Abstract
Hepatitis C virus (HCV) is an important human pathogen, persistently infecting more than 170 million individuals worldwide. Studies of the HCV life cycle have become possible with the development of cell culture systems supporting the replication of viral RNA and the production of infectious virus. However, the exact functions of individual proteins, especially of nonstructural protein 4B (NS4B), remain poorly understood. NS4B triggers the formation of specific, vesicular membrane rearrangements, referred to as membranous webs, which have been reported to represent sites of HCV RNA replication. However, the mechanism of vesicle induction is not known. In this study, a panel of 15 mutants carrying substitutions in the highly conserved NS4B C-terminal domain was generated. Five mutations had only a minor effect on replication, but two of them enhanced assembly and release of infectious virus. Ten mutants were replication defective and used for selection of pseudoreversions. Most of the pseudoreversions also localized to the highly conserved NS4B C-terminal domain and were found to restore replication competence upon insertion into the corresponding primary mutant. Importantly, pseudoreversions restoring replication competence also restored heterotypic NS4B self-interaction, which was disrupted by the primary mutation. Finally, electron microscopy analyses of membrane alterations induced by NS4B mutants revealed striking morphological abnormalities, which were restored to wild-type morphology by the corresponding pseudoreversion. These findings demonstrate the important role of the C-terminal domain in NS4B self-interaction and the formation of functional HCV replication complexes.
INTRODUCTION
Hepatitis C virus (HCV) is an important human pathogen persistently infecting 130 to 170 million individuals worldwide and increasing the risk for steatosis, fibrosis, liver cirrhosis, and hepatocellular carcinoma (28). Owing to high variability, HCV is classified into seven genotypes and more than 100 subtypes (44). The HCV genome is an ∼9.6-kb single-stranded uncapped RNA molecule of positive polarity containing a single open reading frame (ORF) that is flanked by 5′ and 3′ untranslated regions (UTRs). Both UTRs form complex secondary and higher order pseudoknot structures (reviewed in reference 3). The ORF encodes a polyprotein that is co- and posttranslationally processed by cellular and viral proteases (3), giving rise to three structural proteins (core and envelope protein 1 [E1] and E2), the viroporin p7, and six nonstructural (NS) proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B). P7 and NS2 are required for virion assembly (20, 21, 46), whereas NS3 to NS5B constitute the minimal viral replicase (32). As for all other positive-strand RNA viruses (8, 36), HCV RNA replication occurs in close association with cellular membranes (12, 15, 40, 47). Structural and genetic data (5, 11, 16, 17, 26, 30, 51), as well as ultrastructural analyses (10, 12, 15), provide evidence that NS4B is a key organizer of the viral replication complex by inducing the formation of membranous vesicles, which accumulate in large cytosolic clusters, referred to the membranous web. In addition, genetic studies suggest that NS4B might contribute to assembly (22).
NS4B is thought to contain two N-terminal amphipathic helices, spanning amino acids (aa) 6 to 29 (AH1) (11) and aa 42 to 66 (AH2) (16), a highly hydrophobic central core domain (aa 75 to 191) that contains four putative transmembrane segments (35), and a highly conserved C-terminal domain (aa 192 to 261) that is thought to harbor two α-helices (aa 201 to 213 [H1] and aa 228 to 254 [H2]) (reviewed in reference 18). The three-dimensional structure of the second helix has been solved recently (17), whereas the first α-helix has only been predicted thus far. The N-terminal NS4B domain was shown to translocate posttranslationally at least in part into the endoplasmic reticulum (ER) lumen, and this change in topology might contribute to the induction of membranous vesicles (16, 34). In addition, C-terminal palmitoylation was shown by chemical cross-linking experiments to be involved in NS4B oligomerization (51). A recent study has demonstrated that NS4B oligomerizes through multiple conserved determinants and that oligomerization may be required for membranous web induction (19). However, the contribution of the highly conserved C-terminal domain in NS4B oligomerization and membranous vesicle induction remains unknown. To address this important issue, we used reverse and forward genetics in combination with an interaction assay and ultrastructural studies of NS4B-induced membrane alterations. We report that the C-terminal domain is crucial for NS4B self-interaction, which in turn is required for the induction of functional membranous vesicles where HCV RNA replication is thought to occur.
MATERIALS AND METHODS
Antibodies.
Mouse monoclonal antibody 9E10 recognizing NS5A domain III of the Con1 HCV isolate was kindly provided by Charles M. Rice (29). Mouse monoclonal antibody against NS3 of the JFH-1 isolate (2E3) was generated in cooperation with Hengli Tang (2). Rabbit polyclonal antibody against NS4B Con1 was generated by immunization with recombinant NS4B purified as described previously (31). The NS5A 9E10 monoclonal antibody, as well as the polyclonal NS4B-specific rabbit antiserum, recognizes both Con1 and JFH1 proteins.
Cell culture.
Huh7 (37), Huh7-Lunet (13), Huh7-Lunet T7 (1), Huh7.5 (6), and U-2 OS (39) cells were maintained in Dulbecco's modified Eagle medium (DMEM; Invitrogen, Karlsruhe, Germany) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 10% fetal calf serum (DMEM cplt). Huh7-Lunet T7 cells were cultured in the presence of 5 μg of zeocin/ml. G418 (Geneticin; Invitrogen) was added at a final concentration of 250 or 500 μg/ml in the replicon selection assays.
Plasmid constructs.
All nucleotide and amino acid numbers refer to the JFH-1 genome (GenBank accession no. AB047639). The plasmids encoding the subgenomic replicons (pFK_i389LucNS3-3′_JFH_δg), the nonreplicative mutant (pFK_i389LucNS3-3′_NS5BΔGDD_JFH_δg), and the full-length genomes (pFK_JFH_δg and pFK_JFHΔE1E2_δg) have been described previously (42, 50). The plasmids used for FRET assays (pCMVJFH4B-CFP, pCMVJFH4B-YFP, pCMVJFH4B40-130-CFP, pCMVJFH4B40-130-YFP, pCMVJFH4B130-261-CFP, and pCMVJFH4B130-261-YFP), as well as control plasmids, were as described in reference 19. Plasmid pTM-NS3-3′_JFH, used for T7-based expression of the HCV replicase proteins (NS3 to NS5B), has been described previously (2). Single amino acid substitutions in the NS4B C-terminal region were introduced by PCR-based site-directed mutagenesis (the primers used are listed in Table S1 in the supplemental material). After restriction of the amplicons with BamHI, they were inserted into pBSK_SacI_JFH containing a SacI restriction fragment (codon 1407-2204) of the JFH-1 genome. The NS5A mutation K139E was generated by PCR-based site-directed mutagenesis and, after restriction with BspEI and SanDI, the fragment was inserted into pBSK_SacI_JFH or the analogous vectors, into which mutations in the NS4B C-terminal region had already been inserted. Replicon constructs containing these mutations were obtained by transferring the SacI DNA fragment from the corresponding pBSK_SacI_JFH vector into pFK_i389LucNS3-3′_JFH_δg. All constructs were verified by nucleotide sequence analysis. Selectable replicons were generated by replacing the first cistron encoding the firefly luciferase gene by the gene encoding for neomycin phosphotransferase by using flanking AgeI and KpnI restriction sites. Full-length JFH-1 genomes and pTM-based replicase constructs (containing an HCV genome fragment from NS3 to the end of the 3′UTR) containing mutations in NS4B were created by transferring an NsiI/RsrII fragment from a given pFK_i389LucNS3-3_JFH_δg mutant into pFK_JFH_δg and pTM-NS3-3′, respectively. NS4B mutations were inserted into pCMV-based FRET vectors by using PCR with the primers f_NS4B_175 (5′-TCCTGTCTCCGGGAGCCC-3′) and r_NS4B_Bam (5′-ATAGGATCCGCATGGGATGGGGCAGTCC-3′) and a given mutant contained in pFK_i389LucNS3-3_JFH_δg as a template and transfer of a PmlI/BamHI restriction fragment into pCMVJFH4B-CFP, pCMVJFH4B-YFP, pCMVJFH4B130-261-CFP, and pCMVJFH4B130-261-YFP.
In vitro transcription.
