Abstract
HIV infections are initiated by a limited number of variants that diverge into a diverse quasispecies swarm. During in utero mother-to-child transmission (IU MTCT), transmitted viral variants must pass through multiple unique environments, and our previously published data suggest a nonstochastic model of transmission. As an alternative to a stochastic model of viral transmission, we hypothesize that viral selection in the placental environment influences the character of the viral quasispecies when HIV-1 is transmitted in utero. To test this hypothesis, we used single-template amplification to isolate HIV-1 envelope gene (env) sequences from both peripheral plasma and the placentas of eight nontransmitting (NT) and nine IU-transmitting participants. Statistically significant compartmentalization between peripheral and placental HIV-1 env was detected in one of the eight NT cases and six of the nine IU MTCT cases. In addition, viral sequences isolated from IU MTCT placental tissue showed variation in env V1 loop lengths compared to matched maternal sequences, while NT placental env sequences did not. Finally, comparison of env sequences from NT and IU MTCT participants indicated statistically significant differences in Kyte-Doolittle hydropathy in the signal peptide, C2, V3, and C3 regions. Our working hypothesis is that the hydropathy differences in Env associated with IU MTCT alter viral cellular tropism or affinity, allowing HIV-1 to efficiently infect placentally localized cells.
INTRODUCTION
According to UNAIDS estimates, in 2007 there were approximately 1.4 million HIV-infected pregnant women in low- and middle-income countries in need of antiretroviral therapy to prevent HIV-1 mother-to-child transmission (MTCT). In populations where replacement feeding is not feasible, it has been estimated that 12% of MTCT occurs in utero (IU) before 36 weeks gestation, 29% occurs in utero between 36 weeks and delivery, 20% occurs during labor and delivery (peripartum), and the remaining 39% occurs during prolonged breastfeeding (postpartum) (39). Although low- and middle-income countries have made progress to increase the use of antiretrovirals to prevent peripartum and postpartum HIV-1 MTCT, in 2007, only 23% of the estimated 73,000 HIV-infected pregnant Malawians had access to the required antiretrovirals (WHO/UNAIDS/UNICEF 2008 progress report). In addition, access to other preventive measures, such as elective cesarean sections and replacement formula feeding, remain unrealized. As the coverage of new intervention strategies to prevent breastfeeding and intrapartum transmissions increases, the overall rate of transmission will decrease, but a greater proportion of the transmissions will occur in utero via largely uncharacterized biological mechanisms (21).
HIV infections are initiated by a single variant or a limited number of variants that diverge and diversify into a complex quasispecies swarm (1, 62, 67, 70, 79, 82, 84). In other words, although a chronically infected pregnant woman has a heterogeneous viral population, the complexity of the viral population is severely restricted during transmission, and a homogeneous virus population is often seen in vertically infected infants. Three mechanisms have been proposed to explain this restriction: (i) transmission of the most abundant viral variant (stochastic model), (ii) transmission of multiple variants followed by selective amplification of host-adapted variants (selective-amplification model), and (iii) limited variant transmission restricted by host and/or viral biology (selection model) (85). We have previously reported a transmission bottleneck of HIV-1 subtype C during in utero MTCT based on a heteroduplex tracking assay of the V1/V2 region of env (42), and these data suggested that the bottleneck was inconsistent with a stochastic model of transmission. Building on these findings, we investigated the possibility that selection of viral lineages underlies transmission of HIV-1 variants in IU MTCT (model 3 above).
There are several opportunities for selective pressure during IU MTCT. The transmitted variants must cross several cell layers in the placenta (17) and sequentially adapt to three unique immune environments: an immunologically developed or depleted host, an immunologically blunted placenta, and finally, an immunologically naïve infant. We hypothesize that the placental environment provides the selective force for the phenotype of the viral quasispecies when HIV-1 is transmitted in utero. To address this hypothesis, we used single-template amplification to characterize the molecular evolution of HIV-1 maternal quasispecies swarms and the individual lineages that transmit and those that do not transmit during in utero MTCT. We report quantitative and qualitative differences in the gp120 region of HIV-1 env in viral populations isolated from peripheral and placental compartments. Furthermore, we report sequence differences in the env signal peptide, C2, V3, and C3 regions that are associated with MTCT status.
(This research was presented in part at the 16th Conference on Retroviruses and Opportunistic Infections, the 17th Annual Meeting on HIV Dynamics and Evolution, and the 2010 Ohio Collaborative Conference on Bioinformatics.)
MATERIALS AND METHODS
Study participants and clinical isolates.
Clinical isolates of virus were obtained from HIV-infected pregnant female participants in the Malaria, HIV, and Pregnancy (MHP) Cohort from 2001 to 2004 (52, 53) in Blantyre, Malawi. Institutional review board (IRB) approval was obtained from Ohio State University, the University of North Carolina at Chapel Hill, and the Malawi College of Medicine Research Ethics Committee; all participants gave written informed consent. HIV-infected mothers and their infants received nevirapine according to the HIVNET 012 protocol, which was the standard of care at the time of this study (27). In the parent MHP study, participants who were seroreactive on two independent HIV tests were deemed HIV positive. Infant HIV-1 status was determined by real-time PCR according to the methods of Luo et al. (47) and classified as nontransmitting (NT) or intrauterine transmission according to the definitions outlined by Bryson et al. (10). HIV-1-exposed but uninfected infants, here called NT, were HIV-1 DNA negative both at birth and at 6 weeks postpartum, while intrauterine-transmission cases were HIV-1 DNA positive within 48 h of birth. Peripheral blood was collected from the pregnant women by venipuncture, and plasma was isolated according to standard protocols. After delivery, an incision was made in the middle of the maternal surface of the placenta, 2 cm long and through half the thickness of the placental tissue, and the placental blood that pooled in the incision was collected, with the presumption that the resulting blood likely represented an admixture of maternal and fetal blood (56). In addition to placental blood, a full-thickness biopsy specimen was taken from the pericentric area of the placenta and snap-frozen in liquid nitrogen. For simplicity, HIV-1 env genes from placental blood and placental biopsy specimens were aggregated into a single category termed “placenta.” Blood from the umbilical cord vein was collected, and plasma was prepared; previously, we demonstrated that HIV-1 variants in umbilical cord blood represented the infant and not the maternal quasispecies (42). Infant blood was collected from heel pricks at birth and at 6 and 12 weeks. Constrained by the availability of matched placental tissue and maternal plasma, we identified 84 NT participants (29 of which were randomly selected) and 15 IU MTCT cases. Only 9 of the 29 NT cases and 9 of the 15 intrauterine-transmission cases supported gp160 nested-PCR amplification from all tissues, which is described below.
Using the quality control tool from the Los Alamos National Laboratory (LANL) (http://www.hiv.lanl.gov), we determined that all env amplicons were HIV-1 subtype C, that there was no contamination with common laboratory isolates, and that there was epidemiological linkage between all mother-placenta-infant pairs, with one exception. Sequences isolated from MHP sample 2466 were not monophyletic on the phylogenetic tree; all maternal samples formed on a single subclade, whereas the matched placental sequences formed another clade that was not a sister group to the maternal clade. There was no comingling of the 2,466 sequences with other sequences, so we excluded sample cross-contamination as the source of this result. This sample was eliminated from further analysis, resulting in a total of eight NT samples. Three env sequences from three different mother-placenta pairs were predicted to be intersubtype recombinants by the LANL quality control tool. In each case, comparison of these predicted recombinant sequences with their nearest neighbors showed sequence differences that could be explained by a single large deletion, and therefore, these sequences are unlikely to be true intersubtype recombinants.
Viral-RNA quantification, extraction, and cDNA synthesis.
