV1(RIPO)-luc replication was somewhat delayed at 39.5°C (Fig. 4A). In contrast, in SK-N-MC cells and especially in L20B cells, HRV2 IRES-mediated translation, and consequently replication, was low, and it decreased with increasing temperature (Fig. 4B and C). Most strikingly, in mouse L20B cells the PV1(RIPO)-luc replicon failed to show any replication activity at 37°C and 39.5°C, likely as a result of the greatly reduced translation activity (Fig. 4C). This finding is consistent with the growth characteristics of the corresponding chimeric virus, PV1(RIPO), in different mouse cell lines (Fig. 3). However, even PV1(M)-Luc translation and replication displayed aspects of a ts phenotype, albeit less severe, in SK-N-MC cells and L20B cells (Fig. 4B and C). We hypothesize that in this case the translation/replication efficiency is still above a critical threshold level to maintain the wt phenotype and maximum virus production of the PV1(M) parent virus. Our data suggest that the HRV2 IRES-mediated translation in mouse cell lines is defective in a temperature sensitive manner, which ultimately affects viral RNA replication.
Fig. 4.
RNA translation and replication of PV1(RIPO)-luc and PV1(M)-luc replicons. Graphs show results for monolayers of HeLa R19 cells (A), SK-N-MC cells (B), and mouse L20B cells (C) transfected with in vitro-transcribed RNA of poliovirus luciferase replicons and incubated at 33°C, 37°C, and 39.5°C in the presence (for translation) or in the absence (for replication) of 2 mM GnHCl. RNA translation and RNA replication were assessed by measuring the luciferase activity (RLU) at different time points posttransfection.
To further support the conclusion that the presence of the HRV2 IRES causes this ts-dependent translation defect, the kinetics of viral gene expression was analyzed by Western blot detection of viral gene products 2BC/2C in PV1(M)- and PV1(RIPO)-infected cells. The infections were carried out at different temperatures in HeLa R19, SK-N-MC, and L20B cells, and the cell lysates were analyzed for the accumulation of poliovirus proteins 2BC/2C 9 h postinfection. At the same time, the cell lysates from parallel infections were collected to analyze virus titers. PV1(RIPO) was as efficient as PV1(M) in expressing 2BC/2C proteins and producing virus in HeLa R19 cells (Fig. 5 A, lanes 2 to 7). In stark contrast, no 2BC/2C proteins were detectable in PV1(RIPO)-infected SK-N-MC or L20B cells at 37°C and 39.5°C (Fig. 5B and C, compare lane 2 with lanes 3 and 4), a finding which correlated with the lack of net virus production at 37°C and 39.5°C in these cell lines compared to the 0-h titer (marked by asterisks). On the other hand, PV1(M) was almost equally efficient in synthesizing 2BC/2C in both SK-N-MC and L20B cells at various temperature (Fig. 5B and C, lanes 5 to 7). These data strongly suggest that the HRV2 IRES in PV1(RIPO) causes a ts-dependent translation defect in the restricted cells.
Fig. 5.
Detection of poliovirus protein 2C and a corresponding increase in virus titers in PV1(RIPO)- and PV1(M)-infected cells. HeLa R19 cells (A), SK-N-MC cells (B), and mouse L20B cells (C) were infected with PV1(RIPO) and PV1(M) at an MOI of 10 for 9 h. The viral protein 2C was detected by Western blot analysis (WB), and increases in virus titers were measured by plaque assays as described in Materials and Methods. The lack of net virus production at 37°C or 39.5°C in the SK-N-MC and L20B cell lines, compared to the 0-h titer, is indicated by asterisks in the graphs.
Genetic variants in the 5′-NTR can adapt PV1(RIPO) to growth in mouse cells and human neuronal cells.
The strong expression of ts phenotypes of PV1(RIPO) in different cells allowed us to select variants that have escaped restriction and to map the mutations. To this end, PV1(RIPO) was serially passaged in SK-N-MC cells and in mouse H20A cells at 37°C. Two variants (R-1 and R-7) were isolated from the SK-N-MC passages and seven variants (R-1235, R-123, R-124, R-234, R-136, R-12, and R-23) from the H20A passages (Fig. 6 A) (see Materials and Methods). These variants showed increased replication in the cell types in which their progenitors were highly restricted in growth. The 5′-NTRs of the adapted isolates were sequenced, and the mutations were mapped with the aim of understanding the molecular determinants of the high-titer growth phenotype (Fig. 6A). The three most common changes that were observed were the following: (i) 12- or 13-nucleotide deletions in spacer I between the cloverleaf and IRES (mutation 1); (ii) a C134G mutation in the bulge of stem-loop II (mutation 2); (iii) an A228U mutation in the unstructured region between stem-loops II and IV (mutation 3). Among these, mutation 1 was observed in both human SK-N-MC cell-adapted and mouse H20A cell-adapted viruses. We found either a deletion of 12 or 13 nucleotides in spacer I between the cloverleaf and the IRES (Fig. 6A) that removed the EcoRI linker insertion (originally derived from the parental PN6 virus and used for cloning of this virus). On the other hand, point mutations 2 to 6 are most likely important for mouse-specific adaptation, because they are only present in the mouse cell-adapted isolates. Variants containing different combinations of mutations (R-1235, R-123, R-12, and R-23) were then tested for growth in L20B cells at various temperatures. All variants grew very well in L20B cells at 33°C and 37°C, the latter being the temperature at which the cells were selected. At 39.5°C, on the other hand, they still showed growth defects similar to PV1(RIPO) (Fig. 6B). All variants were also tested in two other mouse cell lines (NIH 3T3CD155 and N2aCD155), in which they showed a similar growth phenotype (data not shown).
