Abstract
During cell-to-cell transmission of human immunodeficiency virus type 1 (HIV-1), many viral particles can be simultaneously transferred from infected to uninfected CD4 T cells through structures called virological synapses (VS). Here we directly examine how cell-free and cell-to-cell infections differ from infections initiated with cell-free virus in the number of genetic copies that are transmitted from one generation to the next, i.e., the genetic inheritance. Following exposure to HIV-1-expressing cells, we show that target cells with high viral uptake are much more likely to become infected. Using T cells that coexpress distinct fluorescent HIV-1 variants, we show that multiple copies of HIV-1 can be cotransmitted across a single VS. In contrast to cell-free HIV-1 infection, which titrates with Poisson statistics, the titration of cell-associated HIV-1 to low rates of overall infection generates a constant fraction of the newly infected cells that are cofluorescent. Triple infection was also readily detected when cells expressing three fluorescent viruses were used as donor cells. A computational model and a statistical model are presented to estimate the degree to which cofluorescence underestimates coinfection frequency. Lastly, direct detection of HIV-1 proviruses using fluorescence in situ hybridization confirmed that significantly more HIV-1 DNA copies are found in primary T cells infected with cell-associated virus than in those infected with cell-free virus. Together, the data suggest that multiploid inheritance is common during cell-to-cell HIV-1 infection. From this study, we suggest that cell-to-cell infection may explain the high copy numbers of proviruses found in infected cells in vivo and may provide a mechanism through which HIV preserves sequence heterogeneity in viral quasispecies through genetic complementation.
INTRODUCTION
During the course of human immunodeficiency virus type 1 (HIV-1) infection, a progressive accumulation of viral diversity is generated by viral replication, which occurs in CD4+ immune cells. From in vitro studies, it is known that HIV-1 infection of CD4+ cells is initiated via three major mechanisms: by a cell-free virus particle, by contact with an uninfected cell that has captured cell-free virus particles (25), and by direct contact with an HIV-1-expressing cell (6, 15, 16). Recent studies have revealed that large amounts of viral antigen can be translocated from cell to cell through the formation of virological synapses (VS) (6, 15). A key question arising from these studies is whether the average number of viral copies functionally transmitted from cell to cell is different from that which occurs during infection with cell-free viral inoculums. This question is critical because the copy number or ploidy of cells infected with HIV profoundly affects the virus's ability to tolerate genetic mutations and evolve over the course of chronic infection (7, 22).
In vivo, infected cells can be found to harbor 3 or 4 copies of HIV-1 (17) but it is unclear how such high copy numbers of HIV-1 per cell are achieved in the host cell. This is unlikely to be explained by sequential superinfection because HIV-1 downregulates the viral receptor, CD4 (3). Also unlikely is simultaneous multicopy infection with cell-free virus because cell-free infection of cells at this multiplicity would appear to require viral titers higher than those normally found in the body. One possibility is that cells vary significantly in their receptivity to viral infection with cell-free virus or dendritic cell transinfection (4, 8). A related idea suggests that there is a time window that allows superinfection because of the delay between initial infection and subsequent CD4 downregulation (20). These studies found that coinfection frequencies were detectable as a function of the square of the infection frequency, which, if extended to the infection frequencies observed in vivo (12), would lead to low frequencies of multicopy cells. Thus, the proposed mechanisms are insufficient to explain the frequency of cells infected with more than one virus. Recent modeling studies propose that cell-to-cell infection may explain high levels of coinfection without invoking high viral titers or high infection frequencies (10, 29), but this has yet to be tested experimentally. Here, we directly compared cell-free and cell-to-cell infections in vitro and determined how the frequency of coinfection changes in relation to the infection frequency. We sought to understand how cell-to-cell infection contributes to the simultaneous inheritance of multiple copies of HIV-1, which may act to maintain viral quasispecies diversity through genetic complementation and recombination without the need for high viral titers or high infection frequencies.
MATERIALS AND METHODS
Cells and tissue culture.
Human cell lines Jurkat clone E6-1 (provided by Arthur Weiss) and MT4 (provided by Douglas Richman) were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. MT4 and Jurkat T cells were cultured in RPMI 1640 medium with 10% fetal bovine serum, 100 U/ml penicillin, 100 g/ml streptomycin, and 2 mM glutamine (complete RPMI). Human peripheral blood CD4+ T cells were obtained anonymously from seronegative donors through the New York Blood Center. Primary CD4+ T cells were isolated from peripheral blood mononuclear cells using Miltenyi CD4+ T cell isolation kit II, activated with 2 μg/ml phytohemagglutinin and 10 U/ml interleukin-2 (IL-2) for 2 to 3 days before Lonza nucleofection or infection, and cultured in complete RPMI with 10 U/ml IL-2.
Viral constructs.
HIV Gag-iGFP encodes green fluorescent protein (GFP) between the matrix and capsid proteins (14). NLENG1-IRES, NLRX-IRES, and NLENC1-IRES encode a fluorescent protein in place of nef, and nef is expressed from an internal ribosome entry site (IRES) downstream (11, 18, 20).
Transfection of viral constructs and fluorescent cell labeling.
Lonza nucleofection was used to transfect Jurkat and primary CD4+ T cells. Jurkat cells were cultured at a density of <5 × 105 cells/ml, and CD4+ T cells were cultured at a density of 2 × 106 cells/ml. We mixed 7 × 106 cells with 2.2 μg endonuclease-free DNA in 15 μl endonuclease-free buffer Tris-EDTA and 110 μl solution V with supplement. Amaxa nucleofector program S18 was used, and cells were cultured in antibiotic-free medium overnight. A 4:1-μg ratio of NLRX-IRES to NLENG1-IRES DNA or a 4:1:1 ratio of NLRX-IRES to NLENG1-IRES to NLENC1-IRES was used for cotransfections to preserve equal fluorescence ratios in the donor cells. Cells were dye labeled by incubating 1 × 106 cells/ml in phosphate-buffered saline–1 μM CellTrace Far Red for 10 min at 37°C or in serum free medium–20 μM CellTracker Blue for 1 h at 37°C.
