Abstract
The Drosophila brain tumor (brat) gene encodes a member of the conserved NHL family of proteins, which appear to regulate differentiation and growth in a variety of organisms. One of the founding family members, Caenorhabditis elegans LIN-41, is thought to control posttranscriptional gene expression. However, the mechanism by which LIN-41, or any other NHL protein, acts has not been clear. Using a yeast “four-hybrid” interaction assay, we show that Brain Tumor is recruited to hunchback (hb) mRNA through interactions with Nanos and Pumilio, which bind to the RNA to repress its translation. Interaction with the Nanos/Pumilio/RNA complex is mediated by the Brat NHL domain; single amino acid substitutions in this domain compromise quaternary complex assembly in vitro and hb regulation in vivo. Thus, recruitment of Brat is necessary for translational repression and the normal development of posterior embryonic pattern. In addition to regulating abdominal segmentation, previous genetic analysis has shown that Brat, Nanos, and Pumilio govern a variety of developmental processes. We examined the role of Brat in two of these processes—regulation of maternal Cyclin B mRNA in the embryo and regulation of imaginal disc development. The results of these experiments suggest that NHL domain proteins are recruited to various mRNAs by combinatorial protein–protein interactions.
Keywords: Brain tumor, nanos, pumilio, translation, NHL domain
Posttranscriptional regulation plays an important role in the regulation of development and metabolism (Wickens et al. 1996; Gray and Wickens 1998; Preiss and Hentze 1999). In general, this regulation is mediated by cis-acting signals in the 3′ UTR of targeted mRNAs and by proteins that recognize these signals. One well-studied case is the regulation of hunchback (hb) during early embryogenesis in Drosophila. Maternally derived hb mRNA is uniformly distributed throughout the embryo; the mRNA is translationally repressed in the posterior, giving rise to an anterior-to-posterior gradient of Hb protein (Tautz 1988). Failure of this repression results in the abnormal accumulation of Hb in the posterior, which inhibits abdominal segmentation (Hülskamp et al. 1989; Irish et al. 1989; Struhl 1989).
Two conserved RNA-binding proteins, Pumilio (Pum) and Nanos (Nos), are specifically required to repress hb translation (Barker et al. 1992; Wang et al. 1994). Pum, which is distributed uniformly throughout the embryo, is the founding member of a large family of RNA-binding proteins (Murata and Wharton 1995; Zamore et al. 1997; Zhang et al. 1997; Wharton et al. 1998). Pum binds to 32 nucleotide sites in the 3′ UTR of hb (Nos Response Elements, NREs) to regulate its translation (Murata and Wharton 1995; Zamore et al. 1997; Wharton et al. 1998). Nos, which initially is distributed as a gradient emanating from the posterior pole of the embryo, contains a conserved zinc finger that mediates nonspecific RNA binding (Curtis et al. 1997). Nos is selectively recruited into a ternary complex on hb mRNA by NRE-bound Pum (Sonoda and Wharton 1999). The mechanism by which the resulting Nos/Pum/NRE complex regulates translation is not yet understood, although deadenylation is thought to play a role (Wharton and Struhl 1991; Wreden et al. 1997).
Brain Tumor (Brat) is one of three NHL domain proteins found in Drosophila (Adams et al. 2000; Arama et al. 2000). The family name derives from three of the founding members: NCL-1, HT2A, and LIN-41 (Slack and Ruvkun 1998). All three factors have ties to RNA metabolism: the nucleoli in Caenorhabditis elegans ncl-1 mutants are enlarged (Frank and Roth 1998); HT2A was identified by virtue of interaction with the RNA-binding protein HIV Tat (Fridell et al. 1995); and posttranscriptional regulation of lin-29 mRNA is abrogated in lin-41 mutants (Slack et al. 2000). Little is known of the biological roles of other family members, and no direct molecular mechanism has been described previously for any NHL domain protein (including Brat).
In this report, we show that the NHL domain of Brat mediates its recruitment to the 3′ UTR of hb mRNA. Recruitment occurs through protein–protein interactions with RNA-bound Pum and Nos; formation of the resulting quaternary complex is essential for translational control of hb. These results suggest a general mechanism by which other NHL domain proteins may act to control posttranscriptional gene expression.
Results
Pum and Nos jointly recruit Brat
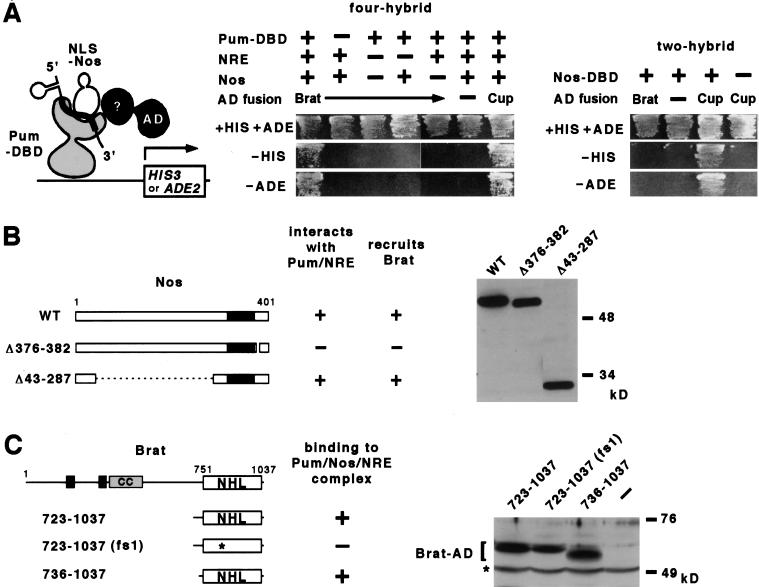
To identify targets or cofactors of the Nos/Pum/NRE ternary complex, we performed a yeast “four-hybrid” experiment, screening a Gal4 activation domain fusion library for proteins that interact with the ternary complex (Fig. 1A). The bait contained the RNA-binding domain of Pum, full-length Nos, and NRE-bearing RNA. As anticipated, we identified factors that interact with individual components in isolation. However, one factor, which proved to be a fragment of Brat, interacts only with the ternary complex and not with either Nos alone, Pum alone, or a Pum/NRE binary complex (Fig. 1A). Deletion analysis revealed that recruitment of Brat is dependent on the conserved carboxy-terminal domain of Nos that mediates its interaction with Pum on hb RNA (Sonoda and Wharton 1999), and not the amino-terminal domain that mediates interaction with Cup during early oogenesis (Fig. 1B) (Verrotti and Wharton 2000). Mutational analysis further showed that a fragment of Brat consisting of little more than the NHL domain is recruited to the ternary complex (Fig. 1C).
Figure 1.
