Abstract
The E2 glycoprotein of hepatitis C virus (HCV) mediates viral attachment and entry into target hepatocytes and elicits neutralizing antibodies in infected patients. To characterize the structural and functional basis of HCV neutralization, we generated a novel panel of 78 monoclonal antibodies (MAbs) against E2 proteins from genotype 1a and 2a HCV strains. Using high-throughput focus-forming reduction or luciferase-based neutralization assays with chimeric infectious HCV containing structural proteins from both genotypes, we defined eight MAbs that significantly inhibited infection of the homologous HCV strain in cell culture. Two of these bound E2 proteins from strains representative of HCV genotypes 1 to 6, and one of these MAbs, H77.39, neutralized infection of strains from five of these genotypes. The three most potent neutralizing MAbs in our panel, H77.16, H77.39, and J6.36, inhibited infection at an early postattachment step. Receptor binding studies demonstrated that H77.39 inhibited binding of soluble E2 protein to both CD81 and SR-B1, J6.36 blocked attachment to SR-B1 and modestly reduced binding to CD81, and H77.16 blocked attachment to SR-B1 only. Using yeast surface display, we localized epitopes for the neutralizing MAbs on the E2 protein. Two of the strongly inhibitory MAbs, H77.16 and J6.36, showed markedly reduced binding when amino acids within hypervariable region 1 (HVR1) and at sites ∼100 to 200 residues away were changed, suggesting binding to a discontinuous epitope. Collectively, these studies help to define the structural and functional complexity of antibodies against HCV E2 protein with neutralizing potential.
INTRODUCTION
Hepatitis C virus (HCV) is a blood-borne hepatotropic virus that infects ∼170 million people worldwide. Approximately 70% of infected individuals progress to chronic liver disease, which carries an increased risk of cirrhosis and hepatocellular carcinoma (7). In general, treatment of chronic HCV infection is complicated by resistance due to extensive genetic diversity. HCV has been classified into seven major genotypes, which differ by ∼30% at the nucleotide level (4), and this positive-sense, single-stranded RNA virus has a capacity for rapid evolution of variant viruses during persistent infection. The current treatment, pegylated α2a interferon (IFN-α2a) and ribavirin, has variable side effects and response rates depending on the virus and host genotype (16). No vaccine is currently available, and preclinical development has been hampered by a lack of understanding of which conserved epitopes on the HCV structural proteins should be targeted.
HCV contains an ∼9.6-kb RNA genome that is translated as a single polyprotein and then cleaved by viral and host proteases into structural proteins (core, E1, and E2), p7, and nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (39). Viral attachment and entry are mediated by the envelope glycoproteins, E1 and E2. Four attachment or entry receptors that are required for infection of hepatocytes have been identified, including CD81 (53), scavenger receptor B1 (SR-B1) (56), and the tight-junction proteins claudin 1 (CLDN1) (14) and occludin (OCLN) (54). The importance of E2 binding to the large extracellular loop of CD81 has been established in vitro (13, 18, 28, 50, 53), and interactions between E2 hypervariable region 1 (HVR1) and SR-B1 have been reported (3, 5, 56). The structural basis of binding of E2 to its cognate cell attachment factors, however, is poorly understood, in part because high-resolution structures of the HCV glycoproteins or intact virion have not been solved.
The role of the humoral immune response in controlling HCV infection in patients remains controversial, as patients with persistent infection develop high-titer antibodies that do not appear to clear infection (reviewed in reference 7). Nonetheless, there are emerging data showing that classes of monoclonal (MAbs) and polyclonal antibodies against HCV have protective activity. Binding to CD81 by soluble forms of E2 (sE2, truncated proximal to the transmembrane domain) is inhibited by antibodies that also neutralize infection of pseudotyped HCV particles (HCVpp) derived from the structural proteins of multiple genotypes (1, 45). Perhaps more convincing, experiments in chimpanzees and chimeric mice have shown that passive transfer of anti-E2 antibodies protects against infection (15, 37, 64), and immunization with E1-E2 virus-like particles (VLPs) and E2 glycoprotein in chimpanzees induces protective antibodies (10, 29, 37). Moreover, in a comprehensive study of neutralizing MAbs derived from infected patients, MAbs that bound regions comprised of amino acid residues 396 to 424, 436 to 447, and 523 to 540 on E2 neutralized HCVpp derived from multiple genotypes (37). Thus, anti-E2 antibodies apparently can restrict HCV infection, although the exact steps (attachment, entry, or fusion) in the viral entry process that are inhibited and the corresponding E2 binding epitopes have not been elucidated.
To gain more insight into the molecular and structural basis of anti-E2 antibody neutralization of HCV infection, we generated a panel of 78 mouse MAbs against soluble, recombinant E2 proteins derived from genotypes 1a (strain H77) and 2a (strain J6) HCV strains. These MAbs were analyzed for inhibitory activity against infectious HCV in cell culture, and their mechanisms of action with respect to inhibition of ligand binding on the cell surface were assessed. By combining this functional analysis with a high-throughput yeast surface display mapping strategy, we identified neutralizing MAbs that bound to distinct regions of E2, including MAbs that recognized determinants with discontinuous epitopes and with primary sequences greater than 100 amino acids apart. These experiments suggest that neutralizing MAbs blocking distinct stages of the HCV cell entry process recognize discontinuous epitopes on the E2 protein.
MATERIALS AND METHODS
Cells and viruses.
Huh-7.5 cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Equitech), nonessential amino acids (Gibco), and antibiotics (penicillin G and streptomycin) at 37°C in a 5% CO2 incubator. SF9 cells were cultured in Grace's insect cell medium (Gibco) supplemented with 10% FBS at 28°C. Hi-5 cells were cultured in Ex-cell medium (Gibco) at 27°C. CHO cells were grown in Ham's F-12 medium (Gibco) supplemented with 10% FBS (HyClone) at 37°C.
The genotype 2 J6/JFH1/JC1 HCV chimera, which expresses luciferase (58), was a generous gift from Apath Inc. The HJ3-5 H77/JFH1 chimera, which expresses the core-NS2 segment of the genotype 1a polyprotein within a genotype 2a background, has been described previously (40, 67). The genotype 1a H77C/JFH1(57), genotype 2a J6/JFH1 (38), genotype 3a S52/JFH1 (23), genotype 4a ED43/JFH1 (57), genotype 5a SA13/JFH1 (32), and genotype 6a HK6a/JFH1 (24) infectious HCV recombinants used in cross-neutralization studies have also been described previously. Cell culture compensatory and adaptive mutations for the HJ3-5, S52/JFH1, ED43/JFH1, SA13/JFH1, and HK6a/JFH1 chimeras have been described previously (24, 40, 67).
To generate virus stocks from infectious cDNA clones, plasmids were linearized and RNA transcription was performed using the T7 DNA-dependent RNA polymerase (Megascript Kit; Ambion). Infectious HCV RNA (2 μg) was electroporated as described previously (38), and virus was harvested at 48, 72, and 96 h, sterile filtered (0.2-mm filter; Corning Inc.), and buffered with 10 mM HEPES, pH 7.2 (Mediatech, Inc.). Virus was stored at 4°C for short-term usage, or aliquots were prepared and stored at −80°C. Virus titration on Huh-7.5 cells was performed by 50% tissue culture infective dose (TCID50) assay as previously described (38).
Generation of CHO cells stably expressing HCV cell entry factors.
Human SR-B1 and CD81 genes were expressed in CHO cells via lentivirus transduction in the context of pTRIP, a self-inactivating lentiviral provirus that expresses no HIV proteins but instead employs an internal cytomegalovirus (CMV) promoter to express cloned genes. An intermediate plasmid, called TRIP-GFP-linker, was generated as a backbone into which SR-B1 and CD81 were cloned (all entry factor templates were kindly provided by C. Rice, Rockefeller University, NY). TRIP-GFP-linker was generated by amplifying the green fluorescent protein (GFP) sequence with the forward oligonucleotide 5′-CGC AAA TGG GCG GTA GGC GTG and the reverse oligonucleotide 5′-CTC GAG CTA GTC GAC TTC GAA ACT AGT GCT AGC CCG CGG CTT GTA CAG CTC GTC CAT GCC. The PCR product was digested with restriction enzymes BamHI and XhoI and ligated into the TRIP-GFP plasmid digested with the same enzymes. The human SR-B1 sequence was amplified with forward oligonucleotide 5′-CCG CGG ATG GGC TGC TCC GCC AAA GCG and reverse oligonucleotide 5′-GCT AGC CAG TTT TGC TTC CTG CAG CAC from the previously described TRIP-hu-SR-B1 plasmid (54) to generate TRIP-GFP-hu-SR-B1-linker. The PCR product was digested with SacII and NheI and ligated into similarly digested TRIP-GFP-linker. The human CD81 sequence was amplified from an expression construct, TRIP-GFP-hu-CD81 (56), with forward oligonucleotide 5′-GCT AGC ATG GGA GTG GAG GGC TGC ACC and reverse oligonucleotide 5′-ACT AGT GTA CAC GGA GCT GTT CCG GAT. The PCR product was digested with NheI and SpeI and ligated into similarly digested TRIP-GFP-linker to generate TRIP-GFP-hu-CD81-linker.
Pseudoparticle production was performed as previously described (54) by cotransfection of three plasmids encoding a TRIP provirus containing a transgene, HIV Gag-Pol, and the vesicular stomatitis virus (VSV) G glycoprotein. 293-T cells were seeded at 1.8 × 106 cells/well into a poly-l-lysine (Sigma)-coated six-well plate. Transfection was performed the next day using a total of 1.5 μg of DNA plasmid, with 6 μl of TransIT-LT1 transfection reagent (Mirus). Supernatants were collected 24, 48, and 72 h posttransfection; filtered (0.45-μm pore size); and mixed with 100 μl of 1 M HEPES buffer. All transductions were performed in the presence of 4 μg/ml Polybrene (Sigma). Receptor expression was verified by flow cytometry using the following protocol. Cells were lifted using phosphate-buffered saline (PBS) supplemented with 4 mM EDTA and 10% FBS, washed, and pelleted in a V-bottom plate. Cells (105) were incubated with 20 μg/ml of either mouse anti-hu-CD81 (BD Biosciences) or rabbit anti-hu-SR-B1 (Ab-Cam) for 30 min on ice, washed, and then incubated with goat anti-mouse IgG or goat anti-rabbit IgG secondary antibody conjugated to Alexa Fluor 647 (Molecular Probes). The cells were washed twice, and receptor expression was analyzed on a FACSArray flow cytometer (Becton-Dickinson) using FloJo software (Tree Star).