In vitro transcripts were generated by using 10 μg of the respective plasmid DNA that had been linearized by 1 h of digestion with MluI. DNA was extracted with phenol and chloroform and, after precipitation with ethanol, dissolved in RNase-free water. In vitro transcription reaction mixtures (total volume, 100 μl) contained 80 mM HEPES (pH 7.5), 12 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol (DTT), 3.125 mM concentrations of each nucleoside triphosphate, 1 U of RNasin (Promega)/μl, 0.1 μg of plasmid DNA/μl, and 0.6 U of T7 RNA polymerase (Promega)/μl. After 2 h of incubation at 37°C, 0.3 U of T7 RNA polymerase/μl of reaction mixture was added, and the reaction mixture was incubated for 2 h at 37°C. Transcription was terminated by the addition of 1.2 U of RNase-free DNase (Promega) per μg of plasmid DNA and 30 min of incubation at 37°C. RNA was extracted with acidic phenol and chloroform, precipitated with isopropanol at room temperature, and dissolved in RNase-free water.
RNA transfection.
Selection assays were conducted with naïve Huh7 cells, whereas transient replication assays were performed with Huh7-Lunet cells that have a higher permissiveness. Single-cell suspensions were prepared by trypsinization and, after being washed with phosphate-buffered saline (PBS), the cells were resuspended at a concentration of 107 cells per ml in Cytomix (49) supplemented with 2 mM ATP and 5 mM glutathione. Next, 5 or 10 μg of subgenomic or genomic HCV in vitro transcripts, respectively, was mixed with 400 μl of the cell suspension and transfected by electroporation using a Gene Pulser system (Bio-Rad, Hercules, CA) and a cuvette with a gap width of 0.4 cm (Bio-Rad) at 975 μF and 270 V. Cells were immediately diluted into 12 ml (transient replication assay) or 20 ml (selection and infectivity release assay) of DMEM cplt and seeded as specified in Results.
Selection for pseudorevertants, amplification of HCV RNA by RT-PCR, and sequence analysis of cloned amplicons.
Huh7 cells transfected with a selectable replicon were resuspended in 20 ml of DMEM cplt and seeded onto two 10-cm-diameter culture dishes. After 24 h, G418 was added to a final concentration of 250 or 500 μg/ml. The medium was exchanged at least twice a week until single cell clones became visible. Two cell clones per 10-cm-diameter dish were isolated and expanded for further analyses. The total RNA of each cell clone was obtained by using a Nucleo Spin RNAII kit (Macherey-Nagel, Düren, Germany) according to the instructions of the manufacturer. A 1-μg portion of total RNA was used for reverse transcription-PCR (RT-PCR), and amplicons were molecularly cloned and used for sequence analysis as described elsewhere (24).
Transient replication assay.
Cells transfected with a subgenomic replicon RNA were resuspended in 12 ml of DMEM cplt and seeded into a six-well plate. Replication was determined by measuring luciferase activity in cells 4, 24, 48, and 72 h after transfection. Cells were washed once with PBS and lysed by adding 350 μl of lysis buffer (0.1% Triton X-100, 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT [pH 7.8]). Cells were frozen immediately at −70°C and, after thawing, the lysates were resuspended by pipetting. For each well, 100 μl of lysate was mixed with 360 μl of assay buffer (25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 2 mM ATP, 15 mM K2PO4 [pH 7.8]) and, after the addition of 200 μl of a luciferin solution (200 μM luciferin, 25 mM glycylglycine [pH 8.0]), were measured for 20 s in a luminometer (Lumat LB9507; Berthold Technologies, Bad Wildbad, Germany). Each well was measured in duplicate. The kinetics of replication were calculated by normalizing the relative light units measured at a given time point to the respective 4-h value.
Infectivity assays.
Huh7-Lunet cells (13) were transfected with viral RNA and resuspended in 20 ml of DMEM cplt. Portions (2 ml) of the cell suspension were seeded into each well of a six-well plate and harvested 4, 24, 48, and 72 h later. Then, a 9-ml portion of the cell suspension was seeded into a 10-cm-diameter dish and used to determine the extra- and intracellular infectivity 48 h later. Supernatants were filtered through a 0.45-μm-pore-size filter and stored at 4°C. The cells were rinsed once with PBS, scraped into 2 ml of PBS, sedimented by 15 min of centrifugation at 750 × g, and resuspended in 1 ml of DMEM cplt. The cell pellets and 1 ml of the corresponding supernatant were subjected in parallel to three freeze-thaw cycles consisting of rapid freezing in liquid nitrogen and thawing at 37°C. The cell debris was removed by centrifugation for 5 min at 10,000 × g, and the infectivity in supernatants and cell lysates was determined by limiting-dilution assay using Huh7.5 target cells and staining of the NS3 protein with the 2E3 antibody, as described elsewhere (24). Virus titers (the 50% tissue culture infective dose [TCID50]/ml) were calculated based on the method of Spearman and Kärber (23, 45).
FRET analysis.
NS4B interaction experiments were measured by using fluorescence resonance energy transfer (FRET) acceptor bleaching as described previously (19). In brief, U-2 OS cells were seeded in a 12-well plate onto glass coverslips at a density of 1.5 × 105 cells per well. About 18 h later, the cells were transfected with 1 μg of each NS4B-CFP and NS4B-YFP expression constructs by using a calcium phosphate transfection kit (Clontech, Mountain View, CA). After 24 h, the cells were washed with PBS and fixed with 4% paraformaldehyde solution in PBS (1.8 mM KH2PO4, 5.9 mM Na2HPO4·2H2O, 2.7 mM KCl, 137 mM NaCl [pH 7.4]) for 10 min at room temperature. After a rinse with PBS, the samples were mounted on glass slides with Fluoromount G (Southern Biotech, Birmingham, AL), and FRET measurements were carried out with a Leica SP2 confocal laser scanning microscope (Leica, Heidelberg, Germany). FRET efficiency was determined according to the following formula: FRETeff = [(EDpost − EDpre)/EDpost] × 100, where ED represents the emitted donor fluorescence before (EDpre) or after (EDpost) photobleaching of the acceptor fluorophore. The FRET efficiency was determined at two different view fields per cell using the Leica Software package RK03032000. Measurements of at least 15 cells from two independent transfections were used to calculate the box plots. Significance values were calculated by using the unpaired t test with the GraphPad Prism 5 software package (GraphPad Software, Inc., La Jolla, CA).
Immunoblot analysis.
Cells were briefly rinsed with PBS, lysed in 2× protein sample buffer (200 mM Tris [pH 8.8], 5 mM EDTA, 0.1% bromophenol blue, 10% sucrose, 3% sodium dodecyl sulfate [SDS], 2% β-mercaptoethanol), and sonicated for 15 s with a Branson Sonifier 450 cup horn (Branson, Danbury, CT). Proteins were separated by SDS-polyacrylamide gel electrophoresis and electrotransferred onto polyvinylidene membranes. After the membranes were blocked with 5% nonfat milk, the proteins were labeled with primary antibodies (NS5A 9E10, 1:10,000; NS4B, 1:1,000) by overnight incubation at 4°C and, after being washed in PBS-0.5% Tween 20, with secondary horseradish peroxidase-conjugated antibodies (Sigma-Aldrich, St. Louis, MO). The membranes were developed by using Western Lightning Plus-ECL (Perkin-Elmer, Waltham, MA), and bands were visualized on BioMax Light films (Kodak, Rochester, MN).
Ultrastructural analysis by transmission electron microscopy.
Huh7-Lunet T7 cells (5 × 104) were seeded onto glass coverslips. On the next day the cells were transfected with pTM-NS3-3′-based expression vectors by using the TransITLT1 transfection reagent (Mirus Bio, LLC, Madison, WI). After 8, 16, or 24 h the cells were washed three times with prewarmed PBS and fixed for 30 min with 2.5% glutaraldehyde in 50 mM sodium cacodylate buffer (pH 7.2) containing 1 M KCl, 0.1 M MgCl2, 0.1 M CaCl2, and 2% sucrose. The cells were washed five times (for 5 min each time) with 50 mM cacodylate buffer and postfixed on ice in the dark with 2% OsO4 in 50 mM cacodylate buffer for 40 min. After the cells were washed overnight with H2O, they were treated with 0.5% uranyl acetate dissolved in H2O for 30 min, rinsed thoroughly with H2O, and dehydrated at room temperature in a graded ethanol series: 40, 50, 60, 70, and 80% for 5 min each time and then 95 and 100% for 20 min each time. The cells were immersed in 100% propylene oxide and immediately embedded in an araldite-Epon mixture (Araldite 502/Embed 812 kit; Electron Microscopy Sciences, Hatfield, PA). After polymerization at 60°C for 2 days, the coverslips were removed, and the embedded cell monolayer was sectioned into 65-nm-thick slices by using a Leica Ultracut UCT microtome and a diamond knife. The slices were counterstained with 3% uranyl acetate in 70% methanol for 5 min, 2% lead citrate in H2O for 2 min, and examined with a Zeiss EM 10 transmission electron microscope at 60 kV (Zeiss, Gottingen, Germany).