Brat interaction with the Nos/Pum/NRE ternary complex in yeast. (A) Schematic of the yeast four-hybrid screen (left). Results of yeast four-hybrid interaction assays (center). Recruitment of a transcriptional activation domain (AD)–Brat fusion to the ternary complex allows growth in the absence of histidine (−HIS) or adenine (−ADE) in the appropriate yeast reporter strain. Growth is dependent on all four fly-derived moieties. The ternary complex also interacts with an AD–Cup fusion protein (last column); however, unlike Brat, Cup interacts with Nos alone in two-hybrid experiments (right), as reported by Verrotti and Wharton (2000). (B) The carboxy-terminal portion of Nos, which contains the conserved Cys/His zinc-binding domain (in black) (Curtis et al. 1997), is sufficient to recruit Brat into a quaternary complex in yeast. The Western blot of yeast extracts (probed with anti-HA antibody) shows that each NLS–HA–Nos derivative is expressed at approximately the same level. Note that residues 1–42 are not required for quaternary complex formation in vitro (see Fig. 3). (C) A mutation in the NHL domain (G774D) prevents recruitment into a quaternary complex in yeast. The Western blot of yeast extracts (probed with α-NHL antibody) shows that each Brat derivative (within the bracket) is expressed at approximately the same level; the asterisk marks a cross-reacting band. In the drawing (to scale) of full-length Brat at the top, the B-box (black boxes), coiled coil (CC) and NHL domains are indicated. The G774D substitution in Bratfs1 is indicated with an asterisk.
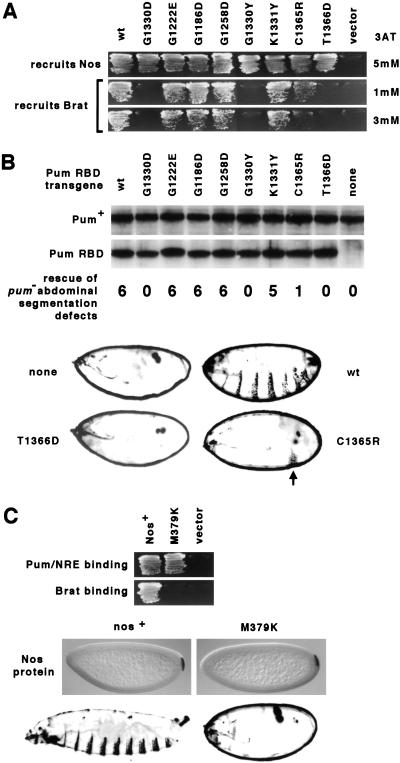
To test the biological significance of the interaction between Brat and the Nos/Pum/NRE ternary complex, we asked whether substitutions in Pum and Nos that interfere with Brat recruitment also abrogate hb regulation in vivo. The impetus for these experiments derives from two properties of the Pum680 mutant that bears the G1330D substitution in the seventh repeat of its RNA-binding domain. First, PumG1330D binds RNA normally and recruits Nos into a ternary complex, but is defective in regulating hb in embryos (Wharton et al. 1998; Sonoda and Wharton 1999). Second, when tested in a yeast four-hybrid experiment, PumG1330D does not recruit Brat (Fig. 2A).
Figure 2.
Correlation between Brat recruitment in yeast and hb regulation in embryos. (A) Activity of Pum mutants in yeast. Genes encoding either the wild-type (wt) Pum RNA-binding domain (RBD) or various mutant derivatives (as indicated) or the empty vector were introduced into yeast to monitor recruitment of Nos into ternary complexes and Brat into quaternary complexes. Transformants were streaked on medium lacking histidine and containing the concentration of the His3 competitor 3-aminotriazole (3AT) indicated on the right. (B) Activity of Pum mutants in embryos. Transgenes encoding each of the indicated RBD derivatives were introduced into flies to monitor complementation of the pum− embryonic phenotype. The Western blot of embryonic extracts prepared from representative transgenic lines shows that each Pum RBD fragment accumulates to essentially the same level in vivo (endogenous full-length Pum serving as a loading control). Below each lane of the blot is shown the typical number of abdominal segments in embryos from pum− females that also bear a single copy of the indicated pum–RBD transgene. Below this list are shown four representative dark-field photographs of embryonic cuticle in which the extent of abdominal segmentation can be assessed. In the absence of pum function (none), hb mRNA is translated in the posterior of the embryo and no abdominal segments form. In this mutant background, expression of the wild-type RBD (wt) results in repression of hb translation sufficient to allow formation of six abdominal segments, on average (Wharton et al. 1998). In this assay, the T1366D derivative is completely inactive, whereas activity of the C1365R derivative typically yields embryos with a single (arrow) abdominal segment. (C) Activity of the NosM379K mutant in yeast and embryos. Above are shown the results of yeast interaction experiments (as in A) that monitor the ability of Nos and NosM379K to enter a ternary complex with Pum and the NRE (Sonoda and Wharton 1999), or to recruit Brat into a quaternary complex, as described in Fig. 1. Transgenes encoding either wild-type or M379K mutant protein were then introduced into flies, and the distribution of encoded protein was examined by whole mount in situ histochemical methods (center) by crossing each transgene into a nosBN background (which otherwise contains no detectable Nos). Dark-field photographs at the bottom show that expression of Nos+ supports development of a normal complement of eight abdominal segments, whereas NosM379K is completely inactive in this assay (although it does rescue the oogenesis defects of nosRC/Df flies and the lethality of nos7117/Df or nosj3B6/Df animals; not shown). The NosM379K embryo (and the embryos in B) is surrounded by the vitelline membrane.
Seven additional Pum mutations were engineered by site-directed mutagenesis into both yeast and Drosophila expression vectors. Residues adjacent to 1330 or at analogous positions in other repeats within the RNA-binding domain were chosen for mutagenesis. The capacity of each Pum mutant to recruit Nos to the NRE or to recruit Brat to the Pum/Nos/NRE complex was assayed in transformed yeast. And the capacity of each Pum mutant to regulate hb translation in embryos (and thereby direct the development of abdominal segmentation) was assayed in transgenic flies. As summarized in Figure 2(A,B), Brat recruitment in yeast correlates with hb regulation in embryos, suggesting that contacts between Pum and Brat are essential in vivo.
To test the role of Nos in Brat recruitment, we screened mutations in its carboxy-terminal domain that were identified originally in a genetic screen for defective nos alleles (Arrizabalaga and Lehmann 1999). Most of the Nos mutants are not recruited by Pum into a ternary complex with the NRE (see Materials and Methods). However, one mutant, NosM379K, is incorporated into a ternary complex normally, but this complex does not interact with Brat (Fig. 2C). When expressed in appropriately engineered transgenic embryos, the M379K derivative is stable but inactive (Fig. 2C), consistent with the idea that contacts between Nos and Brat are also essential for hb regulation in vivo.