Flow cytometry.
The LSRII (BD Biosciences) was used to detect infection and discriminate donor and target cell populations. All cells were discriminated initially by side scatter area versus forward scatter area (FSC-A); doublets were excluded using forward scatter width (FSC-W) versus FSC height. GFP was detected using the fluorescein isothiocyanate channel, DsRed-Express was detected using the phycoerythrin-Texas Red channel, CellTracker Blue was detected using the PacificBlue channel, and CellTrace Far Red was detected using the allophycocyanin channel. All cells within a single experiment were detected using the same voltage settings. Channels were compensated after acquisition using FlowJo, and compensation was always performed equally across all samples of a single experiment.
Cell-free and cell-to-cell infections.
For cell-free infection, HIV supernatant containing 100 ng of p24 based on quantitative enzyme-linked immunosorbent assay or serial 2-fold dilutions thereof were mixed with 3 × 105 MT4 T cells in 400 μl in a 5-ml round-bottom tube for 3 h. The mixture was then transferred to a 6-well tissue culture plate for 27 h. For cell-to-cell infections, donor cells were purified with a Ficoll-Hypaque density gradient at 12 to 18 h posttransfection and then either mixed immediately or dye labeled and then mixed with dye-labeled target cells. The ratio of donors to targets was varied from 3:1 to 1:1,023, with a constant 4 × 105 cells in 400 μl in a 5-ml round-bottom tube. After 3 h, cells were diluted by transfer to a 6-well tissue culture plate for 27 h. Cells were then trypsinized, fixed with 4% paraformaldehyde, and analyzed by flow cytometry. All data points included in the analysis yielded a minimum of 40 infected cells to avoid sampling error at the lowest infection frequencies.
Calculations.
For a random infection, curves for the proportion of infected cells that are multicolored for a given infection frequency were calculated using the Poisson distribution. For two-color experiments, we summed, for a given infection frequency with a multiplicity of infection (MOI) of m, the probability of a cell becoming infected with n particles [(mn × e−m)/n!] multiplied by the probability of a cell that receives n particles will receive at least one HIV(Red) and one HIV(Green) genome (1 − (2·0.5n). The summation equals [1 − (1 − I)1/2]2, where I is the overall infection frequency. This was then divided by the infection frequency (I) to yield the proportion of infected cells that were dually fluorescent (i.e., red and green [RG]). This approach has been previously described as a modified Poisson distribution (10). Alternatively, the probability of coinfection can be determined by the product of the frequency of each virus (8). Solving for the probability of coinfection in terms of the overall infection frequency yields the same equation, [1 − (1 − I)1/2]2, and the proportion of infected cells that are coinfected is given by the same equation divided by I. For a given proportion of infected cells that are coinfected (Y), the expected infection frequency (I), and thus the expected MOI, can be calculated by solving for the variable I [I = 4Y/(1 + Y)2] and using the formula MOI = −ln(1 − I).
For three-color viruses, the summation for the probability of cells receiving two different-colored viruses is 3 × (1 − I) × [1 − (1 − I)−1/3]2 and that of cells receiving at least one of each color is [1 − (1 − I)1/3]3. These values were then divided by the infection frequency, I, to generate the expected proportion of infected cells that would be multicolored for a given infection frequency. Rearranging the equation in terms of the proportion of infected cells that are 3 colors (Y), the expected infection frequency can be determined by the equation
Again, the expected MOI is given by the equation MOI = −ln(1 − I).
For R2, the data for MT4 target cells cocultured with Jurkat donorRG and with Jurkat donorR + donorG was fitted to the expected curve for a Poisson distribution, Y = 100·[100/X]·[1 − (1 − X/100)1/2]2, with minimization of the relative distances squared (1/Y2 weighting). This was performed using Prism software.
Computational model.
We studied an agent-based model that tracks the fate of individual cells over time. The properties of the model are as follows. Assume the existence of n cells, where n = 400,000 in the experiments. Initially, a certain number of these cells will contain red virus only (R), green virus only (G), or both red and green viruses (RG). These numbers are taken from the initial number of cells in the experiments. The cells are allowed to interact for 3 h, after which the number of double-color cells and the total number of multiply infected cells (containing >1 virus) are determined in the simulation (taking into account only the newly produced infected cells). At each time step, the system is randomly sampled m times, where m is the total number of donor cells. The following events can occur based on probabilities. Free cells (not connected to other cells via synapses) have a probability S of forming a synapse with a randomly chosen partner cell. Donor cells that are connected to another cell can pass on one offspring virus to the partner cell with a probability Q. Each infection event is assumed to add one virus particle to a cell. That is, an initially uninfected cell will acquire one virus. A cell infected with i viruses will end up having i + 1 viruses. In addition, there is a probability G that linked cells will break apart. It is assumed that during the time frame considered here, cells do not die. Note that the model considers infection via the synapse only and does not include infection via free virus, as Fig. 1 shows that most of the infection that occurs is likely through a synapse. The model was fitted to the data using least-squares techniques in the following way. The parameter space was randomly sampled 10,000 times. For each parameter combination, the simulation was run 100 times and the average model output was recorded for the different initial numbers of cells used in the experiments. The parameters were chosen from a uniform distribution between zero and one.
Fig. 1.