Cloning, expression, and purification of recombinant HCV E2.
The E2 protein ectodomain of strain H77 (amino acids [aa] 384 to 661) (35) or J6 (aa 385 to 661) was cloned into a baculovirus expression vector (pFastBac derivative) from plasmids containing the structural proteins of H77 (a gift from M. Gale, Jr., University of Washington) or the infectious J6/JFH1/JC1 (58) viral genome (a gift from Apath, Inc.). The baculovirus expression vector adds a honeybee melittin signal peptide at the NH2 terminus and a thrombin-cleavable His6 tag and stop codon at the COOH terminus. Recombinant baculoviruses expressing HCV E2 ectodomains were generated as described previously (31), amplified in SF9 cells, and used for large-scale infection of Hi-5 cells under serum-free conditions. The supernatant was concentrated and buffer exchanged into binding buffer (300 mM sodium citrate, 150 mM sodium chloride, 50 mM sodium phosphate, pH 8.0) using a Centramate tangential-flow concentrator. E2 was purified by sequential nickel affinity and size exclusion chromatography, and monodispersed fractions of monomeric protein were collected and used for subsequent studies.
Generation, purification, and labeling of anti-HCV MAbs.
MAbs were generated by five independent splenocyte-myeloma fusions as described previously (43). Mice were immunized via an intraperitoneal route with sE2 produced from either genotype 1a (H77) or 2a (J6) HCV strains after being complexed with the RIBI Adjuvant System (Corixa Corp.) or complete Freund's adjuvant (Sigma Chemical). Mice were boosted between two and five times with homologous HCV sE2 protein complexed with either incomplete Freund's adjuvant (Sigma), the RIBI Adjuvant System (Corixa), or the Sigma Adjuvant system (Sigma), depending on commercial availability, until adequate titers (>1:2,500 by flow cytometry) were achieved. The mice with the highest serum titers were boosted intravenously with purified sE2 (50 mg) 3 days prior to fusion of splenocytes with P3X63Ag8.53 myeloma cells (12). Hybridomas producing anti-HCV E2 antibodies were identified after binding to Saccharomyces cerevisiae yeast cells expressing sE2 on their surfaces by flow cytometry, subcloned by limiting dilution, and isotyped by enzyme-linked immunosorbent assay (ELISA). For large-scale production, MAbs were generated from ascites or adapted to growth in Hybridoma Serum Free Medium (Gibco) and purified using protein A or G affinity chromatography (Pierce). In some experiments, MAbs were labeled with Alexa Fluor 647 (Molecular Probes) or NHS-FITC (fluorescein isothiocyanate) (Pierce) MAb-labeling kits according to the manufacturer's instructions.
Virus neutralization assays.
Neutralization of HCV infection by viruses containing genotype 1a structural proteins (H77/JFH1) was assessed by a focus-forming unit (FFU) assay. Serial dilutions of HCV-specific MAb, control MAb (WNV E16) (43), anti-human CD81 (clone JS81; BD Biosciences), or anti-human SR-B1 (clone 396; Ab-Cam) were preincubated with 2.4 × 102 FFU of virus for 1 h at 37°C. The virus-MAb mixtures were added to Huh-7.5 cells (1.2 × 104 cells per well) in a 48-well tissue culture plate precoated with poly-l-lysine (Sigma). After 72 h, the cells were fixed with methanol (0°C) and incubated sequentially with a mouse anti-NS5A (APA-1; 40 ng/ml; a generous gift from Apath, Inc.) (38) and secondary goat anti-mouse horseradish peroxidase (HRP) diluted 1:3,000 (Sigma). FFU were visualized using the True Blue Peroxidase Reagent (KPL) and quantitated using an S5 Biospot Macroanalyzer (Cellular Technologies Ltd). The 50% effective concentration (EC50) values were determined using nonlinear regression analysis.
Neutralization of the genotype 2a (J6/JFH1/JC1) HCV was assessed by luciferase assay. Serial dilutions of HCV-specific or control MAbs were preincubated with the J6/JFH1/JC1 virus (102 FFU), which expresses luciferase, for 1 h at 37°C and then added to Huh-7.5 cells (104 cells per well) in a 96-well black flat-bottom polystyrene-treated microplate (Corning). After 48 h, the cells were lysed and luciferase was detected using the Renilla Luciferase Assay System (Promega) according to the manufacturer's instructions. EC50 values were determined using nonlinear regression analysis.
Neutralization of chimeric viruses with genotype 1a- to 6a-specific core-NS2 sequences was assessed by FFU assay as previously described (55). Briefly, 50 to 400 TCID50 of HCV was incubated for 1 h at 37°C with MAb H77.39 or an isotype control and then incubated with cells for 3 h. After 48 h, the cells were immunostained for NS5A as previously described (23). FFU counting was automated using an ImmunoSpot Series 5 UV Analyzer (22). The percent neutralization was calculated by relating FFU counts to the mean of six replicates incubated in the absence of antibody (virus only). Neutralization data were analyzed as variable-slope dose-response curves using GraphPad Prism 4.0, and EC50 values were interpolated by the software.
Pre- and post-virus attachment assays.
To assess the abilities of MAbs to inhibit H77/JFH1 virus at pre- and postattachment steps, FFU assays were modified as follows. For the postattachment assay, prechilled cells were incubated with 4.8 × 102 FFU of virus for 1 h at 4°C. Cells were washed thrice with cold DMEM to remove unbound virus, MAbs (diluted to 50 μg/ml in medium and prewarmed at 37°C) were added, and the cells were shifted to 37°C. After 1 h, a 1:1 MEM-methylcellulose overlay with 4% FBS was added to prevent viral spread. For the preattachment assay, 4.8 × 102 FFU of virus was preincubated with 50 μg/ml of MAb for 1 h at 37°C and then added to preseeded Huh-7.5 cells.
To assess the abilities of MAbs to inhibit J6/JFH1/JC1 at pre- and postattachment steps, the luciferase assay was modified in the following manner. Forty-eight-well tissue culture plates were precoated with poly-l-lysine (Sigma) and seeded with 1.2 × 104 cells per well. For the postattachment assay, 4.8 × 102 FFU of virus was added to prechilled cells and “spinoculated” for 45 min at 400 × g at 4°C, followed by a 15-min incubation at 4°C. The cells were washed, and prewarmed MAbs and methylcellulose were added as described above. For the preattachment assay, 4.8 × 102 FFU of virus was preincubated with 50 μg/ml of MAb for 1 h at 37°C and then added to preseeded Huh-7.5 cells. Cells from both the pre- and postattachment assays were lysed after 48 h and transferred to a 96-well black-bottom plate, and luciferase was detected using the Renilla Luciferase Assay System (Promega) according to the manufacturer's instructions.
Cross-reactivity and mapping analysis of MAbs using yeast surface display.
To assess MAb cross-reactivity with other HCV genotypes, the ectodomain of the E2 genes from genotype 1a (H77; amino acids 384 to 660), genotype 2a (J6; amino acids 385 to 664), genotype 3a (UKN 3A13.6; amino acids 385 to 667), genotype 4a (UKN 4.21.16; amino acids 392 to 663), genotype 5a (SA13 NIH; amino acids 384 to 663), and genotype 6a (UKN 6; amino acids 385 to 668) was amplified by PCR with BamHI and XhoI sites for cloning added at the 5′ and 3′ ends, respectively. The PCR products were cloned as downstream fusion proteins to the Aga2 gene in the pYD1 vector (Invitrogen) for expression on the surface of yeast. To determine the relative binding regions on the E2s of specific MAbs, COOH-terminal truncation constructs, based on previous studies (41), were generated for genotypes 1a and 2a corresponding to regions I (amino acids 384 to 520 in genotype 1a and 384 to 518 in genotype 2a) or I and II (amino acids 384 to 605 in genotype 1a and 384 to 603 in genotype 2a) and displayed on the surface of yeast.
Expression constructs were transformed into Saccharomyces cerevisiae strain EBY100 (17) using the S.c. EasyComp transformation Kit (Invitrogen). Individual yeast colonies were grown to logarithmic phase at 30°C in tryptophan-free yeast selection medium containing 2% glucose. Protein expression was induced by cultivating yeast for an additional 48 to 72 h in tryptophan-free medium supplemented with 2% galactose at 20°C. Yeast cells were washed with PBS containing 1 mg/ml bovine serum albumin (PBS-BSA) and incubated with 40 μl of MAb (neat supernatant or 20 μg/ml purified and diluted in PBS) for 30 min on ice. Yeasts were washed in PBS-BSA, incubated with goat anti-mouse IgG secondary antibody conjugated to Alexa Fluor 647 (Molecular Probes) for 30 min on ice, washed, and analyzed on a FACSArray flow cytometer (Becton-Dickinson) using FloJo software (Tree Star).
Random mutant libraries of E2 were generated from genotype 1a (strain H77) and genotype 2a (strain J6) genes by error-prone PCR using a GeneMorph II random-mutagenesis kit (Stratagene). The libraries were ligated into the pYD1 vector and transformed into XL2-Blue ultracompetent cells (Stratagene) with ∼5.7 × 105 and 5.5 × 105 transformants for genotypes 1a and 2a, respectively. Screening of the libraries for loss-of-binding variants was performed as described previously (43, 59). In brief, yeasts expressing E2 variants that lost specific binding to individual MAbs were sorted using two-color flow cytometry. To eliminate mutations that abolished surface expression of E2, yeasts were stained sequentially with the Alexa Fluor 647-conjugated individual MAb, followed by a FITC-conjugated oligoclonal pool of the cross-reactive MAbs (J6.1, J6.2, J6.16, J6.39, J6.51, and J6.101 for the genotype 1a library and J6.2, J6.14, J6.15, J6.39, J6.51, and J6.99 for the genotype 2a library) on ice for 30 min. Yeasts that stained positively for the oligoclonal pool but negatively for the MAb of interest were collected, cultivated, and iteratively sorted. In some cases, sorting was performed using MACS LS magnetic columns (Miltenyi Biotech). In brief, ∼107 yeast cells were pelleted and resuspended in MACS buffer (PBS plus 0.5% BSA plus 2 mM EDTA) containing a 1:50 dilution of a FITC-labeled MAb of interest for 30 min, washed, and then incubated with 10 μl of anti-FITC microbeads (Miltenyi Biotech) on ice for 15 min. The yeasts were washed and passed over a MACS LS column, and the flowthrough was collected. After four or five rounds, the yeasts were plated, and individual colonies were tested for binding to individual MAbs by flow cytometry. For clones that lost binding to the desired MAb of interest, the plasmid was recovered using a Zymoprep yeast minipreop kit (Zymo Research), transformed into XL1-Blue competent Escherichia coli, purified using a QIAprep spin miniprep kit (Qiagen), and sequenced. In cases where more than one mutation was detected, site-specific mutagenesis using the Quick Change II Mutagenesis kit (Stratagene) was used to generate individual mutations within the E2 protein to define the mutant of interest.
