Abstract
The exogenous and pathogenic Jaagsiekte sheep retrovirus (JSRV) coexists with highly related and biologically active endogenous retroviruses (enJSRVs). The endogenous enJS56A1 locus possesses a defective Gag polyprotein which blocks the late replication steps of related exogenous and endogenous retroviruses by a mechanism known as JSRV late restriction (JLR). Conversely, enJSRV-26, which most likely integrated into the sheep genome less than 200 years ago, is able to escape JLR. In this study, we demonstrate that the ability of enJSRV-26 to escape JLR is due to a single-amino-acid substitution in the signal peptide (SP) of its envelope glycoprotein. We show that enJSRV-26 SP does not localize to the nucleolus, unlike the functional SPs of related exogenous and endogenous sheep betaretroviruses. In addition, enJSRV-26 SP function as a posttranscriptional regulator of viral gene expression is impaired. enJSRV-26 JLR escape relies on the presence of the functional enJS56A1 SP. Moreover, we show that the ratio between enJSRV-26 and enJS56A1 Gag is critical to elude JLR. Interestingly, we found that the domestic sheep has acquired, by genome amplification, several copies of the enJS56A1 provirus. These data further reinforce the notion that transdominant enJSRV proviruses have been positively selected in domestic sheep, and that the coevolution between endogenous and exogenous sheep betaretroviruses and their host is still occurring.
INTRODUCTION
Retroviruses must integrate their genome into the host genomic DNA to replicate successfully. As a consequence of their peculiar replication cycle, retroviruses exist in nature as exogenous retroviruses, transmitted horizontally from infected to uninfected host like any other virus, and endogenous retroviruses (ERVs). ERVs derive from the infection of the host germ line during evolution and are transmitted vertically from generation to generation like any other Mendelian gene (15, 18). ERVs colonize the genome of all vertebrates studied to date, where they represent a significant percentage of the DNA of their host species (e.g., ∼8% of the human and mouse genomes) (15, 18).
During evolution, most ERVs have accumulated genetic defects and lost the ability to express proteins and/or infectious viruses. However, some ERVs have been coopted by their hosts because they fulfill useful functions (4, 5, 12–14, 18). In addition, some ERVs protect the host against the infection of related exogenous pathogenic retroviruses. In chickens and mice, for example, it has been shown that the expression of Env glycoproteins by some ERVs can saturate the receptors used by related exogenous retroviruses to gain entry into the cell (42).
Domestic sheep provide a fascinating model for studying the interplay between retroviruses and their host. The sheep genome harbors at least 27 copies of endogenous betaretroviruses (enJSRVs) that are highly related to the exogenous and pathogenic Jaagsiekte sheep retrovirus (JSRV) (1, 2, 4, 29, 30, 39). enJSRVs have been integrating into the genome of their host throughout the evolution of the Caprinae for the last 5 to 7 million years (i.e., sheep, goats, and related species) (2).
enJSRVs (or at least some of the enJSRV proviruses) can be considered to be in symbiosis with their host, as they play an essential part in the reproductive biology of sheep and interfere with the replication cycle of related exogenous retroviruses (4, 11, 12, 34, 35, 39). A transdominant provirus, enJS56A1, blocks JSRV replication by a unique mechanism that we termed JSRV late restriction (JLR) (25). enJS56A1 possesses a defective Gag protein that does not traffic properly to the pericentriolar area, where newly formed viral particles assemble and use the recycling endosomes to exit from cells (3, 26). Interestingly, the defect of enJS56A1 is transdominant over JSRV as well as other enJSRVs. In other words, enJS56A1 Gag forms multimers with JSRV Gag, which consequently cannot traffic properly and subsequently are degraded by the proteasomal machinery of the cell (3, 26).
The main determinant of JLR is a tryptophan residue (W) at position 21 in enJS56A1 Gag, which replaces an arginine (R) that is well conserved in betaretroviruses (25). We showed that enJS56A1 possessed an arginine residue in Gag at position 21 when originally integrated into the host genome. Subsequently, the transdominant enJS56A1 with the W21 Gag residue appeared in the closest relatives of domestic sheep and then became fixed after domestication (2).
Interestingly, we also have identified five enJSRV proviruses (enJSRV-7, enJSRV-15, enJSRV-16, enJSRV-18, and enJSRV-26) with an intact genomic organization that are able to produce viral particles in vitro (2). These loci are insertionally polymorphic in domestic sheep, in other words, they are present in only some individuals/breeds. These observations suggest that the original integration of these proviruses occurred after domestication (i.e., in the last 10,000 years). In particular, enJSRV-26 probably integrated into the host germ line less than 200 years ago. Remarkably, this virus possesses the unique ability to escape the restriction induced by enJS56A1 (2). enJSRV-26 has been detected in the germ line of a single sheep to date, suggesting that an enJSRV-26-like exogenous retrovirus still circulates within the sheep population.
Sheep betaretroviruses have allowed us to witness sequential counteradaptations between endogenous and exogenous retroviruses and represent an ideal model to study the arms race between virus and host over long evolutionary periods. In this study, we investigated the molecular mechanisms followed by enJSRV-26 to elude the restriction induced by enJS56A1. Using a variety of approaches, we demonstrate that a single amino acid substitution in the signal peptide of the enJSRV-26 Env confers the ability of this virus to escape JLR. We and others have shown previously that the signal peptide (SP) of sheep and mouse betaretrovirus envelope glycoproteins is a multifunctional protein acting as a posttranscriptional regulator of viral gene expression (7, 8, 17). Here, we demonstrate that the SP of enJSRV-26 lacks at least some of these functions. In addition, we show that JLR escape depends on the ratio between enJSRV-26 and enJS56A1 Gag. Interestingly, we also obtain evidence suggesting that the transdominant proviruses are amplified within the genome of domestic sheep.
MATERIALS AND METHODS
Plasmids.
