Abstract
Herpes simplex virus (HSV) pUL34 plays a critical role in virus replication by mediating egress of nucleocapsids from the infected cell nucleus. We have identified a mutation in pUL34 (Y68A) that produces a major defect in virus replication and impaired nuclear egress but also profoundly inhibits cell-to-cell spread and trafficking of gE. Virion release to the extracellular medium is not affected by the Y68A mutation, indicating that the mutation specifically inhibits cell-to-cell spread. We isolated extragenic suppressors of the Y68A plaque formation defect and mapped them by a combination of high-throughput Illumina sequencing and PCR-based screening. We found that suppression is highly correlated with a nonsense mutation in the US9 gene, which plays a critical role in cell-to-cell spread of HSV-1 in neurons. The US9 mutation alone is not sufficient to suppress the Y68A spread phenotype, indicating a likely role for multiple viral factors.
INTRODUCTION
Dissemination of herpes simplex virus (HSV) during recurrent disease in the host is dependent upon efficient viral replication and on the ability of the virus to spread from cell to cell in the face of the host innate and adaptive immune defenses. Cytoplasmic envelopment of HSV-1 virions is followed by vesicular transport of virions to the cell surface and secretion by fusion of the vesicle membrane with the plasma membrane (8, 23, 44). Transport of virions to cell membranes in contact with the extracellular medium results in release of free virions. Transport to surfaces apposed to other cells results in cell-to-cell spread of virus infection. The mechanism by which virions are sorted to junctional or basolateral surfaces in epithelial and fibroblast cells is poorly characterized. About half of the virus-encoded proteins play critical roles in virus replication, but relatively few have been found to have specific functions in cell-to-cell spread of virus. The essential components of the virion entry apparatus, gB, gD, and gH/gL, are required for cell-to-cell spread (7, 17, 30, 52). It is likely that this is because cell-to-cell spread requires interaction of the virus entry proteins with cellular receptors and subsequent fusion of the virion envelope with the plasma membrane of the naïve host cell. A few additional viral proteins have been shown to be required for efficient cell-to-cell spread at least in some cell types. HSV-1 gE and gI form a heterodimeric complex that is required for efficient cell-to-cell spread in the nervous system in vivo (10, 11, 21, 22, 36). The gE/gI complex is also required for spread in cultured neuronal cells and in epithelial and fibroblast cells that form well-defined cell junctions (1, 10, 12, 36). The gE spread phenotype in epithelial cells requires sequences in the cytoplasmic tail of gE and also requires sorting of gE to basolateral cell surfaces and adherens junctions, where it colocalizes with β-catenin (12, 16, 37, 64). Deletion of US9 in pseudorabies virus (PRV) is associated with failure of viral spread in neuronal cultures and in vivo (5, 33). In HSV-1, the effect of US9 deletion on neuronal spread is less clear, and the degree of inhibition of neuronal spread may depend on the experimental system (36, 60). The function of US9 appears to be tied in neurons to sorting of virus components from the neuronal cell body into axons (5, 33, 60, 63).
The HSV-1 UL34 gene, along with its homologs in other herpesviruses, is required for efficient viral replication in all cultured cells tested, presumably because it is required for efficient egress of capsids from the infected cell nucleus (15, 25, 41, 43, 51). The UL34 protein (pUL34) is targeted specifically to the inner nuclear membrane (INM) by a mechanism that requires its interaction with HSV pUL31 (48, 49), and this dependence is a conserved feature of herpesvirus envelopment (18, 26, 48, 49, 55, 56, 65). In addition to their localization at the nuclear envelope in infected cells, pUL31 and pUL34 of HSV and PRV have been shown to be structural components of the perinuclear virion (18, 25, 49). The proteins are lost from the egressing virion upon deenvelopment at the outer nuclear membrane (ONM), and pUL34 and pUL31 and their homologs are not detected in mature virions (15, 18, 19, 25, 49). Localization of these two proteins at the INM results in the recruitment of other proteins, including protein kinase C delta and pUS3, to the nuclear membrane to form a nuclear envelopment complex (NEC) (41, 45, 54). Deletion of the HSV UL34 gene causes failure to disrupt the nuclear lamina and essentially complete failure of nuclear egress, with accumulation of nucleocapsids in the infected cell nucleus (3, 27, 38, 39, 45, 47, 51, 57, 58). The concentration of pUL34 and pUL31 at the nuclear membrane during infection suggests that the nuclear envelope (NE) is likely to be their most important functional site.
Complete deletion of any gene whose product is required at multiple steps in infection will result in arrest of infection at the first of those steps, making identification and analysis of later events impossible. Point mutations in that gene will sometimes result in proteins with full or partial function at early steps and failure of function at later steps, thereby allowing characterization of those later steps. This strategy has been useful in analysis of UL34 gene function, since careful analysis of point mutations has allowed identification of UL34 gene functions in nuclear egress that follow nuclear lamina disruption, including mediation and regulation of membrane curvature around capsids (50). Analysis of point mutations has the additional advantage that extragenic suppressors of the mutant phenotypes can be selected and mapped, allowing identification of functionally important interactions. This genetic approach has had limited use in analysis of herpesvirus morphogenesis, principally because of the difficulty of mapping the extragenic suppressor mutations by marker transfer. Application of the method so far has yielded useful results only when the position of the suppressor could be predicted based on already known or suspected interactions (9, 20). Two recent technical advances allow for more extensive and powerful use of extragenic suppressor analysis. The first of these is the use of high-throughput sequencing methods for rapid and convenient sequencing of whole herpesvirus genomes for identification of single nucleotide polymorphisms (SNPs) between the parental and suppressor mutant viruses (61). The second is the use of molecular clones of herpesvirus genomes in the form of bacterial artificial chromosomes (BACs) as the parent genomes for marker transfer identification of the relevant SNP.