Inhibition of CD81 and SR-B1 binding.
To assess the abilities of neutralizing MAbs to inhibit binding of sE2 to CD81 and SR-B1, 50 μg/ml of purified MAb was preincubated with 20 μg/ml H77 E2 or J6 sE2 for 30 min at 37°C. CHO cells expressing HCV receptors were detached with PBS supplemented with 4 mM EDTA and 10% FBS and washed three times in medium. Cells (105) were pelleted in a V-bottom plate, resuspended with MAb-protein mixture, and incubated on ice for 30 min. The cells were washed and then incubated with a pool of Alexa Fluor 647-labeled anti-E2 MAbs (J6.1, J6.2, J6.39, J6.51, H77.30, and H77.34 for the detection of H77 E2 and J6.2, J6.39, J6.51, J6.60, and J6.101 for the detection of J6 E2) for 20 min on ice. The cells were washed twice, and sE2 binding was analyzed on a FACSArray flow cytometer (Becton-Dickinson) using FloJo software (Tree Star).
Statistical analysis.
All data were analyzed using Graphpad Prism software (version 4.0). For neutralization assays and receptor-binding assays, an unpaired t test was used to determine statistical significance.
RESULTS
MAb generation.
Previous studies have demonstrated that HCV-specific monoclonal and polyclonal antibodies, particularly those that recognize the E2 protein, can control HCV infection in vitro and in vivo (10, 15, 33, 37, 47, 62). However, only a few of these antibodies have been characterized for the ability to inhibit at different stages of HCV infection or mapped to epitopes at the amino acid level. To better define the structural basis of antibody neutralization of HCV, we generated a new panel of anti-HCV MAbs by immunizing BALB/c mice with soluble, recombinant E2 protein that was expressed in insect cells and derived from either genotype 1a (strain H77; amino acids 384 to 664) or genotype 2a (strain J6; amino acids 385 to 664) viruses. After five independent splenocyte-myeloma cell fusions, we subcloned 37 MAbs from genotype 1a-immunized mice and 41 MAbs from genotype 2a-immunized mice, all with reactivity against the E2 structural glycoprotein of HCV (see Table S1 in the supplemental material).
Neutralizing activities of anti-E2 MAbs.
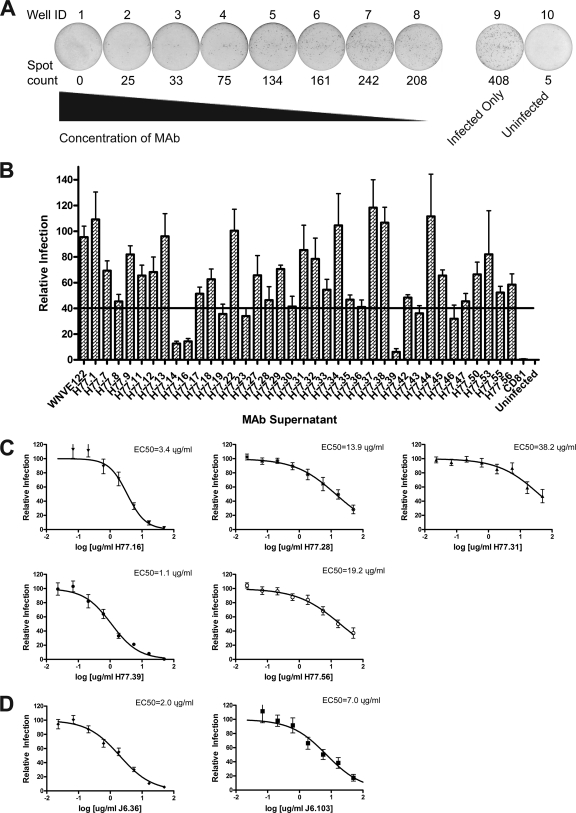
To study the inhibitory capacity of genotype 1a MAbs in cell culture, we utilized an H77-JFH1 chimeric infectious virus that contains genotype 1a core-NS2 sequence in the JFH1 background, with a compensatory Q221L mutation in NS3 (pHJ3-5) (40, 67). For high-throughput screening, we adapted an FFU assay with Huh-7.5 cells so that infectious foci were scored objectively on an enzyme-linked immunospot (ELISPOT) reader, and the reduction in the number of FFU was assessed after preincubation of virus with individual MAbs (Fig. 1A) . We performed a single-endpoint focus reduction neutralization test (FRNT) using neat antibody supernatant (∼10 μg/ml) and identified 9 MAbs that inhibited infection by 40% or greater (Fig. 1B). The candidate neutralizing MAbs identified above and other selected antibodies were purified by immunoaffinity chromatography and tested for inhibitory activity with a more complete dose-response curve (Fig. 1C). We demonstrated that five of these MAbs (H77.16, H77.28, H77.31, H77.39, and H77.56) had reproducible neutralizing activity and determined the concentration of MAb at which 50% of foci were inhibited (the EC50 value) (Fig. 1C). Of these MAbs, H77.16 and H77.39 showed the greatest inhibitory activity, with EC50 values of ∼3.4 μg/ml and ∼1.1 μg/ml, respectively.
Fig. 1.
Identification of neutralizing anti-E2 antibodies against HCV. (A) Examples of MAb neutralization as judged by a reduction in the number of FFU, using the Biospot Macroanalyzer. Spot counts are shown below each well, and well numbers are shown above. Wells 1 through 8 represent decreasing (3-fold) concentrations of the neutralizing MAb H77.39 (starting concentration, 50 μg/ml). Well 9 shows infection in the absence of MAb, and well 10 is an uninfected well. The data are representative of three independent experiments performed in duplicate. (B) MAb supernatant was mixed with the H77-JFH1 chimeric HCV for 1 h at 37°C, and Huh-7.5 cells were infected. Three days later, neutralization was determined by FFU assay. MAb supernatants that decreased the number of FFU to 40% or less (below the solid black line) of the negative-control MAb (anti-WNV E122), as well as additional selected MAbs, were purified for testing in full dose-response analysis. The data are pooled from three independent experiments performed in duplicate. (C) Serial dilutions of genotype 1a-specific purified MAbs were mixed with H77-JFH1 chimeric virus, and neutralization was assessed. Efficient neutralization was observed for five (H77.16, H77.28, H77.31, H77.39, and H77.56) genotype 1a-specific MAbs, but not for the negative-control MAb (data not shown). EC50 values were calculated after nonlinear regression analysis. The data are pooled from at least three independent experiments performed in duplicate. (D) Increasing concentrations of purified genotype 2a-specific MAbs (J6.36 and J6.103) were mixed with J6-JFH1-JC1-luciferase-expressing virus. At 48 h, neutralization was assessed in Huh-7.5 cells by monitoring luciferase expression. EC50 values were calculated after nonlinear regression analysis. The data are pooled from at least three independent experiments performed in duplicate. All error bars represent the standard errors of the mean.
To evaluate the neutralizing activities of MAbs generated against E2 derived from the genotype 2a HCV strain, we utilized a genotype 2a J6/JFH1/JC1 infectious chimera of HCV that contains a Renilla luciferase reporter gene inserted immediately upstream of an NS2A cleavage site (58). All 41 MAbs that bound the genotype 2a E2 protein were purified and assessed for inhibitory activity over a broad range of concentrations to determine the concentration of antibody that reduced luciferase expression by 50% (EC50 value) (data not shown). We identified two antibodies, J6.36 and J6.103, that efficiently neutralized infection, (Fig. 1D), with J6.36 having an EC50 value below 2 μg/ml. Notably, no significant difference in the inhibitory potency of a given neutralizing MAb was observed when the luciferase and FRNT assays were directly compared (data not shown).
Cross-reactivity of anti-E2 MAbs.
HCV is comprised of six epidemiologically important genotypes with ∼70% nucleotide identity (4). A better understanding of the specific epitopes that are conserved and recognized by inhibitory antibodies may facilitate the design of future vaccines. To begin to address this, we assessed how genotype variation affected MAb reactivity using recombinant E2 proteins displayed on yeast (1a, H77; 2a, J6; 3a, UKN 3a; 4a, UKN 4a; 5a, SA13; and 6a, UKN 6) and neutralization capacity with chimeric HCV strains (1a, H77; 2a, J6; 3a, S52; 4a, ED43; 5a, SA13; and 6a, HK6a) containing the nonstructural proteins (NS3 to NS5B) of the genotype 2a JFH1 strain and structural proteins, p7, and NS2 from strains representative of HCV genotypes 1 to 6.
(i) Binding to different HCV genotypes.
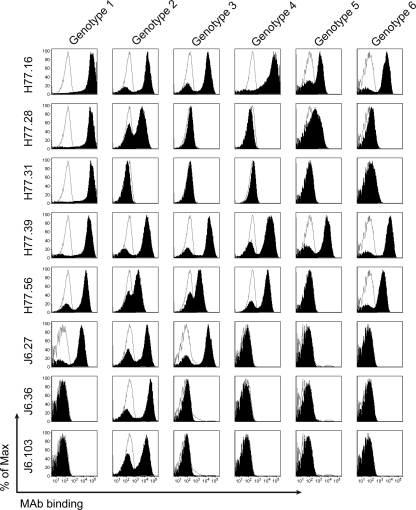
The ectodomain of E2 from individual strains corresponding to HCV genotypes 1 to 6 was expressed on the surface of yeast, incubated with MAbs, and analyzed for binding by flow cytometry. Three of the eight neutralizing MAbs were broadly cross-reactive and recognized all five (H77.16 and H77.39) or four of the five (H77.56) heterologous genotypes (Fig. 2 and Table 1). Three of the neutralizing MAbs (H77.31, J6.36, and J6.103) bound to yeast expressing only the homologous E2.
Fig. 2.
Identification of MAbs that bind heterologous HCV genotypes using yeast display of E2 protein. The E2 ectodomain gene from six strains corresponding to HCV genotypes 1 to 6 was cloned into the PYD1 vector and expressed on the surface of yeast (see Materials and Methods). Yeasts expressing HCV E2 were incubated with MAb supernatants, and binding was assessed by flow cytometry. Representative histograms from all neutralizing MAbs (H77.16, H77.28, H77.31, H77.39, H77.56, J6.27, J6.36, and J6.103; solid black histograms) and negative-control MAb (WNV E16; unfilled gray histograms) are depicted. The data are representative of at least three independent experiments.