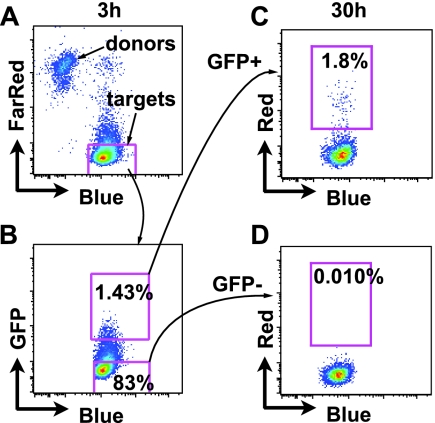
Preferential infection of cells that have internalized high levels of virus following exposure to cell-associated HIV-1. (A) Donor cells are discriminated from target cells using CellTrace Far Red-stained Jurkat HIV(Red)/Gag-iGFP donor cells and CellTracker Blue-stained primary CD4+ T cells (targets). (B) Target cells are flow sorted after 3 h of coculture into GFP+ and GFP− populations. (C) GFP+ target cells expressing HIV(Red) 27 h after sorting. (D) GFP− target cells expressing HIV(Red) 27 h after sorting.
As an alternative, the log10 of the parameters was chosen from a uniform distribution between −6 and 0. To measure how well the model fits the data, a modified sum of squares was recorded for each parameter combination. Each data point consists of a pair of values (fraction of double-color cells and overall fraction of infected cells). We designated the observed data points Xi and Yi and the predicted data points xi and yi. The quantity that was minimized in order to determine the best fit is given by
This expression aims to make the two different measures (the fraction of doubly infected cells and the overall fraction of infected cells) commensurate, as they can have different scales. This procedure gave rise to a number of parameter combinations that result in comparably good fits. We plotted the closest fit to the average number of infected MT4 cells that were dually fluorescent (see Fig. 3G). This predicted that about 21% the infected cells harbored more than one copy and was achieved with the following parameter combination: S = 8.25e−2, G = 0.36, and Q = 9.3e−2. Probabilities are given per minute.
Fig. 3.
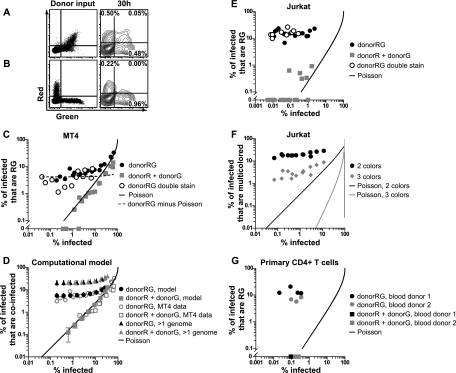
The cofluorescence frequency of cell-to-cell infection is proportional to the overall infection frequency and does not follow a Poisson distribution. (A) Cell-to-cell infection with Jurkat donor cells coexpressing red and green fluorescent HIV, i.e., Jurkat donorRG and MT4 target cells. Flow cytometry dot plots of donor input (left) and log contour plots of target cells after 30 h (right) are shown. (B) Cell-to-cell infection with a mixture of donor cells expressing HIV(Red) or HIV(Green), i.e., Jurkat donorR, Jurkat donorG, and MT4 target cells. Flow cytometry dot plots of donor input (left) and log contour plots of target cells after 30 h (right) are shown. (C) Titration of Jurkat donor cells onto MT4 target cells. Graphs show percentages of infected MT4 target cells that are red and green (RG) (y axis) versus the overall infection frequency (x axis) following infection with cells coexpressing red and green virus, donorRG cells, or separately expressing red or green virus, i.e., donorR and donorG cells. DonorRG double stain indicates experiments where donorRG cells were additionally discriminated from target cells by using a second CellTracker dye. Poisson line indicates the expected curve if coinfection follows a Poisson distribution. DonorRG minus Poisson indicates subtraction of the expected Poisson distribution from the donorRG data points. (D) Computer simulation of infections of Jurkat donor cells and MT4 target cells to estimate the percentage of target cells that carry more than one genome. Plot of average percentages of infected MT4 cells that are coinfected versus the overall infection frequency (see Materials and Methods for details and parameters). Plots are shown with computer simulation data next to actual MT4 data. When provided with donor cell cofluorescence information, the model accurately outputs target cell cofluorescence values for the donorRG experiment and the donorR + donorG experiment and predicts that the percentages of infected cells that have more than one genome will be similar for both donorRG and donor plus donorG experiments. Error bars show standard deviations. (E) Titration of Jurkat donor cells onto Jurkat target cells. (F) Titration of Jurkat donorRGB cells and Jurkat target cells. A graph of the percentage of infected Jurkat target cells expressing only two different fluorescent proteins or all three different fluorescent proteins versus the overall infection frequency is shown. (G) Titration of infected primary CD4+ T cells onto primary CD4+ T cell targets. Shown is a graph of the percentage of infected primary CD4+ T cells that are RG (y axis) versus the overall infection frequency (x axis) after infection with donorRG or donorR + donorG cells for two different donors. Abbreviations: R, HIV(Red); G, HIV(Green); B, HIV(Blue).
In addition to the model fitting, we also performed statistical calculations to predict the total percentage of multiply infected cells based on the initial fraction of donorRG cells and the resulting percentage of double-color cells observed. For these purposes, we focused on experiments performed at a low MOI, where we may assume that the infected target cells were not infected by more than one donor cell. We therefore used only the experiments that resulted in a <5% total infection frequency. We also assume that the duration of the experiment is long relative to the duration of the synapse, which should be true in our experimental system. Based on these assumptions, the total fraction of multiply infected cells is given by F = 2/[RGd/v + 1], where RGd is the fraction of donor RG cells that were used in the experiment and v is the resulting fraction of double-color cells. Taking the average over all data points that resulted in a <5% total infection frequency (10 data points fell into this category), we obtained a predicted total percentage of multiply infected cells of 21% ± 6.2%. While these calculations could be performed only for the low-MOI data points, the predicted percentage of multiply infected cells coincides well with the results of the model-fitting procedure.