The HIV-1 RNA concentrations in peripheral and placental plasma isolates were determined with the Roche Amplicor HIV-1 Monitor test (version 1.5) (53), which had a lower threshold of detection of 400 copies/ml; any samples with viral loads below the lower limit of detection were assigned a value of 200 copies/ml. Differences in matched viral loads (i.e., peripheral versus placental viral load) were tested with the Wilcoxon signed-rank test, and differences between unmatched viral loads (i.e., placental plasma of NT versus placental plasma of intrauterine transmitters) were tested with the two-sample Wilcoxon rank sum (Mann-Whitney) test; both tests were implemented with STATA version 10.1 (STATA Corp., College Station, TX). RNA was extracted from 140 μl centrifuged plasma using the QIAamp viral-RNA minikit that was used for other plasma samples. If the plasma viral concentration was <10,000 RNA copies/ml, samples were concentrated by ultracentrifugation at 24,500 × g for 1.5 h at 4°C. To extract RNA from placental biopsy specimens, approximately 100 mg tissue was preincubated for 3 to 4 min in Qiazol lysis reagent, placed in a QIA shredder column, and centrifuged for 2 min at 21,130 × g. The eluent was processed with the RNeasy Lipid Tissue minikit (Qiagen), and the RNA was eluted into 50 μl of RNase-free water. RNA was reverse transcribed into cDNA with SuperScript III (Invitrogen Life Technologies, Carlsbad, CA) as follows: reaction mixtures containing 50 μl RNA, 0.5 mM deoxynucleoside triphosphates (dNTPs), and 0.25 μM HIV-1 subtype C-specific primer (OFM19) were heated for 5 min at 65°C and then quenched on ice. cDNA synthesis was performed in 100 μl with 1× reverse transcriptase buffer, 5 mM dithiothreitol, 80 units RNaseOut, and 1,000 units SuperScript III (Invitrogen Life Technologies, Carlsbad, CA). The reaction mixture was sequentially incubated at 50°C and 55°C for 1 h each, inactivated at 70°C for 15 min, incubated with 2 units RNase H (Invitrogen Life Technologies, Carlsbad, CA), and then digested at 37°C for 20 min. To ensure that only HIV-1 RNA was isolated from placental biopsy specimens, placental tissue extracts underwent mock cDNA synthesis without SuperScript III; this reaction never supported PCR amplification, indicating that HIV-1 RNA and not HIV-1 DNA in the placental biopsy specimen served as the PCR template (data not shown).
Single-template amplification.
It has been demonstrated that PCR amplification of a related yet heterogeneous mixture of templates is prone to PCR-induced template recombination, which can be mitigated by performing PCR on single templates (59, 77). Single-template amplification was performed according to published protocols (35, 57, 65, 73). Briefly, serial dilutions of cDNA were made until less than 30% of 8 to 10 replicate PCRs generated amplicons. All PCRs used High Fidelity Platinum Taq (Invitrogen) according to the manufacturer's instructions and the subtype C-optimized outer primers VIF and OFM19 and inner primers ENVA* and ENVN (the sequences are listed in Table S1 in the supplemental material).
DNA sequencing.
Prior to being sequenced, PCR amplicons were incubated with exonuclease I (New England BioLabs) and alkaline phosphatase (Roche) at 37°C for 30 min, followed by heat inactivation at 95°C for 5 min (6). Full-length gp160 was sequenced using eight HIV-1 subtype C-optimized primers (Genewiz, Inc., NJ) (see Table S1 in the supplemental material), which provided a minimum of 2-fold coverage. The sequencing chromatograms were manually examined for multiple peaks, and those with multiple concurrent peaks were discarded. env sequences were trimmed, assembled, and manually edited using Geneious software. All sequences were examined with the HIV Sequence Quality Control Tool on the Los Alamos National Laboratory HIV database website (http://www.hiv.lanl.gov). The results from this tool allowed us to exclude potential contamination with laboratory isolates and to conclude that all env sequences are most closely related to HIV-1 subtype C (data not shown). The features of gp41 will be described elsewhere.
Sequence alignment and phylogenetic-tree construction.
Cleaned gp160 sequences were first aligned, using HMMER (19), to the Env protein model provided by the LANL. This alignment was manually adjusted in less than 10 instances to correct the alignment of sequence landmarks and to maintain the reading frame. The adjusted alignment was used to train a new subtype C-specific Env protein model, followed by alignment of the sequences to this new model using HMMER. Finally, the alignment was used for a tree search in RAxML (75) with a general time-reversible substitution model with gamma-distributed rate heterogeneity and a proportion of invariant sites (GTR-gamma-I; determined using Findmodel [http://www.hiv.lanl.gov]). The best-scoring tree under the maximum-likelihood criterion after 100 replicates is shown in Fig. S1 in the supplemental material.
Assessment of recombination.
Genetic algorithm recombination detection (GARD) (38), a standard recombination detection method included in hypothesis testing using phylogenies (HYPHY) (58), was used to screen all epidemiologically linked maternal and placental sequences for evidence of recombination. Significant breakpoints are reported at a P value of <0.05, as reported by HYPHY. The phylogenetic tree associated with each segment identified by GARD was used for the positive-selection analyses described below.
Compartmentalization testing.
Using the tree produced by RAxML, Slatkin-Maddison (74) P values were calculated using the standard analysis packaged in HYPHY (58). A second test of compartmentalization, the Hudson nearest-neighbor method (31) was also used; this method is a robust measure of compartmentalization when there are unequal numbers of sequences in the two compartments (83), as is the case for some of the pregnancies in these data. The analysis was performed both with and without duplicate sequences.
Sequence characteristics.
Evidence of APOBEC-induced sequence hypermutation was measured with Hypermut (http://www.hiv.lanl.gov). Hypervariable loops from the gp120 sequences were extracted with the Gene Cutter program (http://www.hiv.lanl.gov). Potential N-linked glycosylation sites (PNLGs) on both gp120 and the individual hypervariable loops were enumerated with the N-glycoside program (http://www.hiv.lanl.gov). Coreceptor usage of the subtype C gp120 genes was predicted from the extracted V3 sequences with the Web position-specific scoring matrix tool trained for HIV-1 subtype C (PSSM-C), which is maintained by the University of Washington (32).
Alignment position quality controls and multiple-hypothesis correction.
Because multiple amino acid positions are considered simultaneously, it is necessary to perform a multiple-hypothesis correction. An amino acid position was included in the analysis if it was a consensus deletion in no more than 2 (of 17) participants and no more than 12 of the participants shared the same plurality amino acid, excluding those participants for whom the Shannon entropy at that amino acid position (among different variants) was greater than 0.4. This minimum diversity requirement excludes amino acid positions that are too conserved to ever rise above the multiple-hypothesis correction (see below) based on estimates from Fisher's exact test (P < 0.001, with fewer than five mothers showing the rare variant amino acid; a Shannon entropy of 0.4 roughly corresponds to a rare-variant frequency of 22%, which should be detected in 95% of bootstrap replicates.) After filtering, 118 amino acid positions and their corresponding codons were included in the analyses described below. To correct for multiple-hypothesis testing, the Benjamini-Hochberg method was used with a primary false-discovery rate of less than 0.05; in order to reduce the probability of false-negative results, we also report a secondary false-discovery rate of 0.25 (7).
Positive and purifying selection.
Positions under positive selection were identified by differences in relative synonymous and nonsynonymous substitution rates (dN − dS) as reported by the standard analysis packaged with HYPHY (58), including an adjustment for any recombination detected by GARD. HIV-1 sequences derived from each volunteer were treated separately, and the observed and expected values for synonymous and nonsynonymous substitution rates were combined across volunteers after stratification by their transmission status. The P value for the sum of these substitution rates was obtained using the same binomial-approximation calculation employed by HYPHY (58). This analysis was performed only for the 118 codons corresponding to amino acids used in other analyses, and the same multiple-hypothesis correction was applied.
Bootstrapping method to identify sequence differences associated with in utero MTCT.