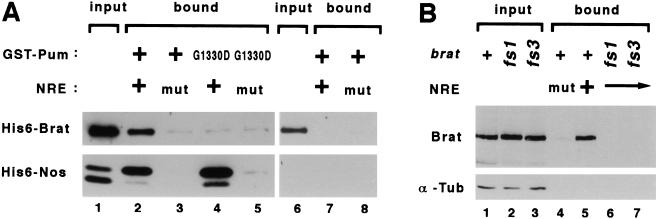
All of the protein–protein interaction experiments described above involve indirect assays performed in yeast. To determine whether the interaction between Brat and the ternary complex is direct and independent of yeast factors, we performed binding experiments in vitro using purified components. As described earlier (Sonoda and Wharton 1999), ternary complexes containing the Pum RNA-binding domain, the carboxy-terminal domain of Nos and the wild-type NRE can be captured on glutathione agarose beads (which bind to the GST moeity attached to Pum). Under the same reaction conditions, Brat is recruited into a quaternary complex that, by three criteria, has the same properties as the complex detected in yeast experiments (Fig. 3A). First, retention of either Nos or Brat is substantially reduced (∼10-fold and 6-fold, respectively) if the NRE bears a mutation that abrogates Nos binding (lanes 2,3). Second, Brat is not detectably retained by a binary Pum/NRE complex in the absence of Nos (lanes 6,7). Third, the PumG1330D mutant captures Nos but not Brat (lane 4). Taken together, these results demonstrate that Nos and Pum act jointly and directly to recruit Brat to the NRE.
Figure 3.
Brat interaction with the Nos/Pum/NRE ternary complex in vitro. (A) The Brat NHL domain binds specifically to wild-type ternary complexes in vitro. The figure is a Western blot probed with anti-Xpress antibody to detect the tag present on both the recombinant His6–Nos and His6–Brat molecules. Lane 1 contains ∼5% of input protein for the experiments of lanes 2–5, and lane 6 contains the same amount of input protein (i.e., Brat only) for the experiments of lanes 7 and 8. After incubation and capture on glutathione agarose beads, bound proteins were eluted and visualized by Western blot. Approximately 1% of the input Brat is bound in the presence of wild-type components (lane 2). The mutant (mut) RNA in the reactions applied to lanes 3, 5, and 8 bears the AA17CC mutation, which specifically prevents Nos binding to the Pum/NRE complex (Sonoda and Wharton 1999). The G1330D mutant Pum (lanes 4,5) is described in the text. (B) Substitutions in the NHL domains of the Bratfs mutant proteins prevent binding to the Pum/Nos/NRE complex. The figure is a Western blot probed with α-CC to detect proteins in embryonic extracts (genotype indicated above) that bind to ternary Pum/Nos/NRE complexes assembled in vitro with purified components. The blot was reprobed with antibodies to α-tubulin, which serves as a loading control. Lanes 1–3 contain ∼5% of input protein, and the reaction loaded in lane 4 contained the NRE AA17CC mutant.
Brat required for repression of hb
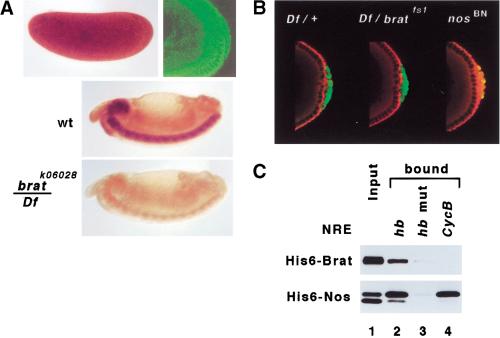
All of the recessive brat alleles are associated to a greater or lesser extent with a variety of phenotypes, including: a dramatic (≤10-fold) overgrowth of the larval brain (Hankins 1991; Arama et al. 2000), early oogenesis defects (Schüpbach and Wieschaus 1991), metastasis of transplanted brain and imaginal tissue (Woodhouse et al. 1998), and a maternal effect on embryonic viability (Hankins 1991). This last class of “female sterile” (fs) alleles appear to interfere preferentially with function in the female germ line, although they also exhibit the other brat phenotypes (Hankins 1991). Unlike lethal alleles that encode truncated proteins, bratfs1 and bratfs3 encode proteins with single amino acid substitutions at conserved residues within the NHL domain (this study; Arama et al. 2000).
We then wanted to determine whether the substitutions in the Bratfs1 and Bratfs3 mutant proteins interfere with recruitment to the Nos/Pum/NRE ternary complex. To this end, ternary complexes were assembled in vitro with purified GST–Pum, Nos, and NRE-bearing RNA; these were subsequently incubated with embryonic extracts prepared from embryos derived from either wild-type or bratfs mutant females (henceforth referred to as bratfs mutant embryos). Complexes were captured on glutathione-agarose beads, and bound proteins displayed on a Western blot probed with Brat-specific antibodies (described below).
As shown in Figure 3B, ∼5% of full-length Brat+ is retained under the conditions of the experiment, but neither Bratfs1 nor Bratfs3 binds appreciably to the ternary complex. The mutant proteins accumulate to normal levels and are stable in vivo. Thus, the single amino acid substitutions in the Bratfs mutant proteins prevent efficient recruitment to the Nos/Pum/NRE complex.
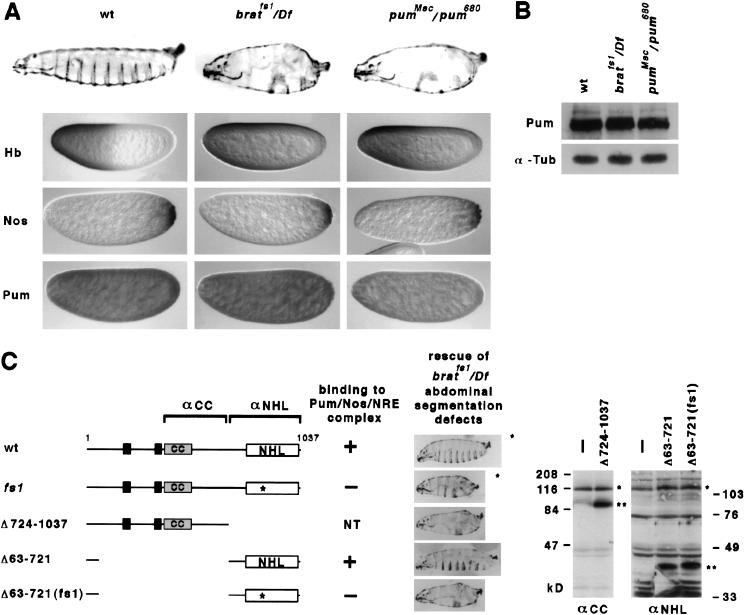
We then examined the consequences of altered Brat function on embryonic development. As shown in Figure 4A and Table 1, bratfs mutant embryos have defects in abdominal segmentation that are essentially indistinguishable from those in embryos with reduced nos or pum function (Lehmann and Nüsslein-Volhard 1987, 1991; Wharton et al. 1998). These defects arise as a result of incomplete translational repression of hb in the posterior of the preblastoderm embryo, as is the case for nos or pum mutants (Fig. 4A). The level and distribution of Nos, Pum, and hb mRNA appear to be normal in bratfs mutant embryos (Fig. 4A,B and data not shown), suggesting that Brat does not act indirectly to regulate abdominal segmentation. Taken with the interaction data of Figures 1–3, we conclude that recruitment of Brat jointly by Nos and Pum to the NRE is required for the normal regulation of hb mRNA in the early embryo.