RESULTS
Highly conserved residues in the NS4B C-terminal region are crucial for HCV RNA replication.
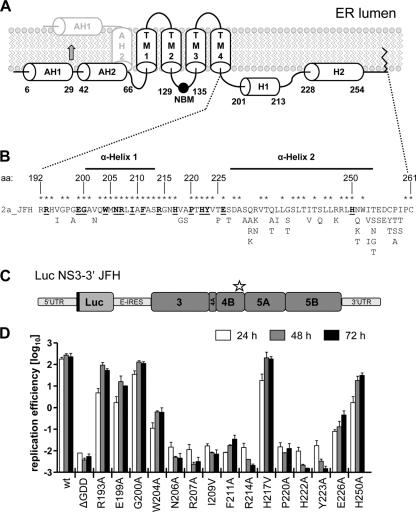
Structure predictions and experimental data indicate two N-terminal amphipathic α-helices spanning aa 6 to 29 (11) and aa 42 to 66 (16), respectively, and a highly hydrophobic central region (aa 75 to 191) containing four putative transmembrane segments (35) (Fig. 1A). The highly conserved C-terminal, cytoplasmic NS4B domain (aa 192 to 261) is thought to contain two α-helices, of which the most C-terminal spanning aa 228 to 254 (α-helix 2) has been confirmed experimentally (17), whereas the α-helix 1 spanning aa 201 to 213 has only been predicted. Given the high sequence conservation of the complete C-terminal NS4B domain, especially of the predicted H1, we conducted a reverse genetics study, targeting mainly residues in H1 and the region in between both α-helices, in order to determine the role of this domain in viral RNA replication (Fig. 1B). Single amino acid substitutions were created in most cases by introducing two nucleotide exchanges affecting the same codon in order to avoid rapid reversion to wild type (see Table S1 in the supplemental material). Highly conserved residues were replaced in most cases by alanine in the context of a subgenomic JFH-1 luciferase replicon (Fig. 1C), and the mutations were analyzed for their effect on viral RNA replication in transient-transfection assays. As shown in Fig. 1D, substitutions R193A, E199A, G200A, H217V, and H250A had only a minor or no effect on RNA replication and moderately slowed down replication kinetics, which is most visible with the 24-h values. A strong impairment of RNA replication was observed for mutants W204A and E226A, whereas substitutions N206A, R207A, I209V, F211A, R214A, P220A, H222A, and Y223A completely abrogated HCV RNA replication, as inferred from the comparison with the nonreplicative ΔGDD polymerase mutant. Interestingly, four of five mutations affecting the extremely conserved α-helix 1 completely abolished RNA replication, arguing for its important role in the formation or activity of the viral replicase.
Fig. 1.
Residues in the C-terminal domain of NS4B are important for HCV RNA replication. (A) Predicted NS4B membrane topology. The schematic representation shows the N-terminal amphipathic α-helices AH1 and AH2, the four transmembrane segments (TM1 to TM4), the nucleotide binding motif (NBM), the C-terminal α-helices H1 and H2, and the very C-terminal palmitoylation site(s). The reported posttranslational flip of the N terminus into the ER lumen is indicated by the arrow. (B) HCV JFH1 protein sequence (2a_JFH) of the NS4B C-terminal region. NS4B amino acid (aa) positions are given. Fully conserved amino acids are marked with asterisks. Alternative amino acid residues found in other genotypes are indicated below. Residues targeted in this mutagenesis study are underlined and highlighted with bold letters. (C) Schematic representation of the subgenomic luciferase reporter replicon. The luciferase is expressed as an N-terminal fusion with 16 aa of the N-terminal region of the core protein (black line) and is translated under the control of the HCV IRES contained in the 5′UTR. The second cistron (NS3 to NS5B) is translated via the internal ribosome entry site of the encephalomyocarditis virus (E-IRES). The star indicates the region in NS4B targeted by the mutations. (D) Huh7 cells were transfected with in vitro transcribed luciferase replicon RNAs specified in the bottom. Cells were lysed 4, 24, 48, and 72 h after transfection, and the luciferase activity in cell lysates was determined. The data were normalized to the 4-h value that reflects transfection efficiency. The background of the assay is determined with the active-site NS5B polymerase mutant (ΔGDD). The mean values of two independent experiments are shown. Error bars indicate standard deviations.
Impact of mutations in the NS4B C-terminal domain on assembly and release of infectious HCV particles.
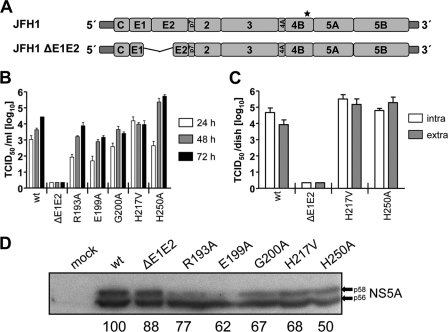
Given the minor effect of mutations R193A, E199A, G200A, H217V, and H250A on HCV RNA replication, we wondered whether these amino acid substitutions might influence the production of infectious HCV, as described earlier (22). The mutations were therefore introduced into the JFH-1 full-length genome (Fig. 2A), and in vitro transcripts derived therefrom were transfected into Huh7-Lunet cells. The wild-type (wt) genome and a mutant that due to a deletion in the E1-E2 coding region was unable to produce infectious virus particles served as positive and negative controls, respectively. Release kinetics of infectious HCV progeny, as well as the ratios of intra- and extracellular infectivity, were determined by limiting-dilution assay (Fig. 2B and C). Mutants R193A, E199A, and G200A produced lower amounts of infectivity compared to the wt (Fig. 2B), probably reflecting their lower replication efficiency, as determined with the subgenomic replicons (Fig. 1D) and the quantification of NS5A amounts produced in cells 72 h after transfection with the corresponding JFH-1 genome (Fig. 2D). We therefore concluded that these mutations did not affect virus production. However, cells transfected with mutant H217V released ∼10 times more infectious virus particles than the wt 24-h posttransfection, although titers stagnated at later time points and were thus comparable to the wt (Fig. 2B). In contrast, mutant H250A produced less infectious virus particles than the wt at 24 h posttransfection, which correlated with the delayed replication kinetics determined in the subgenomic replicon system (Fig. 1D). Nevertheless, a 10- to 100-fold increase in infectivity titer compared to the wt was detected 48 and 72 h after transfection, suggesting that this mutation enhances the assembly or release of HCV particles (Fig. 2B). This was not due to enhanced replication, as deduced from quantification of NS5A amounts produced in cells at 72 h posttransfection (Fig. 2D).
Fig. 2.
Influence of mutations in the NS4B C-terminal region on production of infectious virus. (A) Schematic representation of the full-length JFH1 genome and the JFH1ΔE1E2 mutant containing a deletion removing most of the envelope glycoprotein coding region (codons 217 to 567 [50]). The asterisk indicates the NS4B region into which the mutations were introduced. (B) Release kinetics of infectious HCV particles. Huh7-Lunet cells were transfected with the HCV RNA specified at the bottom of the panel. Culture supernatants were harvested at given time points, and infectivity titers were determined by limiting-dilution assay and are expressed as TCID50/ml. The mean values of two independent experiments are shown; error bars indicate standard deviations. (C) Efficiency of virus assembly and release. Huh7-Lunet cells were transfected with given full-length HCV RNAs. After 48 h, the cells and supernatants were harvested and subjected to three freeze-thaw cycles, and the infectivity titers contained in the cell lysates (intracellular) and corresponding supernatants (extracellular) were determined by limiting-dilution assay. The mean values of two independent experiments are shown; error bars indicate standard deviations. (D) Expression levels of NS5A at 72 h posttransfection. Arrows indicate the basal (p56) and hyperphosphorylated (p58) forms of NS5A, respectively. Bands were quantified by densitometry, and the relative protein levels normalized to the wt are indicated at the bottom of the panel as percentages.
To confirm these results and to distinguish between assembly and release phenotypes, intra- and extracellular infectivity titers were determined (Fig. 2C). In agreement with the earlier results, mutants R193A, E199A, and G200A produced lower amounts of intra- and extracellular infectivity (data not shown). In the case of mutant H217V, both intra- and extracellular infectivity titers were elevated over the wt, arguing for enhanced assembly. In contrast, only the extracellular titer was significantly elevated in the case of mutant H250A, which might be due to an accelerated release of infectious particles. In summary, these results show that the C-terminal region of NS4B is not only essential for RNA replication but also contributes to the assembly of infectious HCV particles.