Here, we show that a mutation in UL34 that results in substitution of alanine for a highly conserved tyrosine at pUL34 position 68 results in a surprising phenotype. This mutation is associated with a major virus replication defect and inhibition of nuclear egress but also results in a profound defect in virus cell-to-cell spread and in trafficking of gE. We isolated extragenic suppressors of the cell-to-cell spread phenotype and showed by whole-genome sequencing and PCR-based screening that phenotypic suppression is correlated with a nonsense mutation in the US9 gene. This mutation alone is not sufficient to suppress the UL34 Y68A phenotype.
MATERIALS AND METHODS
Cells and viruses.
Vero and HEp-2 cells were maintained as previously described (51). The properties of HSV-1(F), vRR1072(TK+), and UL34(−) BACs have been previously described (14, 51).
Plasmids and cell lines.
pRR1072 and pRR1072Rep were previously described (2, 51). To construct an infection-inducible UL34Y68A-expressing cell line, we built the plasmid pRR1374. To achieve this, the 1,250-bp XbaI/Klenow-BspEI fragment of pRR1293 that contains the Y68A UL34 gene on the pRR1072Rep background was ligated into AseI/Klenow-NgoMIV-cut pTuner-IRES2 (Clontech). The resulting plasmid expresses bicistronic pUL34-IRES2-enhanced green fluorescent protein (EGFP) mRNA from the UL34 promoter/regulatory sequences. Clonal cell line Y68A-DD was constructed by transfection of pRR1374 into Vero cells followed by selection with G418 and isolation of clones by limiting dilution. Expressing cell clones were initially screened by assay for EGFP expression 20 h after infection with HSV-1(F). Cell clones that expressed EGFP were further screened for pUL34 expression by immunofluorescence assay of cells 20 h after infection with the UL34-null virus vRR1072(TK+). Although several Y68A pUL34-expressing cell clones were isolated, only one, Y68A-DD, expressed pUL34 after UL34-null virus infection at a level comparable to that seen in wild-type (WT) virus infection. The Y68A-DD cells are referred to in the remainder of this report simply as Y68A UL34-expressing cells. The wild-type pUL34-expressing cell line, called RepAC, was previously described (50), and these cells are referred to as WT UL34-expressing cells.
Plaque assays.
Six-well tissue culture wells were seeded with 1.8 × 106 Vero cells, WT UL34-expressing cells, or Y68A UL34-expressing cells the day before infection. Infection was initiated by removal of growth medium and the addition of 1 ml of virus diluted in V medium (Dulbecco's modified Eagle medium [DMEM] containing 1% heat-inactivated calf serum). The virus inoculum was removed after 90 min and replaced with 2.5 ml V medium containing a 1:250 dilution of pooled human immunoglobulin as a source of HSV-neutralizing antibody (GamaSTAN S/D; Talecris Biotherapeutics). At the indicated times after infection, monolayers were washed twice with phosphate-buffered saline (PBS) and then fixed by incubation for 15 min in 3.7% formaldehyde in PBS. After fixation, monolayers were washed three times with 2 ml PBS. Plaques were stained by indirect immunofluorescence using a 1:5,000 dilution of mouse monoclonal anti-gD DL6 (gift of Gary Cohen and Rosalyn Eisenberg) as the primary antibody and a 1:1,000 dilution of Alexa Fluor 488 goat anti-mouse IgG (Invitrogen) as the secondary antibody.
Single-step growth measurement.
Measurement of replication and release of HSV-1(F), vRR1072(TK+), and Y68ARev viruses on Vero cells, WT UL34-expressing cells, and Y68A UL34-expressing cells after infection at high multiplicity was performed as previously described (29).
Indirect immunofluorescence.
Immunofluorescence was performed as previously described using either a 1:2,000 dilution of mouse monoclonal anti-gE ascites fluid (gift of Lenore Pereira), a 1:500 dilution of mouse monoclonal anti-emerin clone H-12 (Santa Cruz), or a 1:5,000 dilution of mouse monoclonal anti-gD DL6 (2, 48).
Immunoblotting.
Nitrocellulose sheets bearing proteins of interest were blocked in 5% nonfat milk plus 0.2% Tween 20 for at least 2 h. The membranes were probed either with a previously described chicken polyclonal antibody directed against pUL34 (1:1,000) (48), followed by reaction with alkaline phosphatase-conjugated anti-chicken secondary antibody (Aves Laboratories), or with mouse monoclonal antibody directed against the HSV-1 scaffolding protein (1:2,000) (Serotec), followed by reaction with alkaline phosphatase-conjugated anti-mouse secondary antibody (Sigma).
Transmission electron microscopy (TEM) of infected cells.
Confluent monolayers of Vero or Y68A UL34-expressing cells were infected with vRR1072(TK+) at a multiplicity of infection (MOI) of 10 for 20 h and then fixed by incubation in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 2 h. Cells were postfixed in 1% osmium tetroxide, washed in cacodylate buffer, embedded in Spurr's resin, and cut into 95-nm sections. Sections were mounted on grids, stained with uranyl acetate and lead citrate, and examined with a JEOL 1250 transmission electron microscope.
Sequencing of the Y68ARev genome.
Y68ARev genomic DNA was isolated from C capsids from infected HEp-2 cell nuclei purified on sucrose gradients as previously described (51). Viral DNA was sequenced on an Illumina Genome Analyzer II (GAII), as described previously (61). Briefly, viral nucleocapsid DNA was prepared for sequencing using Illumina's genomic DNA sample prep kit, loaded onto one lane of a flow cell, and sequenced for 75 cycles using standard data acquisition by the Illumina Pipeline software version 1.3. Sequence reads were filtered to remove mononucleotides, and the human aligning sequence derived from the HEp-2 cells was used to prepare viral DNA. Then sequence reads were aligned to the HSV-1(F) viral genome (GenBank identifier GU734771) using the Mapping and Alignment with Qualities (MAQ) software package (28). Default MAQ settings were used to call SNPs in the Y68ARev sequence compared to the HSV-1(F) genome. Default MAQ filtering excludes SNP calls in any region where sequence reads align to more than one location. This situation occurs in the large inverted repeat regions of HSV-1, where sequence reads can align to either the terminal or internal copy of the repeat. For this reason, MAQ filtering was lifted for the repeat regions, and all potential SNP calls were considered in the repeat regions, which includes the RS1, RL1, and RL2 genes.