Figure 4.
Brat is required for normal regulation of hb in the early embryo. (A) The maternal effect on embryonic viability of bratfs1. Shown are photomicrographs of embryos from females of the indicated genotype (above): cuticle (row 1), Hb protein (row 2), Nos protein (row 3), and Pum protein (row 4). Note the absence of most abdominal segments and the accumulation of Hb in the posterior of bratfs1 mutant embryos. In contrast, the distributions of Nos and Pum are indistinguishable from wild type. Note the lower level of Pum antigen in pumMsc/pum680 embryos. (B) Western blot to detect the level of Pum in extracts prepared from embryos of the indicated genotype. The blot was reprobed with antibodies to detect α-tubulin, which serves as a loading control. (C) Rescue of the bratfs1 abdominal segmentation defects. Expression of UAS transgenes encoding either wild-type (wt) Brat or various mutant derivatives (left) was driven in the female germ line by crossing to a nos>GAL4–VP16 driver; the endogenous brat gene in these females was mutant (bratfs1/Df). Rescue of the abdominal segmentation defects correlates with binding of each Brat derivative to the Pum/Nos/NRE complex in various experiments. (NT) not tested. A representative embryonic cuticle is shown for each case; the asterisk indicates these are rare embryos, as a result of uncharacterized oogenesis defects associated with ectopic expression of full-length Brat and Bratfs1. Western blots of embryonic extracts (right) show that Brat fragments (**) are expressed at levels somewhat higher than the endogenous Brat+ protein (*). Blots were probed with antibodies that recognize different regions of Brat, as indicated at the top left.
Table 1.
Abdominal segmentation defects in embryos from various mutant females
|
|
|
No. of abdominal segments
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 1
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
||
| pum | Msc/680 | 4 | 46 | 39 | 10 | 2 | |||
| fs1/Df(2L)TE37C-7 | 13 | 52 | 30 | 4 | 1 | ||||
| fs3/Df(2L)TE37C-7 | 4 | 28 | 28 | 24 | 11 | 4 | |||
| fs1/fs3 | 2 | 45 | 34 | 17 | 3 | ||||
| brat | 1/fs3 | 3 | 11 | 42 | 24 | 8 | 13 | ||
| k06028/fs3 | 18 | 30 | 25 | 27 | |||||
| k06028/k06028a | 7 | 29 | 63 | ||||||
| +/fs3 | 100 | ||||||||
| +/Df(2L)TE37C-7 | 100 | ||||||||
Each entry is the percentage of embryos derived from females of the indicated genotype (left) bearing the indicated number of abdominal segments (above). Forty to one-hundred embryos were scored in each case.
Germ-line clone.
Many NHL domain proteins also share three other motifs: a Ring-finger, one or two B-box motifs, and a coiled-coil (RBCC) (Slack and Ruvkun 1998). All of the evidence described above points to the central role of the NHL domain in mediating Brat activity. Analysis of lin-41 alleles also suggests that the NHL domain plays an important role in Lin-41 function (Slack et al. 2000). However, another report shows that expression of the RBCC domain of human BERP (brain expressed Ring-finger protein) in PC12 cells blocks a response to nerve growth factor (El-Husseini and Vincent 1999), suggesting an essential role for this region of BERP and, by extension, other family members.
To test the role of these other motifs in hb regulation, transgenic flies that express wild-type Brat, the amino-terminal BCC domain (Brat lacking a Ring-finger), and the carboxy-terminal NHL domain were prepared. As controls, similar transgenic flies that express full-length Bratfs1 and NHLfs1 derivatives were also prepared (Fig. 4C). Expression of each transgene is controlled by Gal4-binding sites upstream activating sequence (UAS); by appropriate genetic crosses, each Brat derivative was expressed during oogenesis under the control of a nos>GAL4–VP16 transgene (Van Doren et al. 1998) in a bratfs1 mutant background.
As shown in Figure 4C, expression of full-length Brat+ but not Bratfs1 rescues the abdominal defects of bratfs1 embryos. For reasons we do not understand, overexpression of either protein severely disrupts oogenesis and females produce very few eggs. Although we do not understand the basis of this phenotype, we were able to analyze the segmentation pattern among larvae derived from rare fertilized eggs. Expression of the amino-terminal BCC domain has no effect in wild-type females and does not rescue the defects of bratfs1 embryos, although the protein is stable in vivo and accumulates to a higher level than the endogenous mutant Brat protein (Fig. 4C). Somewhat surprisingly, expression of the wild-type NHL domain almost completely rescues the bratfs1 embryonic phenotype. In contrast, expression of the NHLfs1 mutant domain to essentially the same level, does not. Thus, these experiments suggest the Brat NHL domain is necessary and sufficient to regulate hb translation in the early embryo.
Nos and Pum do not recruit Brat to Cyclin B mRNA
Analysis of mutant phenotypes has revealed that Nos and Pum are required for a variety of processes in addition to the development of abdominal segmentation. nos and pum are expressed in tissues other than the female germ line (Wharton et al. 1998; Wang and Lehmann 1991; M. Asano and R.P. Wharton, unpubl.). More important, nos and pum mutants are subviable, revealing an (unknown) essential function for each factor in somatic cells (Lin and Spradling 1997; Spradling et al. 1999). In the germ line, nos and pum mutants exhibit a number of defects including loss of germ-line stem cells in both sexes, failure of precursor cells to migrate into and populate the somatic gonad, and premature proliferation of precursor cells (pole cells) in the embryo (Kobayashi et al. 1996; Lin and Spradling 1997; Forbes and Lehmann 1998; Asaoka-Taguchi et al. 1999; Bhat 1999; Deshpande et al. 1999). The premature proliferation appears to result from the inappropriate derepression of maternal Cyclin B (CycB) mRNA in the pole cells (Asaoka-Taguchi et al. 1999); in no other case is the molecular basis of Nos or Pum action currently understood.
We wanted to determine whether Nos and Pum also act in conjunction with Brat to regulate maternal Cyclin B mRNA. Using antibodies directed against different regions of the protein, we find that Brat is distributed throughout the syncitial blastoderm stage embryo when hb mRNA is repressed, and is also present in the cytoplasm of the pole cells when maternal Cyclin B mRNA is regulated (Fig. 5A). However, Cyclin B mRNA is repressed normally in the pole cells of bratfs mutant embryos, but not in the pole cells of nos mutant embryos, as reported previously (Fig. 5B) (Asaoka-Taguchi et al. 1999). Thus, Brat does not appear to play a role in repression of Cyclin B, although we cannot rule out the possibility that the residual activity of Bratfs1 is sufficient to regulate Cyclin B but not hb.