The above formula was derived as follows. Assuming that a donor cell has synapsed with a target cell and transmitted a genome, the fraction of cells that received more than one genome is given by the formula
where G and Q are the probabilities defined above. This can be simplified to F = 1 − {G/[G + Q − (G·Q)]} or closely approximated by F = Q/(G + Q) if the term G·Q is ignored, as it is relatively small. According to the same line of reasoning, we can calculate the fraction of double-color cells by using the equation
From these expressions, it follows that F = 2/[RGd/v + 1].
FISH experiments.
For fluorescence in situ hybridization (FISH) experiments, donor cells (stained with CellTrace Far Red) were generated by infection at 100 ng p24 HIV(Green) virus/106 cells and the infection was allowed to spread for 3 to 4 days in a 24-well plate at 2 × 106 cells/ml. Target cells were inoculated with cell-free or cell-associated HIV(Green) as described in the methods above. Up to 500 infected target cells (stained CellTracker Blue) were sorted onto one spot on a glass slide, allowed to dry, fixed in Carnoy's fixative, treated with 5 μg/ml RNase, probed for HIV-1 using the pNL4-3 nick-translated probe, and probed for chromosomes X (green) and 8 (aqua; not shown but visible during scoring). Slides were blinded and scored for HIV-1 hybridization signals and chromosomal probes.
RESULTS
Cells that internalize large amounts of virus are preferentially infected.
Cell-to-cell infection spreads more efficiently in vitro than cell-free infection (9, 26, 27), but it has been difficult to measure the extent to which internalization of HIV-1 through VS leads to productive infection (15). A defining feature of cells that have engaged a VS is the contact-dependent internalization of fluorescently tagged virus particles into a trypsin-resistant compartment that is easily detected by flow cytometry (6). Separation of cells with a Transwell barrier or cocultivation with large excesses of cell-free fluorescent virus does not result in a detectable fluorescent signal in exposed target cells (6). Thus, detection of antigen uptake into a trypsin-resistant compartment by flow cytometry may serve as a marker for cells that underwent a VS. Furthermore, quantitative live-imaging experiments found that cells engaged in synapses internalized large amounts of virus, while those only micrometers away did not accumulate any virus above the detection limit of fluorescence microscopy (15). We thus designed an experiment to compare the infection frequency in cells that internalized large amounts of virus through cell-to-cell contact with that in cells in the same culture that had not. We generated HIV-expressing “donor” Jurkat T cells that were cotransfected with two HIV-1 proviral constructs, one that labels virus particles with GFP, called HIV Gag-iGFP (14), and another construct that contains DsRed-Express in parallel with nef, a viral gene expressed early after infection (NLRX-IRES [11]). Target cells that internalize HIV-1 through VS are rapidly marked with green fluorescence (6), and subsequent infection may then be monitored by measuring red fluorescence. A brief 3-h exposure of target cells to the donor cells was followed by trypsin treatment of cells to remove surface-adsorbed virions and to break apart all VS (Fig. 1A). Following this transient exposure, target cells were flow sorted away from all donor cells into populations that internalized labeled virions (GFP+) or did not (GFP−) (Fig. 1B), and each population was analyzed for infection 27 h later (Fig. 1C and D). Of the GFP+ cells, 1.8% became infected, whereas only 0.010% of GFP− cells became infected, yielding a relative risk of becoming infected of 180 for HIV-1 internalization. Given our previous data that showed detectable internalization requires cell-cell contact, we can conclude that in a 3-h coculture of donor cells and target cells, and following trypsinization, most of the detectable infection occurs in cells laden with virus likely acquired through VS.
Coinfection by cell-free virus follows a random Poisson distribution.
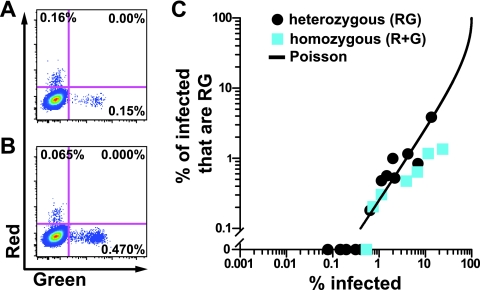
We next set out to measure coinfection frequencies by cell-free and cell-associated viral inoculums utilizing infectious fluorescent molecular clones of HIV-1 called NLRX-IRES, NLENG1-IRES, and NLENC1-IRES, which encode DsRed-Express, enhanced GFP, and enhanced cyan fluorescent protein, respectively (11, 18, 20). For convenience, we refer to these clones as HIV(Red), HIV(Green), and HIV(Blue), respectively. We first generated cell-free virus by cotransfection of molecular clones encoding HIV(Red) and HIV(Green), which would result in individual virions that package two of the same genomes or two different genomes. We refer to these virions arising from cotransfected cells as heterozygous virus. Although each HIV-1 particle contains two RNA genomes, we found that at an MOI of 0.003 (an infection frequency of 0.3%), heterozygous HIV(Red)/HIV(Green) cell-free virus generated exclusively monofluorescent cells (Fig. 2 A). This result is consistent with prior studies which demonstrated that while two genomes are packaged, due to strand transfer events during viral reverse transcription, only a single chimeric genome is integrated into the target cell. This has also been referred to as pseudodiploid inheritance of HIV-1 (5, 13). At a similar infection frequency (0.5%), infection with a mixture of homozygous HIV(Red) and homozygous HIV(Green) also yielded exclusively monofluorescent cells (Fig. 2B). This suggests that dual fluorescence occurs only following infection with multiple viruses. This allowed us to consider each heterozygous virus particle as either HIV(Red) or HIV(Green) and validated the use of multifluorescent cells as a measure of coinfection.
Fig. 2.