For purposes of identifying sequence differences associated with IU MTCT, comparisons were made at individual sites in HIV-1 env sequences derived from IU MTCT pregnancies versus those derived from NT pregnancies. Statistically significant differences were identified using a nested-bootstrapping procedure (16), with a given pregnancy sampled on the outer layer and individual sequences from each pregnancy sampled on the inner layer, for a total of 12 sequences per pregnancy per sample (the fewest for any pregnancy). Allowing this number to match the full number of available sequences did not render any of the results nonsignificant. At each alignment position, in each bootstrap replicate (100,000 replicates), the difference between the average Kyte-Doolittle hydropathy index (43) of the IU MTCT sequences in the sample and the NT sequences in the sample is calculated. At each alignment position, a given bootstrap replicate supports the null hypothesis (similar hydropathy) if the difference is zero or if the difference has a sign opposite to that of the difference observed in the data set as a whole. The P value at each alignment position is the fraction of bootstrap replicates in which the null hypothesis is supported (e.g., the P value drops, becoming more significant, if the difference seen in the data set is also seen in a large fraction of the bootstrap replicates). The bootstrap P values were calculated only for the 118 alignment positions where a significant association could be detected given the available data (see “Alignment position quality controls and multiple-hypothesis correction” above).
Feature mapping on the Env crystal structure.
Amino acid positions of interest were mapped onto the JRFL Env crystal structure (Protein Data Bank identifier [PDB ID] 2B4C) using Jmol, an open-source Java viewer for chemical structures in three dimensions (3D) (http://www.jmol.org/).
Nucleotide sequence accession numbers.
The env sequences from the signal peptide through gp120 (abbreviated sp-gp120) have been deposited in GenBank (http://ncbi.nlm.nih.gov) under accession numbers JF722674 to JF723201.
RESULTS
Comparison of peripheral and placental plasma-associated HIV-1 loads.
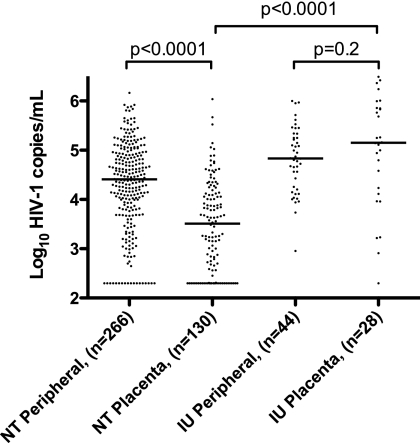
Previous studies from the MHP cohort have reported an association between malaria and the HIV-1 load (53), an association between maternal syphilis seroreactivity and HIV-1 MTCT (52), an association between maternal-infant blood admixture during intrapartum HIV-1 MTCT (40, 41), and a reduction in HIV-1 quasispecies diversity during HIV-1 MTCT (42). In order to better understand the relationship between the HIV-1 load in peripheral and placental plasmas, we stratified viral-load data from the MHP cohort by HIV-1 transmission status. Table 1 shows the clinical characteristics of the 266 NT and 44 in utero-transmitting HIV-infected women whose peripheral viral loads were quantified; a subset of 130 NT and 28 IU MTCT cases had matched placental viral-load data. As shown in Fig. 1, the median placental HIV-1 concentration among NT women was significantly lower than the median HIV-1 concentration in matched peripheral plasma (P < 0.0001), while in contrast, the median placental HIV-1 concentration in intrauterine-transmitting women was equivalent to or slightly higher than the median HIV-1 concentration in matched peripheral plasma (P = 0.2). In comparisons between the two groups, the median placental HIV-1 concentration in intrauterine-transmitting women was significantly higher than in NT women (5.2 log10 copies/ml versus 3.5 log10 copies/ml, respectively; P < 0.0001). While IU MTCT cases were significantly more likely to be coinfected with syphilis (consistent with previous reports [52]), other clinical features did not differ significantly by transmission status. These results show two distinct differences in viral loads that are associated with MTCT status. First, the placental HIV-1 concentration is higher in cases of IU MTCT than in cases of NT. Second, the relationship between peripheral and placental HIV-1 concentrations differs by MTCT status: the vast majority of NT pregnancies had placental viral loads that were on average an order of magnitude lower than matched peripheral concentrations, while the placental viral loads were similar to matched peripheral viral loads among the IU MTCT pregnancies.
Table 1.
Clinical features of HIV-infected women with peripheral and placental HIV-1 viral-load data, stratified by HIV-1 mother-to-child transmission status
| Featurea | NT (n = 266) | In utero transmitter (n = 44) | P value |
|---|---|---|---|
| Median age (IQR) (yr) | 24 (21, 28) | 24 (20, 26) | 0.2b |
| Median CD4+ T-cell count (IQR) | 378 (234, 530) | 293 (202, 439) | 0.2b |
| % Syphilis seroreactivity (n) | 5 (265) | 18 (44) | 0.002c |
| % with placental malaria (n) | 8 (256) | 1 (41) | 0.09c |
IQR, interquartile range.
Kruskal-Wallis equality-of-populations rank test.
Fisher's exact test.
Fig. 1.
The peripheral and placental HIV-1 RNA concentrations vary according to both transmission status and tissue compartment. Each point represents a viral-load measurement from an individual participant, with the horizontal lines representing the median values. P values represent the results of Wilcoxon signed-rank tests of log10 peripheral versus log10 placental viral loads or a two-sample Wilcoxon rank sum test of log10 placental viral loads of NT versus in utero transmitters (IU).
env sequence characteristics.
Previous studies from the same MHP cohort revealed a restriction of HIV-1 env genetic diversity after IU MTCT in a manner inconsistent with a stochastic mechanism of variant transmission (42). Building on these data, we tested an alternative model, natural selection of viral lineages, to explain the genetic restriction observed during IU MTCT. In order to test this hypothesis, we used single-template amplification to isolate HIV-1 env from peripheral plasma (maternal), various placental tissues (placental plasma and placental biopsy specimens), and umbilical cord blood (infant). Table 2 shows the clinical characteristics of the maternal donor, the number of gp160 amplicons sequenced from the peripheral and placental compartments, and the biochemical characteristics of the sequences of these matched samples. A total of 589 gp160 sequences from maternal plasma, placental plasma, a placental biopsy specimen, cord plasma, neonatal plasma, and 12-week infant plasma were obtained. After alignment and in silico translation, 61 of the 589 sequences (∼11%) had multiple stop codons and were excluded from further analysis; none of the sequences showed evidence of APOBEC hypermutation (data not shown). The majority of the sequences with stop codons were isolated from infant tissues (P < 0.05; Fischer's exact test). In total, 528 env sequences, spanning the signal peptide through env gp120 (sp-gp120), from nine intrauterine-transmission and eight NT participants are described.
Table 2.