Figure 5.
Cyclin B mRNA regulated normally in the pole cells of brat mutant embryos. (A) Brat is distributed uniformly throughout syncitial cleavage stage embryos (brown, above left), and is uniform in the cytoplasm of nuclear division cycle 12–13 embryos (green, above right). In particular, the protein is present in the pole cells when translation of Cyclin B mRNA is repressed, as shown in B. The same conditions were used to stain sibling stage 14 wild type (wt) and hemizygous bratk06028 embryos, demonstrating antibody specificity. Note the presence of low levels of residual protein that are also detectable in Fig. 6A,B. (B) Confocal images showing the distributions of Vasa (green) and Cyclin B (red) at the posterior poles of embryos derived from females of the indicated genotype. Translation of maternal Cyclin B mRNA is repressed in the pole cells of wild-type and bratfs1 embryos, but not in nosBN embryos. (C) Brat is not recruited to sequences in the Cyclin B 3′ UTR. In vitro interaction assay using purified factors demonstrates recruitment of Brat to a Nos/Pum complex assembled on the hb NRE (lane 2) but not to the complex assembled with the same concentration of RNA derived from Cyclin B (lane 4). (Lane 3) Control showing that neither Nos nor Brat is recruited efficiently to a complex containing Pum and the AA17CC mutant hb NRE (Sonoda and Wharton 1999). (Lane 1) Approximately 5% of input protein for each reaction.
The cis-acting signals that mediate Nos- and Pum-dependent regulation of Cyclin B have not yet been defined precisely. However, NRE-like sequences are present in the maternal isoform of the mRNA, which is regulated (Dalby and Glover 1993). If indeed Pum, Nos, and NRE-like sequences mediate its regulation, then why would repression of Cyclin B mRNA be Brat independent?
To investigate this issue, we examined the binding of Pum, Nos, and Brat to the Cyclin B NRE-like element in vitro. The RNA used in these experiments contains 136 nucleotides that include all of the NRE homologous elements as well as flanking sequences. Pum binds to this Cyclin B-derived RNA in gel mobility-shift experiments, but not to a derivative bearing mutations in the conserved NRE-like element (Y. Habara and R.P. Wharton, unpubl.), consistent with the idea that similar sequences in Cyclin B and hb are recognized. Bound Pum can recruit Nos into a ternary complex on Cyclin B RNA, much as it does on the hb NRE (Fig. 5C). However, the ternary complex assembled on Cyclin B RNA recruits Brat at least 10-fold less efficiently than the corresponding complex assembled on the hb NRE (Fig. 5C). This surprising observation may in part explain the Brat independence of Cyclin B regulation described above (Fig. 5B). Furthermore, it suggests that the RNA sequence specifies the geometry of the Pum/Nos complex, which in turn determines whether Brat is recruited or not (see Discussion and Fig. 7).
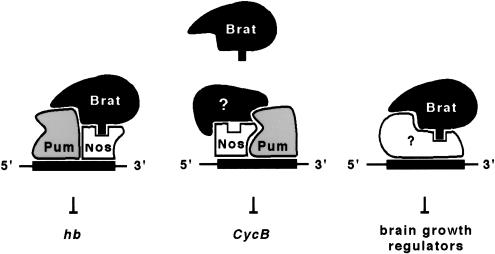
Figure 7.
Models for combinatorial regulation of various mRNAs by Brat, Nos, and Pum. Regulation of the hb and Cyclin B mRNAs occurs at the level of translation. In the brain (and the imaginal discs), the targets of Brat action are unknown. Brat activity is probably not mediated by Pum in either tissue. See discussion for details.
Growth regulation mediated by the NHL domain of Brat
Brat acts as a growth suppressor in the larval brain (Hankins 1991; Arama et al. 2000). Whether Brat acts by a similar molecular mechanism in the brain and in the early embryo (in regulating hb) is unclear; however, the observation that single amino acid substitutions in the NHL domain of the Bratfs mutant proteins disrupt both processes is consistent with such an idea. The role of Brat in the brain is not yet clear, as the phenotype has not been characterized in detail and regulatory targets have yet to be identified.
The role of Brat in the imaginal discs has been even less clear. Loss of brat function leads to no obvious defects in imaginal development (Arama et al. 2000), and rare escaper homozygous brat− flies appear morphologically normal. One role for Brat was revealed by experiments in which imaginal disc tissue was transplanted into the body cavity of adult hosts; brat− but not wild-type discs metastasize and kill the fly (Woodhouse et al. 1998). This observation suggests that Brat is expressed in the discs, which led us to consider the possibility that loss-of-function mutants exhibit no apparent phenotype due to the presence of a redundant activity.
To investigate this possibility, we misexpressed either Brat+ or Bratfs1 using an engrailed (en)–GAL4 driver line and the UAS transgenes described above (see Fig. 4). Flies were examined for phenotypes resulting from the gain of Brat function.
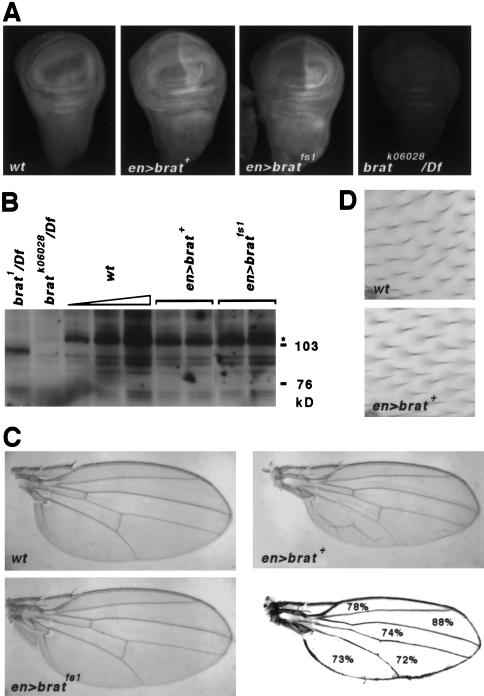
Endogenous Brat accumulates uniformly in the cytoplasm of cells in wing discs from third instar larvae (Fig. 6A). In either UAS>brat+ or UAS>bratfs1 discs that also bear the en>GAL4 driver, a modest excess of protein accumulates in the posterior compartment of the wing disc; analysis of Western blots suggests that the level of overproduction is less than two- to threefold (Fig. 6B). At this stage of development, ectopic expression of either protein does not substantially alter the morphology of the discs.
Figure 6.