BAC construction.
An HSV-1 BAC genome, carrying a UL34 gene deletion and a mutation in the US9 gene creating an R58Stop substitution, was engineered using Red recombineering on the background of a UL34-null BAC as previously described (50, 62). The UL34-null BAC was mutagenized at the US9 locus by insertion and scarless excision of a gentamicin (Gm) resistance (Gmr) cassette.
The Gmr cassette with the mutant US9 flanking sequence was constructed in several steps. First, a Gm resistance cassette containing the Gmr promoter, protein coding sequence, and terminator flanked at the 5′ end with an SceI homing nuclease site was amplified as previously described (50). Second, PCR products containing the 5′ and 3′ halves of the Gm resistance gene were amplified from the Gmr cassette template using the primers US9R58StGm For/Gm mid Rev and Gm mid For/US9R58StGm Rev, respectively. The two resulting PCR products overlap in the Gmr coding sequence. The complete Gm resistance cassette with the US9 flanking sequence was then assembled in a PCR using the overlapping partial genes and the primers US9R58St unique For and US9R58St unique Rev (Table 1). The resulting PCR product was recombined into the UL34-null BAC, Gm-resistant recombinants were picked, and genomes were tested for insertion of the Gm cassette by diagnostic PCR by using the flanking primers US9 test Fwd and US9 test Rev (Table 1). Correct insertion of the Gm cassette was confirmed by direct sequencing of the BAC DNA. Scarless excision of the Gm cassette, leaving an intact US9 gene carrying the R568Stop mutation, was carried out as described previously (50), and Gm-sensitive, kanamycin (Kan)-resistant clones were tested for correct structure at both the US9 and UL34 loci by diagnostic PCR using the UL34 test For, UL34 test Rev, US9 test Fwd, and US9 test Rev primers. Correct structure was confirmed by direct sequencing of the BAC DNA at both loci.
Table 1.
Primers used for construction and analysis of mutant US9 BAC
| Primer name | Primer sequencea |
|---|---|
| US9R58StGm For | 5′-TCCTCGTACGCATGGGCCGCCAACAGTCGGTATTAAGGCGctagCGCAGACGCACCCGCTGCGTGCTCGGATCCTAGGGATAACAGGG-3′ |
| US9R58StGm Rev | 5′-GACAGGCGATCACCATGCCGACGCAGCGGGTGCGTCTGCGctagCGCCTTAATACCGACTGTTGTCTAGAGGCCGCGGCGTTG-3′ |
| Gm mid For | 5′-GGTCGTGAGTTCGGAGACGTAGC-3′ |
| Gm mid Rev | 5′-CACTACGCGGCTGCTCAAACC-3′ |
| R58St unique For | 5′-TCCTCGTACGCATGGGCCG-3′ |
| R58St unique Rev | 5′–GACAGGCGATCACCATGCCGAC-3′ |
| US9 test Fwd | 5′-AGTCACTGCGACCGCAACTTCC-3′ |
| US9 test Rev | 5′-AAGGCTGGGTGCAAATTGCGG-3′ |
Underlined sequences have homology to the US9 gene. Lowercase indicates nucleotides altered to create the R58Stop mutation and an NheI restriction enzyme cleavage site. Sequences not underlined have homology to the Sce-Gmr cassette.
Viruses were rescued from the WT BAC, UL34-null BAC, and UL34-null/US9R58Stop BAC by transfection into UL34-expressing complementing cells. The sequence of the rescued UL34-null/US9R58Stop virus at the US9 locus was verified by sequencing of a PCR product containing the US9 gene.
RESULTS
Y68A mutant pUL34 is deficient in its ability to support virus production and cell-to-cell spread.
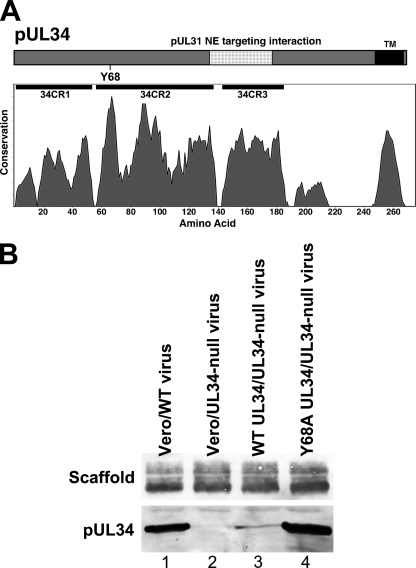
We have previously described a strategy for evaluation of mutant UL34 function based on UL34-null virus infection of cell lines that express mutant pUL34 (50). The Y68A mutant pUL34 was evaluated as part of a larger effort to examine the function of UL34 mutants in which individual conserved amino acids were replaced with alanine. Tyrosine 68 is in the most conserved region of pUL34 and is absolutely conserved among alphaherpesviruses (Fig. 1A). In order to evaluate the function of Y68, we constructed a cell line that expresses Y68A pUL34 under the control of its own promoter regulatory sequences. We compared pUL34 expression relative to a previously constructed WT pUL34-expressing cell line, and to wild-type virus infection, by infecting cells with 10 PFU/cell of either HSV-1(F) or the UL34-null virus vRR1072(TK+) for 18 h. Total protein was determined in each extract, and equivalent amounts were separated on an SDS-PAGE gel, transferred to nitrocellulose, and probed for pUL34 (Fig. 1B). HSV-1 scaffolding protein was used as a loading control. The UL34-null virus-infected Y68A UL34-expressing cells expressed as much pUL34 as WT virus-infected Vero cells (Fig. 1B, compare lanes 1 and 4) and considerably more than UL34-null virus-infected WT UL34-expressing cells (lane 3). We could, therefore, directly compare the activity of Y68A pUL34 to that of WT pUL34 both in WT virus-infected cells and in WT UL34-expressing cells. Y68A mutant phenotypes could not be ascribed to insufficient levels of the mutant protein.