Identification of pseudoreversions rescuing the replication defect caused by mutations in the C-terminal NS4B region.
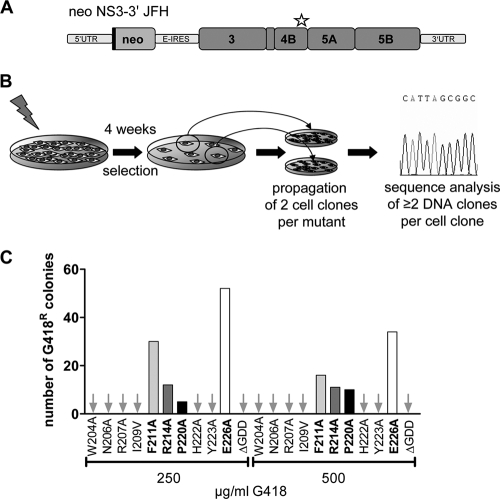
With the aim of selecting pseudoreversions compensating for the replication defect of the NS4B mutations described above, we introduced the single amino acid substitutions of replication-defective NS4B mutants into a selectable subgenomic JFH-1 replicon encoding the neomycin phosphotransferase gene in the first cistron (Fig. 3A). In vitro transcripts derived from these DNA constructs were transfected into Huh7 cells, which were cultured in the presence of the neomycin analogue G418 in order to select for cells containing functional HCV replicons (Fig. 3B). After about 4 weeks of selection, colonies emerging from single cell clones became visible for mutants F211A, R214A, P220A, and E226A, whereas for the other mutants no colonies were obtained, even at low selective pressure (250 μg of G418/ml; Fig. 3C). In order to identify pseudoreversions in selected replicons, total RNA was extracted from single cell clones and used for RT-PCR to amplify a DNA fragment spanning almost the complete nonstructural region. Amplicons were cloned, and at least two DNA clones per cell clone were subjected to nucleotide sequence analysis. Only mutations conserved between the two DNA clones were considered for subsequent analyses as described earlier (25, 30) (Table 1) . In all of the clones, the original NS4B mutation, caused in most cases by at least two nucleotide substitutions (see Table S1 in the supplemental material), had been retained, and at least one additional conserved mutation residing in most cases also in the C-terminal region of NS4B was detected (Fig. 4A). Interestingly, an additional intergenic compensatory mutation residing in NS5A domain I (K139E) was found with replicon mutants F211A and E226A, but this NS5A mutation was only found in combination with a second-site mutation in the NS4B C-terminal region.
Fig. 3.
Identification of pseudoreversions compensating for NS4B defects. (A) Schematic representation of the selectable subgenomic replicon. The neomycin phosphotransferase is expressed as an N-terminal fusion with 16 aa of the N-terminal region of the core protein (black line) and is translated under the control of the HCV IRES contained in the 5′UTR. The second cistron (NS3 to NS5B) is translated via the internal ribosome entry site of the encephalomyocarditis virus (E-IRES). The asterisk highlights the region into which the NS4B mutations were introduced. (B) Experimental approach used to select for pseudoreversions. Huh7 cells were transfected with selectable replicon RNAs, and cells were cultured in the presence of 250 or 500 μg of G418/ml for 4 weeks. For each mutant two cell clones were expanded, the total RNA was isolated, and at least two cDNA clones per cell clone were sequenced. (C) Number of G418-resistant cell colonies. Arrows indicate those mutants for which no colonies could be selected.
Table 1.
Pseudoreversions selected with replication-defective NS4B mutantsa
| Primary mutation | Pseudoreversion | Nucleotide exchangeb |
|---|---|---|
| F211A | L208F | CTT→TTT |
| T221N | ACT→AAT | |
| ±NS5A K139E | AAA→GAAc | |
| R214A | R192G | CGC→GGC |
| P220A | G200E | GGC→GAA |
| I253M | ATA→ATG | |
| E226A | G200S | GGC→AGC |
| A201E | GCG→GAG | |
| +NS5A K139E | AAA→GAAd |
Primary NS4B mutations and corresponding pseudoreversions are given.
That is, the nucleotide substitution detected in the cloned cDNA.
Mutation detected in a fraction of analyzed DNA clones.
Mutation observed in all analyzed DNA clones.
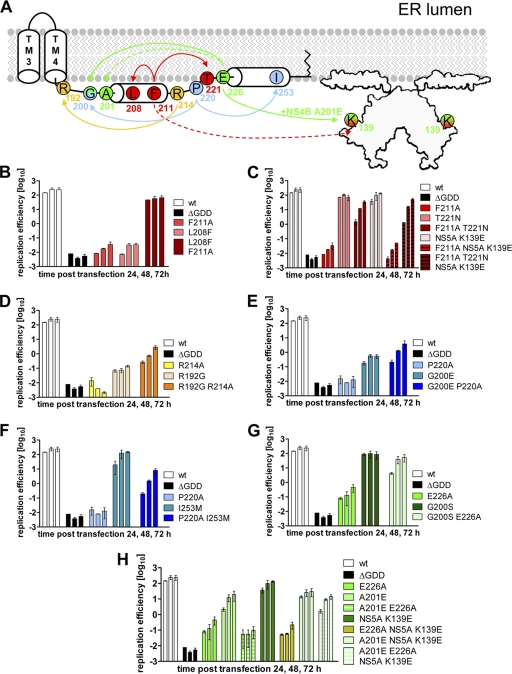
Fig. 4.
Pseudoreversions rescuing the replication defect of NS4B C-terminal mutants. (A) Schematic representation of primary NS4B mutations and the corresponding pseudoreversions. The NS4B C-terminal half (aa 130 to 261) containing TM3+TM4, as well as cytoplasmic helices H1 and H2, is depicted. The structure of the NS5A dimer (48) with the N-terminal amphipathic helix attached to a membrane bilayer is indicated. Primary mutations and corresponding pseudoreversions are labeled with the same color. Arrows originate from the primary mutation and point toward the corresponding pseudoreversion. Solid lines indicate functional rescue; broken lines indicate there was no such rescue. (B to H) Huh7-Lunet cells were transfected with luciferase reporter replicons specified in the right of each panel. Cells were lysed 4, 24, 48, and 72 h after transfection, and the luciferase activity in cell lysates was determined. The data were normalized to the 4-h value that reflects the transfection efficiency. Mean values from two independent experiments are shown; error bars indicate the standard deviations. (B) NS4B mutant F211A and pseudoreversion L208F in NS4B. (C) NS4B mutant F211A and pseudoreversions T221N in NS4B and K139E in NS5A. (D) NS4B mutant R214A and pseudoreversion R192G in NS4B. (E) NS4B mutant P220A and pseudoreversion G200E in NS4B. (F) NS4B mutant P220A and pseudoreversion I253M in NS4B. (G) NS4B mutant E226A and pseudoreversion G200S in NS4B. (H) NS4B mutant E226A and pseudoreversions A201E in NS4B and the intergenic compensatory mutation K139E in NS5A.
We next determined whether the pseudoreversions indeed were responsible for the rescue of HCV RNA replication by introducing them into the original replicon mutant. In addition, the mutations were inserted into the wt replicon in order to determine a general effect on RNA replication. Pseudoreversion L208F rescued RNA replication of mutant F211A but abrogated replication completely when introduced into the wt construct (Fig. 4B). Pseudoreversion T221N also restored replication of the parental F211A mutant, although replication was 5- fold lower compared to the wt (Fig. 4C). This reduction was not compensated for by additional insertion of the NS5A mutation K139E. Moreover, this NS5A mutation had no effect in the context of the wt or the primary NS4B mutant F211A (Fig. 4C). Pseudoreversion R192G rescued RNA replication of the primary NS4B mutant R214A, but this double mutant replicated ∼100-fold less efficient compared to the wt (Fig. 4D). Surprisingly, pseudoreversion R192G dramatically reduced replication of the wt. NS4B mutant P220A was rescued by pseudoreversions G200E (Fig. 4E) and I253M (Fig. 4F). However, while G200E abrogated replication of the wt, I253M had no effect. No impact on HCV RNA replication in the wt context was observed for pseudoreversion G200S, but it was able to rescue the strongly impaired mutant E226A (Fig. 4G). Finally, RNA replication of mutant E226A was also rescued, but only when pseudoreversions A201E and the intergenic compensatory mutation K139E in NS5A were combined, whereas none of them restored replication of the primary E226A mutant individually. Although these data argued for intra- and intermolecular NS4B-specific interactions and possibly also interaction(s) between NS4B and NS5A as a requirement for efficient HCV RNA replication, it was possible that the mutations affected NS4B stability.