Table 1.
Binding of MAbs to HCV E2 from different HCV genotypes
| MAb | Binding to genotypea: |
|||||
|---|---|---|---|---|---|---|
| 1a(H77) | 2a(J6) | 3a(UKN 3) | 4a(UKN4) | 5a(SA13) | 6a(UKN6) | |
| J6.1 | +++ | +++ | +++ | +++ | +++ | +++ |
| J6.2 | + | +++ | +++ | +++ | − | − |
| J6.6 | + | +++ | + | − | − | − |
| J6.7 | +++ | +++ | +++ | + | +++ | +++ |
| J6.8 | + | +++ | − | − | − | − |
| J6.9 | +++ | +++ | − | − | +++ | − |
| J6.12 | +++ | +++ | +++ | +++ | − | − |
| J6.13 | − | +++ | − | − | − | − |
| J6.14 | +++ | +++ | +++ | + | + | +++ |
| J6.15 | − | +++ | +++ | +++ | +++ | +++ |
| J6.16 | +++ | +++ | +++ | − | +++ | − |
| J6.21 | +++ | +++ | − | − | − | − |
| J6.23 | + | +++ | − | − | − | − |
| J6.25 | − | +++ | − | − | − | − |
| J6.27 | +++ | +++ | +++ | − | − | − |
| J6.30 | +++ | +++ | +++ | +++ | +++ | +++ |
| J6.33 | +++ | +++ | +++ | +++ | +++ | +++ |
| J6.34 | +++ | +++ | + | + | +++ | + |
| J6.36 | − | +++ | − | − | − | − |
| J6.39 | +++ | +++ | +++ | − | − | − |
| J6.40 | − | +++ | + | − | − | − |
| J6.42 | + | +++ | +++ | +++ | − | − |
| J6.48 | − | +++ | − | − | − | − |
| J6.49 | − | +++ | − | − | − | − |
| J6.51 | + | +++ | + | +++ | − | − |
| J6.56 | +++ | +++ | +++ | + | +++ | +++ |
| J6.58 | +++ | +++ | +++ | +/− | +++ | − |
| J6.60 | − | +++ | − | − | − | +++ |
| J6.62 | +++ | +++ | +++ | + | +++ | + |
| J6.67 | +++ | +++ | + | − | +++ | − |
| J6.68 | + | +++ | − | − | − | − |
| J6.75 | +++ | +++ | +++ | +++ | +++ | +++ |
| J6.76 | − | +++ | − | − | − | − |
| J6.81 | +++ | +++ | +++ | − | +++ | − |
| J6.85 | − | +++ | +++ | +++ | +++ | +++ |
| J6.86 | + | +++ | +++ | +++ | − | − |
| J6.91 | + | +++ | − | − | − | − |
| J6.98 | − | +++ | + | +++ | − | − |
| J6.99 | − | +++ | − | − | − | + |
| J6.101 | +++ | +++ | + | +++ | − | +++ |
| J6.103 | − | +++ | − | − | − | − |
| H77.1 | +++ | − | − | − | − | − |
| H77.7 | +++ | +++ | +++ | +++ | +++ | +++ |
| H77.8 | +++ | − | − | − | − | − |
| H77.9 | +++ | − | − | − | − | − |
| H77.11 | +++ | − | − | − | − | − |
| H77.12 | +++ | +++ | − | − | +++ | − |
| H77.13 | +++ | − | − | − | − | − |
| H77.14 | +++ | − | − | − | − | − |
| H77.16 | +++ | +++ | +++ | +++ | +++ | +++ |
| H77.17 | +++ | − | − | − | − | − |
| H77.18 | +++ | − | − | − | + | − |
| H77.19 | +++ | − | − | − | − | − |
| H77.22 | +++ | − | − | − | − | − |
| H77.23 | +++ | − | − | − | − | − |
| H77.27 | +++ | +++ | − | − | + | − |
| H77.28 | +++ | +++ | − | − | + | − |
| H77.29 | +++ | +++ | − | − | + | − |
| H77.30 | +++ | − | − | − | − | − |
| H77.31 | +++ | − | − | − | − | − |
| H77.32 | +++ | + | +++ | − | +++ | − |
| H77.33 | +++ | − | − | +++ | − | − |
| H77.34 | +++ | − | − | − | − | − |
| H77.35 | +++ | − | − | − | − | − |
| H77.36 | +++ | +++ | +++ | +++ | +++ | +++ |
| H77.37 | +++ | − | − | − | +++ | − |
| H77.38 | +++ | − | − | − | − | − |
| H77.39 | +++ | +++ | +++ | +++ | +++ | +++ |
| H77.42 | +++ | − | − | − | + | − |
| H77.43 | +++ | − | − | − | − | − |
| H77.44 | +++ | − | − | − | +++ | − |
| H77.45 | +++ | − | − | − | − | − |
| H77.46 | +++ | − | − | − | +++ | − |
| H77.47 | +++ | +++ | − | − | − | − |
| H77.50 | +++ | − | − | − | − | − |
| H77.53 | +++ | − | − | − | +++ | − |
| H77.55 | +++ | − | − | − | +++ | − |
| H77.56 | +++ | +++ | +++ | +++ | − | +++ |
+++, strong binding (40 to 100%) to yeast expressing E2; +, weak binding (15 to 40%) to yeast expressing E2; −, no appreciable binding detected. The data are a summary of 3 to 5 independent experiments.
(ii) Cross-neutralizing potential of MAbs.
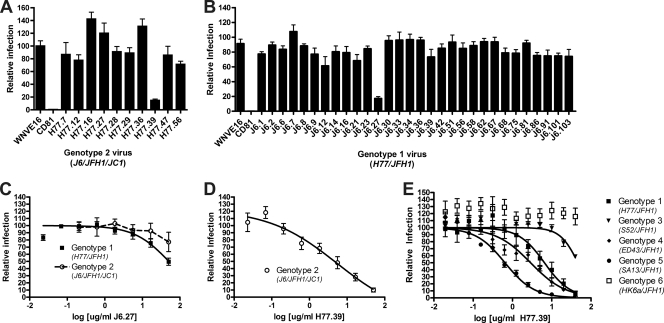
As the MAb binding capacity to recombinant viral structural proteins does not always directly correlate with the neutralizing potential (8), we evaluated the inhibitory activities of several of the cross-reactive MAbs against HCV of other genotypes. Initially, single-endpoint focus reduction assays were performed with high concentrations (50 μg/ml) of purified MAbs generated against genotype 1a or genotype 2a that cross-reacted with genotype 2a or genotype 1a E2, respectively (Fig. 3A and B). Of the cross-reactive MAbs generated against genotype 1a E2, only H77.39 neutralized the genotype 2a virus. Of the cross-reactive MAbs generated against genotype 2a E2, only J6.27 inhibited genotype 1a HCV infection (Fig. 3B and C). This was surprising, because J6.27 lacked neutralizing activity against the genotype 2a strain against which it was generated (Fig. 3C); this pattern of enhanced neutralizing activity of cross-reactive antibodies against the heterologous virus has also been observed with MAbs against distantly related flaviviruses (2, 44). H77.39 inhibited the genotype 2a virus with an EC50 of ∼5 μg/ml (Fig. 3D), which was comparable to that observed with the genotype 1a virus (Fig. 1C). We subsequently tested whether H77.39 neutralized infection of a panel of chimeric viruses that expressed structural proteins from the remaining heterologous HCV genotypes. H77.39 dose-dependently inhibited HCV infection of genotypes 3a, 4a, and 5a but showed reduced activity against a virus containing structural proteins of genotype 6a (Fig. 3E).
Fig. 3.
MAb neutralization of heterologous HCV genotypes. (A and B) MAbs that were generated against genotype 1a (A) or genotype 2a (B) E2 proteins were tested for the ability to neutralize infection by virus from the heterologous genotype. Purified J6 or H77 MAbs (50 μg/ml) were preincubated at 37°C with H77-JFH1 (genotype 1a) or J6-JFH1-JC1 (genotype 2a) virus, respectively, and neutralization was assessed as described in the legend to Fig. 1. (C to E) EC50 analysis was performed with J6.27 MAb and H77-JFH1 virus (▪) or J6-JFH1-JC1 virus (○) (C), H77.39 MAb and J6-JFH1-JC1 virus (○) (D), or H77.39 MAb and H77C/JFH1 (▪), S52/JFH1 (▾), ED43/JFH1 (⧫), SA13/JFH1 (•), and HK6a/JFH1 (□) chimeric viruses (E). The graphs represent pooled data from at least three independent experiments performed in duplicate (A to D) or two independent experiments performed in triplicate (E), and the error bars represent the standard errors of the mean.
Mechanism of MAb neutralization.
Antibody neutralization may involve different stages of viral infection, including attachment, internalization, and fusion (51). To begin to understand how our inhibitory MAbs blocked infection, we performed pre- and postattachment neutralization assays and studies of binding to the CD81 and SR-B1 receptors.
(i) Pre- and postattachment assays.
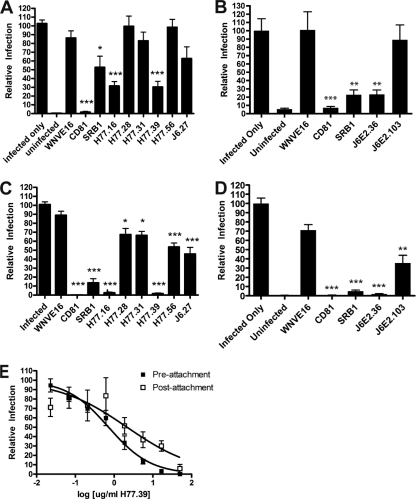
To identify the stage of infection at which MAbs neutralize infection, we adapted a pre- and postattachment inhibition assay originally developed for flaviviruses (42, 60, 66). Purified anti-E2 MAb was incubated with virus before or after attachment at 4°C to Huh-7.5 cells, and infection was measured by a single-endpoint focus reduction assay. Of the nine neutralizing MAbs tested, three (H77.16, H77.39, and J6.36) significantly reduced infection compared to the negative-control MAb (WNV E16) when added after viral absorption to a cell monolayer, suggesting blockade of a postattachment step (Fig. 4A to D). Interestingly, both anti-CD81 and anti-SR-B1 MAbs also inhibited infection after viral adsorption, confirming previous results in Huh-7.5 cells which suggested that HCV binds to CD81 and SR-B1 after initial attachment (6, 25). Inhibition of infection at a postattachment step by H77.39 was confirmed by performing more complete dose-response curve analyses (Fig. 4E).