Fig. 1.
Position of the Y68A mutation and expression of pUL34 by cell lines. (A) Schematic diagrams of pUL34 showing the locations of relevant sequence features. Protein sequence of pUL34 is indicated as a bar with the N terminus at the left. Sequences in pUL34 that mediate nuclear envelope targeting of the NEC are indicated as a stippled region. Position of Y68 is indicated below the bar. Positions of conserved regions are indicated immediately below each of the bars. The conservation plot shows conservation of biochemical properties of amino acids using all available herpesvirus sequences aligned by using the program MUSCLE (13). (B) Expression of wild-type and Y68A mutant pUL34 by stable cell lines. Digital images of Western blots are shown. Vero cells (lanes 1 and 2) or cells stably expressing WT pUL34 (lane 3) or Y68A pUL34 (lane 4) were infected with WT HSV-1(F) (lane 1) or UL34-null vRR1072(TK+) virus (lanes 2 to 4). Blotted infected cell proteins were probed for either scaffolding protein (top) or pUL34 (bottom).
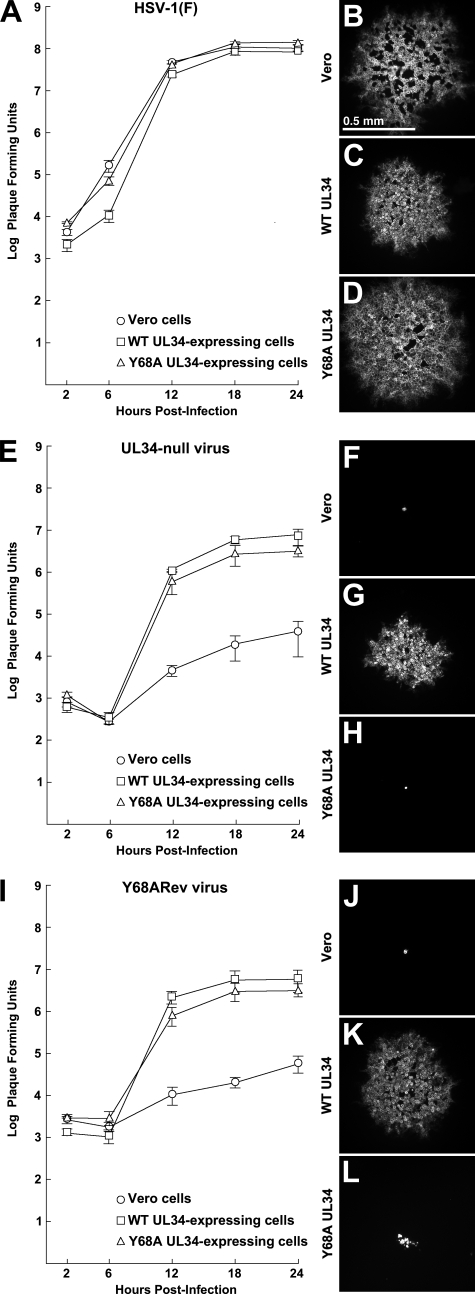
Virus replication and spread were measured in single-step growth and plaque formation assays (Fig. 2 and 3) with several interesting results. Figure 2 shows single-step growth kinetics and the sizes of representative plaques. Figure 3 shows the results of measurement of plaque areas (note that the y axis of Fig. 3 has a logarithmic scale). As expected, wild-type virus grew efficiently and formed robust plaques on all cell lines tested (Fig. 2A to D and 3A). This indicates that the Y68A UL34-expressing cells show no UL34-independent defect in virus growth and no dominant negative effect of the UL34 mutant on virus growth. UL34-null virus replication on Y68A pUL34-expressing cells was less efficient than WT virus replication on any of the cell types, producing about 60-fold-less virus at peak times (compare curves in Fig. 2A with the triangle marker curve in Fig. 2E). Since roughly equivalent amounts of pUL34 were expressed in both cases, this suggests that Y68A pUL34 is deficient in its ability to support single-step growth of HSV-1. This inhibition of single-step growth was, however, much less than that seen in the absence of pUL34 (compare triangle and circle marker curves in Fig. 2E).
Fig. 2.
Single-step growth and plaque formation on Vero, WT pUL34-expressing, or Y68A mutant pUL34-expressing cell lines. For single-step growth replicate, cultures of Vero cells, WT UL34-express-ing cells, or Y68A UL34-expressing cells were infected at an MOI of 5 with HSV-1(F) (A), the UL34-null virus vRR1072(TK+) (E), or Y68ARev (I). Residual virus was removed or inactivated with a low-pH wash, and at the indicated times total culture virus was titrated on WT UL34-expressing cells. Virus yields are expressed as PFU per milliliter. Each data point represents the mean of results from three independent experiments. Error bars indicate the range of values. For plaque formation assays, digital micrographs of infected cell monolayers stained for glycoprotein D are shown. The cell line infected is indicated to the left of each panel. In panels B to D, the infecting virus was HSV-1(F). In panels F to H, the infecting virus was the UL34-null virus vRR1072(TK+). In panels J to L, the infecting virus was Y68ARev. All plaques were fixed and stained at 2 days after infection. All plaque images are shown at the same magnification.