pCMV4JS21, pCMV5-enJS26, pCMV5-enJS18, and pCMV2en56A1 express the full-length JSRV21 molecular clone and the endogenous enJSRV-26, enJSRV-18, and enJS56A1, respectively. These plasmids have been described previously (2, 28, 30). All of the chimeras/mutants employed in this study were derived from the plasmids listed above and are schematically represented in Fig. 1. Specific details for the cloning procedures of any of the plasmids described below are available upon request. Mutants were obtained by site-directed mutagenesis using the QuikChange kit (Stratagene), as suggested by the manufacturer. Chimeras were derived by swapping, respec- tively, the full-length (enJS26-Env18 and enJS18-Env26), the 5′-end (enJS26-5′Env18 and enJS18-5′Env26), or the 3′-end (enJS26-3′Env18 and enJS18-3′Env26) env between enJSRV-26 and enJSRV-18. Mutants enJSRV-26ΔEnv and enJSRV-18ΔEnv contain two nonsense mutations in the first and third methionines of their respective Env glycoproteins. Mutant enJS56A1ΔGag contains two nonsense mutations replacing the first and third methionines of the enJS56A1 Gag. enJS56A1-4CTE contains four copies of the constitutive transport element (CTE) of Mason-Pfizer monkey virus (M-PMV) at the 3′ end of env. The M-PMV CTE was derived from pSarm4, as already described, and was a gift from Eric Hunter (32). Single mutants enJSRV26-EnvD6A, enJSRV18-EnvA6D, and JSRV-EnvA6D express the full-length proviruses with a single point mutation in their Env glycoprotein at position 6. Plasmids penJSEnv26 and penJSEnv18 express the Env of enJSRV-26 and enJSRV-18, respectively. These mutants were obtained by deleting gag, pro, pol, and orf-x from the plasmids encoding their respective full-length proviruses. pSP26-HA, pSP18-HA, and pSP56-HA encode the signal peptide of enJSRV-26, enJSRV-18, and enJS56A1, respectively, tagged with the hemagglutinin (HA) epitope. In penJS18Env-PPT, the signal peptide of enJSRV-18 Env was replaced by the signal peptide of the human preprotrypsin, followed by the FLAG epitope fused at the N terminus of the enJSRV-18 surface domain (SU) of Env. pNLgagSty330 and pRev were kindly provided by Barbara Felber and have been described already (16, 21). pNLgagSty330 is a Rev- and Tat-dependent HIV-1 Gag-Pol expression plasmid and, for simplicity, is termed pHIV1-RRE in this study. pRev is an expression plasmid for HIV-1 Rev. The expression plasmid for HIV-1 Tat was a gift from Mauro Giacca and has been described elsewhere (37). pHIV-SPRE26, pHIV-SPRE18, and pHIV-SPRE56 were obtained by replacing the HIV-1 Rev-responsive element (RRE) in pHIV1-RRE with the signal peptide-responsive elements (encompassing env and the 3′ untranslated region [UTR]) of enJSRV-26, enJSRV-18, and enJS56A1, respectively.
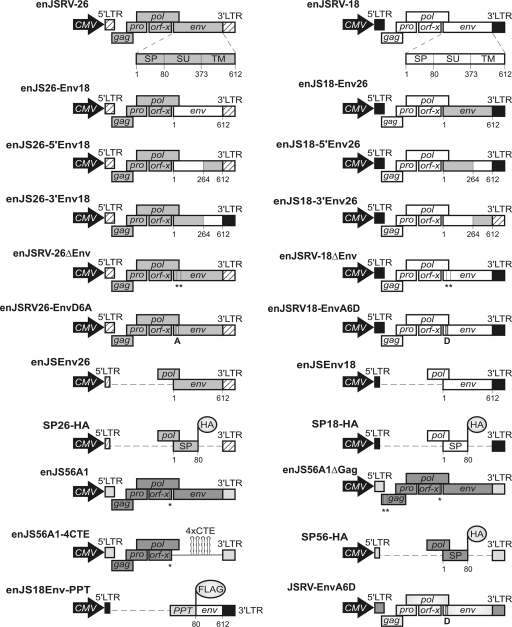
Fig. 1.
Schematic representation of the plasmids employed in this study. All of the mutants/chimeras used in this study were derived from expression plasmids encoding the full-length enJSRV-26 and enJSRV-18 proviruses (top). Numbers indicate amino acid residues of Env. Premature termination codons are indicated with vertical lines and asterisks. LTR, long terminal repeat; CMV, cytomegalovirus.
Cell cultures, transfections, and viral preparations.
293T and COS cells were cultured in Dulbecco's modified Eagle medium (Gibco) and supplemented with 10% fetal bovine serum at 37°C, 5% CO2, and 95% humidity. Virus preparations were obtained by transient transfections of 293T cells with the appropriate plasmids using the Calphos mammalian transfection kit (Clontech). The empty vector pcDNA3.1 (Invitrogen) was used to calibrate the amount of DNA used in each experiment. Cell supernatants were collected at 48 h posttransfection, and viral particles were concentrated by ultracentrifugation as already described (8, 30, 31). For the analysis of intracellular proteins, cells were lysed by standard techniques, as described previously (38).
Western blotting and immunoprecipitation.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed on concentrated viral particles and cell lysates (200 μg of protein extracts) as previously described (8, 30). enJSRV Gag was detected using a rabbit polyclonal serum against the JSRV major capsid protein (CA) (26). A rabbit polyclonal serum toward the JSRV transmembrane protein (TM) was employed to detect Env proteins. Signal peptides tagged with the HA epitope were detected with a mouse monoclonal anti-HA antibody (Abcam), while γ-tubulin was detected with a rabbit polyclonal antibody (Sigma). Membranes were exposed to the appropriate peroxidase-conjugated secondary antibodies and further developed by chemiluminescence using ECL Plus (Amersham). Levels of CA associated with viral particles released in the supernatants were quantified by measuring chemiluminescence in a Molecular Dynamics Storm 840 imaging system using ImageQuant TL software (Molecular Dynamics). Each experiment was repeated independently at least three times, and results are presented as the mean values for each sample (± standard errors). Env expression by enJS18Env-PPT was assessed by immunoprecipitation using a mouse monoclonal anti-FLAG antibody (Sigma) as already described (38).
Confocal microscopy.
293T and COS cells were plated onto two-well chambered glass slides (Lab-Tek; Nalge Nunc International) and transfected with the appropriate plasmids using Lipofectamine (Invitrogen) according to the manufacturer's instructions. Twenty-four h after transfection the cells were washed with phosphate-buffered saline (PBS) and fixed with 3% formaldehyde for 15 min. After fixation, cells were processed essentially as already described (25, 33). SP proteins tagged with the HA epitope were detected with a mouse monoclonal anti-HA (Abcam) antibody. Rabbit polyclonal antibody to fibrillarin (Abcam) was used as a marker for the nucleolus. Goat anti-mouse and anti-rabbit immunoglobulin G conjugated with Alexa Fluor 488 and Alexa Fluor 594, respectively (Molecular Probes), were used as secondary antibodies. Slides were mounted with medium containing DAPI (4′,6-diamidino-2-phenylindole; Vectashield, Vector Laboratories) and analyzed with a Leica TCS SP2 confocal microscope. Single sections from confocal optical sections along the z axis were analyzed.
Gag ELISA.
For enzyme-linked immunosorbent assay (ELISA), 293T cells were transfected with the appropriate plasmids (1 μg) in the presence or absence of SP26-HA, SP18-HA, SP56-HA (or pRev, as a control), and HIV-1 Tat (0.2 μg). At 48 h posttransfection, cell supernatants were assessed for the presence of HIV-1 Gag proteins using a Murex HIV antigen monoclonal antibody (MAb) kit (Abbot Murex) according to the manufacturer's instructions. All experiments were repeated independently at least three times.
qPCR.