Ectopic Brat suppresses growth of the wing. (A) The distribution of Brat in third instar imaginal discs of the indicated genotypes. In addition to endogenous protein, the discs in the center bear ectopic Brat in the posterior compartment (to the right). Analysis by confocal microscopy reveals that Brat is cytoplasmic (not shown). The hemizygous bratk06028 disc (right) shows a very low level of residual staining, demonstrating specificity of the antibody. (B) Western blot of wing imaginal disc extracts prepared from flies of the genotypes indicated above. (Lanes 3–5) Three different amounts of wild-type extract (1×, 2×, 4×); (lanes 4,–7) Extracts from two different UAS transgenic lines. (C) Adult wings from flies of the indicated genotype. Also shown is a tracing of the en>brat+ wing showing the average intervein area for this genotype as a percentage of wild type. (D) High magnification views of wild-type and en>brat+ wings. Each cell secretes a small hair that is visible in the micrographs; note that the cell density is the same.
However, misexpression of Brat+ causes an intriguing growth suppression phenotype that is evident in the wings of adults. Three observations stand out. First, the en>brat+ wings are 24% smaller than control wings (P < 0.0001 by Student's t test) (Fig. 6C). They are also usually deformed, probably as a result of poor adhesion between the dorsal and ventral surfaces. Second, the reduction in wing size appears to be due to a reduction in the number of cells contributing to the wing rather than a reduction in the size of the cells. This conclusion is based on a measurement of the density of epidermal hairs, each secreted by a single cell (Fig. 6D). Third, the phenotype is nonautonomous, extending into the anterior compartment where the en promoter is not active. For example, the anterior-most sector of en>brat+ wings (bounded by the first and second longitudinal veins) is on average 22% smaller than the corresponding region of control wings.
Significantly, none of the phenotypes associated with misexpression of Brat+ is caused by misexpression of similar levels of Bratfs1. This supports the idea that Brat acts by a similar molecular mechanism to regulate abdominal segmentation in the embryo and growth of the wing imaginal disc.
Discussion
Our current model of how Nos, Pum, and Brat act to regulate gene expression is shown in Figure 7. The model involves combinatorial interactions among cis-acting sequences in regulated mRNAs, proteins that recognize these sequences, and the NHL domain of Brat. Below we discuss the evidence that supports these ideas as well as the possibility that other NHL domain proteins act by a similar mechanism.
Recruitment of Brat to the NRE jointly by Nos and Pum is essential for regulation of hb. Three lines of evidence show that the NHL domain plays a key role in this process. First, the NHL domain is sufficient to mediate interaction with the Nos/Pum/NRE complex, thereby targeting Brat to hb mRNA (Figs. 1,3). Second, single amino acid substitutions within the NHL domain attenuate interaction with the ternary complex (Figs. 1,3) and regulation of hb in vivo (Fig. 4). Third, maternal expression of the wild-type NHL domain alone is sufficient to restore hb regulation in bratfs mutant embryos (Fig. 4). This result suggests that the NHL domain contains intrinsic translation regulatory activity. However, activity of the isolated NHL domain is (necessarily) assayed in the presence of Bratfs mutant protein, and thus, we cannot rule out the possibility that the amino-terminal BCC domain participates somehow in hb regulation.
Brat appears to play no role in regulating Cyclin B mRNA in the pole cells (Fig. 5), although Nos and Pum are required for this process (Asaoka-Taguchi et al. 1999). This observation is perhaps not surprising, as translation of Cyclin B mRNA in the posterior region of the syncitial cleavage stage embryo appears to be uninhibited, even in the presence of Nos, Pum, and Brat. Only in the pole cells, which extrude from the posterior extreme of the embryo, is Cyclin B mRNA repressed. Perhaps the specialized pole plasm incorporated into these cells contains a Cyclin B-specific corepressor that acts in conjunction with Nos and Pum (as hypothesized in Fig. 7). Alternatively, the Nos/Pum complex on Cyclin B mRNA may be sufficient to regulate translation without a cofactor in the pole cells.
In either case, the experiments of Figure 5 reveal an unanticipated complexity: the Nos/Pum complexes assembled on Cyclin B and hb mRNAs apparently have different conformations, as revealed by their ability to interact with Brat. Perhaps the RNA sequence acts as a scaffold, bringing Nos and Pum together on the RNA in different relative orientations in the two cases, as suggested in Figure 7. Alternatively, the RNA might act as an allosteric effector, altering the conformation of Pum to allow interaction with different cofactors. A recent study of the structure and function of the Pum RNA-binding domain suggests how the quaternary complex assembles on the hb NRE (T.A. Edwards and A.K. Aggarwal, pers. comm). Extension of these studies to complexes containing Cyclin B mRNA should reveal how the RNA sequence specifies cofactor identity.
Brat acts as a growth suppressor in the larval brain and, upon modest overexpression, in the wing imaginal disc (Fig. 6). Current evidence suggests that, in these tissues, Brat likely acts with cofactors other than Pum or Nos (as indicated in Fig. 7), although the supporting evidence is relatively weak. The brains of mutant larvae bearing the strongest extant alleles of pum do not exhibit a tumorous brat phenotype (not shown), consistent with the idea that some other factor acts in conjunction with Brat in this tissue. We attempted to test the role of Pum in mediating the en>brat+ phenotype, but were unable to recover flies of the appropriate genotype (presumably due to the subviability of both pum− and en>brat+ flies). The role of Nos in mediating Brat action is less easily assessed, as larvae bearing lethal nos alleles die before the third instar (not shown) when both the brat− and en>brat+ phenotypes are evident. Weaker alleles, such as nosRC, have substantial residual activity (Verrotti and Wharton 2000).
Perhaps the most striking aspect of the en>brat+ phenotype is that ectopic Brat suppresses growth nonautonomously. This is in contrast to the action of other growth regulators that have been the focus of recent research in flies. Regulation of cell size and cell number in the imaginal discs is complex. One class of regulators are the extracellular signals of the EGF, TGF-β, and Wg pathways that coordinately control pattern and growth. Another class consists of signals mediated by molecules such as Ras, Myc, TOR, and members of the insulin receptor pathway that primarily control cell size or number, but not pattern (Weinkove et al. 1999; Oldham et al. 2000; Stocker and Hafen 2000; Zhang et al. 2000; and references therein). For many of these “pure” growth regulators, ectopic expression enhances (or suppresses) growth of the imaginal discs to an extent similar to that observed for Brat in Figure 6. However, in none of these cases is the effect on growth transmitted to surrounding cells, as is true for Brat. Thus, Brat appears to regulate either novel pathways or novel combinations of pathways that generate extracellular signals.
All three Drosophila NHL proteins regulate some aspect of growth, suggesting this may be a common role for NHL proteins in general. Mutations in dappled and mei-P26 reveal that the proteins encoded by these loci suppress growth of melanotic and ovarian tumors, respectively (Rodriguez et al. 1996; Page et al. 2000); Mei-P26 is also required for a normal frequency and distribution of genetic exchange during meiosis. Given the structural similarity among the three fly proteins, we assume that Dappled and Mei-P26, like Brat, act by regulating translation or some other aspect of mRNA metabolism. Little else is currently known of the cell biological processes controlled by Brat, Dappled, or Mei-P26.