Clinical and virological features of the participants contributing env sequencesa
| MHP ID | MTCT status | Peripheral VL (no. of copies/ml) | Placental VL (no. of copies/ml) | CD4+ T cells (no. of cells/ml) | No. of env sequences |
Avg no. of substitutions/position |
Median V1 length (amino acids) (minimum, maximum) |
Median V2-C5 length (amino acids) |
Median predicted no. of PNLGs |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | P | M | P | M | P | M | P | M | P | |||||
| 1639 | NT | 160,259 | NA | 375 | 7 | 26 | 0.004 | 0.003 | 21 (21) | 21 (17, 21) | 369 | 369 | 30 | 30 |
| 1669 | NT | 81,178 | NA | 485 | 5 | 13 | 0.015 | 0.014 | 14 (10, 14) | 14 (10, 14) | 356 | 356 | 27 | 27 |
| 1702 | NT | NA | NA | 273 | 12 | 7 | 0.017 | 0.027 | 17 (17, 33) | 19 (13, 33) | 357 | 354 | 27 | 27 |
| 2437 | NT | 13,529 | NA | 445 | 10 | 10 | 0.019 | 0.036 | 19 (19) | 19 (17, 19) | 361 | 359 | 26 | 26 |
| 2502 | NT | 154,972 | NA | 38 | 12 | 15 | 0.070 | 0.061 | 20 (13, 20) | 20 (14, 20) | 372 | 370 | 28 | 28 |
| 2512 | NT | 23,301 | NA | 222 | 7 | 6 | 0.040 | 0.054 | 21 (20, 23) | 22 (20, 29) | 358 | 361 | 24 | 25 |
| 2544 | NT | 587,702 | NA | NA | 19 | 19 | 0.018 | 0.024 | 17 (17) | 17 (17) | 360 | 358 | 25 | 25 |
| 3274 | NT | NA | NA | 103 | 13 | 18 | 0.073 | 0.063 | 15 (9, 25) | 15 (9, 18) | 361 | 361 | 26 | 26 |
| 1468 | IU | 36,453 | 87,276 | 274 | 17 | 6 | 0.035 | 0.040 | 12 (12, 17) | 12 (12, 17) | 356 | 355 | 26 | 25 |
| 1472 | IU | 51,551 | 24,116 | 397 | 16 | 12 | 0.028 | 0.048 | 17 (13, 19) | 13 (13, 19) | 370 | 360 | 28 | 26 |
| 1485 | IU | 125,813 | 708,115 | 329 | 7 | 34 | 0.051 | 0.014 | 21 (10, 31) | 10 (10, 21) | 364 | 361 | 28 | 19 |
| 1646 | IU | 902,778 | NA | 95 | 14 | 9 | 0.050 | 0.040 | 17 (16, 24) | 16 (16, 23) | 364 | 364 | 26 | 26 |
| 1851 | IU | NA | NA | 761 | 6 | 6 | 0.015 | 0.040 | 8 (8, 11) | 9 (8, 9) | 355 | 356 | 23 | 27 |
| 2080 | IU | 165,000 | NA | 91 | 14 | 7 | 0.052 | 0.037 | 15 (14, 38) | 14 (14) | 365 | 365 | 26 | 25 |
| 2400 | IU | 98,793 | NA | 439 | 16 | 43 | 0.043 | 0.022 | 17 (17, 22) | 22 (17, 22) | 349 | 353 | 24 | 26 |
| 2797 | IU | NA | NA | 399 | 14 | 18 | 0.041 | 0.002 | 23 (17, 27) | 23 (23) | 345 | 344 | 26 | 25 |
| 3321 | IU | NA | NA | 220 | 8 | 11 | 0.065 | 0.047 | 24 (11, 38) | 35 (16, 35) | 357 | 357 | 26 | 27 |
M, maternal plasma; P, placental tissue; VL, viral load; V, variable; C, conserved; NA, no data available.
The CD4+ T-cell count and HIV concentration have been correlated with viral population diversity and IU MTCT; therefore, it is plausible that the observed sequence differences between intrauterine transmission and NT samples (see below) might reflect differences in progression or immunological status between the intrauterine-transmission and NT groups. To address this concern, we calculated the mean viral sequence diversity for each pregnancy and measured its correlation with the viral load or CD4+ T-cell counts. Maternal viral population diversity increased in both intrauterine transmission and NT, with declining CD4+ T-cell counts (P < 0.003; Pearson's R2), but not with rising viral load (Table 2). No significant relationship was observed between transmission status and viral population diversity in either compartment or between maternal sequences stratified by transmission status (0.04 ± 0.01 substitutions per nucleotide in the intrauterine-transmission maternal sequences and 0.03 ± 0.03 substitutions per nucleotide in the NT maternal sequences). Therefore, the sequence differences observed in the sequencing subcohort are unlikely to be a proxy for disease progression or immunological status.
Regardless of transmission status, there was variability in the env length within the nine intrauterine-transmission and eight NT isolates (Table 2). First, we compared the median lengths of the hypervariable regions of gp120 (V1 to V5) isolated from matched placental and maternal plasmas. Within single participants, insertions and deletions occurred most frequently in env V1. The median env V1-V5 lengths between epidemiologically linked sp-gp120 sequences were similar in six of eight NT isolates, but in seven of the nine IU MTCT isolates, the env V1-V5 loop lengths were different (either longer or shorter). These differences were most robust in the env V1 loop, where differences as great as 12 amino acids between epidemiologically linked samples were observed (Table 2). On average, env V1 loops from matched IU MTCT maternal-peripheral and placental samples differ in length by 4 amino acids in spite of the epidemiological linkage. This is equal to the average difference between env V1 loop lengths from different mothers (not epidemiologically linked) and much greater than the average of 1 amino acid change in average env V1 loop lengths seen between NT maternal-peripheral and placental sequences. Finally, this is in strong contrast to other regions of gp120; for example, the entire region from the end of env V1 to the end of env C5 (V2-C5 length in Table 2), which is over 20 times as long as env V1 on average, shows an average length change of 8 amino acids between different mothers but an average change of only 3 amino acids in length when matched IU MTCT maternal-peripheral sequences were compared to placental sequences. The absolute change in average env V1 loop lengths is significantly greater in IU MTCT pregnancies than in NT pregnancies (P < 0.02; Wilcoxon's rank sum test), but the absolute change in average env C2-V5 length did not differ significantly by transmission status (P = 0.5; Wilcoxon's rank sum test). The contrast between env V1 length differences (high between tissues from the same participant and low between mothers) and env C2-V5 length differences (low between tissues from the same participant and high between mothers) indicates that the env V1 length differences observed are not simply an indirect consequence of overall sequence differences associated with a placental compartment. This suggests that over a short evolutionary time frame, the env V1 loop among IU MTCT isolates varies tremendously between the periphery and the placenta.
Consistent with the env V1 loop length data, differences in the numbers of PNLGs in epidemiologically linked plasma/placental isolates were observed in eight of nine IU MTCT samples, while NT pairs had similar numbers of PNLGs in seven of eight samples (Table 2). The average number of PNLGS differed between the maternal-peripheral and placental compartments in each transmitting participant, but not in nontransmitting participants (P < 0.008 by the Wilcoxon rank test). However, the change in PNLGs was closely correlated with the change in overall loop length (Pearson R, 0.81; significant to a P value of <0.005). In one extreme case, the placental variants from one IU MTCT pair (MHP1485) had nine additional PNLGs.
Overall, env V3 loop lengths were consistent between placental and peripheral variants, and evidence of predicted CXCR4 (X4) tropism was identified in 100 of the 528 sequences (19%) and 10 of the 17 (59%) epidemiologically linked isolates: four of eight NT and six of nine intrauterine transmission mother-placenta pairs. In all cases, if predicted X4 tropism was found in the peripheral blood, it was also found in the placental tissue. Infant samples were available for five cases of IU MTCT, and two infants had env sequences with a predicted X4-tropic phenotype.
Next, we used GARD to identify significant recombination breakpoints in the sp-gp120 sequences. The eight NT mothers had 3.0 ± 2.0 recombination breakpoints significant at a P value of <0.05, while the nine intrauterine-transmtting mothers had 3.4 ± 1.7 recombination breakpoints at the same significance threshold. It is possible that recombination might interfere with the detection of a real compartment (discussed below). In this case, however, the intrauterine-transmitting mothers (in whom significantly more HIV-1 compartmentalization is detected) also show more recombination in sp-gp120, although the difference is not significant.
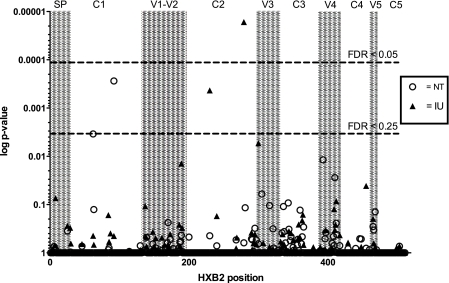
The phylogenetic trees produced by GARD were used to calculate dN − dS values and the dN and dS values were pooled among volunteers according to transmission status to identify amino acid positions/codons that are under statistically significant positive (dN − dS > 0) or purifying (dN − dS < 0) selection. Positions 279 and 230 in the intrauterine transmitters were under significant purifying selection, with dN − dS values of −1.41 and −1.17 (HXB2 coordinates; false-discovery rates, <0.05 and 0.25, respectively). In the NT group, positions 62 and 92 were also under purifying selection, with dN − dS values of −0.72 and −1.28 (HXB2 coordinates; false-discovery rate, <0.25). Figure 2 shows that the intrauterine-transmission positions under purifying selection are clustered in env C2 and that the NT positions under purifying selection are in env C1.
Fig. 2.