Fig. 4.
Pre- or postattachment neutralization. (A to D) To determine whether MAbs neutralize HCV infection at a postattachment step, Huh-7.5 cells were prechilled at 4°C, and 480 FFU of genotype 1a (H77-JFH1) (A) or genotype 2a (J6-JFH1-JC1) (B) virus was added to each well for 1 h at 4°C. After three washes with 4°C DMEM, saturating concentrations of MAbs (50 μg/ml) were added for 1 h at 37°C, and the neutralization assay was completed. In comparison, a standard preincubation neutralization test was performed at 37°C, in which genotype 1a virus (C) or genotype 2a virus (D) and MAb were preincubated at 37°C prior to addition to cells. The data shown are the averages of three independent experiments, with the error bars representing standard errors of the mean. Statistically significant differences in neutralization are compared to infection in the presence of a negative-control MAb (WNV E16): *, P < 0.05; **, P < 0.01; and ***, P < 0.001. (E) To confirm the ability of H77.39 to neutralize infection at both pre- and postattachment steps, a dose-response curve was performed under both pre- and postattachment conditions, as described above, using H77/JFH1 virus. The graphs represent pooled data from at least three independent experiments performed in duplicate, and the error bars represent the standard errors of the mean.
(ii) MAb inhibition of sE2 binding to receptors.
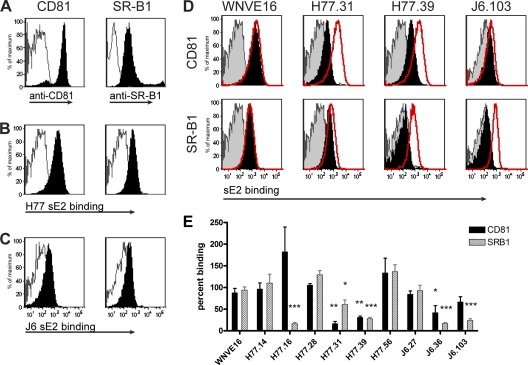
Given that anti-CD81, anti-SR-B1, and several anti-E2 MAbs all blocked after HCV attached to Huh-7.5 cells, it was difficult to discern whether some antibodies blocked binding to individual HCV receptors. To address this, we developed an assay for sE2 binding to CHO cells that ectopically expressed human CD81 or SR-B1. CHO cells were transduced with a lentiviral vector encoding CD81 or SR-B1 fused to GFP. Surface staining of intact cells with anti-CD81 and anti-SR-B1 MAbs confirmed high-level receptor expression (Fig. 5A), as did analysis of cells for GFP fluorescence (data not shown). Binding of genotype 1a (Fig. 5B) and genotype 2a (Fig. 5C) sE2 to CD81- and SR-B1-expressing CHO cells (solid histograms), but not control CHO cells (outlined histograms), was confirmed by flow cytometry. To determine whether sE2-CD81/SR-B1 receptor interactions could be disrupted by anti-E2 MAbs, neutralizing or control (anti-WNV E16) MAbs were preincubated with sE2 and added to wells containing CHO cells expressing CD81 or SR-B1, and loss of binding was assessed by flow cytometry (Fig. 5D). The neutralizing MAb H77.39 significantly blocked (>70%; P < 0.01) sE2 binding to both CD81 and SR-B1. In comparison, H77.31 also reduced binding of sE2 to both receptors, although inhibition of SR-B1 binding was more modest (∼40%; P = 0.04) than that seen with CD81 (>80%; P = 0.003). Conversely, J6.36 efficiently inhibited sE2–SR-B1 binding (>80%; P = 0.0002) yet only modestly (∼50%; P < 0.05) diminished sE2-CD81 binding. H77.16 and J6.103 blocked sE2 binding to only a single receptor, with both efficiently reducing (>75%; P = 0.0005) binding to SR-B1 (Fig. 5D). Three neutralizing MAbs, H77.28, H77.56, and J6.27, did not significantly inhibit sE2 attachment to either CD81 or SR-B1, suggesting that they may block an alternate attachment or entry step (Fig. 5E).
Fig. 5.
Inhibition of sE2 binding to CD81 and SR-B1 by neutralizing MAbs. (A) Verification of ectopic CD81 and SR-B1 receptor expression on CHO cells. CHO-CD81 or CHO–SR-B1 cells were incubated with either mouse anti-hCD81 or rabbit-anti-hSR-B1 (black histograms) or an irrelevant MAb (unfilled gray histograms) for 30 min on ice. The cells were washed, incubated with the appropriate secondary antibodies, and processed by flow cytometry. (B and C) Binding of genotype 1a (H77) E2 (B) or genotype 2a (J6) E2 (C) to CHO-CD81 and CHO–SR-B1, but not wild-type (WT) CHO cells. CHO-CD81 or CHO–SR-B1 (solid black histograms) or WT CHO (unfilled gray histograms) cells were incubated with sE2, and binding was assayed by flow cytometry. The data are representative of at least three independent experiments. (D) Assessment of inhibition of sE2 binding to CHO-CD81 or CHO–SR-B1 cells by neutralizing MAbs. sE2 was preincubated with neutralizing MAbs and added to CHO cells, and binding was detected by flow cytometry. Examples of MAbs that inhibit sE2 binding to CD81 preferentially (H77.31), to both CD81 and SR-B1 (H77.39), or only to SR-B1 (J6.103), as well as a negative-control MAb (WNV E16), are shown. The histograms are representative of three individual experiments. The solid black histograms represent sE2 binding in the presence of MAb, the red histograms represent sE2 binding in the absence of MAb, and the shaded gray histograms represent sE2 binding to CHO WT cells. (E) Graphical representation of sE2 binding to CHO-CD81 and CHO–SR-B1 cells in the presence of neutralizing MAbs. The values were determined by dividing the fluorescence quotient (mean fluorescence intensity × percent positive cells) for E2 binding in the presence of a neutralizing MAb by the fluorescence quotient of sE2 binding to either CHO-CD81 or CHO–SR-B1 cells alone. The asterisks represent statistically significant differences in sE2 binding compared to the negative-control MAb, WNV E16: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. The error bars represent the standard errors of the mean. The data are pooled from three independent experiments.
Epitope localization of MAbs.
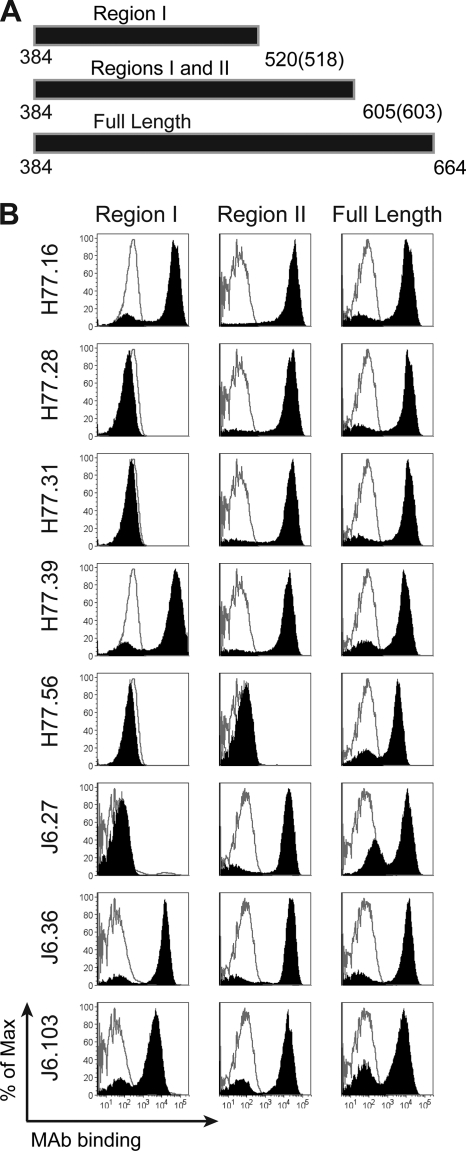
To correlate the functions of the anti-E2 MAbs with the structure of the HCV E2 protein, we localized their epitopes using a previously validated yeast surface display mapping assay (43, 44, 61). Initially, COOH-terminal truncated versions of E2, based on those described previously (41), were displayed on the surface of yeast, and MAbs were tested for immunoreactivity by flow cytometry (Fig. 6) (see Table S1 in the supplemental material). Neutralizing MAbs showed different requirements for binding. H77.16, H77.39, J6.36, and J6.103 bound to a region bracketed by amino acids 384 to 520 of genotype 1a and 384 to 518 of genotype 2a E2 (designated “region I”), whereas H77.28, H77.31, and J6.27 required amino acids 521 to 605 of genotype 1a or 519 to 603 of genotype 2a E2 (designated “region II”) for binding. In contrast, MAb H77.56 required the full E2 ectodomain (amino acids 1 to 664), suggesting that it interacts with amino acids 606 to 664 alone or requires a conformation of E2 that this region stabilizes. MAbs that neutralized efficiently at a postattachment step, H77.16, H77.39, and J6.36, all bound to region I of E2.
Fig. 6.
Mapping of anti-E2 antibodies using COOH-terminal truncation mutants. (A) Scheme of E2 truncations used for mapping. cDNA containing region I (aa 384 to 520 and aa 384 to 518 in E2 of genotypes 1a and 2a, respectively) or I and II (aa 384 to 605 and 384 to 603 in E2 of genotypes 1a and 2a, respectively) and the full-length ectodomain (aa 384 to 664) were displayed on the surface of yeast. (B) MAb supernatants were incubated with yeast and assessed for binding by flow cytometry. Neutralizing MAbs binding to regions I (H77.16, H77.39, J6.36, and J6.103) and II (H77.28, H77.31, and J6.27) and the full-length E2 ectodomain (H77.56) are shown. The solid black histograms depict binding of HCV-specific MAbs, and the gray unfilled histograms represent binding of a negative-control MAb (WNV E16). The histograms are representative of three independent experiments.