Cofluorescence during cell-free HIV-1 infection follows a random Poisson distribution for homozygous or heterozygous virus. (A) Flow cytometry plots of cell-free infection of MT4 cells from supernatants of HIV(Red)/HIV(Green)-cotransfected 293T cells (heterozygous virus). (B) Flow cytometry plots of cell-free infection of MT4 cells using a mixture of HIV(Red) and HIV(Green) pooled from separate transfections (homozygous virus). (C) Plot of the proportion of infected cells that are coinfected (y axis) versus the overall infection frequency (x axis). See Materials and Methods for Poisson curve calculations.
To evaluate how the cofluorescence frequency changes as a function of the overall infection frequency for a cell-free infection, we titrated heterozygous virus or a mixture of homozygous virus onto MT4 T cells and analyzed cells at 30 h postinfection (Fig. 2C). Infection with cell-free virus resulted in random infection of cells, where the frequency of dually fluorescent cells followed a Poisson distribution, scaling as the product of each fluorescent color frequency, as previously observed (20).
Each HIV-1-expressing cell can infect a target cell with multiple copies of HIV-1.
To study cell-to-cell infections, we mixed donor Jurkat T cells cotransfected with HIV(Red) and HIV(Green) plasmids (donorRG) with target MT4 T cells (Fig. 3 A) at a low donor-to-target cell ratio. After 30 h of coculture, target cells were analyzed for HIV(Red) and HIV(Green) expression by flow cytometry. This short duration of infection ensured a single round of infection (1, 23, 28), and the low donor-to-target cell ratio minimized the chances of a target cell becoming infected with viruses from different donor cells. When donor cells were mixed with MT4 target cells at a ratio of 1:127, 1.03% of the MT4 cells were infected and 0.05% (about 5% of the infected cells) expressed both HIV(Red) and HIV(Green). In comparison, an infection with cell-free virus that infected the same percentage of target cells yielded a 16-fold lower cofluorescence frequency (Fig. 2C). To test if coinfection occurred by a single donor cell or by multiple donor cells, Jurkat cells were singly transfected with either HIV(Red) or HIV(Green), pooled (donorR + donorG), and mixed with MT4 cells for 30 h (Fig. 3B). At a similar infection frequency (1.18%), dually fluorescent target cells were rarely detected and comprised about 0.25% of the infected cells.
If single cell-cell interactions give rise to multicopy infection, then as one titrates toward lower infection frequencies, one may predict that the cofluorescence frequency will approach a constant fraction of the overall infection frequency. We tested this by varying the donor-to-target cell ratio and found that as the infection frequency decreased, the fraction of infected cells that were coinfected did not fall in proportion to the square of the overall infection frequency (Fig. 3C, black circles; see Fig. S1 in the supplemental material). Instead, the cofluorescence frequency was maintained as a constant fraction of the infection frequency as the donor-to-target ratios were decreased. The presence of doublets or fused cells did not explain this phenomenon because doublets were excluded using FSC-W versus FSC-A. Additionally, doublets and fused cells were also excluded by staining both donor and target cells with CellTrace Far Red and CellTracker Blue, respectively, which yielded similar results (Fig. 3C, white circles).
To determine the frequency of coinfection caused by multiple donor cells, Jurkat cells were individually transfected with HIV(Red) or HIV(Green), pooled, and mixed with MT4 target cells at different donor-to-target ratios. Infection with Jurkat donorR + donorG cells yielded cofluorescent cells, suggesting that viruses from two different donors could infect a single cell when sufficiently high numbers of donor cells existed. Notably, the cofluorescence frequency was approximated by the Poisson distribution (R2 = 0.8090) compared to donorRG (R2 = −8787).
To estimate the cofluorescence frequency resulting purely from single donorRG cells, the expected Poisson cofluorescence frequency was subtracted from the donorRG cofluorescence frequency. This gave rise to a constant fraction of infected MT4 cells that were dually fluorescent, with an average of 4.3% (Fig. 3C, dashed line, slope not significantly different from zero [P = 0.41, linear regression]), suggesting that there was a 4.3% chance of at least two different genomes transmitted per synapse infectious event. In comparison, to achieve a 4.3% fraction of infected cells that are dually fluorescent, an MOI of 0.17 would be required, which is equivalent to a 15.6% infection frequency. In a cell-to-cell infection, however, this steady level of cofluorescence occurs at infection frequencies that are lower by several orders of magnitude and thereby obviates the need for high viral titers to observe coinfection.
Using multifluorescence as a surrogate for coinfection underestimates true coinfection because cells that are infected with multiple copies of the same reporter virus will become monofluorescent. Furthermore, not all transfected donor cells in our donorRG system express both HIV(Red) and HIV(Green), which may further underestimate the true coinfection frequency. To estimate the coinfection frequency of a cell-to-cell infection, we used a computational, stochastic, agent-based model (29) to simulate a cell-to-cell infection (Fig. 3D and Materials and Methods; see Fig. S2 in the supplemental material). The model received input data from experiments between Jurkat donor cells and MT4 target cells, which included the numbers of single- and dual-expressing donor cells, as well as the resulting single- and dual-expressing target cells at the end of the infection. The model then simulated cell-to-cell infection testing a range of probabilities for synapse formation, virus transfer, and synapse breakage. Iterative computational simulations identified the parameter values that best fit the empirical data, both the infection frequency and the cofluorescence frequency (Fig. 3D). The best-fit model replicated the observed data when donorR + donorG cells were used (Fig. 3D, grey squares) or when donorRG cells were used (Fig. 3D, black circles). The computational approach illustrates elegantly that a simple cell-cell infection model can accurately fit the cofluorescence and infection frequency data from both donorRG and donorR + donorG experiments. Furthermore, based on the parameter values of the best fit, the model then provided an estimate of the coinfection frequency without regard to cofluorescence status, predicting that 21% of the infected cells were coinfected when either donorRG or donorR + donorG cells were used (Fig. 3D, black and grey triangles). To complement the model fitting, we also used a statistical calculation to predict the average total percentage of multiply infected cells that is based on a subset of data points that resulted from low-MOI infection (see Materials and Methods). According to this calculation, the average percentage of multiply infected cells is 21% (standard deviation = 6.2%), in line with the model-fitting results. Thus, this model predicts a plausible range by which cofluorescence may underestimate the true coinfection frequency.