Several positions in Env show statistically significant selection. The vertical axis shows the P value for each codon among all HIV-1 sp-gp120 sequences stratified by transmission status; all significant positions are under purifying selection. The P values correspond to the probability of at most the observed number of nonsynonymous substitutions given the synonymous substitutions and amino acids observed (as estimated using HYPHY). These results account for any recombination breakpoints identified by GARD. The horizontal axis shows amino acid positions within sp-gp120, which are numbered according to the HXB2 reference sequence. The dashed lines indicate the thresholds for statistical significance at false-discovery rates (FDR) of 0.05 and 0.25. The shaded areas indicate the hypervariable (V) or conserved (C) domain of env, also according to the HXB2 reference sequence.
env sequence differences associated with transmission status.
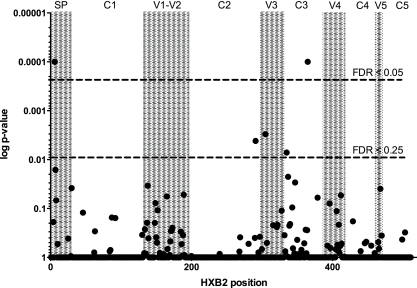
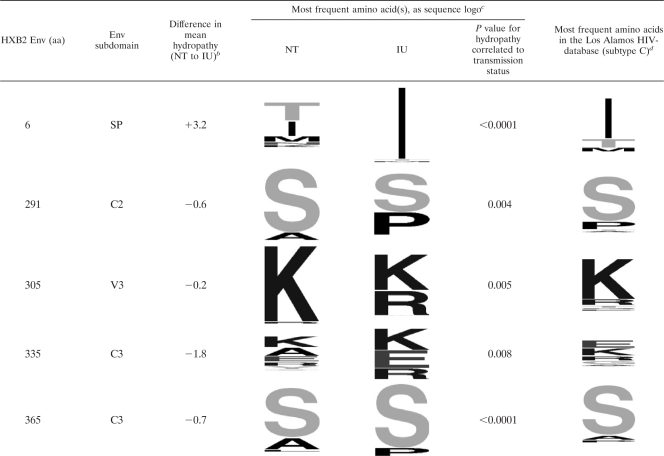
We identified several characteristic sequence differences associated with MTCT, which are shared by the viral variants present in the IU MTCT participants regardless of their tissue of origin. In order to better understand these differences and determine the statistical significance of the observed differences, we converted the observed amino acids to a quantitative measurement and used a nested-bootstrapping method to measure differences in Kyte-Doolittle hydropathy at individual gp-120 amino acid positions, stratified by mother-to-child transmission status. Hydropathy was chosen over other measures of amino acid physical properties because it is especially sensitive to amino acid changes that alter the energetics of ligand-binding interactions, which are dominated by interactions between the protein and the solvent (23). Among the 120 amino acid positions with significant amino acid variability, statistically significant differences in hydropathy as a function of transmission status were identified at gp-120 amino acid positions 6 and 365 (HXB2 coordinates; false-discovery rate, 0.05) and amino acid positions 291, 305, and 335 (false-discovery rate, 0.25) (Fig. 3). The most frequent amino acids at the individual positions, segregated by transmission status, are displayed in Table 3 using sequence logos (13, 68). For comparison, the most frequent amino acids among HIV-1 subtype C env genes currently deposited in the Los Alamos HIV-1 database are shown in the final column (only one sequence per patient was included in the analysis). Note that although amino acids at low frequency can produce a statistically significant result, the nested-bootstrapping method requires those rare amino acids to be present in all or most participants in order to produce a significant P value. Using the same false-discovery thresholds, no significant differences in hydropathy were found between sequences in the same transmission type but from different tissues; this is in notable contrast to the V1 loop length differences described above.
Fig. 3.
Several positions in Env are associated with in utero mother-to-child transmission. Shown are amino acid positions where differences in Kyte-Doolittle hydropathy values at the individual positions of HIV-1 sp-gp120 are associated with transmission status. The P values (vertical axis) shown correspond to the fraction of bootstrap replicates in which the sign of the observed hydropathy difference is maintained. The dashed lines indicate the thresholds for statistical significance at false-discovery rates of 0.05 and 0.25. The shaded areas indicate the hypervariable (V) or conserved (C) domains of env, also according to the HXB2 reference sequence (horizontal axis).
Table 3.
Changes in hydropathy at several Env positions are correlated with in utero mother-to-child transmissiona
Shown are the individual amino acid (aa) positions, according to HXB2 numbering, and the corresponding subdomain of Env where that position is found (SP, signal peptide; V, variable; C, conserved).
Difference in mean hydropathy indicates the difference in the bootstrap mean estimate of the Kyte-Doolittle hydropathy at the position, stratified by transmission status. A positive value indicates more hydrophobic residues in in utero mother-to-child transmission (IU); a negative value indicates more hydrophobic residues in the nontransmitters (NT).
The most frequent amino acids are displayed using sequence logo, where the size of the letter is proportional to its frequency. The P values represent the raw (not multiple-hypothesis-corrected) correlation between hydropathy at the position and the transmission status.
The most frequent amino acids in 757 HIV-1 subtype C sequences in the Los Alamos HIV database.
Placental compartmentalization.
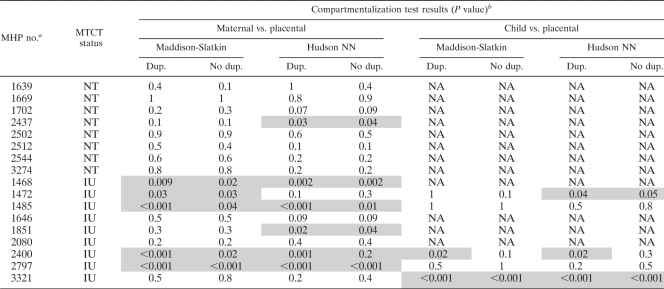
The quantitative data presented in Fig. 1 indicate that the viral load in the placenta is not equivalent to that in the periphery. This result could indicate an independent compartment of HIV-1 replication. In order to test the hypothesis that HIV-1 is compartmentalized in the placenta during IU MTCT, both a tree-based (Slatkin-Maddison [74]) and a distance-based (Hudson fixation index [FST] [31]) test of compartmentalization were performed on the sp-gp120 sequences isolated from the placenta and peripheral plasma (phylogenetic trees and highlighter plots are shown in Fig. S2 in the supplemental material). In seven of the eight NT pregnancies, both tests failed to support statistically significant compartmentalization (defined as a P value of <0.05) between the placenta and the maternal periphery (Table 4). In contrast, six of the nine intrauterine-transmission pregnancies showed significant compartmentalization (P < 0.05) by either the tree-based Slatkin-Maddison test or the Hudson FST nearest-neighbor test. The discordant results between these tests of compartmentalization could be a result of the different numbers of sequences sampled in the two potential compartments, in which case the Hudson FST is reportedly a more sensitive test of compartmentalization (83). Exclusion of duplicate sequences attenuated the significance of the compartmentalization tests, but it did not alter the general association between compartmentalization and IU MTCT. In addition, five of the nine groups of intrauterine-transmission isolates (MHP ID 1468, 1485, 1851, 2400, and 2797) and one of the NT isolates (MHP ID 2437) showed evidence of local viral replication in the placenta (see Fig. S2 in the supplemental material). Overall, the intrauterine-transmission pregnancies showed more compartmentalization, as assessed by a Wilcoxon sum-of-ranks test on the P values associated with each individual volunteer: P < 0.0006 for Slatkin-Maddison with duplicates; P < 0.02 for Slatkin-Maddison without duplicates; P < 0.02 for Hudson nearest neighbor with duplicates; and P < 0.12 for Hudson nearest neighbor without duplicates.
Table 4.
Evidence for placental compartmentalization of HIV-1 env sequences
The MHP number is the participant's unique number.
Results of a tree-based (Slatkin-Maddison) and distance-based (Hudson) nearest-neighbor (NN) test of compartmentalization. Statistically significant compartmentalization (P < 0.05) is shaded. Dup. and No dup. indicate whether duplicate sequences were included in the analysis. NA, not available.