Influence of mutations in the C-terminal region on NS4B protein stability.
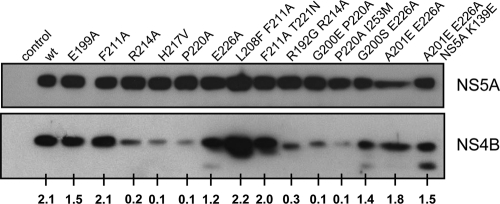
To address the possibility that mutations in NS4B affected its stability, we introduced a subset of mutations and the corresponding pseudoreversions into a construct directing the expression of an NS3 to NS5B polyprotein fragment corresponding to the minimal viral replicase (32). This approach allowed the analysis of NS4B protein stability also for the mutants abrogating viral replication, which would not be possible in a replicon system. Huh7-Lunet T7 cells were transfected with the respective constructs and steady-state levels of NS4B and NS5A were determined by Western blotting (Fig. 5). Although some of the primary NS4B mutations (R214A, H217V, and P220A) severely reduced NS4B abundance, these reductions did not correlate with RNA replication, e.g., mutant H217V replicated to wt level even though this NS4B was much less stable (for a complete summary, see Table 2). Moreover, stability of mutants F211A and E226A was not affected, even though they were severely impaired in RNA replication. Finally, pseudoreversions restoring replication ability of R214A and P220A did not raise NS4B protein stability but restored RNA replication, albeit to levels below the wt. We therefore concluded that the defects in RNA replication caused by the primary NS4B mutations are mainly due to mechanism(s) other than reduced NS4B stability.
Fig. 5.
Impact of mutations in the C-terminal region on NS4B protein stability. Huh7-Lunet T7 cells were transfected with the expression vector (control) or the same vector encoding the NS3-5B polyprotein fragment corresponding to wt NS4B or a NS4B mutant specified in the top. Cells were harvested 24 h posttransfection and subjected to Western blot analysis by using monospecific antibodies for NS4B or NS5A. The numbers below each lane refer to the relative amounts of NS4B proteins as determined by densitometry. After subtraction of the background, the NS4B signals were normalized to those of the corresponding NS5A. One result of two independent experiments is shown.
Table 2.
Properties of NS4B mutantsa
| Mutant | Replication | Virus production | Heterotypic interaction | Membrane alteration | Protein stability |
|---|---|---|---|---|---|
| R193A | + | + | ++ | ND | ND |
| E199A | + | + | ++ | Regular | ++ |
| G200A | ++ | + | ++ | ND | ND |
| W204A | − | ND | − | ND | ND |
| N206A | − | ND | − | ND | ND |
| R207A | − | ND | − | ND | ND |
| I209V | − | ND | − | ND | ND |
| F211A | − | ND | − | Aberrant | ++ |
| L208F + F211A | + | ND | ++ | Regular | ++ |
| F211A + T221N | + | ND | ++ | Regular | ++ |
| R214A | − | ND | − | Aberrant | − |
| R192G + R214A | + | ND | ++ | Regular | − |
| H217V | ++ | +++ | ++ | Regular | − |
| P220A | − | ND | − | Aberrant | − |
| G200E + P220A | + | ND | ++ | Regular | − |
| P220A + I253M | + | ND | ++ | Regular | − |
| H222A | − | ND | − | ND | ND |
| Y223A | − | ND | − | ND | ND |
| E226A | − | ND | − | Aberrant | ++ |
| G200S + E226A | + | ND | ++ | Regular | ++ |
| A201E + E226A | − | ND | + | Aberrant | ++ |
| A201E + E226A + NS5A K139E | + | ND | + | Regular | ++ |
| H250A | + | +++ | ++ | ND | ND |
+++, ∼10 times more than the wild type (wt); ++, same as the wt; +, ∼5 times less than the wt; -, at least 10 times less than the wt, significantly reduced (heterotypic interaction). ND, not determined.
Impact of mutations in the NS4B C-terminal region on homo- and heterotypic interactions.
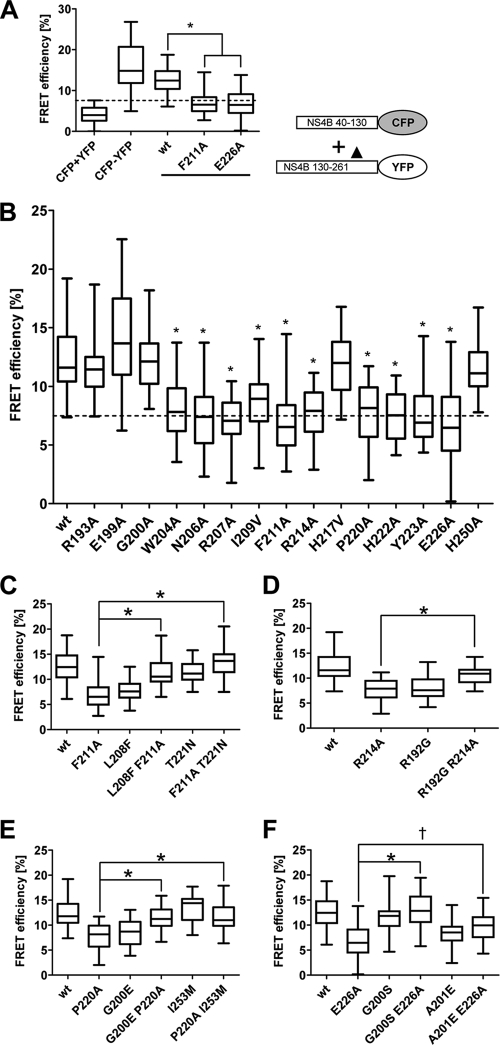
Since NS4B can oligomerize (19) and since this process is probably required to establish functional HCV replication complexes, we hypothesized that the primary NS4B mutations might have affected oligomerization. To address this hypothesis, we used a FRET-based assay (19) to determine the impact of the primary NS4B mutations and the corresponding pseudoreversions on NS4B oligomerization. As described recently (19), NS4B oligomerization can be mediated by interactions between identical protein fragments such as two C-terminal NS4B fragments (homotypic interactions) or by interactions between two different protein fragments such as an N-terminal and a C-terminal NS4B fragment (heterotypic interactions). In the initial set of experiments we tested a small subset of mutations (F211A and E226A) in order to determine which of these interactions might be affected. The mutations were inserted into expression constructs encoding for full-length NS4B or the C-terminal half (aa 130 to 261), each fused C-terminally to CFP or YFP. When using full-length NS4B proteins, mutations did not disrupt homodimerization to an extent detectable in the FRET assay (see Fig. S1A in the supplemental material). The same was found in FRET assays probing the homotypic interaction between the C-terminal NS4B fragments (see Fig. S1B and C). In contrast, when NS4B mutations were probed for their impact on heterotypic interaction, i.e., with the wt N-terminal fragment and a C-terminal NS4B fragment containing either of the mutations F211A or E226A, interaction was profoundly reduced (Fig. 6A). In fact, FRET efficiencies were significantly decreased arguing for a strong impairment of the NS4B heterotypic interaction (wt, 12.6 ± 0.4; F211A, 6.9 ± 0.5; E226A, 6.9 ± 0.6; means ± the standard errors of the mean [SEM]). Therefore, all other NS4B mutants were analyzed with this assay. Intriguingly, mutations that severely interfered with RNA replication also disrupted heterotypic NS4B interaction (Fig. 6B). In contrast, mutations R193A, E199A, G200A, H217V, and H250A that had no or only a very moderate effect on RNA replication also did not affect FRET between the NS4B N- and C-terminal fragments.
Fig. 6.
Mutations in the C-terminal region of NS4B and their impact on interaction with the NS4B N-terminal region. (A) Heterotypic interaction between the N-terminal and the C-terminal NS4B fragment. U2-OS cells were transfected for 24 h with expression plasmids encoding for NS4B CFP and YFP fusion proteins depicted in the lower right prior to FRET acceptor photobleaching. The C-terminal NS4B fragment corresponds either to the wt sequence or contains a mutation (▴) specified in the bottom of the diagram. Coexpression of CFP and YFP (CFP+YFP) and expression of a CFP-YFP fusion protein (CFP-YFP) served as negative and positive controls, respectively. Box-and-whisker diagrams show the median FRET efficiency (middle line) from 32 measurements. The bottom and top lines in the boxes represent the 25th and 75th percentiles, respectively. Minimum and maximum values are indicated by vertical lines. The dashed line indicates the background, as determined by the negative control. (B) Analogous to panel A but showing a larger panel of primary NS4B mutants. (C to F) Pseudoreversions restoring heterotypic NS4B interaction. U2-OS cells were transfected with constructs as described in the text, and interaction was determined by FRET analysis. The results are shown for mutants F211A (C), R214A (D), P220A (E), and E226A (F). For each data set, the statistical significance was calculated by using the unpaired Student t test as described in Materials and Methods (*, P < 0.0001; †, P = 0.0017).