Fig. 6.
Genetic analyses of the 5′-NTR nucleotide sequences of the adapted isolates of PV1(RIPO). (A) Isolate names and the mutation(s) they carry. The mutations found were numbered according to their position in the 5′-NTR, starting from the 5′ end. The numbers in parentheses show the number of isolates among total isolates containing any particular mutation. The sequences of the deletions for mutation 1 are shown. The nucleotide numbers were assigned according to their position in the human rhinovirus 2 IRES. (B) Growth of PV1(RIPO) and selected mouse L cell-adapted isolates in mouse cell line L20B. The virus titers were determined by plaque assay on monolayers of HeLa R19 cells, as described in Materials and Methods.
To identify the most effective point mutation(s) in variants of PV1(RIPO) that lead to efficient replication in mouse cells (in addition to the deletion mutation), several of the mouse cell-adapted isolates were reconstructed by introducing the most commonly observed point mutation(s) into the 5′-NTR of PV1(RIPO). When the deletion either alone or in combination with three point mutations was introduced into the parental PV1(RIPO), the resulting reconstructed viruses, named R-1r and R-1235r, respectively, showed growth kinetics similar to that of the adapted isolates R-1 and R-1235 in L20B cells (data not shown). This observation indicates that all important adaptive mutations were confined to the 5′-NTR and that no significant second-site reversions existed elsewhere in the genome, e.g., in viral proteins or in the genome downstream of the IRES, the 3′-NTR included. Some additional mutants were constructed in vitro by introducing mutations 2, 3, and 5 either singly (R-2r, R-3r, and R-5r) or in combination (R-123r, R-12r, and R-35r). The reconstructed mutants were then compared with the original adapted isolates for their growth phenotypes in L20B cells (Table 1). The data support the conclusion that the replication of the variants roughly increases with the number of adaptive mutations in the 5′-NTR and that all significant adaptation mutations are localized in the 5′-NTR.
Table 1.
Neurovirulence study results in CD155 tg mice
| Virus | PLD50a (PFU) | Relative increase in PLD50b | Fold increase in virus titer in L20B cellsc |
|---|---|---|---|
| PV1(RIPO) | >108d | 1 | 0.5 |
| R-1 | >108d | ≤1 | 960 |
| R-1r | NDe | ND | 400 |
| R-2r | >108 | 1 | 140 |
| R-3r | >107 | <10 | 0.3 |
| R-5r | >107 | <10 | 5.7 |
| R-35r | 107 | 10 | 7,500 |
| R-12r | 106 | 100 | 8,400 |
| R-23 | 105.5 | 316 | 12,000 |
| R-123 | 105 | 1,000 | 22,000 |
| R-123r | 105.3 | 501 | 30,000 |
| R-1235 | 103 | 100,000 | 130,000 |
| R-1235r | 103.2 | 63,096 | 100,000 |
| PN6 | 104.7 | 1,995 | 16,000 |
| PV1(M) | 102 | 10,00,000 | 250,000 |
Groups of four mice were infected intracerebrally with a given amount of virus. PLD50 values were calculated by using the method of Reed and Muench (49).
The relative increase in the PLD50 was calculated by considering the PLD50 of PV1(RIPO) as 1.
Fold increase in virus titer in L20B cells at 24 h postinfection at an MOI of 0.01, compared with the titer at 0 h postinfection.
The LD50 could not be determined because no mice died at the highest dose inoculated.
ND, not determined.
Covariation between mouse cell adaptive mutations and mouse neurovirulence.