Do other NHL proteins act in a manner similar to Brat? Relatively little is known about the molecular mechanism by which these factors act in vivo, and thus it is not clear whether they regulate translation or some other aspect of posttranscriptional gene expression. Nevertheless, an argument for analogous function can be made for three of the family members, based on current knowledge. First, the HT2A human protein interacts with the site-specific RNA-binding Tat protein (Fridell et al. 1995), much as Brat interacts with Nos and Pum. Second, C. elegans NCL-1 appears to regulate growth, although the mutant worms have larger cells rather than more cells (Frank and Roth 1998). The most striking analogy with Brat function comes from the third case, C. elegans LIN-41, which acts in the penultimate step of the heterochronic pathway (Reinhart et al. 2000; Slack et al. 2000). Like Brat, LIN-41 is a posttranscriptional regulator. And like Brat, which acts in concert with Nos and Pum, LIN-41 appears to play a role in the switch from sperm to oocyte production in hermaphrodites that is governed by homologs of Nos and Pum (Zhang et al. 1997; Kraemer et al. 1999; Subramaniam and Seydoux 1999). Thus, it seems likely that LIN-41 and Brat act by a similar mechanism, interacting with RNA-bound factors to repress translation.
Materials and methods
Strains and reagents
Transgenes were constructed in pCasPeR derivatives and introduced into w1118 flies by standard methods. The pum transgenes used in Figure 2 were mutant derivatives of pum(Δ2‘) (Wharton et al. 1998). At least three independent transgenic lines were crossed into a pumMsc/pumFC8 background to test activity in the absence of endogenous pum function (by counting the number of abdominal segments). The nos transgenes are as described by Verrotti and Wharton (2000); their activity was tested in the embryonic null background nosBN/Df(3R)DlFX1. The following mutants were obtained from either the Bloomington or Umea Stock Centers: nos7117, nosj3B6, bratfs1, bratfs3, brat1, bratk06028, Df(2L)TE37C-7; as were the en>GAL4, act5C>GAL4, and nos>GAL4-VP16 (Van Doren et al. 1998) drivers. Germ-line clones of bratk06028 were prepared by standard methods by heat shocking P{ry +t7.2=hsFLP}22; bratk06028 P{ry +t7.2=neoFRT40A}/P{w+mC=ovoD1–18}12L P{ry +t7.2=neoFRT40A} females. An UAS-brat transgene was prepared using the UASp vector (Rørth 1998), fusing nucleotides 626–3940 (accession no. AB022432) of brat, which encodes part of the 5′ UTR and the entire protein-coding region, to sequences encoding the αtub84B 3′ UTR. Antibodies to Nos and Pum have been described (Sonoda and Wharton 1999), and antibodies to Cyclin B were a gift from Christian Lehner (University of Bayreuth, Germany). Antibodies to Brat were raised against GST fusion proteins bearing residues 376–722 and 723–1037 yielding αCC and αNHL sera (Fig. 4). Similar results were obtained using both sera for whole mount in situ histochemistry. Antibodies against α-tubulin were from Sigma, and against the Xpress tag from Invitrogen.
Yeast four-hybrid screen
Yeast strain PJ69–4A (James et al. 1996) was transformed with three types of plasmids. One is a derivative of pGBT9 (TRP1 selection) that encodes not only a Gal4 DNA-binding domain PumRBD (amino acids 1093–1426) fusion, but also a chimeric nuclear RNA bearing two tandem copies of the 32-nucleotide NRE described by Wharton and Struhl (1991) (each bearing a UUUU>UUAU substitution to prevent premature transcriptional termination) driven by the Pol III promoter present on pIIIEx424 RPR (Good and Engelke 1994). A BamHI–SalI fragment encoding the RNA expression cassette was inserted at the NaeI site of pGBT9 to generate pJ2110. The second plasmid, a derivative of pRS424 (URA3 selection), encodes a fusion of the SV40 NLS, an HA epitope tag, and full-length Nos driven by the PGK promoter (Mellor et al. 1983) (pJ2701). The third plasmid is a member of a library encoding transcriptional activation domain fusions to early embryo cDNAs in pGAD10 (LEU2 selection, a gift of A. Dahanukar, Yale University, CT). Approximately 2 × 106 potential colony-forming units were plated on selective medium containing 3 mM 3-aminotriazole but no histidine, and subsequently replica-plated onto medium lacking adenine. PJ69–4A bears both HIS3 and ADE2 transcriptional reporters that are activated upon recruitment of the AD moiety (in this case, fused to Brat) to the DNA-bound bait. Plasmids were recovered from 165 colonies. Of these, 83 retested true; these are derived from 16 different genes, including Cup and Brat (amino acids 707–1037). With the exception of the Brat clone, all others interacted with a ternary complex containing the PumG1330D mutant fusion protein.
For the experiments of Figure 2C, the activities of 11 Nos derivatives (Arrizabalaga and Lehmann 1999) were examined in yeast. The S337L, V338E, R339Q, P348S, L350R, R351Q, R351H, G362E, and A365T mutants do not form a ternary complex in yeast with Pum/NRE and thus, were not tested further. The V354M substitution has no apparent effect on Nos activity in yeast or in the embryo.
Quaternary complex assay in vitro
The experiments of Figures 4A and 5C were performed essentially as described by Sonoda and Wharton (1999), with the addition of purified His6-tagged Brat NHL domain (residues 723–1037) to a concentration of 0.5 μM. Estimates of the fraction of retained Nos or Brat were derived from ECL detection and comparison with multiple autoradiographic exposures of a dilution series of input protein. In the experiments of Figure 4B, ternary complex was incubated with embryonic extracts prepared as described by Murata and Wharton (1995). In Figure 5C, the Cyclin B RNA contains nucleotides 2249–2384 (accession no. M33192) from the 3′ UTR. In control experiments, Pum and Nos do not bind to RNAs of similar size derived from the 3′ UTRs of pum, nos, or tor.