Fig. 3.
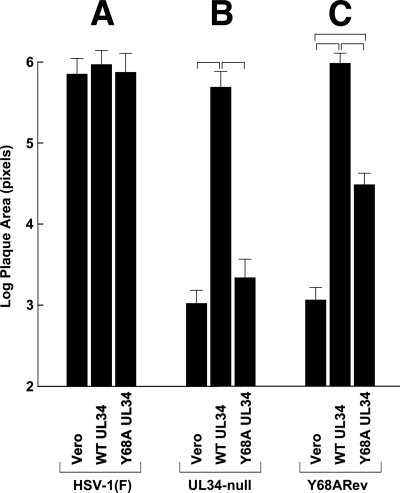
Quantitation of plaque formation on wild-type and mutant UL34-expressing cell lines. Histograms of mean plaque sizes on Vero, WT pUL34-expressing, and Y68A pUL34-expressing cells are shown. Plaques stained 2 days after infection, as described in the legend to Fig. 2, were photographed, and plaque areas in image pixels were determined using ImageJ. For each bar, 20 randomly selected plaques from two independent experiments (40 plaques total) were measured. Brackets indicate pairwise statistical comparisons performed using a Student t test. All of the indicated comparisons showed a highly significant difference (P < 0.001). Note that the y axis has a logarithmic scale.
Interestingly, the single-step growth of UL34-null virus on Y68A UL34-expressing cells (triangle marker curve in Fig. 2E) was very similar to that obtained on WT UL34-expressing cells (square marker curve in Fig. 2E), probably because of the low level of WT pUL34 expression (Fig. 1B). The latter observation was in striking contrast to the result of plaque formation assays. While UL34-null virus formed robust plaques on WT pUL34-expressing cells, it formed only tiny foci of one to a few infected cells on Y68A pUL34-expressing cells (compare Fig. 2G and H and 3B). Y68A pUL34-expressing cells supported plaque formation no better than cells that express no pUL34 at all (compare Fig. 2F and H and 3B). Since infected cultures for plaque formation assays were incubated in the presence of HSV-neutralizing antibody to make plaque formation exclusively dependent on cell-to-cell spread, this result suggests that the Y68A mutation in UL34 results in a severe defect in cell-to-cell spread. Spread of WT virus in Y68A UL34-expressing cells was just as efficient as that in Vero cells (Fig. 2D and 3A), indicating that Y68A UL34-expressing cells have no UL34-independent defect in virus spread and showing that the Y68A UL34 has no dominant negative effect on virus spread.
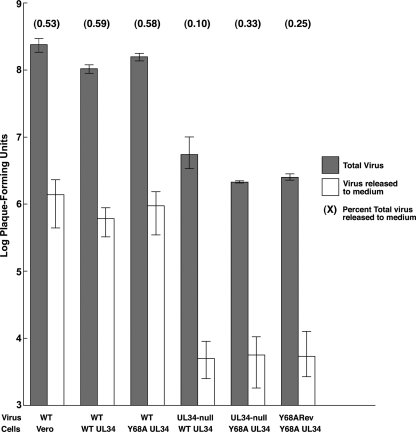
Failure in plaque formation could be due to specific inhibition of virion trafficking to cell junctions, or it could reflect inhibition of virion trafficking to any cell surface. In order to determine whether Y68A pUL34 is associated with a general virion release defect, amounts of total infectious virus and infectious particles released to the medium at 18 h after infection were measured in cells that express WT or Y68A pUL34 following infection with either UL34-null virus or Y68ARev (Fig. 4). Again, virus replication was slightly depressed in Y68A-expressing cells compared to that in cells that express WT pUL34 (Fig. 4, gray bars). As is typical for Vero cells, only a small fraction of total virus was released to the medium (white bars). The amounts of virus released from UL34-null virus-infected cells were the same regardless of whether WT or Y68A pUL34 was expressed, indicating that the Y68A mutation does not interfere with release of mature virus from the cell. Furthermore, the efficiency of release (i.e., the fraction of total virus produced that is released to the medium) from UL34-null virus-infected Y68A UL34-expressing cells (0.33%) is similar to that seen in wild-type virus infection of either Vero (0.53%) or Y68A UL34-expressing (0.58%) cells. This indicates that there is no UL34-independent inhibition of virus release occurring in the UL34-expressing cell lines. Given the severe inhibition of plaque formation, these data suggest that the Y68A mutation is associated with a major defect in cell-to-cell spread.
Fig. 4.
Virus release to the medium mediated by wild-type and mutant pUL34. Replicate cultures of Vero, wild-type pUL34-expressing, or Y68A mutant pUL34-expressing cells were infected at an MOI of 5 with wild-type virus HSV-1(F), UL34-null virus vRR1072(TK+), or Y68ARev. Residual virus was removed or inactivated with a low-pH wash, and at 18 h after infection total culture virus (gray bars) or culture supernatant (white bars) was titrated on WT UL34-expressing cells. Virus yields are expressed as PFU/ml. Each data point represents the mean of results from three independent experiments. Error bars indicate the range of values. The values in parentheses above each pair of bars indicate the percentages of total virus released to the medium calculated as the mean amount of released virus divided by the mean amount of total virus times 100. Low pH wash completely removed residual extracellular virus, as the titer of culture supernatant at 2 h postinfection was undetectable.
Y68A mutant pUL34 induces exaggerated disruption of the nuclear envelope and a nuclear egress defect.