Quantitative PCR (qPCR) assays were designed to estimate the dosage of target genes (enJS56A1, enJSRV-6, enJSRV-18, and SOX9) compared to that of the β-actin gene (used as reference gene) in genomic DNA of domestic and wild sheep. The genomic DNA tested was collected from various breeds (Dorset, Suffolk, Texel, Jacob, Red Maasai, Merino, Xalda, Rambouillet, Soay, Norway, and Finsheep) of domestic sheep (Ovis aries), wild sheep (O. dalli, O. canadensis, O. ammon, and O. vignei), Mediterranean mouflon (O. orientalis musimon), and members of the genera Budorcas (B. taxicolor) and Pseudois (P. nayaur). All DNA samples were obtained and used in a previous study (9). Standard curve efficiency was 99.2% for enJS56A1, 96.6% for enJSRV-6, 99% for enJSRV-18, 100% for β-actin, and 100% for SOX9. PCR assays were performed using a reverse primer complementary to the genomic 3′ flanking region for each provirus (enJS56A1, 5′-GGA AGG ATC TGA AAC GTG GA-3′; enJSRV-6, 5′-CAG GGG AAT AAC TGG TGC TAC CT-3′; and enJSRV-18, 5′-CAA GTG CCA GAG CCC AGA GCC A-3′) and a forward primer designed in a conserved region in env (5′-ATA AAG AGA GGG GAG CTG CG-3′). Primers for β-actin (forward, 5′-ATC ATG TTT GAG ACC TTC AAC ACC CC-3′; reverse, 5′-CCA GGA AGG AAG GCT GGA AGA GAG C-3′) and SOX9 (forward, 5′-CCT AGC TTT TCT TGC AGC C-3′; reverse, 5′-GCA TTC CCC AGA CAG ATT TC-3′) were designed on highly conserved regions of both genes. qPCR assays were carried out in triplicate in a total volume of 25 μl and performed in a Mx30005P (Stratagene) thermocycler, using the Brilliant II SYBR green QPCR low ROX master mix (Stratagene) and the Brilliant SYBR green QPCR core reagent kit (Stratagene) according to the manufacturer's instructions. The reaction mixture contained 20 ng of sheep genomic DNA. The reaction mixture was subjected to a denaturation step at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 30 s, primer annealing at the temperature appropriate for each primer (55°C to 60°C for 30 s), and elongation at 72°C for 30 s, ending with a melting-curve analysis to validate the specificity of the PCR products. Results were expressed as the ratio between the estimated number of molecules in the target and reference genes in each sample.
Genotyping enJS56A1 proviruses.
The presence of the codon encoding an arginine or tryptophan residue in enJS56A1 (and enJS56A1-like proviruses) at position 21 in Gag was assessed by PCR. Genomic DNA samples collected from various breeds of domestic sheep (Texel, n = 2; Merino, n = 1), wild sheep (O. dalli, n = 1; O. canadensis, n = 1; O. ammon, n = 1), and the Mediterranean Mouflon (Ovis orientalis musimon, n = 1) were amplified by PCR using a forward primer complementary to the genomic 5′-flanking region of the enJS56A1 or enJSRV-20 provirus and a reverse primer complementary to their gag gene as previously described (2). PCR products then were cloned into a pCR4-TOPO vector (Invitrogen), and 40 individual clones for each PCR product were completely sequenced.
FISH.
Fluorescent in situ hybridization (FISH) analysis was carried out essentially as already described (9). Briefly, sheep peripheral blood cells were cultured at 37°C in RPMI 1640 medium (Invitrogen) supplemented with 15% fetal bovine serum, 1.5% concanavalin A (Sigma), and penicillin-streptomycin (Invitrogen). Cells then were synchronized with 300 μg/ml thymidine (Sigma) and, after 18 h, washed and resuspended in medium containing 15 μg/ml bromodeoxyuridine (Sigma) and 30 μg/ml Hoechst 33258 (Invitrogen). Cells then were incubated for 6 h at 37°C (the last hour in the presence of 0.1 μg/ml colcemid; Sigma) and then treated with a hypotonic solution and washed three times with methanol-acetic anhydride. Cell suspensions then were plated onto slides, incubated overnight at 50°C, and stained for 10 min with 25 μg/ml Hoechst 33258. Slides were further probed with biotin-labeled bacterial artificial chromosome (BAC) clones containing the appropriate provirus. Hybridization, chromosome staining, signal detection, and image processing were performed as already described (9) in at least 30 metaphases for each probe. Chromosome identification was carried out using the R-banding karyotype by adding fluorescein avidin DCS and biotinylated anti-avidin (Vectors Laboratories), as recommended by the manufacturer. Chromosome identification and band nomenclature followed the International System for Chromosome Nomenclature of Domestic Bovids (10).
RESULTS
The 5′ portion of the enJSRV-26 env is the main determinant of JLR escape.
The first aim of this study was to identify the molecular determinants of enJSRV-26 that are necessary to escape JLR. At the nucleotide level, enJSRV-26 is 98% identical to enJSRV-18 along the entire genome. enJSRV-18 is another insertionally polymorphic provirus present in the sheep genome. Interestingly, enJSRV-26 and enJSRV-18 Gag and Env are 100 and 99.3% identical, respectively. However, enJSRV-26 and enJSRV-18 display different phenotypes in the presence of enJS56A1. enJSRV-18 is restricted by the transdominant enJS56A1 (like the exogenous JSRV), whereas enJSRV-26 escapes JLR (2) (Fig. 2A). Because the major differences between enJSRV-26 and enJSRV-18 are found in env, we reasoned that this region contains the main determinants for JLR escape. The enJSRV-26 and enJSRV-18 env genes differ for only 10 nucleotides, resulting in six synonymous and four nonsynonymous mutations.
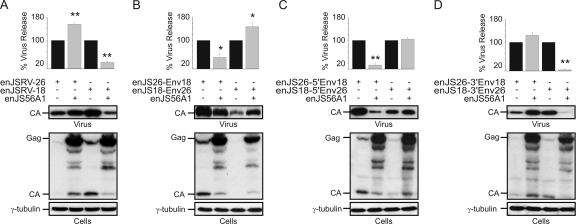
Fig. 2.
enJSRV-26 env 5′ end contains the determinants necessary to escape JLR. Shown are Western blots of concentrated supernatants (virus) and cell extracts (cells) of 293T cells transfected with the plasmids indicated in each panel. Membranes were incubated with antibodies against the major capsid protein of JSRV (CA) or γ-tubulin as a loading control. Levels of CA associated with viral particles released in the supernatants were quantified by chemifluorescence using ImageQuant TL software (Molecular Dynamics). Graphs represent data obtained from three independent experiments. The values obtained by each chimera expressed in the absence of enJS56A1 (black bars) were arbitrarily set as 100%. Error bars indicate standard errors; statistically significant differences are indicated with one (P < 0.05) or two (P < 0.01) asterisks. (A) Interference assays with wild-type enJSRV-26 and enJSRV-18. (B, C, and D) Interference assays using various enJSRV-26/enJSRV-18 chimeras as indicated in each panel. Only those chimeras containing the 5′ end of the env gene of enJSRV-26 are able to elude enJS56A1 restriction.
To identify the determinants of JLR escape, we generated a series of chimeras between enJSRV-18 and enJSRV-26 (Fig. 1) and carried out interference assays with the transdominant enJS56A1 (Fig. 2). As shown in Fig. 2B, the release of enJS26-Env18 viral particles was restricted in the presence of enJS56A1, while enJS18-Env26 escaped JLR. Furthermore, chimeras enJS18-5′Env26 and enJS26-3′Env18 also were able to escape JLR, while enJS56A1 was able to inhibit enJS26-5′Env18 and enJS18-3′Env26 viral particle release (Fig. 2C and D). Collectively, these results indicated that the 5′ end of enJSRV-26 env contains the main determinants of JLR escape.
Amino acid residue D6 in the enJSRV-26 Env is the main determinant of JLR escape.