DISCUSSION
This study was designed to test the hypothesis that selected HIV-1 variants cross the placental barrier and cause IU MTCT. After genotypically characterizing 528 env genes from matched peripheral and placental tissues of pregnant women with documented HIV-1 vertical-transmission status, we observed the following: (i) tissue-specific differences in viral loads, which varied by HIV-1 transmission status; (ii) discordant env V1 lengths between peripheral and placental HIV-1 isolated from cases of IU MTCT; (iii) phylogenetic reconstruction consistent with the presence of a placental compartment; and (iv) differences in env hydropathy within the env-CD4 binding interface that are associated with IU MTCT.
There are several limitations to these findings, including the following: a small sample size at the participant level, which limits the generalizability of our findings, and a potential bias toward the characterization of the most abundant viral variants, which is an inherent limitation of single-template amplification. In addition, HIV-1 isolated from the placenta can be a proxy for infant HIV present in the fetal vasculature within the placenta (56). Maternal-fetal blood admixture could confound the interpretation of the placenta-specific observations, but the following observations argue against the placenta representing only infant blood. First, viral diversity in the placental biopsy specimens did not differ by transmission status (P = 0.8; Student's t test). Second, infant sequences are generally homogeneous and therefore unlikely to contribute significantly to viral diversity (54, 61). Third, compartmentalization tests comparing infant env sequences to placental env sequences indicated the presence of both a placental and an infant compartment in three of five mother-infant pairs. Nevertheless, even if these findings were to represent the characteristics of HIV-1 in the infant and not the placenta, these data are still important because they suggest features of env that could be associated with enhanced viral fitness in the immunologically naïve newborn.
Although we observed genotypic differences between peripheral and placental HIV-1 isolates (varied env V1 loop lengths) and codon-specific differences in hydropathy between isolates from in utero transmitters versus nontransmitters, the environmental pressures that selected for these changes in env are unknown. Immunological pressures on viral replication are a likely selective force, and several of the observed differences in env in this report have previously been associated with immune escape. For example, extension of the V1 loop length has been associated with neutralizing-antibody escape through a hypothesized “glycan shield” masking mechanism (64, 81). Our data do not show a consistent increase or decrease in env V1 loop length between anatomical compartments; rather, we observed that the env V1 loops are frequently different during IU MTCT. Amino acid 230, which is under purifying selection in the intrauterine-transmission cases, is an established glycosylation site, and amino acid 279 is within the N-linked glycosylation site pattern and could potentially influence the efficiency of glycosylation. In addition, one of the positions with different hydropathy values is located on the α2 helix of env C3 (HXB2 position 335), and this position is a known target of autologous neutralizing antibodies (51) and likely relevant to neutralizing-antibody escape (25, 63). Although the sensitivity of the env isolates from this study to antibody neutralization has not been tested, another study using samples from the MHP cohort failed to detect altered sensitivity of in utero-transmitted Env variants to autologous, heterologous, or the well-characterized monoclonal antibodies (63a). It is currently unknown if alterations in these Env positions allow HIV-1 to escape other antibody-directed immune reactions, such as antibody-dependent cellular cytotoxicity (ADCC).
In addition to selective pressure from the immune system, the differences in env associated with either placental localization or in utero MTCT could also be a result of viral adaptation to the tissues of the placenta, which has a unique repertoire of cellular targets for viral infection. The maternal-fetal placental barrier is comprised of several cell layers, including syncytiotrophoblast and cytotrophoblasts, and in the absence of placental lesions, it is presumed that HIV-1 crosses through these cells during in utero MTCT. To date, there are no published data that support maternal-fetal blood admixture as a mechanism of in utero HIV-1 mother-to-child transmission (40, 41, 46). Thus, our working assumption is that HIV-1 must cross an intact maternal-fetal barrier during in utero MTCT. Several studies have demonstrated HIV-1 infection of trophoblasts and syncytiotrophoblast in both immortalized and primary trophoblasts (4, 5, 15, 50, 71). Paradoxically, it has also been reported that syncytiotrophoblast express either a low level of CD4 or CCR5 molecules or none (18, 44), although this finding is inconsistent (14, 50). Trophoblasts are known to express various C-type lectins, like gelatins, DC SIGN, globoside, and syndecans, on their surfaces (12, 33, 34), many of which can serve as attachment receptors for HIV-1 in CD4-negative cell lines (8, 20, 28, 45, 80). Because env V1 length and glycosylation are important in several aspects of the viral life cycle, including replication competence (76), tropism (9, 26), and cytopathology (37, 55, 76), it is possible that the observed differences in env V1 loop lengths between peripheral and placental compartments in cases of in utero transmission are a manifestation of placental adaptation, such as an increased adherence to C-type lectins.
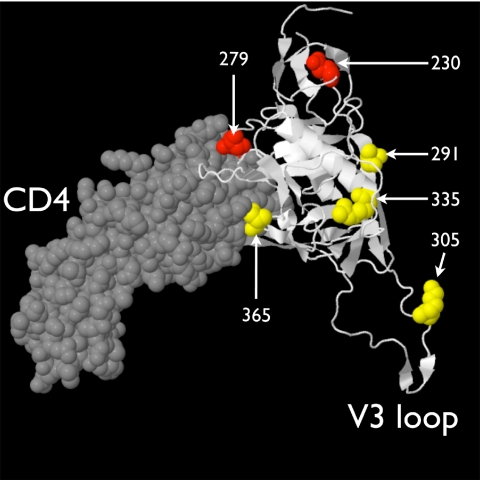
After comparing sequences from participants stratified by transmission status, we found significant differences in hydropathy between intrauterine-transmission and NT isolates at several amino acid positions. Hydrophobicity was chosen for this application in order to be maximally sensitive to mutations between polar and nonpolar residues. However, the mutations identified (Table 3) include two relatively conservative K-to-R mutations. Although less conservative mutations are expected to have a stronger impact on the biochemistry of the protein product, this is not always the case—even relatively conservative amino acid substitutions may have significant biological effects (36). Although they were not associated with placental localization, it is possible that the sp-gp120 env positions associated with in utero MTCT are also a marker of placental adaptation. One of the five hydrophobicity changes (position 365) and one of the positions under purifying selection during intrauterine transmission (position 279) are likely in direct contact with the CD4 molecule (Fig. 4). In addition to its role in binding the CD4 molecule, it has been shown that the amino acid sequence of env V3 through C3 is similar to those of the chemokines that bind either CCR5 or CXCR4 (72). Perhaps the observed changes in env associated with in utero mother-to-child transmission expand the repertoire of coreceptors available to HIV-1 for entry into cells in the placenta or infant.
Fig. 4.
Locations of the amino acid positions associated with in utero mother-to-child transmission. The HIV-1 JR-FL gp120 core (PDB 2B4C) is displayed in white in a cartoon format complexed with a fragment of the CD4 ligand, which is drawn in gray in a space-filling format. Four of the 5 amino acids correlated with in utero transmission are colored yellow (position 6, in the signal peptide, is not included in the structure); the two positions with evidence of purifying selection during in utero transmission are displayed in red. Amino acid positions are indicated in white text.
Finally, the observation that during in utero MTCT the placenta shows phylogenetic evidence of viral compartmentalization is consistent with the hypothesis that HIV-1 has adapted to the placental environment. HIV-1 compartmentalization has been described in semen (2, 11) and brain (69), kidney (49), and metastatic (66) tissue, but limited, if any, HIV-1 compartmentalization has been observed in breast milk (29, 30). Sites of HIV-1 infection in the placenta that could produce a distinct compartment include placental macrophages, trophoblasts, decidual mononuclear cells, and T-cell infiltrates in the intervillous space and decidua (3, 4, 48). Placental macrophages (PMs) express lower levels of the CD4, CXCR4, and CCR5 receptors than monocyte-derived macrophages (MDMs) in the peripheral blood compartment (22, 78). Though CD4 and coreceptor expression is low, viral integration does not differ between PMs and MDMs, (24), suggesting that alternate coreceptors or enhanced affinity may be involved in infection of placental macrophages.