We next examined the Jurkat T cell line as a target cell line. When Jurkat donorRG cells were used to infect Jurkat target cells at different donor-to-target cell ratios (see Fig. S3 in the supplemental material), we again observed that the cofluorescence frequency approached a constant proportion of infected cells as the donor-to-target ratio was decreased (Fig. 3E, black circles). Multicolor analyses that further excluded doublets or fused cells again yielded similar results (Fig. 3E, white circles). Interestingly, the average proportion of infected cells that were coinfected was consistently higher than that observed with MT4 target cells (16.0% versus 4.3%). In comparison to cell-free virus, an MOI of 0.64 would be required to reach a magnitude of 16% of infected cells that are dually fluorescent. When a pooled mixture of Jurkat donorR and donorG cells was used (Fig. 3E, grey squares; see Fig. S3 in the supplemental material), cofluorescence was rarely detected after the analysis of 1 × 105 to 5 × 105 target cells. However, when cofluorescence was detected, a slightly higher-than-random frequency of cofluorescence was observed, perhaps indicative of a fraction of target cells that are more susceptible to infection (8).
Since dually fluorescent cells were coinfected with at least two genotypes, we asked if cells could be coinfected with at least three genotypes following cell-to-cell infection. To do this, we cotransfected Jurkat T cells with HIV(Red), HIV(Green), and HIV(Blue) (donorRGB). We mixed these cells with CellTrace Far Red-stained Jurkat target cells and analyzed cells 30 h later (Fig. 3F; see Fig. S4 in the supplemental material). At low infection frequencies (<5%), we again found that dilution of donor cells yielded a function that approached a constant proportion of infected cells that were doubly or triply fluorescent (18.3% and 2.8%, respectively), illustrating that donor cells can also deliver three or more infectious units to target cells. The level of triple fluorescence was vastly greater than the fraction of three-color-infected cells predicted by random infection (Fig. 3F). Given that many triply infected cells may have been infected with a combination of viruses that express only two different fluorescent proteins, it is likely that the trifluorescence frequency also underestimates the number of cells infected with three copies.
We next tested if multicopy infection occurs in primary human CD4+ T cells (Fig. 3G). To do this, we cotransfected activated primary CD4+ T cells with HIV(Red) and HIV(Green), mixed them with activated, autologous primary CD4+ T cells stained with CellTracker Blue, and analyzed cells by flow cytometry 30 h later (see Fig. S5 in the supplemental material). Similar to the results obtained with T cell lines, dilution of donor cells did not have a strong effect on the fraction of infected cells that were coinfected (average of 11.2%), despite low infection frequencies (Fig. 3G, black and grey circles). When pooled donorR and donorG cells were used, we did not detect coinfected target cells (Fig. 3G, black and grey squares; see Fig. S5 in the supplemental material). These data suggest that in primary cells, coinfection from cell-associated HIV-1 persists even at low ratios of donor to target cells because a single synapse can transmit multiple HIV-1 genomes that coinfect a target cell at a constant frequency.
Increased nuclear HIV-1 proviral copy number in a cell-to-cell infection compared to a cell-free infection.
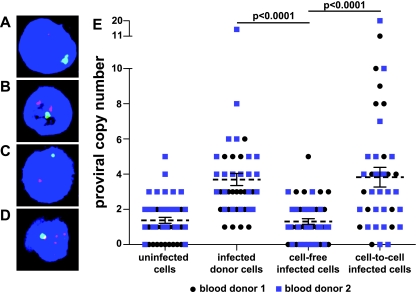
The fraction of infected cells that are coinfected does not provide complete information about the distribution of viral copies per cell. To examine this directly, we measured the proviral copy number on a single-cell basis following cell-free or cell-to-cell infection using FISH. Activated primary CD4+ target T cells were inoculated with cell-free HIV(Green) or with autologous donor CD4+ T cells that had been infected for 3 to 4 days with HIV(Green). Infected target cells were flow sorted 30 h postinfection and analyzed by FISH using an HIV-1 genomic DNA probe and control chromosomal markers (Fig. 4 A to D). Blinded scoring of nuclei for HIV-1 hybridization signals obtained an average of 3.82 (standard error of the mean [SEM] = 0.56) proviral copies in cells following cell-to-cell infection, compared to 1.30 (SEM = 0.16) following cell-free infection (Fig. 4E). Costaining for chromosomal probes ruled out dividing cells or syncytia as a source of multiple proviral copies (Fig. 4A to D, green). Interestingly, donor cells alone, which were infected initially by cell-free virus but then were allowed to propagate the infection over 3 to 4 days, had proviral copy numbers similar to those of cell-to-cell-infected cells. This may suggest that high-copy cell-to-cell infection propagates the infection in these cells, despite being initiated by cell-free virus in vitro. It should be noted that the background of the FISH assay was relatively high, with similar levels of FISH signal detected in the cell-free infected cells and the uninfected controls. While this high background obscured our ability to identify cells with low HIV-1 proviral copy numbers, the assay was sufficiently sensitive to show that cell-to-cell infections generate significantly higher copy numbers than cell-free infections.
Fig. 4.