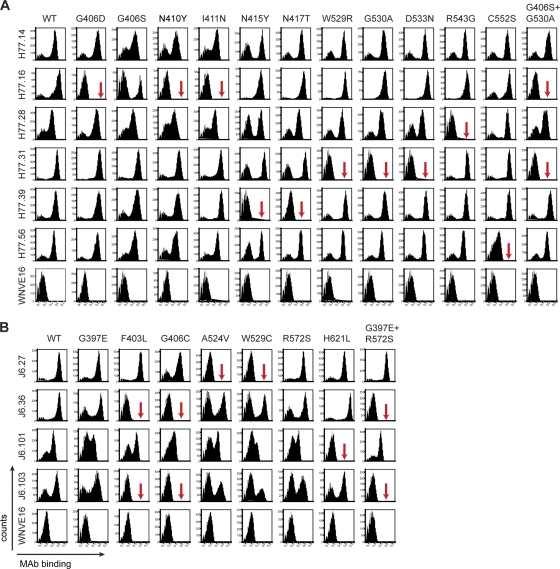
To localize MAb epitopes more clearly, we used error-prone PCR mutagenesis and yeast surface display to create a library of H77 and J6 E2 variants to define individual amino acid binding residues of neutralizing and nonneutralizing MAbs. Yeasts that lost expression of individual MAb epitopes were sorted by flow cytometry, and plasmids were recovered, sequenced, and tested for reactivity against a select panel of MAbs (Fig. 7 and Tables 2 and 3).
Fig. 7.
Epitope localization of anti-HCV MAbs. Binding of neutralizing MAbs to yeast expressing E2 protein variants. (A) Flow cytometry histograms of wild-type and loss-of-binding genotype 1a E2 variants (G406D, G406S, N410Y, I411N, N415Y, N417T, W529R, G530A, D533N, R543G, C552S, and G406S plus G530A). Representative histograms are shown for the MAbs H77.14, H77.16, H77.28, H77.31, H77.39, H77.56, and WNV E16 (negative control) with WT H77 E2 and each of the variants. The data shown are representative of three independent experiments. The red arrows indicate >80% loss of binding of a specific MAb for a given variant. (B) Flow cytometry histograms of wild-type and loss-of-function genotype 2a E2 variants (G397E, F403L, G406C, A524V, W529C, R572S, H621L, and G397E plus R572S) with individual neutralizing MAbs. Representative histograms are shown for the MAbs J6.27, J6.36, J6.101, J6.103, and WNV E16 (negative control) with the wild-type E2 and each of the variants. The data shown are representative of three independent experiments. The arrows indicate >80% loss of binding of a specific MAb for a given variant.
Table 2.
Summary of MAb binding to genotype 1 mutants expressed on the surface of yeast
| MAb | Bindinga |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G406D | G406S | N410Y | I411N | N415Y | T416I | N417T | G470A | W529R | G530A | G530D | D533N | R543G | C552S | G406S + G530A | G406D + G530D | |
| H77.14 | 100 | 100 | 100 | 100 | 67 | 100 | 58 | <1 | 100 | 91 | 100 | 100 | 66 | 100 | 100 | 100 |
| H77.16 | <1 | 58 | <1 | <1 | 76 | 97 | 100 | 100 | 100 | 97 | 100 | 100 | 100 | 100 | <1 | <1 |
| H77.23 | 100 | 100 | 100 | 100 | 75 | 100 | 61 | <1 | 100 | 100 | 100 | 100 | 89 | 100 | 100 | 100 |
| H77.27 | 77 | 91 | 100 | 97 | 64 | 100 | 80 | 81 | <1 | <1 | 11 | 27 | 84 | 100 | <1 | <1 |
| H77.28 | 96 | 100 | 100 | 74 | 38 | 96 | 57 | 74 | 100 | 61 | 100 | 76 | <1 | >500 | 100 | 100 |
| H77.31 | 77 | 100 | 100 | 100 | 96 | 100 | 65 | 100 | <1 | <1 | 24 | <1 | 100 | 100 | <1 | <1 |
| H77.36 | 97 | 88 | 100 | 92 | 100 | 100 | 98 | 100 | 1.2 | 26 | <1 | 80 | 100 | 100 | 21 | <1 |
| H77.39 | 100 | 100 | 100 | 100 | <1 | 81 | <1 | 61 | 100 | 100 | 100 | 100 | 50 | 100 | 100 | 100 |
| H77.56 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 92 | 100 | 9.5 | 100 | 100 |
The values shown were obtained by dividing the total fluorescence product (percent positive population × mean fluorescence intensity) of a mutant for a given MAb by the total fluorescence product of the wild-type E2 for a given MAb. This value was then divided by the total fluorescence product of a mutant for an oligoclonal MAb pool and by the total fluorescence product of WT E2 for the same oligoclonal pool (to control for E2 binding) and multiplied by 100. The values in boldface indicate complete loss of binding, with reductions in MAb binding greater than or equal to 80% for a given mutation. The underlined values indicate a partial reduction in binding, between 50 and 79%. The italicized, boldface, and underlined value shows enhancement of binding greater than 500%. The results are the averages of three independent experiments for each mutant and each antibody.
Table 3.
Summary of MAb binding to genotype 2a mutants expressed on yeast
| MAb | Bindinga |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| G397E | F403L | G406C | S440P | Y443C | A524V | W529C | E531V | R572S | H617L | G397E + R572S | |
| J6.1 | 71 | 97 | 100 | 100 | 83 | 83 | 100 | 88 | 100 | 100 | 61 |
| J6.2 | 71 | 100 | 16 | 100 | 93 | 73 | 20 | 68 | 42 | 6 | 56 |
| J6.6 | 64 | 100 | 26 | 41 | 100 | 6 | 4 | 69 | 100 | 100 | 100 |
| J6.15 | 100 | 100 | 100 | 75 | 85 | 66 | 13 | 34 | 100 | 100 | 79 |
| J6.16 | 100 | 100 | 100 | 76 | 100 | 80 | 100 | 86 | 100 | 100 | 100 |
| J6.27 | 74 | 67 | 100 | 90 | 95 | <1 | <1 | 77 | 83 | 100 | 80 |
| J6.30 | 85 | 100 | 100 | 100 | 75 | 58 | 88 | 82 | 100 | 3 | 100 |
| J6.36 | 100 | <1 | <1 | 60 | 95 | 80 | 100 | 100 | 100 | 100 | <1 |
| J6.39 | 94 | 91 | 100 | 100 | 83 | 100 | 3 | 63 | 100 | 100 | 81 |
| J6.40 | 98 | 100 | 32 | 32 | 100 | 75 | 63 | 65 | 40 | 15 | 80 |
| J6.60 | 51 | 58 | 100 | <1 | <1 | 65 | 90 | 81 | 70 | 73 | 77 |
| J6.75 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| J6.85 | 100 | 100 | 100 | 100 | 91 | 72 | 10 | 27 | 100 | 100 | 80 |
| J6.101 | 83 | 100 | 43 | 100 | 100 | 100 | 57 | 91 | 82 | 3 | 50 |
| J6.103 | 100 | 2 | <1 | 26 | 100 | 100 | 100 | 100 | 100 | 100 | <1 |
The values shown were obtained by dividing the total fluorescence product (percent positive population × mean fluorescence intensity) of a mutant for a given MAb by the total fluorescence product of the wild-type E2 for a given MAb. This value was then divided by the total fluorescence product of a mutant for an oligoclonal pool of MAbs and by the total fluorescence product of WT E2 for the oligoclonal pool (to control for E2 binding) and multiplied by 100. The values in boldface indicate complete loss of binding, with reductions in MAb binding greater than or equal to 80% for a given mutation. The underlined values show partial loss of binding, with a reduction between 50 and 79%. The results are the averages of three independent experiments for each mutant and each antibody. Polyprotein amino acid numbering was determined by alignment with the H77 strain using the Sequence Location tool on the Los Alamos HCV database (http://hcv.lanl.gov/cgi-bin/LOCATE/locate.cgi).
H77.39, the most potent and highly cross-neutralizing MAb, showed markedly reduced binding when residues N415 and N417 of E2 were changed (Fig. 7A and Table 2). Two neutralizing MAbs (J6.36 and J6.103) showed significant loss of binding when a pair of mutations was introduced. J6.36 and J6.103 lost binding with changes in HVR1 and a more distal region of E2; mutation of residues G406 and F403 or a combined mutation at residues G397 and R572 abrogated MAb binding. Single mutations of G397 and R572, however, did not affect binding (Fig. 7B and Table 3). Similarly, H77.16 showed weakly reduced binding when a serine was introduced at residue G406 (Fig. 7A) but complete loss of binding when residue G530 was altered in combination with G406S. However, complete loss of H77.16 binding also was observed when residue G406 was mutated to an aspartic acid residue.
The neutralizing MAbs that were quantitatively weaker in our neutralization assays, H77.31 and J6.27, showed decreased binding when residues in the putative CD81 binding region (amino acids 523 to 535) (48) were changed. H77.31 binding to E2 on yeast was lost when residues W529, G530, and D533 were mutated, whereas J6.27 binding was abolished when amino acids A524 and W529 were altered. The remaining two weakly neutralizing MAbs (H77.28 and H77.56) showed reduced binding with changes at residues R543 and C552, respectively (Fig. 7A and Tables 2 and 3).
Some nonneutralizing MAbs also were mapped. Several nonneutralizing MAbs (H77.27, H77.36, J6.2, J6.6, J6.15, J6.39, and J6.85) shared residues that impacted binding of H77.31 or J6.27 (Tables 2 and 3), and a few (J6.2, J6.6, J6.40, and J6.101) had total or partial loss of binding to residue G406, which was identified as an important recognition residue for the neutralizing MAbs H77.16, J6.36, and J6.103. In addition to G406, J6.2, J6.40, and J6.101 recognition was also affected by mutation of residue H621, thus defining another discontinuous epitope, albeit one that is not apparently involved in neutralization (Fig. 7 and Tables 2 and 3). Additional residues that uniquely affected binding by nonneutralizing MAbs included G470 (H77.14 and H77.23), S440 (J6.60), Y443 (J6.60), and H621 (J6.30).
DISCUSSION
In this study, we generated a novel panel of 78 MAbs against the E2 proteins of HCV genotypes 1a and 2a, analyzed them functionally for inhibition of HCV infection, and localized epitopes using yeast surface display of truncated and substituted forms of the E2 protein. We defined MAbs that mapped to distinct regions of E2, neutralized infection at different stages, and differentially affected CD81 and SR-B1 engagement. Our mapping data also suggest a tertiary interaction between HVR1 and the COOH-terminal membrane-proximal regions of E2, which provides new insight into the quaternary structural aspects of neutralization by functionally relevant antibodies.