Acknowledgments
We thank the Fehon laboratory, H. Bogerd, B. Cullen, and C. Lehner for advice and reagents; Z. Lev for helpful discussions; C. Gardner and S. Pyle for technical assistance; S. Boyles for administrative help; and G. Johnson for media preparation. Drosophila stocks were obtained from the Bloomington and Umea Stock Centers, and information from the Berkeley Drosophila Genome Project (BDGP). R.P.W. is an Assistant Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rwharton@duke.edu; FAX (919) 681-8984.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.870801.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Arama E, Dickman D, Kimchie Z, Shearn A, Lev Z. Mutations in the β-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene. 2000;19:3706–3716. doi: 10.1038/sj.onc.1203706. [DOI] [PubMed] [Google Scholar]
- Arrizabalaga G, Lehmann R. A selective screen reveals discrete functional domains in Drosophila Nanos. Genetics. 1999;153:1825–1838. doi: 10.1093/genetics/153.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat Cell Biol. 1999;1:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- Barker DD, Wang C, Moore J, Dickinson LK, Lehmann R. Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes & Dev. 1992;6:2312–2326. doi: 10.1101/gad.6.12a.2312. [DOI] [PubMed] [Google Scholar]
- Bhat KM. The posterior determinant gene nanos is required for the maintenance of the adult germline stem cells during Drosophila oogenesis. Genetics. 1999;151:1479–1492. doi: 10.1093/genetics/151.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Treiber DK, Tao F, Zamore PD, Williamson JR, Lehmann R. A CCHC metal-binding domain in Nanos is essential for translational regulation. EMBO J. 1997;16:834–843. doi: 10.1093/emboj/16.4.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby B, Glover DM. Discrete sequence elements control posterior pole accumulation and translational repression of maternal cyclin B RNA in Drosophila. EMBO J. 1993;12:1219–1227. doi: 10.1002/j.1460-2075.1993.tb05763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Calhoun G, Yanowitz JL, Schedl PD. Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Cell. 1999;99:271–281. doi: 10.1016/s0092-8674(00)81658-x. [DOI] [PubMed] [Google Scholar]
- El-Husseini AE, Vincent SR. Cloning and characterization of a novel RING finger protein that interacts with class V myosins. J Biol Chem. 1999;274:19771–19777. doi: 10.1074/jbc.274.28.19771. [DOI] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- Frank DJ, Roth MB. ncl-1 is required for the regulation of cell size and ribosomal RNA synthesis in Caenorhabditis elegans. J Cell Biol. 1998;140:1321–1329. doi: 10.1083/jcb.140.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell RA, Harding LS, Bogerd HP, Cullen BR. Identification of a novel human zinc finger protein that specifically interacts with the activation domain of lentiviral Tat proteins. Virology. 1995;209:347–357. doi: 10.1006/viro.1995.1266. [DOI] [PubMed] [Google Scholar]
- Good PD, Engelke DR. Yeast expression vectors using RNA polymerase III promoters. Gene. 1994;151:209–214. doi: 10.1016/0378-1119(94)90658-0. [DOI] [PubMed] [Google Scholar]
- Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- Hankins GR. Analysis of a Drosophila neuroblastoma gene. PhD Thesis. Charlottesville, VA: University of Virginia; 1991. [Google Scholar]
- Hülskamp M, Schröder C, Pfeifle C, Jäckle H, Tautz D. Posterior segmentation of the Drosophila embryo in the absence of a maternal posterior organizer gene. Nature. 1989;338:629–632. doi: 10.1038/338629a0. [DOI] [PubMed] [Google Scholar]
- Irish V, Lehmann R, Akam M. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature. 1989;338:646–648. doi: 10.1038/338646a0. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Yamada M, Asaoka M, Kitamura T. Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature. 1996;380:708–711. doi: 10.1038/380708a0. [DOI] [PubMed] [Google Scholar]
- Kraemer B, Crittenden S, Gallegos M, Moulder G, Barstead R, Kimble J, Wickens M. NANOS-3 and FBF proteins physically interact to control the sperm/oocyte switch in C. elegans. Curr Biol. 1999;9:1009–1018. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Nüsslein-Volhard C. Involvement of the pumilio gene in the transport of an abdominal signal in the Drosophila embryo. Nature. 1987;329:167–170. [Google Scholar]
- ————— The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Mellor J, Dobson MJ, Roberts NA, Tuite MF, Emtage JS, White S, Lowe PA, Patel T, Kingsman AJ, Kingsman SM. Efficient synthesis of enzymatically active calf chymosin in Saccharomyces cerevisiae. Gene. 1983;24:1–14. doi: 10.1016/0378-1119(83)90126-9. [DOI] [PubMed] [Google Scholar]
- Murata Y, Wharton RP. Binding of Pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes & Dev. 2000;14:2689–2694. doi: 10.1101/gad.845700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, McKim KS, Deneen B, Van Hook TL, Hawley RS. Genetic studies of mei-P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila. Genetics. 2000;155:1757–1772. doi: 10.1093/genetics/155.4.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T, Hentze MW. From factors to mechanisms: Translation and translational control in eukaryotes. Curr Opin Genet Dev. 1999;9:515–521. doi: 10.1016/s0959-437x(99)00005-2. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Zhou Z, Tang ML, Meller S, Chen J, Bellen H, Kimbrell DA. Identification of immune system and response genes, and novel mutations causing melanotic tumor formation in Drosophila melanogaster. Genetics. 1996;143:929–940. doi: 10.1093/genetics/143.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Ruvkun G. A novel repeat domain that is often associated with RING finger and B- box motifs. Trends Biochem Sci. 1998;23:474–475. doi: 10.1016/s0968-0004(98)01299-7. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes & Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H, Hafen E. Genetic control of cell size. Curr Opin Genet Dev. 2000;10:529–535. doi: 10.1016/s0959-437x(00)00123-4. [DOI] [PubMed] [Google Scholar]
- Struhl G. Differing strategies for organizing anterior and posterior body pattern in Drosophila embryos. Nature. 1989;338:741–744. doi: 10.1038/338741a0. [DOI] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- Tautz D. Regulation of the Drosophila segmentation gene hunchback by two maternal morphogenetic centres. Nature. 1988;332:281–284. doi: 10.1038/332281a0. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Verrotti AC, Wharton RP. Nanos interacts with Cup in the female germline of Drosophila. Development. 2000;127:5225–5232. doi: 10.1242/dev.127.23.5225. [DOI] [PubMed] [Google Scholar]
- Wang C, Lehmann R. Nanos is the localized posterior determinant in Drosophila. Cell. 1991;66:637–647. doi: 10.1016/0092-8674(91)90110-k. [DOI] [PubMed] [Google Scholar]
- Wang C, Dickinson LK, Lehmann R. Genetics of nanos localization in Drosophila. Dev Dyn. 1994;199:103–115. doi: 10.1002/aja.1001990204. [DOI] [PubMed] [Google Scholar]
- Weinkove D, Neufeld TP, Twardzik T, Waterfield MD, Leevers SJ. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class I(A) phosphoinositide 3-kinase and its adaptor. Curr Biol. 1999;9:1019–1029. doi: 10.1016/s0960-9822(99)80450-3. [DOI] [PubMed] [Google Scholar]
- Wharton RP, Struhl G. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell. 1991;67:955–967. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- Wharton RP, Sonoda J, Lee T, Patterson M, Murata Y. The Pumilio RNA-binding domain is also a translational regulator. Mol Cell. 1998;1:863–872. doi: 10.1016/s1097-2765(00)80085-4. [DOI] [PubMed] [Google Scholar]
- Wickens M, Kimble J, Strickland S. Translational control of developmental decisions. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 411–450. [Google Scholar]
- Woodhouse E, Hersperger E, Shearn A. Growth, metastasis, and invasiveness of Drosophila tumors caused by mutations in specific tumor suppressor genes. Dev Genes Evol. 1998;207:542–550. doi: 10.1007/s004270050145. [DOI] [PubMed] [Google Scholar]
- Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124:3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the drosophila target of rapamycin dTOR. Genes & Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]