To further assess the relationship between the high-titer growth phenotype of the mouse cell-adapted PV1(RIPO) variants (and their equivalent recombinant reconstructions) and the neurovirulence in CD155 tg mice, groups of 4 CD155 tg mice were inoculated via the intracerebral route with 102 to 108 PFU (30 μl/mouse), as previously described (15). All mice were observed and scored daily for at least 21 days postinoculation for symptoms of poliomyelitis and/or death. Using the data obtained, the PLD50 was determined using the method of Reed and Muench (49) (Table 1). As was observed before, PV1(RIPO) was at least 106 times less pathogenic in CD155 tg mice than wild-type PV1(M) (15, 16). In fact, a PLD50 for PV1(RIPO) could not be established (Table 1), as the virus did not kill at the highest tested dose, whereas the PLD50 for PV1(M) was 102 PFU. This result correlates with the inability of PV1(RIPO) to replicate in mouse L20B cells at 37°C (Table 1). In contrast, the mutant virus PN6 with the insertion of 11 nt between the cloverleaf and IRES is attenuated in CD155 tg mice, with a PLD50 of 104.7 [which is still higher by orders of magnitude compared to PV1(RIPO)]. The reduced neurovirulence of PN6 is presumably due to the slight replication defect in mouse cells. It should be noted that the replication defect of PN6 in mouse cells was more pronounced when infections were performed at a low MOI (0.01) (see also data posted at http://ms.cc.sunysb.edu/∼smueller/Jahan/FigureS3.html), a scenario more akin to the situation a virus might find upon encounter of a host cell in an infected animal.
PV1(RIPO) isolates, adapted to grow in mouse cells, exhibited different degrees of neuropathogenicity in CD155 tg mice whereas, importantly, the human SK-N-MC cell-adapted isolate R-1 was still as neuroattenuated as PV1(RIPO) (Table 1). Thus, rescuing the replication in neuron-type cells did not restore neurovirulence in the mouse model. The mouse fibroblast-adapted PV1(RIPO) isolates R-1235 and R-123, on the other hand, showed significantly increased neurovirulence over that of the parental PV1(RIPO), which supported the fact that the mutations are not only able to control the high-titer replication phenotype in mouse cell tissue culture but also improve neurovirulence in the CD155 tg mice. When reconstructed, the PV1(RIPO) recombinant mutants R-1235r and R-123r showed the same trend in neuropathogenicity (within the margin of error of the experiment), an observation indicating the importance of these 5′-NTR mutations as the sole determinants of the increased mouse neurovirulence (Table 1). Reconstructed mutants R-12r and R-35r, harboring a combination of two mutations, were still capable of inducing paralysis and/or death in CD155 tg mice, but this required a relatively higher dose of virus. Mutants R-1r, R-2r, R-3r, and R-5r, which have a single mutation, are incapable of increasing the neuropathogencity of PV1(RIPO). We conclude that the important mutations in the 5′-NTR have a cumulative effect in neurovirulence in CD155 tg mice.
Adaptation of PV1(RIPO) in human neuronal cells cannot relieve the host restriction in murine cells.
To distinguish between a tissue-specific (neuronal versus nonneuronal) and/or host species-specific (mouse versus human) block, the SK-N-MC- and H20A-adapted isolates were evaluated in a crosswise comparison of their growth restrictions in L20B and SK-N-MC cells, respectively. It should be noted that H20A and L20B cells, both mouse L cells expressing human CD155, were used interchangeably. The mouse cell-adapted PV1(RIPO) isolate R-1235, with a replication phenotype nearly identical to wt PV1(M) in mouse cells, also replicated with kinetics similar to wt PV1(M) in SK-N-MC cells (Fig. 7 A), whereas the human SK-N-MC-adapted PV1(RIPO) isolate (R-1) still showed an attenuated phenotype in L20B cells (Fig. 7B). This result indicates that the deletion in the spacer region alone is sufficient for the complete restoration of replication in human neuronal cells but is not sufficient to restore a high-titer growth phenotype in mouse cells. Additional point mutations identified in the HRV2 IRES are essential for overcoming the mouse-specific host restriction.
Fig. 7.
Evaluation of species-specific growth restriction of mouse cell-adapted and human neuronal cell-adapted isolates of PV1(RIPO). Human neuronal cell-adapted (R-1) and mouse fibroblast-adapted (R-1235) variants of PV1(RIPO) were used to infect human SK-N-MC cells (A) or mouse L20B cells (B) at an MOI of 10 and incubated at 37°C. The virus titers at various time points were determined by plaque assay on monolayers of HeLa R19 cells, as described in Materials and Methods. Mouse-adapted variant R-1235 completely restored replication competency for human neuronal cells, while human neuron-adapted variant R-1 did not.
While deletion of the EcoRI linker alone restored replication in human neuronal cells, it did not suffice to restore neurovirulence in CD155 tg mice, as such virus still could not grow well in any mouse cells per se. This was best illustrated by the phenotype of the resulting virus (R-1), which retained the mouse-specific defect, which can be overcome with additional adaptive mutations within the HRV2 IRES (Table 1). This indicates that the neuron-specific defect of PV1(RIPO) is not a result of the choice of the HRV2 IRES to drive translation. It is rather a result of suboptimal spacer length or disruption of an unknown sequence signal or host factor binding site due to the EcoRI linker between the cloverleaf and IRES.