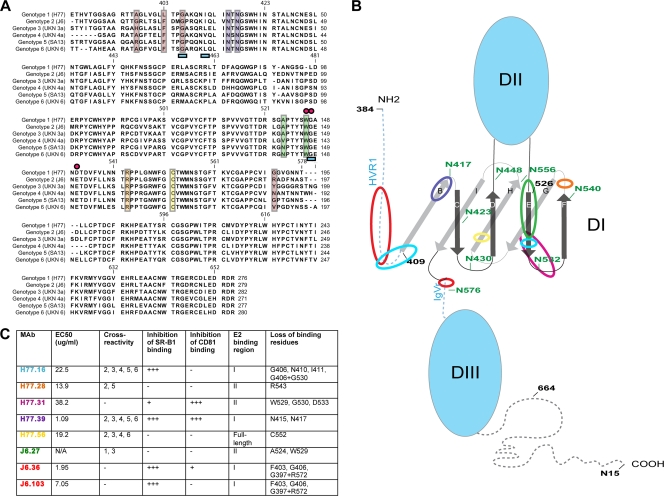
Prior mapping studies of anti-E2 MAbs have utilized peptide binding (9, 18), phage display (65), alanine-scanning mutagenesis of recombinant E1-E2 (33, 37, 49, 62) or E2 (34), or generation of neutralization escape mutants (19) to localize antibody binding sites. In comparison, we used a forward genetic mutational approach coupled with yeast surface display to identify mutants in the context of the entire ectodomain of E2 protein in an unbiased manner. Three of our eight neutralizing MAbs showed loss-of-binding phenotypes with paired amino acid mutations greater than 100 amino acids apart in the linear sequence, suggesting that discontinuous regions of E2 come together to create functionally important antibody epitopes. H77.16 showed a loss-of-binding phenotype when mutations in HVR1 (e.g., G406S) and the more COOH-terminal residue (G530A) were paired, suggesting that H77.16 binds a conformational epitope. Although complete loss of binding could be achieved with a single less conserved mutation (G406D), the more conserved G406S change required a second mutation at a discontinuous site (G530) for loss of binding. This finding, which suggests that HVR1 interacts with more COOH-terminal residues, is consistent with MAb competition studies with recombinant proteins that suggested that amino acids 396 to 424, 436 to 447, and 523 to 540 comprise an antigenic region (designated “antigenic region 3”) within E2 (37) and with sequencing results of MAb AP33 escape variants, which identified noncontiguous amino acid residues (N415 and E655) as factors in the loss-of-neutralization phenotype (19). Additionally, these data support the recently described model of HCV E2 based on the three-domain structure of class II E proteins in Flaviviridae and Togaviridae, which predicts that HVR1 proximally apposes the proposed HCV domain I (D1) (36) (Fig. 8B).
Fig. 8.
Localization of MAb binding residues on E2. (A) Alignment of E2 sequences from HCV genotypes 1 to 6 with superimposed mapping of MAb binding residues. The sequences of E2 from strains representative of the different genotypes (genotype 1a, H77; genotype 2a, J6; genotype 3a, UKN 3; genotype 4a, UKN 4a; genotype 5a, SA513; genotype 6a, UKN 6) used in the yeast-mapping studies (Fig. 2) were aligned. Colored boxes and symbols were used to highlight neutralizing MAb binding residues as follows: red boxes, J6.36 and J6.103; purple boxes, H77.39; blue underscoring, H77.16; green boxes, J6.27; pink circles, H77.31; orange box, H77.28; yellow box, H77.56. (B) Putative model of the structure of the E2 protein with MAb binding regions highlighted. A scheme depicting a possible E2 structure was adapted from Krey et al. (36) to highlight regions involved in MAb recognition. N-linked glycosylation residues are labeled in green, and amino acids are numbered in black at intervals; β-sheets in D1 are labeled as previously described (36). MAb binding regions are highlighted with colored circles as follows: red circles, J6.36 and J6.103; purple circle, H77.39; light-blue circles, H77.16; green circle, J6.27; pink circle, H77.31; orange circle, H77.28; yellow circle, H77.56. (C) Summary of neutralizing MAbs described in this study. EC50 values (neutralization against homologous virus), cross-reactivity to E2 from different genotypes, inhibition of binding to CD81 and SR-B1, reactivity with different regions of E2, and loss-of-binding residues are listed. MAb names are color coded to correspond to panels A and B.
Two other neutralizing MAbs, H77.31 and J6.27, also recognized residues within the third segment of antigenic region 3 (A524, W529, G530, and D533) but did not show a loss-of-binding phenotype when amino acids within segment 1 (396 to 424) were changed. These two MAbs less potently neutralized infection and were less cross-reactive. In comparison, human anti-HCV MAbs (A8, 1:7, and CBH5) that share epitopes in this region (26, 33) have been characterized as inhibitory and cross-reactive (Table 4). Although further analysis is required, the differences in function of the mouse and human MAbs could be related to affinity or possibly to the fact that the human MAbs bind additional sites and do not exclusively recognize the linear epitope centered at residues G523 to D535, as was suggested in previous studies (1, 26).
Table 4.
Previously characterized anti-E2 MAbs with available mapping information
| MAb name | Source of MAb | Binding residues | Neutralization of HCVpp | Neutralization of HCVcc | References |
|---|---|---|---|---|---|
| AP33 | Mouse | L413, N415, G418, W420 | Yes; all genotypes | Yes; >80% neutralization with 50 μg/ml MAb | 19, 45, 46, 62 |
| 3/11 | Rat | N415, W420, H421 | Yes; genotypes 1, 2, 4, 5, and 6 by >50% | NDa | 18, 30, 45, 62 |
| 9/27 | Rat | 396–407 | Yes; genotype 1 by >99% | ND | 18, 30, 45 |
| 11/20 | Rat | 436–447 | Yes; genotype 1 by >99% | ND | 18, 30 |
| 7/16b | Rat | 436–447 | Yes; neutralized HCVpp representative of genotype 1 by >99% | ND | 18, 30 |
| 2/69a | Rat | 432–443 | Yes; genotype 1 by >99% | ND | 18, 30 |
| 2/64a | Rat | 524–531 | Yes; genotype 1 by 40% | ND | 18, 30 |
| 1/39 | Rat | 432–443 | Yes; genotype 1 by 20% | ND | 18, 30 |
| CBH-5 | Human | G523, P525, G530, D535, N540 | Yes; all genotypes >40% | Yes | 26, 47 |
| CBH-7 | Human | N540, W549 | Yes; genotypes 1, 2 and 4 by >20% | Yes | 26, 47 |
| HCV-1 | HuMAb miceb | L413, W420 | Yes; genotypes 1, 2, 3, and 4 | ND | 9 |
| 95-2 | HuMAb mice | L413, W420 | Yes; genotypes 1, 2, 3, and 4 | ND | 9 |
| 1:7 | Human | G523, T526, Y527, W529, G530, D535 | Yes; all genotypes | Yes | 33 |
| A8 | Human | G523, T526, Y527, W529, G530, D535 | Yes; all genotypes | Yes | 33 |
| AR2A | Human | N540A | Yes; genotypes 1, 2, 4, and 5 | No | 37 |
| AR3A | Human | S424, G523, P525, G530, D535, N540 | Yes; genotypes 1, 2, 4, and 5 | Yes | 37 |
| AR3B | Human | S424, G530, D535 | Yes; genotypes 1, 2, 4, and 5 | Yes | 37 |
| AR3C | Human | S424, P525, G530, D535, N540 | Yes; genotypes 1, 2, 4, and 5 | Yes | 37 |
| AR3D | Human | S424, G530 | Yes; genotypes 1, 2, 4, and 5 | Yes | 37 |
| e137 | Human | T416, W420, W529, G530, D535 | Yes; genotypes 1, 2, and 4 | Yes | 49 |
ND, not done.
HuMAb mice are transgenic for human μ, γ1, and κ germ line genes.
The neutralizing MAbs J6.36 and J6.103 also mapped to a discontinuous epitope, requiring residues within HVR1 (G397, F403, and G406) and the more COOH-terminal residue R572. Although neutralizing MAbs (9/27 [18, 30] and AP213 [65]) have been mapped to HVR1, to our knowledge, MAbs that bind residues at or near R572 have not been identified. MAb 9/27 does not block binding of sE2 to CD81 (18, 30), although it did inhibit HCV VLP interaction with CD81 (45), suggesting that it also may recognize a conformational or possibly oligomeric epitope.
The MAb in our study with the greatest inhibitory activity, H77.39, localized to 2 amino acids, N415 and N417, that are highly conserved among all HCV genotypes (48, 65). N415 and N417 were defined previously as possible binding residues for MAbs AP33 and 3/11 (11, 19, 62) (Table 4). Residue N417 comprises part of a highly conserved N-linked glycosylation site (20, 21) that is implicated in obscuring antibody-mediated neutralization (27). H77.39, as well as AP33 and 3/11, is thus unique in mapping to an N-linked glycan that is paradoxically hypothesized to impair antibody recognition.
To relate binding epitopes to function, MAbs were tested for the ability to inhibit sE2 engagement with the HCV cognate receptors CD81 and SR-B1. The MAbs J6.36, J6.103, and H77.16, which recognized residues within HVR1, as well as the more COOH-terminal region, blocked sE2–SR-B1 binding. These results are consistent with data suggesting HVR1 participates in SR-B1 binding (3, 5, 56) and that the HVR1-specific MAb 9/27 inhibits sE2–SR-B1 interactions (5, 56). Although J6.36 did not map to any of the predicted CD81 binding residues (48), it partially inhibited sE2 binding to CD81. J6.36 could map to additional amino acid residues (within the CD81 binding site) not identified in our study, or steric hindrance could mediate this partial inhibition. In the recently modeled E2 structure (36), the J6.36-interacting residues lie in proximity to the HCV D1, which is predicted to contain key CD81 binding residues (36, 48) (Fig. 8B). Conversely, H77.31, which potently inhibited CD81 binding and maps to residues (W529 and G530) involved in CD81 binding (48), partially inhibited SR-B1 engagement despite a lack of contact residues in HVR1. The inability of J6.103 to inhibit binding to CD81 despite localizing to the same residues as J6.36 could be explained by overlapping but not identical MAb footprints or perhaps differences in affinities of interaction.
Only one MAb, H77.39, potently inhibited sE2 binding to both CD81 and SR-B1. Interestingly, H77.39 did not map to residues within known SR-B1 or CD81 binding regions, suggesting that it may recognize a site that, once occupied, can sterically prevent receptor engagement. This concept is supported by studies showing that N415 and N417 can obscure the CD81 and SR-B1 binding sites (11, 27). Finally, the E2 model recently proposed by Krey et al. predicts that residues N415 to N417 lie at the junction of HVR1 and D1 (Fig. 8B), in proximity to both HVR1 and the CD81 binding residues located within the C and D loops of D1 (36, 48).