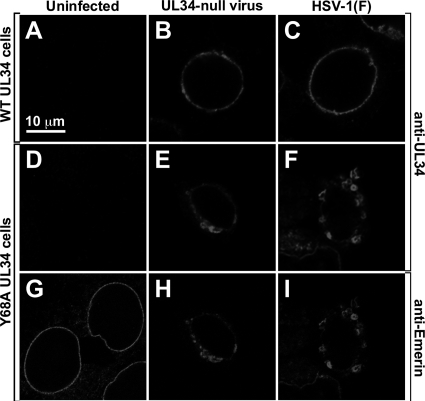
The Y68A mutation is outside the region of pUL34 that is necessary and sufficient for nuclear envelope targeting (29). To test whether the Y68A mutation interferes with proper localization of pUL34, WT UL34-expressing cells and Y68A UL34-expressing cells that were mock infected or infected with 10 PFU/cell of HSV-1(F) or UL34-null viruses for 16 h were fixed and assayed for localization of pUL34 or the host cell nuclear envelope protein emerin by immunofluorescence (Fig. 5). As we have seen with other cell lines that express pUL34 from its own promoter/regulatory sequences, pUL34 expression in both wild-type UL34- and Y68A UL34-expressing cells is strictly dependent upon infection and therefore cannot be detected in uninfected cells (Fig. 5A and D). As previously reported, wild-type pUL34 was tightly localized to the nuclear rim in cells infected with either UL34-null virus or HSV-1(F) (Fig. 5B and C). Y68A pUL34 was also tightly localized to the nuclear rim, but the appearance of the rim was changed such that there were numerous, sometimes large blebs on the outer nuclear envelope, indicating an exaggerated disruption of the architecture of the NE (Fig. 5E). Interestingly, the same effect was observed when Y68A pUL34-expressing cells were infected with wild-type virus (Fig. 5F), indicating that the mutant phenotype dominates even when WT pUL34 is present. The blebs formed in Y68A cells include the host NE protein emerin (Fig. 5H and I), and their induction depends upon infection since the appearance of the nuclear envelope is normal in uninfected cells (Fig. 5G). The exaggerated NE disruption due to the Y68A mutation is not likely to contribute significantly to the virus production or cell-to-cell spread defects, since WT HSV-1(F) replicated normally on Y68A UL34-expressing cells (Fig. 2A) and formed plaques normally (Fig. 2D and 3), even though the same degree of NE disruption was induced.
Fig. 5.
Localization of WT pUL34, Y68A mutant pUL34, and emerin in infected cells. Digital confocal images of WT UL34-expressing cells (A to C) or Y68A UL34-expressing cells (D to I) are shown. Cells were mock infected (A, D, and G) or infected for 16 h with UL34-null virus (B, E, and H) or with HSV-1(F) (C, F, and I) and then fixed and immunofluorescently stained for pUL34 (A to F) or emerin (G to I). All images are shown at the same magnification.
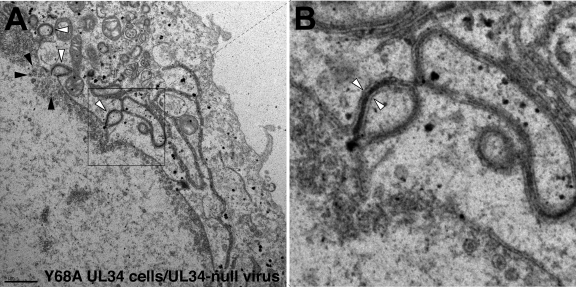
Examination of Y68A pUL34-expressing cells that were infected with UL34-null virus by TEM revealed two interesting phenotypes. First, as suggested by the immunofluorescence localization experiments above, there was blebbing of the nuclear membrane into the cytoplasm (Fig. 6A, white arrowheads). These blebs were formed by distension of both inner and outer nuclear membranes into the cytoplasm. In some cases, we also observed multilamellar structures containing multiple thicknesses of nuclear envelope separated by an electron-dense layer (Fig. 6B, white arrowheads). In all these cytoplasmic structures, the inner and outer nuclear envelopes appeared to be properly spaced, suggesting distortion of the nuclear envelope as a whole rather than just the inner or outer nuclear membranes. This phenotype is in distinct contrast to the behavior of UL34-null mutants in cells that express no pUL34, where the nuclear envelope is generally of uniform shape (27, 51). Second, in most Y68A UL34-expressing cells infected with UL34-null virus, we observed no cell surface virions or cytoplasmic egress intermediates despite the presence of numerous A, B, and C capsids in the infected cell nucleus (C capsids are indicated in Fig. 6A, black arrowheads). In this regard, Y68A-expressing cells were similar in appearance to UL34-null mutant-infected cells that express no UL34 at all, suggesting that the Y68A mutation causes a major defect in nuclear egress. The lack of surface virions and cytoplasmic egress intermediates was unsurprising given the magnitude of the single-step growth defect associated with the Y68A mutation (Fig. 2). Even in EM examination of wild-type virus-infected Vero cells, we would ordinarily observe less than 50 such structures per cell. Our failure to observe extranuclear virus here was consistent with the 60-fold single-step growth defect conferred by the Y68A mutation and suggests that the single-step growth defect is due largely or entirely to a defect in nuclear egress.
Fig. 6.
TEM analysis of cells that express Y68A UL34. Digital micrographs show Y68A UL34-expressing cells infected with the UL34-null virus vRR1072(TK+) for 20 h. White arrowheads in panel A point to examples of blebbing of the nuclear membrane into the cytoplasm. Black arrowheads point to examples of C capsids in the nucleus. The boxed area in panel A is enlarged (×4) in panel B. The white arrowheads in panel B point to an instance of a bleb with two thicknesses of nuclear envelope separated by an electron-dense layer.
pUL34 function is required for proper localization of glycoprotein E.