Finally, luciferase replicons of R-1r and R-1235r were used to examine whether these mutations can restore the translation and replication activities of the replicon RNA in mouse L20B cells. The activities of these replicons were compared with those of PV1(RIPO)-luc, PN6-luc, and PV1(M)-luc (Fig. 8). As expected, the PV1(RIPO)-luc replicon failed to show translation and replication activities at higher temperatures (37°C and 39.5°C) at both early (5-h) and late (11-h) time points posttransfection (Fig. 8), while PV1(M)-luc exhibited the highest level of luciferase activity among all replicons at all the temperatures. The presence of the linker in PV1(M) in PN6-luc reduced translation and, consequently, replication at elevated temperature in L20B cells, but the Luc signal was still much higher than with PV1(RIPO)-luc. Interestingly, removal of the linker in R-1r-luc did not lead to a significant improvement in translation/replication activities in L20B cells, an observation suggesting that the linker in combination with the HRV2 IRES does not contribute significantly to the translation/replication defect. This conclusion is supported by the activity of the R-1235r-luc replicon, which possesses three point mutations in the HRV2 IRES in addition to the linker sequence deletion. Convincingly, the R-1235-luc replicon exhibited a similar luciferase activity as the PV1(M)-luc replicon 37°C, indicating that translation in mouse cells was restored by the adaptive mutations in the 5′-NTR (Fig. 8). This result correlated well with the restoration of the higher growth titer of R-1235r virus in mouse L20B cells (Table 1 and Fig. 7B) and improved neurovirulence of this virus in the CD155 tg mice (Table 1).
Fig. 8.
RNA translation and replication of luciferase replicons in L20B cells. Monolayers of mouse L20B cells were transfected with in vitro-transcribed RNA of luciferase replicons and incubated at 33°C, 37°C, and 39.5°C in the presence (for translation) or in absence (for replication) of 2 mM guanidine hydrochloride (GnHCl). RNA translation and RNA replication were assessed by measuring the luciferase activity (RLU) at 5 h posttransfection (A) or 11 h posttransfection (B).
DISCUSSION
The experiments reported here were designed to find an explanation for the remarkable neuro-attenuation phenotype in CD155 tg mice (15, 18) and in nonhuman primates (7) of PV1(RIPO), a chimera in which the IRES of PV has been exchanged with that of HRV2 (15, 18).
The question of whether the IRES is an important determinant of PV pathogenesis led to 2-decades-long studies of poliovirus attenuation and neurovirulence by different groups. Early studies linked a single mutation in domain V of the IRES of each of the three PV vaccine genomes to attenuation (60); this mutation in the Sabin 3 strain (C472U) was of particular interest because it reverted (U472C) rapidly in vaccine recipients (11, 60). The importance of the C472U mutations was supported by studies in experimental animals (30) and in tissue culture (31) and biochemical experiments (55). After replacing the IRES of PV1(M) with that of coxsackie virus B or hepatitis C virus or the Sabin virus type 3 PV, however, Kauder and colleagues (26) originally concluded that the tropism of wild-type and vaccine strains of poliovirus is determined in a step after IRES-mediated translation. Using a different experimental strategy, they subsequently suggested that IRES-derived translation plays an important role in neurovirulence of a chimeric virus (P1/HRV2) that is nearly identical to PV1(RIPO) (25).
The HRV2 IRES, just like the PV IRES, is a type I IRES (36, 61), yet the 5′-NTR of PV1(RIPO) is quite different from that of PV. First, there is an insertion of a “foreign” sequence (EcoRI linker) in a previously unrecognized highly conserved region in spacer I of the enterovirus 5′-NTRs (see data posted at http://ms.cc.sunysb.edu/∼smueller/Jahan/FigureS1.html). This linker insertion has previously been thought of as innocuous for poliovirus, since a poliovirus mutant, referred to as PN6, exhibited a wt phenotype in HeLa cells (58). As we have shown here, the PN6 mutant is associated with a phenotype in murine cell lines and CD155 tg mice. Second, the poliovirus 5′-NTR has in its 3′ terminus the mysterious spacer II (61), a long seemingly unused segment of mostly nonconserved RNA (124 nt, measured from the end of domain VI to the initating AUG) that human rhinovirus genomes lack altogether. Nevertheless, PV1(RIPO) replicates in HeLa cells with kinetics similar to PV1(M).
We observed a very strong species-specific ts phenotype of PV1(RIPO) in all mouse cell lines tested (L, NIH 3T3, and N2a expressing CD155); there was virtually no proliferation at 37°C and 39.5°C but, remarkably, nearly wt PV1(M) proliferation of PV1(RIPO) at 33°C. It should be noted that the chimera described by Kauder et al. (PV/HRV2) (25) did replicate in HeLa cells at 37°C, albeit with a peculiar delay of 10 to 15 h postinfection. The difference may relate to the EcoRI insertion in the 5′-NTR of PV1(RIPO) that is lacking in the construct described by Kauder et al. (25). Moreover, slight changes in the temperature during growth may also contribute to changes in virus yield.