Increased nuclear HIV-1 proviral copy number in cell-to-cell infections compared to that in cell-free infections. (A to D) Z projection of confocal images of FISH of primary CD4+ T cells. Red, HIV-1 provirus; blue, 4′,6-diamidino-2-phenylindole (DAPI); green, X chromosome. (A) Uninfected cells alone. (B) HIV-1-infected donor cells alone. (C) Target cells infected with cell-free HIV-1. (D) Target cells infected cell to cell with HIV-1. (E) Distributions of nuclear HIV-1 proviral copies from two different blood donors. Combined means ± SEM are shown. Blood donor 1: cell-free mean = 1.11, n = 26; cell-to-cell mean = 3.72, n = 25, P = 0.0003. Blood donor 2: cell-free mean = 1.55, n = 20; cell-to-cell mean = 3.95, n = 20, P = 0.0138. Combined blood donors: donor cell mean = 3.71, n = 45, cell-free mean = 1.30, n = 46, cell-to-cell mean = 3.82, n = 45; P values are displayed. Significance was tested using the two-tailed Mann-Whitney U test with an alpha level of 0.05.
DISCUSSION
The theoretical framework traditionally used to study viral infections has relied heavily on the expectation that viral infections behave with Poisson statistics. Partly because animal models of viral infections are most often initiated by cell-free virus, it is also tacitly assumed that viral infections in vivo behave in a manner similar to that of cell-free virus in vitro. Our study directly examined how cell-to-cell transmission alters the genetic inheritance of HIV-1. The results suggest that the unit of inheritance is dramatically altered when we directly compare cell-to-cell infections with cell-free viral infections. As we can distinguish the two modes of infection in vitro based upon ploidy of infection, we may now have the ability to experimentally examine the extent to which each mode of infection operates during physiological infection in vivo.
A number of studies have measured the relative increase in efficiency of cell-to-cell HIV infection compared to cell-free infection, often finding increases ranging from 100- to 10,000-fold. Our own previous studies illustrated that, on a population basis, the amount of green fluorescence that could be transmitted from a donor population to a target population could be thousands-fold greater when one normalizes to the amount of cell-free virus typically produced during a maximally productive infection in vitro (6). Here we consider a direct functional test of those cells that have taken up this large amount of virus and physically sorted those cells that internalized high versus undetectable amounts of virus. We found that those cells that internalized large doses of viral antigen were hundreds of fold enriched in infected cells, consistent with the magnitude of the advantage of cell-associated infection reported in previous studies (9, 21, 27). In our studies, given the proximity of cells in a coculture, it may be difficult to conclude that the preferential antigen uptake is all driven by bona fide VS, particularly when highly permissive MT4 cells are used as target cells. However, when considered with our direct imaging studies that suggest that mere proximity in vitro cannot result in significantly increased levels of antigen transfer into target cells (15), it is likely that most infection is secondary to VS.
The experiments in this study are among the first to test the extent to which each individual HIV-infected donor cell can infect a target cell with multiple copies of HIV-1. Interestingly, when the cofluorescence frequency of pooled donor cells that expressed only HIV(Red) or HIV(Green) was measured, cell-to-cell HIV-1 infections followed random, single-hit kinetics, behaving like cell-free virus. That is, donor cells are distributed randomly onto target cells and follow a Poisson distribution. However, the distribution of HIV-1 genomes onto target cells does not follow a Poisson distribution, which is revealed when testing HIV-1 donor cells that simultaneously express two distinct copies of HIV-1. Thus, during cell-to-cell infection, a single infectious event, likely defined by the VS, can transmit a variable number of genomes to the target cell. At low infection frequencies, this results in a constant fraction of infected cells becoming coinfected with multiple genomes. In contrast to previous coinfection studies, which suggested that coinfection occurs more frequently than random at high infection frequencies due to target cell heterogeneity (4, 8), our data suggest that coinfection occurs more frequently than random at low infection frequencies because a single donor cell can infect a target cell with multiple copies of HIV-1. Surprisingly, given the amount of viral antigen uptake observed in this study and previous studies (6, 15, 16), a relatively small fraction of infected cells was coinfected. If every virus that was internalized by target cells led to an infectious event, the frequency of dually fluorescent cells would likely be near 100%; instead, 4 to 20% of infected cells were dually fluorescent, depending on the type of target cell, suggesting that only a small fraction of the internalized viral particles successfully infect the target cell. Interestingly, the fraction of infected target cells that were coinfected was reproducible for each target cell type but differed greatly between the types of target cells (4 to 20%). This suggests that in vivo, the average number of infectious virions transmitted from donor to target cell may be influenced by cellular factors in the target cell such as lineage, state of differentiation, or activation, which may control the efficiency of the synapse in a cell population.
Cofluorescence in our studies provided direct evidence for multiple viral genomes being functionally expressed in single cells. It is clear, however, that the level of cofluorescence is likely to underestimate the frequency of multicopy infection (Fig. 3D). To estimate the coinfection frequency of cell-to-cell infection, we found, rather compellingly, that our experimental data sets could be accurately fitted to a probabilistic computer model that allowed only cell-to-cell infection using three simple parameters. The model suggested that the experiments could underestimate the true level of coinfection by 4-fold. This assumed that the number of virus particles transmitted per synapse followed a random distribution. Alternatively, it is plausible that most dually fluorescent cells harbored even more than two genomes, and thus cofluorescence may have accurately approximated the true coinfection frequency.
The use of FISH to detect proviral copies suggested that there is a broad distribution of proviral copy numbers found in infected cells. The discrepancy between this high copy number and the lower frequency of multifluorescent cells in the previous experiments could be explained by the ability of FISH to detect unintegrated and/or unexpressed genomes, or the presence of many monofluorescent donor cells used to initiate the infections in Fig. 3G (see Fig. S5 in the supplemental material). What is most clear from the FISH data is that the proviral copy number was higher when the infection was initiated with cell-associated virus. Also interesting was the transition of cells infected by cell-free virus from a culture where infected cells had low proviral copy numbers at 24 h to one with high copy numbers by day 3 as the infection spread. This result is consistent with prior studies which suggest that cells are initially infected with cell-free virus and that cell contact is required to sustain and amplify the infection in vitro (27). Notably, the proviral copy number detected following cell-to-cell infection is similar to that published in a previous study of splenocytes from HIV-1 patients, 3 or 4 proviral copies per cell (17), and may lend some mechanistic credence to the theory that cell-to-cell spread of HIV-1 is prevalent in vivo.