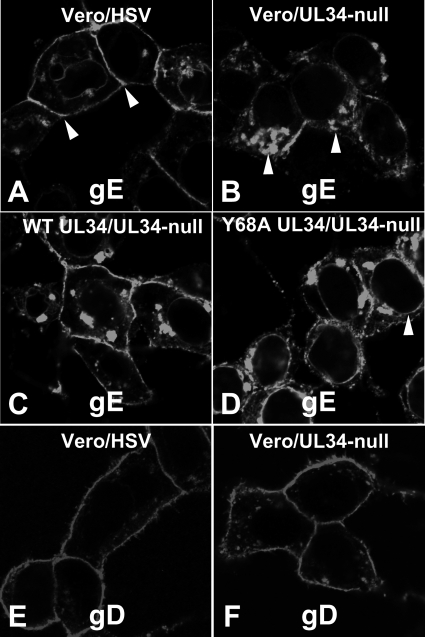
Cell-to-cell spread of HSV-1 is correlated with trafficking of glycoprotein E, and probably virions, to cell junctions. While gE is not required for cell-to-cell spread in Vero cells, we reasoned that alterations in its localization might reflect defects in proper trafficking of viral or cellular components required for cell-to-cell spread. To determine whether pUL34 function is necessary for proper gE sorting, we infected Vero and WT or mutant pUL34-expressing cells and assayed for gE and gD localization at 16 h after infection by immunofluorescence (Fig. 7). In Vero cells infected with wild-type virus (Fig. 7A), gE was localized on intracellular membranes, including the nuclear membrane in some cells. It was most prominent, however, on the plasma membrane, especially at junctional surfaces (Fig. 7A, arrowheads). In Vero cells infected with UL34-null virus, plasma membrane staining was much less prominent. Instead, gE accumulated in large membrane aggregates in the cytoplasm (Fig. 7B, arrowheads), and in most cells, junction staining was not evident. Plasma membrane and junction staining was restored when Vero cells that express WT pUL34 were infected with UL34-null virus (Fig. 7C), indicating that abnormal gE trafficking was due to lack of pUL34 expression. Infection of cells that express Y68A pUL34 with UL34-null virus resulted in an intermediate phenotype (Fig. 7D). gE accumulated more on cytoplasmic membranes than in a wild-type infection, but there was more cell surface and junctional staining than what was seen in infections with no pUL34. In addition, Y68A pUL34 expression resulted in more pronounced nuclear rim staining (Fig. 7D, arrowhead) than that seen in infections with wild-type or with no pUL34. These effects on gE trafficking did not reflect general disruption of cell surface trafficking of viral glycoproteins, since gD localized normally to the plasma membrane regardless of pUL34 expression (Fig. 7E and F). gB and gH also did not differ in localization in WT and UL34-null virus-infected cells (not shown).
Fig. 7.
Wild-type pUL34 expression is required for normal localization of gE but not gD. Vero cells (A, B, E, and F), WT UL34-expressing cells (C), and Y68A UL34-expressing cells (D) were infected with HSV-1(F) (A and E) or UL34-null virus (B to D and F) for 16 h and then fixed and immunofluorescently stained for gE (A to D) or gD (E and F). Arrowheads in panel A point to junctional surfaces, where gE concentrates in wild-type infection. Arrowheads in panel B point to cytoplasmic membranes, where gE concentrates during UL34-null infection. Arrowhead in panel D points to the nuclear envelope.
Isolation and characterization of an extragenic suppressor of the Y68A mutation.
Plaque formation defects allow selection of extragenic suppressors of the growth defect. In order to isolate suppressors, 10 replicate cultures of Y68A UL34-expressing cells were infected with 1 × 107 PFU of the UL34-null recombinant vRR1072(TK+) for 24 h. At the end of the infection, virus stocks were prepared and serial dilutions were plated onto Y68A UL34-expressing cells. Suppressors were rare; after 3 days, only one of the selection stocks gave rise to minute plaques on the mutant pUL34-expressing cells. The suppressor virus (designated Y68ARev) was plaque purified twice on Y68A UL34-expressing cells and then amplified to a high titer stock on WT pUL34-expressing cells. The behavior of the Y68ARev virus in single-step growth and plaque formation are shown in Fig. 2I and J and in Fig. 3C. In single-step growth, Y68ARev replicated only slightly less well in Y68A UL34-expressing cells than in the WT UL34-expressing cells and much less well in cells that express no pUL34 at all (Fig. 2I). In this respect, Y68ARev behaves exactly the same as its UL34-null virus parent, indicating that the suppressor mutation(s) do not improve virus replication.
Y68ARev could form plaques on Y68A UL34-expressing cells, but those plaques were minute (Fig. 2L and 3C). The plaques formed by the Y68ARev virus in Y68A UL34-expressing cells were about 15-fold larger than those formed by UL34-null virus in the same cells but about 25 times smaller than those formed by WT virus. These results indicate that the Y68A plaque formation phenotype is suppressed, albeit inefficiently, by mutations in the Y68ARev virus. The Y68ARev virus formed robust plaques in WT pUL34-expressing cells (Fig. 2K and 3), indicating that the genetic changes leading to suppression do not interfere with the function of WT pUL34. We further compared virus production and extracellular release by Y68ARev and found levels comparable to those of its UL34-null parent strain (Fig. 4). Taken together, these data suggest that genetic changes in the Y68ARev strain might provide insight specifically into the cell-to-cell spread (plaque formation) defect of the Y68A UL34 mutation.
Extragenic suppression of the Y68A mutation correlates with a mutation in US9.