What is the block in PV1(RIPO) proliferation in SK-N-MC cells or in mouse cells? Evidence presented here (Fig. 8) strongly suggests that inefficient translation is the major block for efficient replication. Assays with reporter genes, however, call for cautious interpretation of the data. Campbell et al. (4) reported that assays with IRES-driven luciferase reporter constructs that consisted only of the HRV-2 IRES and the reporter gene did express Luc well (or even better) in HEK-293 or SK-N-MC cells than the equivalent PV IRES-driven reporter constructs. Those authors commented that the results indicated that “translation in a reporter context does not recapitulate the neuron-specific functional deficit of the HRV2 IRES in the context of the PV genome” (4). If so, the results of the IRES studies of Kauder et al. (25, 26) using dicistronic reporter mRNA produced by an adenovirus might not have yielded results that can be interpreted to reflect IRES tropism or attenuation. In our experiments, luciferase replicons were used, which are intact, full-length, replicating poliovirus genomes, with the exception that the viral capsid-encoding region has been replaced with the luciferase gene and the poliovirus IRES was replaced with the HRV2 IRES element. These replicons therefore mirror very closely the circumstances faced by their infectious virus counterpart.
Interestingly, 6 out of 9 isolates from separate passages in human SK-N-MC cells and mouse H20A cells lost the EcoRI linker insertion and surrounding sequences in spacer I (mutation 1). Whereas the striking neuroattenuation of PV1(RIPO) in CD155 tg mice correlated with its inability to replicate in the SK-N-MC neuroblastoma cell line (Fig. 7), variant R-1 grew well in SK-N-MC cells but still replicated only poorly in mouse L20B cells (Fig. 7); fittingly, it retained a high degree of neuroattenuation in CD155 tg mice (Table 1). Replication in a human neuronal cell line (SK-N-MC) and neurovirulence in CD155 tg mice, therefore, do not covary. Further mutations within the HRV2 IRES itself were necessary to adapt the virus to mouse cells. As has been pointed out before by Campbell et al. (4), assays in neuronal tissue culture cells alone may not be a reliable indicator of neurovirulence.
The selection of R-1 variants indicates that the inserted linker sequence is not neutral in SK-N-MC or in mouse cells. In the context of poliovirus propagated in HeLa cells, however, the insertion appears to be stable, presumably because the advantage of deleting it is small under these conditions. Interestingly, the deletion restores both the sequence and the length of a highly conserved region (GTAACTTAGAAG) in spacer I of entero- and rhinovirus genomes (see data posted at http://ms.cc.sunysb.edu/∼smueller/Jahan/FigureS1.html). Similar observations by De Jesus et al. (8) indicated an abundance of conserved nucleotides among different PV serotypes and human C-cluster coxsackie A viruses in this region, although the significance of these conserved regions is not known. As mentioned earlier, spacer I harbors important signals for RNA synthesis (56) and neurovirulence in CD155 tg mice (57).
Mutation 2 (R-2), a C134G transversion, was observed in 6 out of 7 H20A-adapted isolates (Fig. 6). It maps just downstream of a highly conserved sequence of 6 nt (CAATAG), overlapping with a prominent bulge in domain II of all entero- and rhinovirus IRES elements (see data posted at http://ms.cc.sunysb.edu/∼smueller/Jahan/FigureS1.html). Interestingly, a transition mutation, A133G, was observed at a similar position in the domain II bulge by Shiroki et al. (53) when a heat-resistant mutant of PV was isolated by serial passage of wt PV1(M) at 40°C in L cells expressing the PV receptor CD155. Remarkably, Toyoda et al. (57) also reported an A133G transition when a poliovirus carrying the cis-acting element cre in the spacer region (mono-crePV) was either passaged in mouse N2aCD155 cells or isolated from a mouse tumor (neuroblastoma). Again, the mutation at nt 133 was responsible for the increased replication of this A133Gmono-crePV1 variant, compared to mono-crePV in mouse N2aCD155 cells. These observations, together with the R-2 mutation reported here, indicate that a G residue in position 133/134 [in PV1(M) and HRV2, respectively] favors replication in mouse cells and is, thus, a host range mutation. It can be speculated that the bulge of domain II interacts with a host protein (perhaps the host ribosome).
Mutation 3 (A228U) is also frequent and maps to a position in the HRV2 IRES, but the significance of this is unknown. It should be noted that an A at position 228 has not been observed in any other natural HRV2 isolate or other highly homologous HRV serotypes (HRV 49, 30, 23, 74, or 9) but rather is unique to the sequence of the infectious HRV2 cDNA used in this study (GenBank accession number X02316).
Other mutations mapping to domain V or thereafter are sparse. Previous studies (15, 16) showed that the PV1(RIPO) IRES-containing domains V and VI of the PV1(M) IRES exhibited the same neurovirulence as PV1(M) in CD155 tg mice. This observation suggested that both of these domains of the PV1(M) IRES are required for mouse neurovirulence. Subsequently, using chimeric IRES constructs and human HEK-293 cells as indicator cells for neurovirulence, domain VI of the IRES was found to be dispensable for the expression of attenuation, but the entire domain V was required for a growth phenotype of PV1(RIPO) in HEK-293 cells (4). It is possible that some of the mutations in domain V allow this domain to function in neurovirulence. This may include interactions with the 3′-NTR that may augment the neurovirulent phenotype (9).