While cell-free HIV-1 infection is aptly described as pseudodiploid (5, 13), we suggest that infection by cell-to-cell routes provides HIV-1 with a functionally multiploid inheritance pathway, which has several consequences. First, spontaneous point mutations generated during replication are likely to be coinherited with nonmutant copies a set fraction of the time, allowing the virus to accumulate recessive negative mutations without incurring a fitness cost. Indeed, complementation by multicopy infection can preserve less fit mutants in vitro (11). Second, multicopy infection may enhance the opportunity for recombination by allowing rare cells that are coinfected with divergent viruses to recombine over multiple generations. Related quasispecies variants would also be iteratively shuffled by repeated coinheritance and recombination. Third, the simultaneous entry of multiple viruses may saturate endogenous restriction factors and enhance viral infection (2, 24). Fourth, multicopy cell-to-cell infection can explain the viral quasispecies concept, which suggests that selection acts not on individual genomes but rather on a group of different genomes that make up a consensus sequence (19). We suggest that multiploid inheritance following cell-to-cell infection alters how we may describe and model HIV-1 inheritance, diversification, and evolution.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Chen lab and A. Sigal and D. Baltimore for helpful discussions.
This work was supported in part by NIAID 5F31 A1075570 to A.D.P.; NIAID R01 A1078783 to D.N.L.; and NIAID A1074420, NIDA DP1DA028866, a Hirschl Award, and a Burroughs Wellcome Fund grant to B.K.C.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Barbosa P., Charneau P., Dumey N., Clavel F. 1994. Kinetic analysis of HIV-1 early replicative steps in a coculture system. AIDS Res. Hum. Retroviruses 10:53–59 [DOI] [PubMed] [Google Scholar]
- 2. Besnier C., Takeuchi Y., Towers G. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. U. S. A. 99:11920–11925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen B. K., Gandhi R. T., Baltimore D. 1996. CD4 down-modulation during infection of human T cells with human immunodeficiency virus type 1 involves independent activities of vpu, env, and nef. J. Virol. 70:6044–6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen J., et al. 2005. Mechanisms of nonrandom human immunodeficiency virus type 1 infection and double infection: preference in virus entry is important but is not the sole factor. J. Virol. 79:4140–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen J., et al. 2009. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. U. S. A. 106:13535–13540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen P., Hübner W., Spinelli M. A., Chen B. K. 2007. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J. Virol. 81:12582–12595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coffin J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483–489 [DOI] [PubMed] [Google Scholar]
- 8. Dang Q., et al. 2004. Nonrandom HIV-1 infection and double infection via direct and cell-mediated pathways. Proc. Natl. Acad. Sci. U. S. A. 101:632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dimitrov D. S., et al. 1993. Quantitation of human immunodeficiency virus type 1 infection kinetics. J. Virol. 67:2182–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dixit N. M., Perelson A. S. 2004. Multiplicity of human immunodeficiency virus infections in lymphoid tissue. J. Virol. 78:8942–8945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gelderblom H. C., et al. 2008. Viral complementation allows HIV-1 replication without integration. Retrovirology 5:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haase A. T. 1999. Population biology of HIV-1 infection: viral and CD4+ T cell demographics and dynamics in lymphatic tissues. Annu. Rev. Immunol. 17:625–656 [DOI] [PubMed] [Google Scholar]
- 13. Hu W. S., Temin H. M. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. U. S. A. 87:1556–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hübner W., et al. 2007. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J. Virol. 81:12596–12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hübner W., et al. 2009. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science 323:1743–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jolly C., Kashefi K., Hollinshead M., Sattentau Q. J. 2004. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J. Exp. Med. 199:283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jung A., et al. 2002. Recombination: multiply infected spleen cells in HIV patients. Nature 418:144. [DOI] [PubMed] [Google Scholar]
- 18. Kutsch O., Benveniste E. N., Shaw G. M., Levy D. N. 2002. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 76:8776–8786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lauring A. S., Andino R. 2010. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 6:e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levy D. N., Aldrovandi G. M., Kutsch O., Shaw G. M. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl. Acad. Sci. U. S. A. 101:4204–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin N., et al. 2010. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J. Virol. 84:3516–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Overbaugh J., Bangham C. R. 2001. Selection forces and constraints on retroviral sequence variation. Science 292:1106–1109 [DOI] [PubMed] [Google Scholar]
- 23. Perelson A. S., Neumann A. U., Markowitz M., Leonard J. M., Ho D. D. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582–1586 [DOI] [PubMed] [Google Scholar]
- 24. Richardson M. W., et al. 2008. Mode of transmission affects the sensitivity of human immunodeficiency virus type 1 to restriction by rhesus TRIM5alpha. J. Virol. 82:11117–11128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rowland-Jones S. L. 1999. HIV: The deadly passenger in dendritic cells. Curr. Biol. 9:R248–R250 [DOI] [PubMed] [Google Scholar]
- 26. Sato H., Orenstein J., Dimitrov D., Martin M. 1992. Cell-to-cell spread of HIV-1 occurs within minutes and may not involve the participation of virus particles. Virology 186:712–724 [DOI] [PubMed] [Google Scholar]
- 27. Sourisseau M., Sol-Foulon N., Porrot F., Blanchet F., Schwartz O. 2007. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J. Virol. 81:1000–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vandegraaff N., Kumar R., Burrell C. J., Li P. 2001. Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA. J. Virol. 75:11253–11260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wodarz D., Levy D. N. 2011. Effect of different modes of viral spread on the dynamics of multiply infected cells in human immunodeficiency virus infection. J. R. Soc. Interface 8:289–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.