We investigated whether the enJSRV-26 Env protein per se or cis-acting regions within the env gene were involved in JLR escape. We derived enJSRV-26 and enJSRV-18 mutants containing two premature termination codons in Env (enJSRV-26ΔEnv and enJSRV-18ΔEnv) to prevent its expression while maintaining the intact full-length viral genome. As expected, we found that enJSRV-26ΔEnv was able to escape JLR when the enJSRV-26 Env was provided in trans, whereas it was restricted in the presence of the enJSRV-18 Env (Fig. 3A). On the other hand, enJSRV18ΔEnv was restricted by enJS56A1 in the presence of the enJSRV-18 Env but escaped JLR when coexpressed with the enJSRV-26 Env (Fig. 3B). These data confirmed that the enJSRV-26 Env protein per se is necessary to escape JLR.
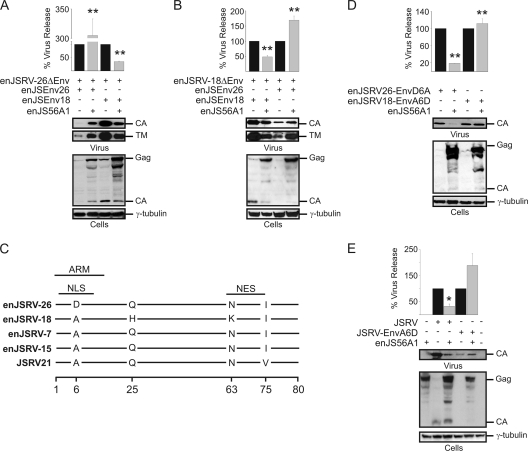
Fig. 3.
Amino acid residue D6 in the enJSRV-26 Env plays a major role in JLR escape. (A, B, D, and E) Western blots of concentrated supernatants (virus) and cell extracts (cells) of 293T cells transfected with the plasmids indicated in each panel. Membranes were incubated with antibodies against the major capsid protein (CA) of JSRV or γ-tubulin as a loading control. Levels of CA associated with viral particles released in the supernatants were quantified by chemifluorescence using ImageQuant TL software (Molecular Dynamics). Graphs represent data obtained from three independent experiments. The values obtained by each mutant expressed in the absence of enJS56A1 (black bars) were arbitrarily set as 100%. Error bars indicate standard errors; statistically significant differences are indicated with one (P < 0.05) or two (P < 0.01) asterisks. Env expression was controlled by incubating membranes with antibodies against the transmembrane (TM) domain of JSRV. (C) Graphic representation of the alignment of the amino acid sequences of the signal peptides of four insertionally polymorphic enJSRV proviruses and the exogenous JSRV. Lines represent identical residues in the sequences, while letters indicate differences in the amino acid residues. Numbering corresponds to amino acid residues in Env. ARM, arginine-rich motif; NLS, nuclear localization signal; NES, nuclear export signal.
The N-terminal region of the retroviral Env includes the signal peptide. The SPs of enJSRV-26 and enJSRV-18 Env differ in only three amino acid residues (Fig. 3C). However, the SP of enJSRV-7 and enJSRV-15 (two enJSRV loci that, like enJSRV-18, are restricted by enJS56A1) differ by only a single amino acid residue (residue 6) from the SP of enJSRV-26 (2). The alanine (A) residue in position 6 is well conserved in the SP of JSRV and all the insertionally polymorphic enJSRVs, with the exception of enJSRV-26, where it is replaced by an aspartic acid (D) residue (Fig. 3C). Thus, we hypothesized that the D6 residue plays a critical role in the ability of enJSRV-26 to escape JLR. To this end, we derived the full-length single mutants enJSRV26-EnvD6A and enJSRV18-EnvA6D. As shown in Fig. 3D, the D6A mutation conferred susceptibility to JLR to enJSRV-26, while the reciprocal mutation (A6D) allowed enJSRV-18 to escape enJS56A1-induced restriction. Similar results were obtained for the exogenous JSRV, where the A6D mutation allowed JSRV to escape JLR (Fig. 3E). These data show conclusively that a single amino acid substitution in the SP of the Env glycoprotein allows enJSRV-26 to escape JLR.
SPs of the enJSRV-26 and enJSRV-18 Env localize in different cellular compartments.
We investigated whether the SP of the enJSRV-26 Env possessed a different biological activity from that of SPs of enJSRV-18 and enJS56A1. We and others have shown that the SPs of sheep betaretroviruses are Rev-like multifunctional proteins that localize in the nucleoli and favor full-length viral RNA nuclear export and enhance Gag synthesis and viral particle release (8, 17). We have shown above that the A6D substitution allows enJSRV-26 to escape JLR. This residue lays within a predicted nuclear localization signal (NLS) and an arginine-rich RNA binding motif (ARM) of the SP (Fig. 3C). Thus, we investigated whether the A6D substitution affected the intracellular localization of the enJSRV-26 SP.
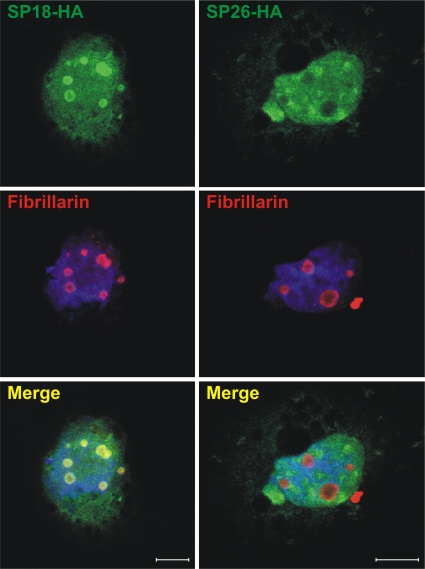
By confocal microscopy, we observed that both the enJSRV-18 and enJSRV-26 SPs localize in the cytoplasm and in the nucleus of transfected cells (Fig. 4). The enJSRV-18 SP colocalized with nucleolar markers, such as fibrillarin, as we previously observed for the JSRV SP (8). On the other hand, the enJSRV-26 SP displayed a diffuse nuclear staining pattern with no accumulation in the nucleoli. The relative number of cells expressing the enJSRV-18 SP with nucleolar localization was about 80-fold higher than those expressing the enJSRV-26 SP (data not shown). We also determined that the enJSRV-26 SP did not relocalize in the nucleoli in the presence of enJS56A1, indicating that its defect was not rescued by the functional SP of enJS56A1 (data not shown). We performed these assays in both COS and 293T cells, obtaining essentially the same results (data not shown).
Fig. 4.
Signal peptide (SP) of enJSRV-26 does not localize in the nucleoli. The intracellular localization of the signal peptide of enJSRV-18 and enJSRV-26 is shown. COS cells were transfected with expression plasmids for the SPs of enJSRV-18 or enJSRV-26 (tagged with the HA epitope), fixed 24 h posttransfection, and incubated with anti-HA (top) and fibrillarin (middle) antibodies. Nuclei are shown in blue. Both SPs display nuclear localization, but only the enJSRV-18 SP shows a strong colocalization with nucleolar markers such as fibrillarin. Bars correspond to 10 μm.
The phenotype of enJSRV-26 can be attributed to a relative lack of function of its SP.