Principally, the more adapting mutations there are in the IRES, the better the growth phenotype in mouse cells (Fig. 6B; Table 1) and the more neurovirulent the corresponding isolate in CD155 tg mice (Table 1). As mentioned before, placing the escape mutations into the IRES by site-directed mutagenesis led to the recovery of the growth and neurovirulence phenotype; again, the more mutations that are reintroduced, the better the loss of restriction. This indicates that mutations outside the IRES are not required for the growth and neurovirulence phenotype.
The replication of human rhinoviruses is broadly restricted in mouse cells. However, host range variants of HRV2 (62) and HRV 39 (34) that bypass the block in mouse L cells have been reported to produce 2C protein with altered electrophoretic mobility. Harris et al. (19, 20) showed more recently that these host cell restrictions can be overcome by specific mutations in proteins mapping to the P2/P3 nonstructural region of the genome. However, in the background of a full-length rhinovirus genome, no mouse adaptive mutations have ever been reported to localize to the IRES. It should be noted that the HRV2 mouse cell adaptation experiments reported by others (34, 62) were most likely carried out at 33°C (the optimal growth temperature for rhinoviruses). According to our data, at this temperature no IRES defect would be expected, and consequently no mouse adaptation mutations localizing to the IRES should be isolated. The mouse-specific mutations in the HRV2 IRES observed here suggest to us that the poliovirus replication machinery may interact poorly with the PV1(RIPO) 5′-NTR at elevated temperatures at some stage during the viral life cycle. This may also explain why the remarkable host restriction is only seen in the context of a PV genome (full length or replicon) but apparently not in IRES-driven reporter constructs (4, 26).
Our work demonstrates that changes in the 5′-NTR alone, particularly the mutations in the HRV2 IRES, are sufficient to rescue HRV2 IRES-mediated translation in mouse cells and, consequently, RNA replication in mouse L cells. Most importantly, these mutations were not only able to rescue the defective growth of PV1(RIPO) in mouse cells but also to produce a highly neurovirulent virus in CD155 tg mice. Because of this covariation, neurovirulence tests of chimeric polioviruses, and perhaps other recombinant polioviruses, in CD155 tg mice may not adequately predict their neurovirulence potential in humans, thus indicating the continued need for nonhuman primate tests in the assessment of live attenuated poliovirus vaccines.
ACKNOWLEDGMENTS
We are deeply indebted to Aniko Paul for help and suggestions during the preparation of the manuscript. We thank Yixin Fu, Sean Coleman, and Xiaoyu Li, who contributed to some aspects of this work over the years.
This work was supported by NIH grants 5R01AI03948508 and 5R37AI01512233 to Eckard Wimmer.
Footnotes
Published ahead of print on 11 May 2011.
REFERENCES
- 1. Alexander L., Lu H. H., Wimmer E. 1994. Polioviruses containing picornavirus type 1 and/or type 2 IRES elements: genetic hybrids and the expression of a foreign gene. Proc. Natl. Acad. Sci. U. S. A. 91:1406–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bernhardt G., Bibb J. A., Bradley J., Wimmer E. 1994. Molecular characterization of the cellular receptor for poliovirus. Virology 199:105–113 [DOI] [PubMed] [Google Scholar]
- 3. Caliguiri L. A., Tamm I. 1968. Action of guanidine on the replication of poliovirus RNA. Virology 35:408–417 [DOI] [PubMed] [Google Scholar]
- 4. Campbell S. A., Lin J., Dobrikova E. Y., Gromeier M. 2005. Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J. Virol. 79:6281–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cello J., Paul A., Wimmer E. 2002. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297:1016–1018 [DOI] [PubMed] [Google Scholar]
- 6. Cello J., et al. 2008. Growth phenotypes and biosafety profiles in poliovirus-receptor transgenic mice of recombinant oncolytic polio/human rhinoviruses. J. Med. Virol. 80:352–359 [DOI] [PubMed] [Google Scholar]
- 7. Chumakov K., et al. 2001. Inactivated vaccines based on alternatives to wild-type seed virus. Dev. Biol. (Basel) 105:171–177 [PubMed] [Google Scholar]
- 8. De Jesus N., Franco D., Paul A., Wimmer E., Cello J. 2005. Mutation of a single conserved nucleotide between the cloverleaf and internal ribosome entry site attenuates poliovirus neurovirulence. J. Virol. 79:14235–14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dobrikova E., Florez P., Bradrick S., Gromeier M. 2003. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′ nontranslated region. Proc. Natl. Acad. Sci. U. S. A. 100:15125–15130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dolan T. M., Fenters J. D., Fordyce P. A., Holper J. C. 1968. Rhinovirus plaque formation in WI-38 cells with methylcellulose overlay. Appl. Microbiol. 16:1331–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunn G., Begg N. T., Cammack N., Minor P. D. 1990. Virus excretion and mutation by infants following primary vaccination with live oral poliovaccine from two sources. J. Med. Virol. 32:92–95 [DOI] [PubMed] [Google Scholar]