In summary, this study supports viral compartmentalization in the placenta during IU MTCT, and it identified changes in the CD4-gp120 binding interface that correlate with IU MTCT. Our working hypothesis is that these mutations alter viral tropism, allowing HIV-1 to efficiently infect cells in the placenta. Whether this model is relevant for all HIV-1 subtypes or just subtype C, which has been shown to be transmitted in utero more frequently than HIV-1 subtypes A and D (60), remains to be evaluated. Future studies addressing the phenotypic consequences of these findings could provide insight into the mechanism of in utero HIV-1 mother-to-child transmission.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful for the participation of the Malawian women and their newborns; the Malaria, HIV, and Pregnancy Cohort staff running the MHP study; Debbie Kamwendo for managing the MHP study; Paul Wilson for the viral-load measurements; Debbie Knight for her logistical support; Joe Verducci of the OSU Department of Statistics, for assistance with the statistics; and the anonymous reviewers for their helpful suggestions. We thank the Medical Center Information Services team of OSU and the Ohio Supercomputer Center for hosting computing clusters used in this study.
This project was supported in part by NIH grants K99-HD056586 and R00HD056586 to J.J.K. and R01-AI49084 and R21-AI065369 to S.R.M. S.J.R. was supported by Wellcome Trust Senior Research Fellowship number 063215. We acknowledge that this material is based upon work supported by, or in part by, the U.S. Army Research Laboratory and Office under grant number W911NF-05-1-0271 (D.J.). This material is also based upon work partially supported by the OSU Mathematical Biosciences Institute (National Science Foundation grant 0635561 to S.K.H.).
The content of this article is solely the responsibility of the authors, and it does not necessarily represent the official views of the funding agencies.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Ahmad N., Baroudy B. M., Baker R. C., Chappey C. 1995. Genetic analysis of human immunodeficiency virus type 1 envelope V3 region isolates from mothers and infants after perinatal transmission. J. Virol. 69:1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson J. A., et al. 2010. HIV-1 populations in semen arise through multiple mechanisms. PLoS Pathog. 6:e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arias R. A., Muñoz L. D., Muñoz-Fernández M. A. 2003. Transmission of HIV-1 infection between trophoblast placental cells and T-cells take place via an LFA-1-mediated cell to cell contact. Virology 307:266–277 [DOI] [PubMed] [Google Scholar]
- 4. Backé E., et al. 1992. Demonstration of HIV-1 infected cells in human placenta by in situ hybridisation and immunostaining. J. Clin. Pathol. 45:871–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bácsi A., et al. 2001. Pseudotypes of vesicular stomatitis virus-bearing envelope antigens of certain HIV-1 strains permissively infect human syncytiotrophoblasts cultured in vitro: implications for in vivo infection of syncytiotrophoblasts by cell-free HIV-1. J. Med. Virol. 64:387–397 [DOI] [PubMed] [Google Scholar]
- 6. Bell J. 2008. A simple way to treat PCR products prior to sequencing using ExoSAP-IT. Biotechniques 44:834. [DOI] [PubMed] [Google Scholar]
- 7. Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289–300 [Google Scholar]
- 8. Boily-Larouche G., et al. 2009. Functional genetic variants in DC-SIGNR are associated with mother-to-child transmission of HIV-1. PLoS One 4:e7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boyd M. T., Simpson G. R., Cann A. J., Johnson M. A., Weiss R. A. 1993. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J. Virol. 67:3649–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bryson Y. J., Luzuriaga K., Sullivan J. L., Wara D. W. 1992. Proposed definitions for in utero versus intrapartum transmission of HIV-1. N. Engl. J. Med. 327:1246–1247 [DOI] [PubMed] [Google Scholar]
- 11. Butler D. M., et al. 2010. The origins of sexually transmitted HIV among men who have sex with men. Sci. Transl. Med. 2:18re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crescimanno C., et al. 1999. Expression pattern alterations of syndecans and glypican-1 in normal and pathological trophoblast. J. Pathol. 189:600–608 [DOI] [PubMed] [Google Scholar]
- 13. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. David F. J., et al. 1992. Human trophoblast cells express CD4 and are permissive for productive infection with HIV-1. Clin. Exp. Immunol. 88:10–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. David F. J., et al. 1995. HIV infection of choriocarcinoma cell lines derived from human placenta: the role of membrane CD4 and Fc-Rs into HIV entry. Virology 208:784–788 [DOI] [PubMed] [Google Scholar]
- 16. Davison A. C., Hinkley D. V. (ed.). 1997. Bootstrap methods and their application. Cambridge series in statistical and probabilistic mathematics no. 1. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 17. Douglas G. C., Thirkill T. L. 2001. Chemokine receptor expression by human syncytiotrophoblast—a review. Placenta 22(Suppl. A):S24–S28 [DOI] [PubMed] [Google Scholar]
- 18. Douglas G. C., et al. 2001. Chemokine receptor expression by human syncytiotrophoblast. J. Reprod. Immunol. 49:97–114 [DOI] [PubMed] [Google Scholar]
- 19. Eddy S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755–763 [DOI] [PubMed] [Google Scholar]
- 20. Fantini J., Cook D. G., Nathanson N., Spitalnik S. L., Gonzalez-Scarano F. 1993. Infection of colonic epithelial cell lines by type 1 human immunodeficiency virus is associated with cell surface expression of galactosylceramide, a potential alternative gp120 receptor. Proc. Natl. Acad. Sci. U. S. A. 90:2700–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farquhar C., et al. 2010. Illness during pregnancy and bacterial vaginosis are associated with in-utero HIV-1 transmission. AIDS 24:153–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fear W. R., Kesson A. M., Naif H., Lynch G. W., Cunningham A. L. 1998. Differential tropism and chemokine receptor expression of human immunodeficiency virus type 1 in neonatal monocytes, monocyte-derived macrophages, and placental macrophages. J. Virol. 72:1334–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Friesner R. A. 2009. Development of scoring functions for computing protein-ligand binding affinities, abstr. CINF64. Am. Chem. Soc. Div. Chem. Inform. Natl. Meet. [Google Scholar]
- 24. García K., Garcia V., Perez Laspiur J., Duan F., Melendez L. M. 2009. Characterization of the placental macrophage secretome: implications for antiviral activity. Placenta 30:149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gnanakaran S., et al. 2007. Clade-specific differences between human immunodeficiency virus type 1 clades B and C: diversity and correlations in C3-V4 regions of gp120. J. Virol. 81:4886–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Groenink M., et al. 1993. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science 260:1513–1516 [DOI] [PubMed] [Google Scholar]
- 27. Guay L. A., et al. 1999. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet 354:795–802 [DOI] [PubMed] [Google Scholar]
- 28. Harouse J. M., et al. 1991. Inhibition of entry of HIV-1 in neural cell lines by antibodies against galactosyl ceramide. Science 253:320–323 [DOI] [PubMed] [Google Scholar]
- 29. Heath L., et al. 2010. Restriction of HIV-1 genotypes in breast milk does not account for the population transmission genetic bottleneck that occurs following transmission. PLoS One 5:e10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Henderson G. J., et al. 2004. HIV-1 populations in blood and breast milk are similar. Virology 330:295–303 [DOI] [PubMed] [Google Scholar]
- 31. Hudson R. R. 2000. A new statistic for detecting genetic differentiation. Genetics 155:2011–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jensen M. A., Coetzer M., van't Wout A. B., Morris L., Mullins J. I. 2006. A reliable phenotype predictor for human immunodeficiency virus type 1 subtype C based on envelope V3 sequences. J. Virol. 80:4698–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jokimaa V., et al. 1998. Expression of syndecan-1 in human placenta and decidua. Placenta 19:157–163 [DOI] [PubMed] [Google Scholar]
- 34. Jordan J. A., DeLoia J. A. 1999. Globoside expression within the human placenta. Placenta 20:103–108 [DOI] [PubMed] [Google Scholar]
- 35. Keele B. F., et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kennedy A. P., Walsh D. A., Nicholson R., Adams J. G., III, Steinberg M. H. 1986. Influence of HbS levels upon the hematological and clinical characteristics of sickle cell trait. Am. J. Hematol. 22:51–54 [DOI] [PubMed] [Google Scholar]