We investigated whether the altered localization of the enJSRV-26 SP was correlated with its altered function and JLR escape. We cotransfected 293T cells with expression plasmids for the enJSRV-26ΔEnv mutant and the SPs of enJSRV-26 (pSP26-HA) or enJSRV-18 (pSP18-HA), and we used Western blotting to analyze the amount of virus produced in the presence or absence of enJS56A1 (Fig. 5A). We found that enJSRV-26ΔEnv was able to escape JLR if the enJSRV-26 SP was provided in trans (Fig. 5A, lanes 6 and 7), while it was impaired in the presence of the enJSRV-18 SP (Fig. 5A, lanes 8 and 9). enJSRV-26ΔEnv also was able to escape enJS56A1 restriction when expressed by itself (Fig. 5A, lanes 4 and 5). Accordingly, we also noticed that the release of viral particles from enJSRV26ΔEnv was much more strongly enhanced by the enJSRV-18 SP than by the enJSRV-26 SP (Fig. 5A, lanes 4, 6, and 8). Similar results were obtained with the enJSRV-18ΔEnv mutant (data not shown).
Fig. 5.
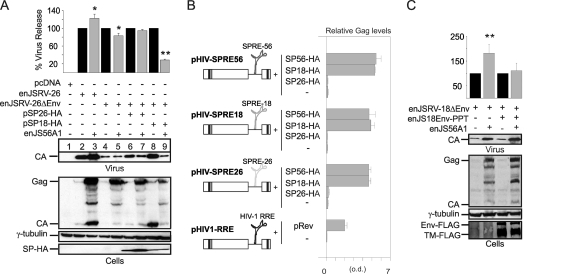
Signal peptide of enJSRV-26 does not function as a posttranscriptional regulator of viral gene expression and is essential to escape JLR. (A and C) Western blots of concentrated supernatants (virus) and cell extracts (cells) of 293T cells transfected with the plasmids indicated in each panel. Membranes were incubated with antibodies against the JSRV major capsid protein (CA), the HA or FLAG epitopes (to detect SPs or Env, respectively), or γ-tubulin as a loading control. Levels of CA associated with viral particles released in the supernatants were quantified by chemifluorescence using ImageQuant TL software (Molecular Dynamics). Graphs represent data obtained from three independent experiments. The values obtained by enJSRV-26 or related mutants in the absence of enJS56A1 (black bars) were arbitrarily set as 100%. Error bars indicate standard errors; statistically significant differences are indicated with one (P < 0.05) or two (P < 0.01) asterisks. (B) HIV Gag ELISAs were performed on supernatants of 293T cells transfected with HIV-1 Gag-Pol expression plasmids (pHIV-SPRE26, pHIV-SPRE18, and pHIV-SPRE56) in the presence or absence of expression plasmids for the enJSRV-26, enJSRV-18, and enJS56A1 SPs (SP26-HA, SP18-HA, and SP56-HA). Controls included supernatants of cells transfected with the HIV-1 Gag-Pol expression plasmid containing HIV-1 RRE (pHIV1-RRE) in the presence or absence of the HIV-1 Rev (pRev).
Note that in a previous study we showed that enJS56A1 also expresses a defective Env glycoprotein (2). Betaretroviruses assemble in the pericentriolar area, and their Env facilitate intracellular Gag trafficking and viral particle release (3, 33). Thus, interference assays involving enJSRV-26ΔEnv can be more difficult to interpret in the absence of functional Env. However, the data described above overall suggest that the A6D mutation in the enJSRV-26 Env impairs at least some of the functions played by its SP, resulting in JLR escape. To test this hypothesis, we used a Rev-RRE-dependent HIV-1 Gag-Pol expression vector (pNLgagSty330; termed pHIV1-RRE in this study) (16, 21) and derived constructs where the HIV-1 RRE was replaced with the signal peptide responsive elements (SPREs) of enJS56A1 (pHIV-SPRE56), enJSRV-18 (pHIV-SPRE18), and enJSRV-26 (pHIV-SPRE26). These constructs where cotransfected with either pSP18-HA, pSP26-HA, or pSP56-HA (encoding the enJSRV-18, enJSRV-26, and enJS56A1 SPs, respectively), and the release of HIV particles in the supernatants was measured by ELISA (Fig. 5B). We observed that in the presence or absence of the enJSRV-26 SP, the levels of Gag in the supernatants of cells transfected with pHIV-SPRE56, pHIV-SPRE18, or pHIV-SPRE26 did not change, while they increased substantially in the presence of the SP of either enJSRV-18 or enJS56A1 (Fig. 5B).
Collectively, the data presented so far suggested that the ability of enJSRV-26 to escape JLR was due to the relative lack of function of its SP. To experimentally prove this point, we derived a mutant of the enJSRV-18 Env expression plasmid in which the SP was replaced with the heterologous SP of the human preprotrypsin protein (penJS18Env-PPT). Interestingly, we found that enJSRV-18ΔEnv escaped JLR when enJS18Env-PPT was provided in trans (Fig. 5C). These data suggest that JLR escape is due to a lack of function, rather than a gain of function, of the SP of the enJSRV-26 Env.
enJS56A1 Env plays a key role in enJSRV-26 JLR escape.
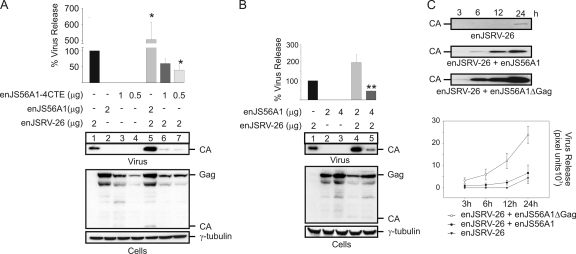
The data obtained so far suggested that the SP of the transdominant enJS56A1 is able to enhance Gag expression as well as enJSRV-26 viral particle release. Thus, our next aim was to assess whether the expression of the enJS56A1 Env SP was a requirement for JLR escape. To this end, we generated an enJS56A1 mutant (enJS56A1-4CTE) in which the Env glycoprotein (including its SP) was replaced by four repeats of the M-PMV CTE. Gag expression of enJS56A1-4CTE therefore is SP independent, but it relies on the M-PMV CTE, which functions in cis (6). As expected, we found that enJSRV-26 viral particle release was blocked by enJS56A1-4CTE, indicating that the expression of the enJS56A1 Env is necessary for enJSRV-26 to escape JLR (Fig. 6A, lanes 1, 6, and 7).
Fig. 6.
enJS56A1 Env expression is critical to escape JLR. (A and B) Western blots of concentrated supernatants (virus) and cell extracts (cells) of 293T cells transfected with the plasmids indicated in each panel. Membranes were incubated with antibodies against the JSRV major capsid protein (CA) and γ-tubulin as a loading control. Note that enJS56A1-4CTE lacks the viral Env and is able to block enJSRV-26. Levels of CA associated with viral particles released in the supernatants were quantified by chemifluorescence using ImageQuant TL software (Molecular Dynamics). Graphs represent data obtained from three independent experiments. The values obtained by enJSRV-26 were arbitrarily set as 100%. Error bars indicate standard errors; statistically significant differences are indicated with one (P < 0.05) or two (P < 0.01) asterisks. (C) Western blot analysis of concentrated supernatants of 293T cells transfected with the indicated plasmids at 3, 6, 12, and 24 h posttransfection. The bottom panel represents the quantification of blots by chemifluorescence as described for panels A and B. Values are expressed as arbitrary pixel units derived from three independent experiments.