- 12. Ehrenfeld E., Teterina N. L. 2002. Initiation of translation of picornavirus RNAs: structure and function of the internal ribosome entry site, p. 159–169 In Semler B. L., Wimmer E. (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 13. Goetz C., Everson R. G., Zhang L. C., Gromeier M. 2010. MAPK signal-integrating kinase controls cap-independent translation and cell type-specific cytotoxicity of an oncolytic poliovirus. Mol. Ther. 18:1937–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goetz C., Gromeier M. 2010. Preparing an oncolytic poliovirus recombinant for clinical application against glioblastoma multiforme. Cytokine Growth Factor Rev. 21:197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gromeier M., Alexander L., Wimmer E. 1996. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl. Acad. Sci. U. S. A. 93:2370–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gromeier M., Bossert B., Arita M., Nomoto A., Wimmer E. 1999. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J. Virol. 73:958–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gromeier M., Lachmann S., Rosenfeld M. R., Gutin P. H., Wimmer E. 2000. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc. Natl. Acad. Sci. U. S. A. 97:6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gromeier M., et al. 1997. Determinants of poliovirus neurovirulence. J. Neurovirol. 3(Suppl. 1):S35–S38 [PubMed] [Google Scholar]
- 19. Harris J. R., Racaniello V. R. 2003. Changes in rhinovirus protein 2C allow efficient replication in mouse cells. J. Virol. 77:4773–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harris J. R., Racaniello V. R. 2005. Amino acid changes in proteins 2B and 3A mediate rhinovirus type 39 growth in mouse cells. J. Virol. 79:5363–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ida-Hosonuma M., et al. 2005. The alpha/beta interferon response controls tissue tropism and pathogenicity of poliovirus. J. Virol. 79:4460–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iwasaki A., et al. 2002. Immunofluorescence analysis of poliovirus receptor expression in Peyer's patches of humans, primates, and CD155 transgenic mice: implications for poliovirus infection. J. Infect. Dis. 186:585–592 [DOI] [PubMed] [Google Scholar]
- 23. Jacobson M. F., Baltimore D. 1968. Morphogenesis of poliovirus. I. Association of the viral RNA with coat protein. J. Mol. Biol. 33:369–378 [DOI] [PubMed] [Google Scholar]
- 24. Jang S. K., et al. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kauder S., Kan S., Racaniello V. R. 2006. Age-dependent poliovirus replication in the mouse central nervous system is determined by internal ribosome entry site-mediated translation. J. Virol. 80:2589–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kauder S. E., Racaniello V. R. 2004. Poliovirus tropism and attenuation are determined after internal ribosome entry. J. Clin. Invest. 113:1743–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kohara M., et al. 1985. In vitro phenotypic markers of a poliovirus recombinant constructed from infectious cDNA clones of the neurovirulent Mahoney strain and the attenuated Sabin 1 strain. J. Virol. 53:786–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koike S., et al. 1991. Transgenic mice susceptible to poliovirus. Proc. Natl. Acad. Sci. U. S. A. 88:951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koike S., et al. 1990. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 9:3217–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. La Monica N., Almond J. W., Racaniello V. R. 1987. A mouse model for poliovirus neurovirulence identifies mutations that attenuate the virus for humans. J. Virol. 61(9):2917–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. La Monica N., Racaniello V. R. 1989. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol. 63:2357–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li X., Lu H. H., Mueller S., Wimmer E. 2001. The C-terminal residues of poliovirus proteinase 2A(pro) are critical for viral RNA replication but not for cis- or trans-proteolytic cleavage. J. Gen. Virol. 82:397–408 [DOI] [PubMed] [Google Scholar]
- 33. Loddo B., Ferrari W., Brotzu G., Spanedda A. 1962. In vitro inhibition of infectivity of polio viruses by guanidine. Nature 193:97–98 [DOI] [PubMed] [Google Scholar]
- 34. Lomax N. B., Yin F. H. 1989. Evidence for the role of the P2 protein of human rhinovirus in its host range change. J. Virol. 63:2396–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu H. H., Wimmer E. 1996. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 93:1412–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martínez-Salas E., Ryan M. D. 2010. Translation and protein processing, p. 141–161 In Ehrenfeld E., Domingo E., Roos R. P. (ed.), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 37. Mendelsohn C. L., Wimmer E., Racaniello V. R. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56:855–865 [DOI] [PubMed] [Google Scholar]
- 38. Molla A., Paul A. V., Wimmer E. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647–1651 [DOI] [PubMed] [Google Scholar]
- 39. Mueller S., Wimmer E. 2003. Recruitment of nectin-3 to cell-cell junctions through trans-heterophilic interaction with CD155, a vitronectin and poliovirus receptor that localizes to alpha (v) beta3 integrin-containing membrane microdomains. J. Biol. Chem. 278:31251–31260 [DOI] [PubMed] [Google Scholar]