Pseudoreversions restore heterotypic NS4B interaction.
Next, we wanted to know whether pseudoreversions that rescue RNA replication also restore heterotypic NS4B interaction. Pseudoreversions were therefore introduced into the construct encoding the C-terminal NS4B fragment (aa 130 to 261) and corresponding to either the wt sequence or containing the primary NS4B mutation, and interaction was measured by FRET assay. Interestingly, the heterotypic NS4B interaction that was disrupted by F211A was restored by the corresponding pseudoreversion L208F or T221N, whereas L208F, but not T221N, significantly reduced heterotypic NS4B interaction in the absence of the primary NS4B mutation (Fig. 6C). Moreover, pseudoreversion R192G significantly increased heterotypic NS4B interaction that was disrupted by the primary mutation R214A, whereas the same pseudoreversion decreased interaction with the N-terminal NS4B fragment when this primary mutation was absent (Fig. 6D). The impaired interaction of NS4B mutant P220A was restored by pseudoreversion G200E and I253M, whereas the G200E, but not the I253M substitution impaired heterotypic NS4B interaction when expressed in the wt construct (Fig. 6E). Finally, interaction of mutant E226A was significantly increased by pseudoreversion G200S or A201E, whereas A201E, but not G200S impaired the interaction between the NS4B N-terminal and C-terminal fragment when analyzed in the wt context (Fig. 6F). We note, however, that the increase in FRET efficiency achieved with pseudoreversion A201E in the context of the primary E226A substitution was considerably lower compared to other pseudoreversions. This observation might explain, why pseudoreversion A201E alone did not rescue RNA replication of mutant E226A (Fig. 4H) and required in addition the intergenic NS5A mutation K139E. We therefore assume that the primary NS4B mutation might have disrupted both NS4B dimerization and interaction with NS5A. In conclusion, as summarized in Table 2, the results reveal a direct correlation between HCV RNA replication and heterotypic NS4B interaction.
Expression of the HCV replicase proteins induces ER membrane alterations, including double membrane vesicles.
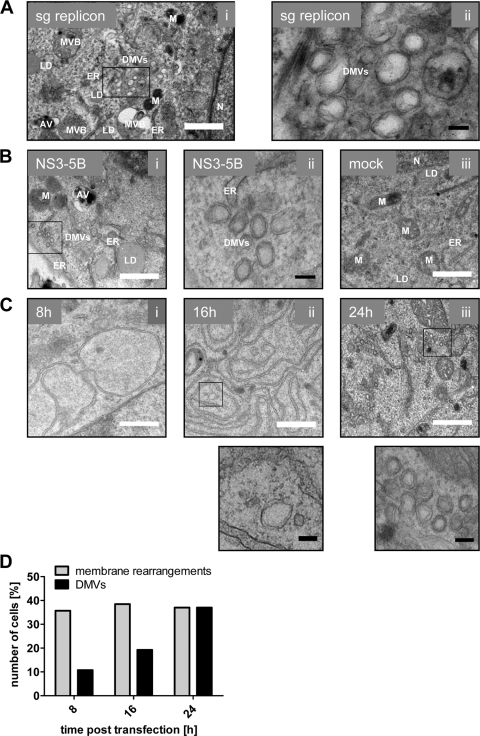
HCV infection (38), subgenomic replicons (12, 15), and (over-)expression of NS4B (10) have been reported to induce ER membrane alterations, in particular membranous vesicles that accumulate in the cytoplasm and form a heteromorphic structure designated “membranous web.” Since NS4B is believed to be the scaffold protein of the HCV replication complex and the key player in the induction of membrane remodeling, we sought to analyze our set of NS4B mutants and pseudorevertants for their capacity to induce membrane rearrangements. Given the possible contribution of other HCV proteins on membranous web formation, and to allow analysis of NS4B mutants disrupting RNA replication, we utilized the expression system described above (Fig. 5). In the initial set of experiments, we compared the membrane alterations induced by expression of the NS3-5B polyprotein fragment or by an actively replicating subgenomic JFH-1 replicon by using transmission electron microscopy (TEM). Clusters of double-membrane vesicles (DMVs) were consistently found in cells containing the subgenomic HCV RNA (Fig. 7A) or expressing NS3 to NS5B (Fig. 7Bi and ii) 24 h after transfection of the replicon RNA or the DNA expression construct, respectively, but not in mock-transfected cells (Fig.7Biii). DMVs, the presumed sites of RNA replication (12, 15), of mostly regular and circular shape, with closely apposed inner and outer membrane and a diameter of 100 to 200 nm were prominently found in close proximity to ER membranes, mitochondria, and lipid droplets. Time course experiments revealed the appearance of dilated ER membranes forming large circular structures 8 h after DNA transfection, and these rearrangements accumulated thereafter (Fig. 7Ci and ii, respectively). DMVs also became first detectable 8 h after transfection. Their abundance increased dramatically and 24 h posttransfection they were the predominant structures induced by the viral proteins (Fig. 7Ciii and D). Interestingly, we observed ER membrane alterations at all time points in ca. 30% of the cells, reflecting transfection efficiency (Fig. 7D). In contrast, the number of cells containing DMVs increased over time arguing that formation of these structures might require prior membrane rearrangements (Fig. 7D).
Fig. 7.
Ultrastructural analysis of membrane rearrangements induced by the HCV replicase proteins. (A) Huh7-Lunet cells were transfected with in vitro transcribed subgenomic replicon RNA. After 24 h, the cells were fixed, processed, and analyzed by TEM as described in Materials and Methods. The boxed area in the left panel is shown enlarged in the right panel. (B) Huh7-Lunet T7 cells were transfected with the expression vector encoding nonstructural proteins NS3 to NS5B (i and ii) or empty vector (iii). After 24 h, the cells were fixed, processed, and analyzed by TEM. The boxed area in the left panel (i) is shown enlarged in the middle panel (ii). (C) Time-dependent membrane alterations induced by expression of the HCV replicase proteins (NS3 to NS5B). Huh7-Lunet T7 cells were fixed 8 h (i), 16 h (ii), and 24 h (iii) after transfection and analyzed by TEM. The boxed areas in the middle and right panels are shown enlarged below. (D) Percentage of cells displaying membrane rearrangements and/or DMVs at given time points after transfection. Note the time-dependent increase of DMVs. For each time point, ∼30 cells were examined. White scale bar, 1 μm; black scale bar, 100 nm. AV, autophagic vacuole; DMVs, double membrane vesicles; ER, endoplasmic reticulum; LD, lipid droplet; M, mitochondrium; MVB, multivesicular body; N, nucleus.
Morphological alterations of intracellular membranes induced by NS4B mutants are restored by pseudoreversions.
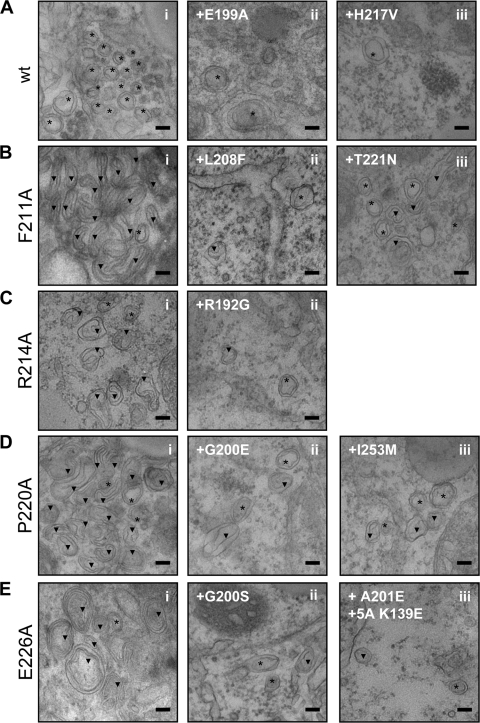
To examine the effect of the NS4B mutations and the corresponding pseudoreversions on induction of membrane alterations, especially DMV formation, mutations were introduced into the NS3-5B expression construct, transfected into Huh7-Lunet T7 cells, and analyzed by TEM as described above. Cells were fixed 24 h after transfection, because at this time point DMVs were most prominent. Mostly regular DMVs of wt morphology were observed in case of mutants E199A and H217V that only moderately reduced RNA replication (Fig. 8A and Table 2). However, DMV abundance was lower in the case of the two mutants, and the vesicles were only rarely found in clusters. In case of H217V this reduced frequency might be due to lower stability of this NS4B protein (Fig. 5 and Table 2). In contrast, mutations F211A, R214A, P220A, and E226A dramatically altered the morphology of membrane rearrangements (Fig. 8B to E), giving rise to long tubular structures (mutants F211A and P220A), U-shaped tubular structures (R214A), or predominantly multimembrane vesicles (MMVs) (mutant E226A). Induction of membrane rearrangements with an aberrant morphology did not correlate with stability of the respective NS4B protein (Fig. 5 and Table 2). Although mutants R214A and P220A were less stable compared to the wt, mutants F211A and E226A were not, and yet these proteins expressed in the context of the NS3-5B polyprotein induced different morphotypes.