- 37. Koito A., Harrowe G., Levy J. A., Cheng-Mayer C. 1994. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J. Virol. 68:2253–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kosakovsky Pond S. L., Posada D., Gravenor M. B., Woelk C. H., Frost S. D. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098 [DOI] [PubMed] [Google Scholar]
- 39. Kourtis A. P., Lee F. K., Abrams E. J., Jamieson D. J., Bulterys M. 2006. Mother-to-child transmission of HIV-1: timing and implications for prevention. Lancet Infect. Dis. 6:726–732 [DOI] [PubMed] [Google Scholar]
- 40. Kwiek J. J., et al. 2006. Maternal-fetal microtransfusions and HIV-1 mother-to-child transmission in Malawi. PLoS Med. 3:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kwiek J. J., et al. 2008. Maternal-fetal DNA admixture is associated with intrapartum mother-to-child transmission of HIV-1 in Blantyre, Malawi. J. Infect. Dis. 197:1378–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kwiek J. J., et al. 2008. The molecular epidemiology of HIV-1 envelope diversity during HIV-1 subtype C vertical transmission in Malawian mother-infant pairs. AIDS 22:863–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kyte J., Doolittle R. F. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132 [DOI] [PubMed] [Google Scholar]
- 44. Lairmore M. D., et al. 1993. Cellular localization of CD4 in the human placenta. Implications for maternal-to-fetal transmission of HIV. J. Immunol. 151:1673–1681 [PubMed] [Google Scholar]
- 45. Larkin M., et al. 1989. Oligosaccharide-mediated interactions of the envelope glycoprotein gp120 of HIV-1 that are independent of CD4 recognition. AIDS 3:793–798 [DOI] [PubMed] [Google Scholar]
- 46. Lee T. H., Chafets D. M., Biggar R. J., McCune J. M., Busch M. P. 2010. The role of transplacental microtransfusions of maternal lymphocytes in in utero HIV transmission. J. Acquir. Immune Defic. Syndr. 55:143–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Luo W., Yang H., Rathbun K., Pau C.-P., Ou C.-Y. 2005. Detection of HIV-1 DNA in dried blood spots using a duplex real-time PCR assay. J. Clin. Microbiol. 43:1851–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marlin R., et al. 2009. Antigen-presenting cells represent targets for R5 HIV-1 infection in the first trimester pregnancy uterine mucosa. PLoS One 4:e5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marras D., et al. 2002. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat. Med. 8:522–526 [DOI] [PubMed] [Google Scholar]
- 50. Mognetti B., et al. 2000. HIV-1 co-receptor expression on trophoblastic cells from early placentas and permissivity to infection by several HIV-1 primary isolates. Clin. Exp. Immunol. 119:486–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Moore P. L., et al. 2008. The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J. Virol. 82:1860–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mwapasa V., et al. 2006. Maternal syphilis infection is associated with increased risk of mother-to-child transmission of HIV in Malawi. AIDS 20:1869–1877 [DOI] [PubMed] [Google Scholar]
- 53. Mwapasa V., et al. 2004. The effect of Plasmodium falciparum malaria on peripheral and placental HIV-1 RNA concentrations in pregnant Malawian women. AIDS 18:1051–1059 [DOI] [PubMed] [Google Scholar]
- 54. Nowak P., et al. 2002. The selection and evolution of viral quasispecies in HIV-1 infected children. HIV Med. 3:1–11 [DOI] [PubMed] [Google Scholar]
- 55. O'Brien W. A., et al. 1990. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature 348:69–73 [DOI] [PubMed] [Google Scholar]
- 56. Othoro C., et al. 2006. Evaluation of various methods of maternal placental blood collection for immunology studies. Clin. Vaccine Immunol. 13:568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Palmer S., et al. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pond S. L., Frost S. D., Muse S. V. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679 [DOI] [PubMed] [Google Scholar]
- 59. Qiu X., et al. 2001. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Renjifo B., et al. 2004. Preferential in-utero transmission of HIV-1 subtype C as compared to HIV-1 subtype A or D. AIDS 18:1629–1636 [DOI] [PubMed] [Google Scholar]
- 61. Renjifo B., et al. 2003. In-utero transmission of quasispecies among human immunodeficiency virus type 1 genotypes. Virology 307:278–282 [DOI] [PubMed] [Google Scholar]
- 62. Ritola K., et al. 2004. Multiple V1/V2 env variants are frequently present during primary infection with human immunodeficiency virus type 1. J. Virol. 78:11208–11218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rong R., et al. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J. Virol. 81:1350–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63a. Russell E. S., et al. The genetic bottleneck in vertical transmission of subtype C HIV-1 is not driven by selection of especially neutralization-resistant virus from the maternal viral population. J. Virol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sagar M., Wu X., Lee S., Overbaugh J. 2006. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J. Virol. 80:9586–9598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Salazar-Gonzalez J. F., et al. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Salemi M., et al. 2009. Distinct patterns of HIV-1 evolution within metastatic tissues in patients with non-Hodgkins lymphoma. PLoS One 4:e8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Samleerat T., et al. 2008. Characteristics of HIV type 1 (HIV-1) glycoprotein 120 env sequences in mother-infant pairs infected with HIV-1 subtype CRF01_AE. J. Infect. Dis. 198:868–876 [DOI] [PubMed] [Google Scholar]
- 68. Schneider T. D., Stephens R. M. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schnell G., Price R. W., Swanstrom R., Spudich S. 2010. Compartmentalization and clonal amplification of HIV-1 variants in the cerebrospinal fluid during primary infection. J. Virol. 84:2395–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shankarappa R., et al. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489–10502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sheikh A. U., Polliotti B. M., Miller R. K. 2000. In situ PCR detection of HIV expression in the human placenta. Methods Mol. Biol. 137:75–86 [DOI] [PubMed] [Google Scholar]
- 72. Shimizu N., Gojobori T. 2000. How can human and simian immunodeficiency viruses utilize chemokine receptors as their coreceptors? Gene 259:199–205 [DOI] [PubMed] [Google Scholar]
- 73. Simmonds P., et al. 1990. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J. Virol. 64:864–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Slatkin M., Maddison W. P. 1989. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics 123:603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stamatakis A., Hoover P., Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57:758–771 [DOI] [PubMed] [Google Scholar]
- 76. Sullivan N., Thali M., Furman C., Ho D. D., Sodroski J. 1993. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J. Virol. 67:3674–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tanabe K., et al. 2002. In vitro recombination during PCR of Plasmodium falciparum DNA: a potential pitfall in molecular population genetic analysis. Mol. Biochem. Parasitol. 122:211–216 [DOI] [PubMed] [Google Scholar]
- 78. Torres G., et al. 2001. Expression of the HIV-1 co-receptors CCR5 and CXCR4 on placental macrophages and the effect of IL-10 on their expression. Placenta 22(Suppl. A):S29–33 [DOI] [PubMed] [Google Scholar]
- 79. Verhofstede C., et al. 2003. Diversity of the human immunodeficiency virus type 1 (HIV-1) env sequence after vertical transmission in mother-child pairs infected with HIV-1 subtype A. J. Virol. 77:3050–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Vidricaire G., Gauthier S., Tremblay M. J. 2007. HIV-1 infection of trophoblasts is independent of gp120/CD4 interactions but relies on heparan sulfate proteoglycans. J. Infect. Dis. 195:1461–1471 [DOI] [PubMed] [Google Scholar]
- 81. Wei X., et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 82. Wolinsky S. M., et al. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134–1137 [DOI] [PubMed] [Google Scholar]
- 83. Zárate S., Pond S. L., Shapshak P., Frost S. D. 2007. Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. J. Virol. 81:6643–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang L. Q., et al. 1993. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J. Virol. 67:3345–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhu T., et al. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179–1181 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.