- 40. Mueller S., Papamichail D., Coleman J. R., Skiena S., Wimmer E. 2006. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J. Virol. 80:9687–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mueller S., Wimmer E., Cello J. 2005. Poliovirus and poliomyelitis: a tale of guts, brains, and an accidental event. Virus Res. 111:175–193 [DOI] [PubMed] [Google Scholar]
- 42. Ochiai H., et al. 2004. Treatment of intracerebral neoplasia and neoplastic meningitis with regional delivery of oncolytic recombinant poliovirus. Clin. Cancer Res. 10:4831–4838 [DOI] [PubMed] [Google Scholar]
- 43. Ohka S., et al. 2007. Establishment of a poliovirus oral infection system in human poliovirus receptor-expressing transgenic mice that are deficient in alpha/beta interferon receptor. J. Virol. 81:7902–7912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paul A. V. 2002. Possible unifying mechanism of picornavirus genome replication, p. 227–246 In Semler B. L., Wimmer E. (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 45. Pelletier J., Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320–325 [DOI] [PubMed] [Google Scholar]
- 46. Pfister T., Wimmer E. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992–7001 [DOI] [PubMed] [Google Scholar]
- 47. Pilipenko E. V., Viktorova E. G., Guest S. T., Agol V. I., Roos R. P. 2001. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J. 20:6899–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pipkin P. A., Wood D. J., Racaniello V. R., Minor P. D. 1993. Characterisation of L cells expressing the human poliovirus receptor for the specific detection of polioviruses in vitro. J. Virol. Methods 41:333–340 [DOI] [PubMed] [Google Scholar]
- 49. Reed L. J., Muench M. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. (Lond.) 27:493–497 [Google Scholar]
- 50. Ren R., Costantini F., Gorgacz E. J., Lee J. J., Racaniello V. R. 1990. Transgenic mice expressing a human poliovirus receptor: a new model for poliomyelitis. Cell 63:353–362 [DOI] [PubMed] [Google Scholar]
- 51. Ren R., Racaniello V. R. 1992. Human poliovirus receptor gene expression and poliovirus tissue tropism in transgenic mice. J. Virol. 66:296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shaw G., Morse S., Ararat M., Graham F. L. 2002. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 16:869–871 [DOI] [PubMed] [Google Scholar]
- 53. Shiroki K., et al. 1995. A new cis-acting element for RNA replication within the 5′ noncoding region of poliovirus type 1 RNA. J. Virol. 69:6825–6832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Solecki D., Schwarz S., Wimmer E., Lipp M., Bernhardt G. 1997. The promoters for human and monkey poliovirus receptors. Requirements for basic and cell type-specific activity. J. Biol. Chem. 272:5579–5586 [DOI] [PubMed] [Google Scholar]
- 55. Svitkin Y. V., Pestova T. V., Maslova S. V., Agol V. I. 1988. Point mutations modify the response of poliovirus RNA to a translation initiation factor: a comparison of neurovirulent and attenuated strains. Virology 166:394–404 [DOI] [PubMed] [Google Scholar]
- 56. Toyoda H., Franco D., Fujita K., Paul A. V., Wimmer E. 2007. Replication of poliovirus requires binding of the poly(rC) binding protein to the cloverleaf as well as to the adjacent C-rich spacer sequence between the cloverleaf and the internal ribosomal entry site. J. Virol. 81:10017–10028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Toyoda H., Yin J., Mueller S., Wimmer E., Cello J. 2007. Oncolytic treatment and cure of neuroblastoma by a novel attenuated poliovirus in a novel poliovirus-susceptible animal model. Cancer Res. 67:2857–2864 [DOI] [PubMed] [Google Scholar]
- 58. Trono D., Andino R., Baltimore D. 1988. An RNA sequence of hundreds of nucleotides at the 5′ end of poliovirus RNA is involved in allowing viral protein synthesis. J. Virol. 62:2291–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van der Werf S., Bradley J., Wimmer E., Studier F. W., Dunn J. J. 1986. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 83:2330–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wimmer E., Hellen C. U. T., Cao X. M. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353–436 [DOI] [PubMed] [Google Scholar]
- 61. Wimmer E., Paul A. 2010. The making of a picornavirus genome, p. 33–55 In Ehrenfeld E., Domingo E., Roos R. P. (ed.), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 62. Yin F. H., Lomax N. B. 1983. Host range mutants of human rhinovirus in which nonstructural proteins are altered. J. Virol. 48:410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yin J., Paul A. V., Wimmer E., Rieder E. 2003. Functional dissection of a poliovirus cis-acting replication element [PV-cre(2C)]: analysis of single- and dual-cre viral genomes and proteins that bind specifically to PV-cre RNA. J. Virol. 77:5152–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yoshikawa T., et al. 2006. Role of the alpha/beta interferon response in the acquisition of susceptibility to poliovirus by kidney cells in culture. J. Virol. 80:4313–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]