Fig. 8.
Effects of mutations in the C-terminal NS4B region and pseudoreversions on HCV-induced membrane rearrangements. Lunet-T7 cells were transfected with the NS3-5B expression vector containing either wt NS4B or given mutation(s). After 24 h, the cells were fixed, processed, and analyzed by TEM as described in Materials and Methods. (A) DMV morphology induced by replication-competent NS3 to NS5B containing wt NS4B (i) or the replication-competent NS4B mutants E199A (ii) or H217V (iii). (B to E) DMV morphology induced by replication-incompetent NS4B mutants specified in the left of each panel (i) and the corresponding pseudorevertants (subpanels ii and iii in each panel). Regular (*) and aberrant (▾) DMVs are indicated. The scale bar in each panel represents 100 nm.
We next examined whether pseudoreversions would restore altered morphology of the membrane rearrangements. Pseudoreversions L208F and T221N indeed restored DMV morphology when combined with the primary NS4B mutation F211A (Fig. 8Bii and iii, respectively). Likewise, pseudoreversion R192G rescued the DMV morphology of the primary mutant R214A (Fig.8Cii), and the aberrant vesicular structures induced by the P220A mutant were restored to wt DMV morphology by pseudoreversions G200E and I253M (Fig. 8Dii and iii, respectively). Importantly, the pseudoreversions did not increase diminished NS4B protein stability in the case of primary NS4B mutants R214A and P220A (Fig. 5 and Table 2) but rescued the morphology of membrane alterations and RNA replication. Finally, the aberrant MMV phenotype observed in case of mutant E226A was rescued by pseudoreversions G200S (Fig.8Eii) or by the combination of pseudoreversion A201E in NS4B and the intergenic mutation K139E in NS5A (Fig.8Eiii), whereas A201E alone did not restore the DMVs of wt morphology (data not shown). In addition, drastic alterations of ER membrane rearrangements induced by primary mutation E226A were only reverted to the wt, when mutation K139E in NS5A was present (see Fig. S2 in the supplemental material). These results argue for a possible NS4B-NS5A interaction required for the establishment of functional HCV replication complexes.
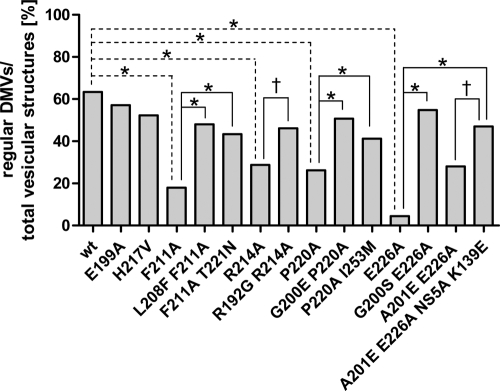
For a more quantitative analysis of the various phenotypes, the observed differences in DMV morphology were classified into regular DMVs and aberrant DMVs or MMVs and used to calculate a ratio (Fig. 9). In agreement with our previous observations, ∼60% of total vesicular structures induced by the wt replicase were regular DMVs, and a similar ratio of ∼50 to 55% was detected in the case of replication-competent NS4B mutants (E199A and H217V). In contrast, the relative amounts of regular DMVs were considerably decreased by replication-inhibiting mutations (F211A, R214A, P220A, and E226A) but elevated to wt levels by the corresponding pseudoreversions (Fig. 9 and Table 2). In summary, these results reveal a direct correlation between NS4B self-interaction, RNA replication, and the induction of DMVs. We conclude that the heterotypic NS4B self-interaction described here is essential for the formation of a functional replication complex.
Fig. 9.
Quantitative analysis of DMV morphotypes. At least 100 vesicular structures per sample from at least three different cells were counted. For each construct the ratio of regular DMVs per total HCV-induced vesicular structures (regular DMVs, aberrant DMVs, and MMVs) is given. The significance of the differences was calculated by using the two-sided Fisher's exact test and is highlighted for the wt and the primary mutants (dashed lines) or the primary mutant and corresponding pseudorevertants (black lines) (*, P < 0.0001; †, P < 0.01).
DISCUSSION
In this study we provide evidence that heterotypic interaction between the NS4B N-terminal region and the highly conserved cytoplasmic C-terminal domain is required to induce membrane alterations, most notably DMVs. Their presence and a specific morphology correlates with efficient replication, arguing that DMVs might be the site of viral RNA amplification. Clusters of DMVs might morphologically resemble membranous web structures described previously (10, 15). In support of this notion, DMVs were the most prominent structures induced by the HCV replicase (NS3 to NS5B) and were previously reported to contain nonstructural proteins and HCV RNA (12). A very critical determinant for proper DMV formation is the predicted α-helix H1 (aa 201 to 213; Fig. 1A). All mutations affecting this domain abolished RNA replication, and 4 of 7 selected pseudoreversions resided in H1 or the capping N-terminal glycine at position 200. The predicted helical structure of H1 is supported by the fact that pseudoreversion L208F is localized 3 aa residues N-terminal of the primary F211A mutation, a finding consistent with one turn of an α-helix. In a three-dimensional structure, this pseudoreversion will preserve a phenylalanine residue exposed on the same side of the α-helix.
The heterotypic interaction described here appears to be inconsistent with the posttranslational membrane passage of the NS4B N-terminal region from the cytoplasmic side into the ER lumen (34). However, only a fraction of NS4B was shown to undergo this topology switch, suggesting that the majority of NS4B molecules are available for heterotypic N-/C-terminal interaction. Moreover, the membrane passage probably is a highly regulated and dynamic process involving not only viral proteins such as NS5A (34) but eventually also cellular proteins. Supporting this idea, the strict conservation of H1 among all HCV isolates may indicate that this α-helix is engaged in a direct interaction with a cellular partner. In this case, complexes of different composition and NS4B membrane topology might form. Whether such complexes also exert different functions remains to be determined.
Several NS4B proteins containing mutations in the C-terminal region were of low abundance as determined by Western blotting. Although the diminished signals might be due to poor recognition by the NS4B-specific antiserum, this possibility is not very likely because we used a polyclonal antiserum raised against the full-length protein that most likely contains several epitopes. We therefore assume that the mutations in the C-terminal region of NS4B reduced protein stability. Importantly, reduced NS4B amounts did not correlate with RNA replication (e.g., H217V) or induction of DMVs and dimerization (e.g., G200E + P220A; for a complete summary, see Table 2). Moreover, even mutants with unaltered stability induced aberrant DMVs and had lost replication competence (e.g., E226A). Finally, some pseudoreversions restored RNA replication of unstable NS4B mutants without increasing NS4B stability (e.g., R214A and P220A). Thus, NS4B quantity is not a major determinant of HCV RNA replication.
Proteins able to trigger membrane curvature and membranous vesicles are known to oligomerize and form higher-order complexes (41, 43, 52). This is well described for replicase proteins of RNA viruses such as Flock house virus protein 1a (9), which induces vesicular invagination of the outer mitochondrial membranes (27), and mouse hepatitis virus nsp4 that triggers the formation of regularly shaped DMVs (7, 14). Considering previous work (19) and the present study, we assume that the same might hold true for NS4B of HCV, thus reflecting the evolutionary relationship between positive-strand RNA viruses. Interestingly, in mutant E226A we identified a compensatory intergenic mutation in NS5A that is required in addition to the pseudoreversion in NS4B to restore the replication competence of the mutant and regular DMV morphology. This NS5A mutation is exposed on the surface of either of the dimeric complexes of NS5A domain I (33, 48). We therefore assume that, in addition to NS4B oligomerization, its interaction with NS5A is important for formation of fully functional replication complexes.