Taken together, the results obtained suggested that the enJS56A1 and enJSRV-26 SPRE compete for the only functional SP within the cell (i.e., the enJS56A1 SP). Consequently, we hypothesized that by overexpressing enJS56A1, the intracellular levels of enJS56A1 SP would favor the synthesis of the transdominant Gag to levels sufficient to block enJSRV-26. To test this point, we cotransfected 293T cells with different ratios of the enJSRV-26 and enJS56A1 expression plasmids and assessed viral particle release by Western blotting (Fig. 6B). We found that enJSRV-26 was able to escape JLR when the ratio of the transfected expression plasmids was 1 to 1 (Fig. 6B, lane 4), while it was blocked when the ratio was 1 to 2 (Fig. 6B, lane 5), suggesting that enJS56A1 can inherently block enJSRV-26 viral particle release. To confirm these data, we performed cotransfection assays with expression plasmids for enJSRV-26 and either wild-type enJS56A1 or an enJS56A1 mutant deleted of gag (enJS56A1ΔGag) (Fig. 6C). As expected, we found that the levels of enJSRV-26 viral particles were higher in the presence of enJS56A1ΔGag than with the full-length enJS56A1. These data suggest that enJS56A1 is intrinsically able to interfere with enJSRV-26, and that JLR is directly related to the relative ratio between transdominant and functional Gag. By using interference assays, we also established that the enJSRV-26 SP does not possess any dominant-negative role and does not affect, by itself, enJS56A1 Gag synthesis (data not shown).
enJS56A1 is amplified within the genome of domestic sheep.
It is difficult to correlate the data obtained in in vitro experiments with events that in nature lead to the selection of transdominant proviruses and viruses escaping JLR. Our results suggest that the relative ratio between defective transdominant and functional Gag could determine the efficiency of JLR. The sheep genome has an overwhelming majority of enJSRV loci with functional Gag. However, in a previous study, we noticed that the BAC clones containing enJS56A1 were overrepresented in the sheep genomic BAC library used to clone the known enJSRV loci (2). Indeed, 22% of BAC positive for enJSRV sequences contained the enJS56A1 provirus, whereas the expected frequency was 3.7% (2). The overrepresentation of particular clones could be due to artifacts related to the construction, and the screening of the library but also could be due to the amplification of the genomic region containing enJS56A1. Here, using locus-specific primers, we determined the relative gene dosage of the transdominant enJS56A1 in genomic DNA of wild sheep within the genus Ovis (O. dalli, n = 1; O. canadensis, n = 2; O. ammon, n = 4; O. vignei, n = 1), the Mediterranean Mouflon (Ovis orientalis musimon, n = 4), and different breeds of domestic sheep (Dorset, n = 1; Suffolk, n = 1; Texel, n = 10; Jacob, n = 2; Red Maasai, n = 2; Merino, n = 3; Xalda, n = 2; Rambouillet, n = 1; Soay, n = 3; Norway, n = 2; Finsheep, n = 3). DNA from animals within the genera Budorcas (B. taxicolor, n = 2) and Pseudois (P. nayaur, n = 1) was used as additional negative controls. In addition, we estimated the relative gene dosage of enJSRV-6 (fixed in the Ovis genus), the insertionally polymorphic enJSRV-18 (present in most but not all domestic sheep), and the ovine SOX9 gene as additional controls. We expressed the data as the ratio between the estimated number of molecules of target genes and the β-Actin gene, which was used as a reference gene (Fig. 7A).
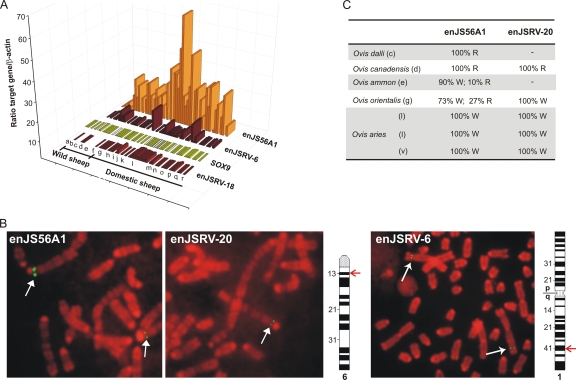
Fig. 7.
enJS56A1 is amplified in the domestic sheep genome. (A) Graph representing qPCR used to estimate the gene dosage, in wild and domestic sheep, of the transdominant proviruses enJSRV-6, enJSRV-18, and SOX9. Each bar represents an individual animal, and each letter represents a different species/breed. Gray boxes indicate assays that could not be performed due to the exhaustion of sample DNA. The absence of a bar indicates no amplification of the corresponding enJSRV locus. Samples tested included genomic DNA collected from B. taxicolor (a), P. nayaur (b), O. dalli (c), O. canadensis (d), O. ammon (e), O. vignei (f), O. orientalis (g), and various breeds of domestic sheep (O. aries), such as Soay (h), Norway (i), Dorset (j), Suffolk (k), Texel (l), Jacob (m), Red Maasai (n), Finsheep (o), Merino (p), Xalda (q), and Rambouillet (r). (B) Fluorescent in situ hybridization of metaphase R-banded chromosomes derived from a Merino sheep (mixed breed). Fluorescent probes were derived from BAC clones containing the enJSRV-20, enJS56A1, or enJSRV-6 proviruses as described in Materials and Methods. The green fluorescent signals, indicated by arrows, are specific for the two transdominant proviruses (both located on chromosome 6 at band 6q13) and the enJSRV-6 locus (situated on chromosome 1 at band 1q41). Ideograms of Ovis aries chromosomes with R-banding patterns also are shown. (C) Relative frequency of the wild-type arginine (R) or the transdominant tryptophan (W) Gag residue at position 21 of the enJS56A1-like proviruses. The 5′ region of gag of enJS56A1 and enJSRV-20 was amplified by PCR from genomic DNA collected from the species indicated in the panel. Note that letters in parentheses (c, d, e, etc.) refer to the code used for panel A. PCR products were cloned into the pCR4-TOPO vector (Invitrogen), and 40 individual clones for each PCR product were sequenced to determine the relative presence of an arginine or trypthophan residue at position 21 in Gag.
As expected, the dosage of the SOX9 gene did not change significantly across domestic and wild sheep. The modern enJSRV-18 provirus also showed low variations between the samples tested, whereas enJSRV-6 (which integrated before the divergence of the genera Ovis and Capra) displayed modest variations in some wild and domestic sheep (Fig. 7A). In contrast, the transdominant enJS56A1 revealed major differences between the samples analyzed, with clear indications of genomic amplification in some domestic sheep breeds (Fig. 7A). FISH analysis on metaphase chromosomes derived from domestic sheep (Fig. 7B) showed that at least one of the copies of the transdominant enJS56A1 provirus (enJSRV-20) maps exactly to the same chromosomal location as enJS56A1 in chromosome 6 (6q13).
In a previous study, we speculated that enJSRV-20 arose by a process of recombination/gene conversion with enJS56A1 (2). enJSRV-20 and enJS56A1 can be distinguished by minor nucleotide sequence differences (23 nucleotides along the entire genome) and by the 5′ flanking region (the env sequences of an enJSRV provirus for enJSRV-20), but they share identical 3′-flanking regions. However, this study suggests that genome amplification has driven the amplification of enJS56A1-like proviruses with identical genomic flanking regions in any given animal.