Pre- and postattachment neutralization studies provided additional insight into the relative potencies of MAbs. Studies with distantly related flaviviruses have shown that MAbs inhibiting at a postattachment step tend to have greater inhibitory activity in vitro and in vivo because they require reduced virion occupancy for neutralization (43, 52, 60, 63, 66). Indeed, our three most potent MAbs, H77.16, H77.39, and J6.36, neutralized infection in the postattachment assay. Nevertheless, J6.103 shared apparent binding epitopes with J6.36 yet did not neutralize efficiently when added after attachment. This discrepancy may be explained by J6.36 having additional amino acid contacts not identified in our study.
MAb binding to conserved residues may not directly predict cross-binding or cross-neutralizing capabilities (8, 59). Despite mapping to highly conserved residues, MAbs H77.31, J.36, and J6.103 failed to cross-react with any other strains tested, and J6.27 was cross-reactive with only two of the strains tested. In comparison, MAb H77.16 was highly cross-reactive but still did not neutralize heterologous strains. In contrast, H77.39 cross-reacted with genotypes 1 to 6 and neutralized chimeric virus representative of all strains except genotype 6. The inability of H77.39 to neutralize the genotype 6 chimeric virus may be explained by the presence of a mutation in one of the recognition residues, N417T (24). This mutation is rare in natural HCV isolates (11, 48) but was required for adaptation of the HK6a/JFH1 chimera in vitro (24). Mutations at N415 are rare (11, 48) and attenuating in the context of HCV infection (19).
Generation of an HCV vaccine has been impeded by the lack of a structural understanding of the epitopes on E2 that should be targeted by inhibitory antibodies. Although direct structural confirmation is necessary, our data suggest the existence of discontinuous epitopes that are recognized by antibodies that inhibit CD81 and SR-B1 binding. The yeast surface display antibody-mapping data also provide support for a recently proposed structural model of E2 in which the residues comprising the CD81 binding region lie within a single domain of β-pleated sheets that contains HVR1 as an N-terminal extension (36). The epitopes defined by the MAbs H77.16, J6.36, and J6.103 suggest that HVR1 might lie in proximity to this domain, creating a conformational epitope (Fig. 8B) that could be a useful target for vaccines and therapeutic antibodies.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the Washington University Institute of Clinical and Translational Science (to M.S.D. and D.H.F.) and an NRSA predoctoral fellowship from NIDDK (F30 DK088385 to M.C.S.). S.E.H. is a predoctoral trainee and was supported in part by a U.S. Public Health Service Institutional Research Training Award (AI07647). M.J.E. is supported in part by the Pew Charitable Funds and the NIAID (R00 AI077800). This study also was supported by a Ph.D. stipend from the Faculty of Health Sciences, University of Copenhagen (J.P.), and research grants from the Lundbeck Foundation (J.B.).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Allander T., et al. 2000. Recombinant human monoclonal antibodies against different conformational epitopes of the E2 envelope glycoprotein of hepatitis C virus that inhibit its interaction with CD81. J. Gen. Virol. 81:2451–2459 [DOI] [PubMed] [Google Scholar]
- 2. Balsitis S. J., et al. 2010. Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS Pathog. 6:e1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bankwitz D., et al. 2010. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J. Virol. 84:5751–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartenschlager R., Frese M., Pietschmann T. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71–180 [DOI] [PubMed] [Google Scholar]
- 5. Bartosch B., et al. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624–41630 [DOI] [PubMed] [Google Scholar]
- 6. Bertaux C., Dragic T. 2006. Different domains of CD81 mediate distinct stages of hepatitis C virus pseudoparticle entry. J. Virol. 80:4940–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bowen D. G., Walker C. M. 2005. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 436:946–952 [DOI] [PubMed] [Google Scholar]
- 8. Brien J. D., et al. 2010. Genotype-specific neutralization and protection by antibodies against dengue virus type 3. J. Virol. 84:10630–10643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Broering T. J., et al. 2009. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J. Virol. 83:12473–12482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choo Q. L., et al. 1994. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 91:1294–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dhillon S., et al. 2010. Mutations within a conserved region of the hepatitis C virus E2 glycoprotein that influence virus-receptor interactions and sensitivity to neutralizing antibodies. J. Virol. 84:5494–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Diamond M. S., Garcia-Aguilar J., Bickford J. K., Corbi A. L., Springer T. A. 1993. The I domain is a major recognition site on the leukocyte integrin Mac-1 (CD11b/CD18) for four distinct adhesion ligands. J. Cell Biol. 120:1031–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drummer H. E., Wilson K. A., Poumbourios P. 2002. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 76:11143–11147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Evans M. J., et al. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805 [DOI] [PubMed] [Google Scholar]
- 15. Farci P., et al. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. U. S. A. 93:15394–15399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feld J. J., Hoofnagle J. H. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436:967–972 [DOI] [PubMed] [Google Scholar]
- 17. Feldhaus M. J., et al. 2003. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat. Biotechnol. 21:163–170 [DOI] [PubMed] [Google Scholar]
- 18. Flint M., et al. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gal-Tanamy M., et al. 2008. In vitro selection of a neutralization-resistant hepatitis C virus escape mutant. Proc. Natl. Acad. Sci. U. S. A. 105:19450–19455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goffard A., et al. 2005. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J. Virol. 79:8400–8409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goffard A., Dubuisson J. 2003. Glycosylation of hepatitis C virus envelope proteins. Biochimie 85:295–301 [DOI] [PubMed] [Google Scholar]
- 22. Gottwein J. M., et al. 2010. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. J. Virol. 84:5277–5293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gottwein J. M., et al. 2007. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology 133:1614–1626 [DOI] [PubMed] [Google Scholar]
- 24. Gottwein J. M., et al. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49:364–377 [DOI] [PubMed] [Google Scholar]
- 25. Haberstroh A., et al. 2008. Neutralizing host responses in hepatitis C virus infection target viral entry at postbinding steps and membrane fusion. Gastroenterology 135:1719–1728 [DOI] [PubMed] [Google Scholar]
- 26. Hadlock K. G., et al. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 74:10407–10416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Helle F., et al. 2010. Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J. Virol. 84:11905–11915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Higginbottom A., et al. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Houghton M., Abrignani S. 2005. Prospects for a vaccine against the hepatitis C virus. Nature 436:961–966 [DOI] [PubMed] [Google Scholar]
- 30. Hsu M., et al. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. U. S. A. 100:7271–7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hüssy P., Schmid G., Mous J., Jacobsen H. 1996. Purification and in vitro-phospholabeling of secretory envelope proteins E1 and E2 of hepatitis C virus expressed in insect cells. Virus Res. 45:45–57 [DOI] [PubMed] [Google Scholar]
- 32. Jensen T. B., et al. 2008. Highly efficient JFH1-based cell-culture system for hepatitis C virus genotype 5a: failure of homologous neutralizing-antibody treatment to control infection. J. Infect. Dis. 198:1756–1765 [DOI] [PubMed] [Google Scholar]
- 33. Johansson D. X., et al. 2007. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 104:16269–16274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Keck Z. Y., et al. 2008. Definition of a conserved immunodominant domain on hepatitis C virus E2 glycoprotein by neutralizing human monoclonal antibodies. J. Virol. 82:6061–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kolykhalov A. A., et al. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570–574 [DOI] [PubMed] [Google Scholar]
- 36. Krey T., et al. 2010. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 6:e1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Law M., et al. 2008. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 14:25–27 [DOI] [PubMed] [Google Scholar]
- 38. Lindenbach B. D., et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 39. Lindenbach B. D., Rice C. M. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933–938 [DOI] [PubMed] [Google Scholar]
- 40. Ma Y., Yates J., Liang Y., Lemon S. M., Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 82:7624–7639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martinez-Donato G., et al. 2006. Expression and processing of hepatitis C virus structural proteins in Pichia pastoris yeast. Biochem. Biophys. Res. Commun. 342:625–631 [DOI] [PubMed] [Google Scholar]
- 42. Nybakken G. E., et al. 2005. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature 437:764–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oliphant T., et al. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11:522–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oliphant T., et al. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J. Virol. 80:12149–12159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Owsianka A., Clayton R. F., Loomis-Price L. D., McKeating J. A., Patel A. H. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877–1883 [DOI] [PubMed] [Google Scholar]
- 46. Owsianka A., et al. 2005. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J. Virol. 79:11095–11104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Owsianka A. M., et al. 2008. Broadly neutralizing human monoclonal antibodies to the hepatitis C virus E2 glycoprotein. J. Gen. Virol. 89:653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Owsianka A. M., et al. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J. Virol. 80:8695–8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Perotti M., et al. 2008. Identification of a broadly cross-reacting and neutralizing human monoclonal antibody directed against the hepatitis C virus E2 protein. J. Virol. 82:1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Petracca R., et al. 2000. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 74:4824–4830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pierson T. C., Diamond M. S. 2008. Molecular mechanisms of antibody-mediated neutralisation of flavivirus infection. Expert Rev. Mol. Med. 10:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pierson T. C., et al. 2007. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe 1:135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pileri P., et al. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941 [DOI] [PubMed] [Google Scholar]
- 54. Ploss A., et al. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Prentoe J., et al. 2011. Hypervariable region 1 differentially impacts viability of hepatitis C virus strains of genotypes 1 to 6 and impairs virus neutralization. J. Virol. 85:2224–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Scarselli E., et al. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Scheel T. K., et al. 2008. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc. Natl. Acad. Sci. U. S. A. 105:997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Scheller N., et al. 2009. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc. Natl. Acad. Sci. U. S. A. 106:13517–13522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shrestha B., et al. 2010. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog. 6:e1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sukupolvi-Petty S., et al. 2010. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J. Virol. 84:9227–9239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sukupolvi-Petty S., et al. 2007. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J. Virol. 81:12816–12826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tarr A. W., et al. 2006. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology 43:592–601 [DOI] [PubMed] [Google Scholar]
- 63. Thompson B. S., et al. 2009. A therapeutic antibody against West Nile virus neutralizes infection by blocking fusion within endosomes. PLoS Pathog. 5:e1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vanwolleghem T., et al. 2008. Polyclonal immunoglobulins from a chronic hepatitis C virus patient protect human liver-chimeric mice from infection with a homologous hepatitis C virus strain. Hepatology 47:1846–1855 [DOI] [PubMed] [Google Scholar]
- 65. Vieyres G., Dubuisson J., Patel A. H. 2011. Characterization of antibody-mediated neutralization directed against the hypervariable region 1 of hepatitis C virus E2 glycoprotein. J. Gen. Virol. 92:494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vogt M. R., et al. 2009. Human monoclonal antibodies against West Nile virus induced by natural infection neutralize at a postattachment step. J. Virol. 83:6494–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yi M., Ma Y., Yates J., Lemon S. M. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.