Aberrant membrane alterations observed for replication-defective NS4B mutants might be a consequence of mutation-induced accumulation of misfolded NS4B proteins, which could trigger the unfolded protein response, leading to cellular degradation processes, including autophagy. However, the typical double membrane autophagic vesicles (AV) observed in cells expressing wt HCV replicase proteins (Fig.7Bi) were bigger (>400 nm) than the aberrant vesicular membrane alterations induced by NS4B mutants and contained very electron-dense material. Therefore, we conclude that mainly intrinsic NS4B mechanisms were affected by the mutations, thus leading to aberrant membrane alterations.
Apart from its major role in RNA replication, we found that the C terminus of NS4B influences the production of infectious HCV progeny. Mutations H217V and H250A significantly enhanced assembly or release of virus particles. Interestingly, a recent study reported N216A as a titer-enhancing mutation (22), emphasizing the role of the C-terminal NS4B domain in the assembly or release process. The underlying mechanism is not known but might be rather indirect. One possibility is an altered interaction of NS4B with NS5A, which is an RNA-binding protein that critically contributes to assembly (reviewed in reference 4). Such an altered interaction might facilitate the transfer of viral RNA from the replicase to the assembly site, thereby stimulating virus production.
In conclusion, we show that heterotypic NS4B self-interactions are required for formation of functional DMVs, which are the likely sites of HCV RNA replication. The results argue for a central role of NS4B in formation of functional replication complexes, a process that most likely requires additional viral and cellular factors. Their identification and molecular characterization will be the next important task.
Supplementary Material
Acknowledgements
We are grateful to Ulrike Herian and Annette Stradtmann for excellent technical assistance and to Cvetelina Coneva, Matthias Reiss, and Maria Xydia for initial help in constructing the NS4B mutants. We thank Takaji Wakita for the gift of the JFH-1 isolate and Charles Rice for Huh7.5 cells and 9E10 antibody. We are grateful to the Electron Microscopy Core Facility of the University of Heidelberg for their continuous support and providing access to their unit.
This study was supported by grants of the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 638 [Teilprojekt A5] and SFB/TRR 83 [Teilprojekt 13]) and the Swiss National Science Foundation (3100A0-122447).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Appel N., Pietschmann T., Bartenschlager R. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J. Virol. 79:3187–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Backes P., et al. 2010. Role of annexin A2 in the production of infectious hepatitis C virus particles. J. Virol. 84:5775–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartenschlager R., Frese M., Pietschmann T. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71–180 [DOI] [PubMed] [Google Scholar]
- 4. Bartenschlager R., Penin F., Lohmann V., Andre P. 2010. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 19:95–103 [DOI] [PubMed] [Google Scholar]
- 5. Blight K. J. 2007. Allelic variation in the hepatitis C virus NS4B protein dramatically influences RNA replication. J. Virol. 81:5724–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blight K. J., McKeating J. A., Rice C. M. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clementz M. A., Kanjanahaluethai A., O'Brien T. E., Baker S. C. 2008. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology 375:118–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. den Boon J. A., Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 64:241–256 [DOI] [PubMed] [Google Scholar]
- 9. Dye B. T., Miller D. J., Ahlquist P. 2005. In vivo self-interaction of nodavirus RNA replicase protein a revealed by fluorescence resonance energy transfer. J. Virol. 79:8909–8919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Egger D., et al. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elazar M., Liu P., Rice C. M., Glenn J. S. 2004. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J. Virol. 78:11393–11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferraris P., Blanchard E., Roingeard P. 2010. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J. Gen. Virol. 91:2230–2237 [DOI] [PubMed] [Google Scholar]
- 13. Friebe P., Boudet J., Simorre J. P., Bartenschlager R. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gadlage M. J., et al. 2010. Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function. J. Virol. 84:280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gosert R., et al. 2003. Identification of the hepatitis C virus RNA replication complex in huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gouttenoire J., et al. 2009. Identification of a novel determinant for membrane association in hepatitis C virus nonstructural protein 4B. J. Virol. 83:6257–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gouttenoire J., Montserret R., Kennel A., Penin F., Moradpour D. 2009. An amphipathic alpha-helix at the C terminus of hepatitis C virus nonstructural protein 4B mediates membrane association. J. Virol. 83:11378–11384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gouttenoire J., Penin F., Moradpour D. 2010. Hepatitis C virus nonstructural protein 4B: a journey into unexplored territory. Rev. Med. Virol. 20:117–129 [DOI] [PubMed] [Google Scholar]
- 19. Gouttenoire J., Roingeard P., Penin F., Moradpour D. 2010. Amphipathic α-helix AH2 is a major determinant for the oligomerization of hepatitis C virus nonstructural protein 4B. J. Virol. 84:12529–12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jirasko V., et al. 2008. Structural and functional characterization of nonstructural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 283:28546–28562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones C. T., Murray C. L., Eastman D. K., Tassello J., Rice C. M. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374–8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones D. M., Patel A. H., Targett-Adams P., McLauchlan J. 2009. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J. Virol. 83:2163–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kärber G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn Schmiedebergs Arch. Pharmakol. Exp. Pathol. 162:480–487 [Google Scholar]
- 24. Kaul A., et al. 2009. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS. Pathog. 5:e1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaul A., Woerz I., Meuleman P., Leroux-Roels G., Bartenschlager R. 2007. Cell culture adaptation of hepatitis C virus and in vivo viability of an adapted variant. J. Virol. 81:13168–13179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koch J. O., Bartenschlager R. 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J. Virol. 73:7138–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kopek B. G., Perkins G., Miller D. J., Ellisman M. H., Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS. Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lavanchy D. 2009. The global burden of hepatitis C. Liver Int. 29(Suppl. 1):74–81 [DOI] [PubMed] [Google Scholar]
- 29. Lindenbach B. D., et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 30. Lohmann V., Hoffmann S., Herian U., Penin F., Bartenschlager R. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lohmann V., Korner F., Herian U., Bartenschlager R. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lohmann V., et al. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 33. Love R. A., Brodsky O., Hickey M. J., Wells P. A., Cronin C. N. 2009. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 83:4395–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lundin M., Lindstrom H., Gronwall C., Persson M. A. 2006. Dual topology of the processed hepatitis C virus protein NS4B is influenced by the NS5A protein. J. Gen. Virol. 87:3263–3272 [DOI] [PubMed] [Google Scholar]
- 35. Lundin M., Monne M., Widell A., Von Heijne G., Persson M. A. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 77:5428–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller S., Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nakabayashi H., Taketa K., Miyano K., Yamane T., Sato J. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858–3863 [PubMed] [Google Scholar]
- 38. Pfeifer U., et al. 1980. Experimental non-A, non-B hepatitis: four types of cytoplasmic alteration in hepatocytes of infected chimpanzees. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 33:233–243 [DOI] [PubMed] [Google Scholar]
- 39. Ponten J., Saksela E. 1967. Two established in vitro cell lines from human mesenchymal tumours. Int. J. Cancer 2:434–447 [DOI] [PubMed] [Google Scholar]
- 40. Quinkert D., Bartenschlager R., Lohmann V. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 79:13594–13605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reynwar B. J., et al. 2007. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature 447:461–464 [DOI] [PubMed] [Google Scholar]
- 42. Schaller T., et al. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 81:4591–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shibata Y., Hu J., Kozlov M. M., Rapoport T. A. 2009. Mechanisms shaping the membranes of cellular organelles. Annu. Rev. Cell Dev. Biol. 25:329–354 [DOI] [PubMed] [Google Scholar]
- 44. Simmonds P., et al. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973 [DOI] [PubMed] [Google Scholar]
- 45. Spearman C. 1908. The method of “right and wrong cases” (“constant stimuli”) without Gauss's formulae. Br. J. Psychol. 2:227–242 [Google Scholar]
- 46. Steinmann E., et al. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS. Pathog. 3:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Targett-Adams P., Boulant S., McLauchlan J. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J. Virol. 82:2182–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tellinghuisen T. L., Marcotrigiano J., Rice C. M. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van den Hoff M. J., Christoffels V. M., Labruyere W. T., Moorman A. F., Lamers W. H. 1995. Electrotransfection with “intracellular” buffer. Methods Mol. Biol. 48:185–197 [DOI] [PubMed] [Google Scholar]
- 50. Wakita T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yu G. Y., Lee K. J., Gao L., Lai M. M. 2006. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 80:6013–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zimmerberg J., Kozlov M. M. 2006. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell. Biol. 7:9–19 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.