Previously, we also showed that the W21 amino acid residue in Gag of enJS56A1 (and enJSRV-20) became fixed during sheep domestication. Considering that the present study showed the presence of multiple copies of enJS56A1, we sought to determine the relative frequency of the codon for the Gag R/W21 residue in the proviruses harbored in the genome of representative wild and domestic sheep. To this end, we amplified the 5′ gag region of enJS56A1-like proviruses (including enJSRV-20) from genomic DNA of wild sheep (O. dalli, n = 1; O. canadensis, n = 1; O. ammon, n = 1), the Mediterranean Mouflon (O. orientalis musimon, n = 1), and two different breeds of domestic sheep (Texel, n = 2; Merino, n = 1). We then cloned the PCR products obtained and sequenced at least 40 individual clones for each sample. We detected the codon corresponding to the Gag R21 residue in 100% of the PCR clones derived from the amplification of DNA collected from O. dalli and O. canadensis, which are the species phylogenetically more distant from the domestic sheep among those analyzed in this study, and for which we had no evidence of genomic amplification (Fig. 7C) (2). In species phylogenetically closer to domestic sheep (O. ammon and O. orientalis), 73 to 90% of the PCR clones sequenced contained the codon corresponding to W21 in Gag. On the other hand, 100% of the clones amplified from domestic sheep (Texel and Merino breeds) contained the codon corresponding to W21 in Gag. These data confirm that enJS56A1-like transdominant proviruses became fixed in the host genome around sheep domestication.
DISCUSSION
In this study, we revealed the molecular mechanisms underlying the ability of the recently integrated enJSRV-26 provirus to elude the late restriction induced by the defective and transdominant enJS56A1. We demonstrated that a single point mutation in the SP of the enJSRV-26 Env allows this virus to escape enJS56A1. Signal peptides mediate the targeting and translocation of membrane and secretory proteins to the endoplasmic reticulum (20, 41). Generally, they are 15 to 25 amino acid residues long and contain the cleavage site for the cellular signal peptidase which, in turn, releases the signal peptide from the rest of the protein (40). Signal peptides usually are degraded by the signal peptide peptidase. However, in some cases signal peptide sequences display other important biological functions. For instance, it has been shown that the mouse mammary tumor virus (MMTV) possesses an SP that is identical for both the Rem regulatory protein (another HIV Rev-like protein) and the envelope glycoprotein (7, 22–24). The MMTV SP requires processing by the cellular signal peptidase and retrotranslocation for nuclear function (7).
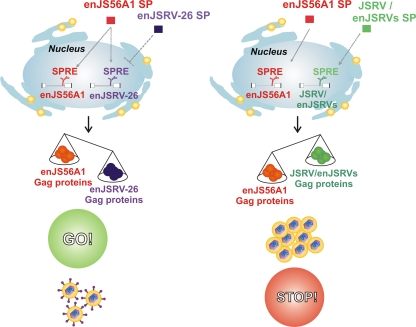
The signal peptides of JSRV and related enJSRVs are unusually long (80 amino acid residues) compared to those of other retroviruses. We and others have shown that the JSRV SP is a multifunctional protein that favors full-length viral RNA nuclear export and enhances Gag synthesis and viral particle release (8, 17). Conversely, in this study, we demonstrated that enJSRV-26 SP function is impaired due to a single point mutation. Interestingly, the A6D substitution lies within two predicted regions of the enJSRV-26 SP, the NLS and the ARM domains, which are important for SP intracellular localization and function (8, 17). Our results showed that the enJSRV-26 SP is excluded from the nucleolus, though it still can enter the nucleus, where it may passively diffuse given the small size of this protein. Interestingly, the staining pattern of the enJSRV-26 SP observed by confocal microscopy resembled what we previously obtained with a JSRV SP mutant deleted of its NLS (8). The HIV-1 Rev is targeted to the nucleolus, and this localization is crucial for mRNA trafficking and viral replication (19). Indeed, our data have shown that, unlike JSRV/enJSRV SPs, the enJSRV-26 SP function is impaired, and this defect allows this virus to elude enJS56A1 restriction. Our study supports a model in which enJS56A1 and enJSRV-26 SPREs compete for the only functional SP (i.e., the enJS56A1 SP), resulting in an increased synthesis of enJSRV-26 and reduced levels of enJS56A1 Gag proteins (Fig. 8). Using a Rev-RRE-dependent HIV-1 Gag-Pol vector, we have shown data suggesting that SPREs of enJS56A1 and enJSRV-26 respond equally well to the SP of enJS56A1. Indeed, we demonstrated that JLR escape depends on the stoichiometry between enJS56A1 and enJSRV-26 Gag, which in turn is regulated by the SPs of these proviruses.
Fig. 8.
Model of enJSRV-26 JLR escape. The ability of enJSRV-26 to elude JLR restriction is dependent on the impaired function of its SP. Consequently, the signal peptide responsive element (SPRE) of enJSRV-26 competes with the SPRE of enJS56A1 for the functional SP of the latter, resulting in the reduced expression of the transdominant Gag.
By studying the evolutionary history of the enJSRV proviruses, we found that enJS56A1 possessed the wild-type R residue at position 21 in Gag when it first entered the host genome. Only subsequently did the transdominant enJS56A1 genotype harboring W21 appear in the host genome, and it became fixed around the time of sheep domestication. With domestication, a relatively large number of animals suddenly were kept in restricted spaces, and this likely facilitated the spread of infectious agents more easily than before. Under these circumstances, it is feasible to hypothesize that sheep with transdominant proviruses had a selective advantage.
In our previous study, we speculated that a second transdominant provirus, enJSRV-20, arose by processes of recombination and/or gene conversion with enJS56A1. The results obtained in this study suggest that the chromosomal location containing enJS56A1 has been amplified several times, especially in some breeds of domestic sheep. Thus, sheep domestication has contributed to the selection and amplification of transdominant proviruses.
The interplay between host and pathogen is a dynamic process. The host has evolved sophisticated mechanisms to block infection by pathogens, which in turn have developed countermeasures to escape host defenses. The endogenous betaretroviruses of sheep represent a unique model to study virus-host coevolution during long evolutionary periods. Sheep, like koalas, harbor several copies of intact and insertionally polymorphic endogenous proviruses (2, 27, 36). The presence of enJSRV-26, in particular, a provirus that we estimated integrated in its host within the last 200 years and escapes restriction by enJS56A1, suggests that betaretroviruses still are invading the sheep genome. The presence of multiple copies of transdominant enJSRV proviruses in modern sheep breeds therefore may be another mechanism adopted by the host to counteract retrovirus infection.
ACKNOWLEDGMENTS
This study was funded by the Wellcome Trust and in part by NIH grant HD052745, by the BBSRC, and by a Strategic Research Developmental Grant by the Scottish Funding Council.
We thank Pietro Parma (Università di Milano), Andrew Shaw, and members of our laboratories for useful suggestions. We also thank Barbara Felber, Eric Hunter, Mauro Giacca, and Jim DeMartini for providing useful reagents.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Arnaud F., et al. 2010. Interplay between ovine bone marrow stromal cell antigen 2/tetherin and endogenous retroviruses. J. Virol. 84:4415–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arnaud F., et al. 2007. A paradigm for virus-host coevolution: sequential counter-adaptations between endogenous and exogenous retroviruses. PLoS Pathog. 3:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnaud F., Murcia P. R., Palmarini M. 2007. Mechanisms of late restriction induced by an endogenous retrovirus. J. Virol. 81:11441–11451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arnaud F., Varela M., Spencer T. E., Palmarini M. 2008. Coevolution of endogenous betaretroviruses of sheep and their host. Cell Mol. Life Sci. 65:3422–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blond J. L., et al. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bray M., et al. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. U. S. A. 91:1256–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Byun H., et al. 2010. Retroviral Rem protein requires processing by signal peptidase and retrotranslocation for nuclear function. Proc. Natl. Acad. Sci. U. S. A. 107:12287–12292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caporale M., et al. 2009. The signal peptide of a simple retrovirus envelope functions as a posttranscriptional regulator of viral gene expression. J. Virol. 83:4591–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chessa B., et al. 2009. Revealing the history of sheep domestication using retrovirus integrations. Science 324:532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cribiu E. P. 2001. International system for chromosome nomenclature of domestic bovids. Cytogenet. Cell Genet. 92:283–299 [DOI] [PubMed] [Google Scholar]
- 11. Dunlap K. A., Palmarini M., Spencer T. E. 2006. Ovine endogenous betaretroviruses (enJSRVs) and placental morphogenesis. Placenta (Suppl. A):S135–S140 [DOI] [PubMed] [Google Scholar]
- 12. Dunlap K. A., et al. 2006. Endogenous retroviruses regulate peri-implantation conceptus growth and differentiation. Proc. Natl. Acad. Sci. U. S. A. 103:14390–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dupressoir A., et al. 2005. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. U. S. A. 102:725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dupressoir A., et al. 2009. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. U. S. A. 106:12127–12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eiden M. V. 2008. Endogenous retroviruses-aiding and abetting genomic plasticity. Cell Mol. Life Sci. 65:3325–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Felber B. K., Hadzopoulou-Cladaras M., Cladaras C., Copeland T., Pavlakis G. N. 1989. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. U. S. A. 86:1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hofacre A., Nitta T., Fan H. 2009. Jaagsiekte sheep retrovirus encodes a regulatory factor, Rej, required for synthesis of Gag protein. J. Virol. 83:12483–12498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jern P., Coffin J. M. 2008. Effects of retroviruses on host genome function. Annu. Rev. Genet. 42:709–732 [DOI] [PubMed] [Google Scholar]
- 19. Kubota S., Furuta R., Maki M., Hatanaka M. 1992. Inhibition of human immunodeficiency virus type 1 Rev function by a Rev mutant which interferes with nuclear/nucleolar localization of Rev. J. Virol. 66:2510–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martoglio B., Graf R., Dobberstein B. 1997. Signal peptide fragments of preprolactin and HIV-1 p-gp160 interact with calmodulin. EMBO J. 16:6636–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mermer B., Felber B. K., Campbell M., Pavlakis G. N. 1990. Identification of trans-dominant HIV-1 rev protein mutants by direct transfer of bacterially produced proteins into human cells. Nucleic Acids Res. 18:2037–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mertz J. A., Chadee A. B., Byun H., Russell R., Dudley J. P. 2009. Mapping of the functional boundaries and secondary structure of the mouse mammary tumor virus Rem-responsive element. J. Biol. Chem. 284:25642–25652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mertz J. A., Lozano M. M., Dudley J. P. 2009. Rev and Rex proteins of human complex retroviruses function with the MMTV Rem-responsive element. Retrovirology 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mertz J. A., Simper M. S., Lozano M. M., Payne S. M., Dudley J. P. 2005. Mouse mammary tumor virus encodes a self-regulatory RNA export protein and is a complex retrovirus. J. Virol. 79:14737–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mura M., et al. 2004. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc. Natl. Acad. Sci. U. S. A. 101:11117–11122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murcia P. R., Arnaud F., Palmarini M. 2007. The transdominant endogenous retrovirus enJS56A1 associates with and blocks intracellular trafficking of the JSRV Gag. J. Virol. 81:1762–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oliveira N. M., Farrell K. B., Eiden M. V. 2006. In vitro characterization of a koala retrovirus. J. Virol. 80:3104–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Palmarini M., et al. 2000. Molecular cloning and functional analysis of three type D endogenous retroviruses of sheep reveal a different cell tropism from that of the highly related exogenous Jaagsiekte sheep retrovirus. J. Virol. 74:8065–8076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palmarini M., Mura M., Spencer T. E. 2004. Endogenous betaretroviruses of sheep: teaching new lessons in retroviral interference and adaptation. J. Gen. Virol. 85:1–13 [DOI] [PubMed] [Google Scholar]
- 30. Palmarini M., Sharp J. M., De las Heras M., Fan H. 1999. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J. Virol. 73:6964–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palmarini M., Sharp J. M., Lee C., Fan H. 1999. In vitro infection of ovine cell lines by Jaagsiekte sheep retrovirus (JSRV). J. Virol. 73:10070–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rhee S. S., Hui H. X., Hunter E. 1990. Preassembled capsids of type D retroviruses contain a signal sufficient for targeting specifically to the plasma membrane. J. Virol. 64:3844–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sfakianos J. N., Hunter E. 2003. M-PMV capsid transport is mediated by Env/Gag interactions at the pericentriolar recycling endosome. Traffic 4:671–680 [DOI] [PubMed] [Google Scholar]
- 34. Spencer T. E., Johnson G. A., Bazer F. W., Burghardt R. C., Palmarini M. 2007. Pregnancy recognition and conceptus implantation in domestic ruminants: roles of progesterone, interferons and endogenous retroviruses. Reprod. Fertil. Dev. 19:65–78 [DOI] [PubMed] [Google Scholar]
- 35. Spencer T. E., Mura M., Gray C. A., Griebel P. J., Palmarini M. 2003. Receptor usage and fetal expression of ovine endogenous betaretroviruses: implications for coevolution of endogenous and exogenous retroviruses. J. Virol. 77:749–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tarlinton R. E., Meers J., Young P. R. 2006. Retroviral invasion of the koala genome. Nature 442:79–81 [DOI] [PubMed] [Google Scholar]
- 37. Vardabasso C., Manganaro L., Lusic M., Marcello A., Giacca M. 2008. The histone chaperone protein nucleosome assembly protein-1 (hNAP-1) binds HIV-1 Tat and promotes viral transcription. Retrovirology 5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Varela M., Chow Y. H., Sturkie C., Murcia P., Palmarini M. 2006. Association of RON tyrosine kinase with the Jaagsiekte sheep retrovirus envelope glycoprotein. Virology 350:347–357 [DOI] [PubMed] [Google Scholar]
- 39. Varela M., Spencer T. E., Palmarini M., Arnaud F. 2009. Friendly viruses: the special relationship between sheep and retroviruses. Proceedings of the New York Academy of Sciences 1178:157–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. von Heijne G. 1990. Protein targeting signals. Curr. Opin. Cell Biol. 2:604–608 [DOI] [PubMed] [Google Scholar]
- 41. Walter P., Johnson A. E. 1994. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 10:87–119 [DOI] [PubMed] [Google Scholar]
- 42. Weiss R. A. 1995. Retrovirus receptors. Cell 82:531–533 [DOI] [PubMed] [Google Scholar]