Suppression of the plaque formation defect of Y68A UL34 was not strong enough to allow mapping of the relevant mutation(s) by marker transfer. Accordingly, we sequenced the whole genome of the Y68ARev virus and compared it to the recently published HSV-1(F) genomic sequence (61). Surprisingly, we found 139 SNPs compared to the sequence of HSV-1(F), 34 of which produce amino acid substitutions in known viral proteins (Table 2). We further reasoned that many of these SNPs would have accumulated during the multistep construction of the UL34-null virus, vRR1072(TK+), which was the immediate parent of Y68ARev (51). Since these were far too many SNPs to test individually by construction of recombinant viruses, we chose to focus on 11 gene products with known roles in virus assembly, egress, or cell-to-cell spread (indicated by boldface in Table 2). We PCR amplified the relevant regions from the genome of vRR1072(TK+) to determine which of the SNPs were found only in the suppressor virus genome and found that eight of the SNPs were suppressor specific (Table 2). Finally, we repeated the suppressor selection procedure in five separate experiments to isolate an additional five Y68A suppressor viruses. We then PCR amplified and sequenced the regions containing suppressor-specific SNPs to see which, if any, were present in most or all suppressor genomes (Table 2). Only one SNP, found in the US9 protein coding sequence, was present in more than one of the suppressor genomes. This SNP, which changes the arginine codon at position 58 to a stop codon, was found in five of the six independently selected suppressors, suggesting that this substitution might be responsible for phenotypic suppression.
Table 2.
Amino acid sequence-altering SNPs in Y68ARev compared to the HSV-1(F) sequence
| Genea | SNPb | Codon changec | Amino acid changed | Suppressor specifice | Frequency in 5 additional Y68A suppressors |
|---|---|---|---|---|---|
| UL8 | C to T (+) | GCG to GTG | A51V | ||
| UL15 | G to A (+) | AGC to AAC | S497N | Y | 0/5 |
| UL15 | G to T (−) | GCC to GTC | A704V | N | |
| UL18 | T to G (−) | ATC to ACC | I160T | Y | 0/5 |
| UL21 | C to T (+) | GCC to GTC | A41V | ||
| UL23 | C to A (−) | GCC to GTC | A272V | ||
| UL26 | G to A (+) | GAA to AAA | E249K | Y | 0/5 |
| UL26/26.5 | G to A (+) | GAT to AAT | D626/319N | Y | 0/5 |
| UL28 | G to A (+) | GTG to ATG | T112 M | N | |
| UL36 | A to G (−) | CTG to CCG | L3099P | N | |
| UL36 | C to T (−) | GCA to ACA | A3082T | N | |
| UL36 | G to A (−) | CGC to TGC | R2646C | Y | 1/5 |
| UL36 | G to A (−) | ACG to ATG | T2584 M | N | |
| UL36 | T to C (−) | ATG to GTG | M2439V | N | |
| UL36 | G to A (−) | TCG to TTG | S2368L | N | |
| UL37 | G to A (−) | CGT to TGT | R651C | N | |
| UL37 | G to A (−) | CGG to TGG | R531W | N | 0/5 |
| UL38 | T to C (+) | TGC to CGC | C35R | ||
| UL39 | C to T (+) | GCT to GTT | A474V | ||
| UL42 | G to A (+) | AGC to AAC | S349N | ||
| UL42 | G to A (+) | AGC to AAC | S358N | ||
| UL48 | T to C (−) | ACG to GCG | T212A | Y | 0/5 |
| UL50 | C to A (+) | CCC to ACC | P134T | ||
| UL51 | G to A (−) | ACG to ATG | T74 M | Y | 0/5 |
| UL52 | C to T (+) | GCT to GTT | A655V | ||
| US8 | C to T (+) | GCG to GTT | A441V | N | |
| US9 | C to T (+) | CGA to TGA | R58Stop | Y | 4/5 |
| US9 | C to T (+) | CTC to TTC | L89F | N | |
| RS1 | A to C (−) | CAG to CCG | Q163P | ||
| RS1 | A to C (−) | GAG to GCG | E266A | ||
| RS1 | G to A (−) | CGC to CAC | R267H | ||
| RS1 | G to A (−) | ACC to GCG | A272T | ||
| RS1 | G to C (−) | GCG to CCG | A273P | ||
| RS1 | A to G (−) | GAC to GGC | D712G |
Genes indicated in boldface have reported functions related to virus assembly, egress, or cell-to-cell spread and were tested to determine whether they were suppressor specific.
All SNPs are listed as nucleotide changes on the top strand of the prototype arrangement of the HSV-1(F) genome. In some cases, this will be the sense strand (+), in other cases, this will be the antisense strand (−), and in the case of RS1, both.
All codons are rendered on the sense strand of the gene.
Amino acids are numbered from the first amino acid encoded by the open reading frame. Amino acid differences indicated in boldface also occur in the published sequence of HSV-1 (17) (GenBank accession NC_001806).
Y, yes; N, no.
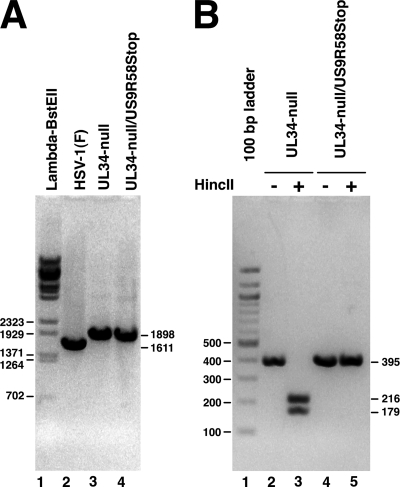
To test this hypothesis, we introduced a stop codon at position 58 in the US9 sequence into our previously characterized UL34-null BAC (50) and rescued mutant virus on WT pUL34-expressing complementing cells. The replacement of the UL34 gene with the kanamycin resistance gene cassette in the UL34-null and UL34-null/US9R58Stop viruses was confirmed by PCR amplification of the UL34 locus, and PCR products of the expected sizes were produced (Fig. 8). To confirm the presence of the expected mutation at the US9 locus and to assess the purity of the rescued virus, the US9 gene was PCR amplified from the rescued virus genome and digested with HincII restriction enzyme. Introduction of the US9R58Stop mutation resulted in elimination of an HincII restriction enzyme site. PCR amplification of the US9 locus from UL34-null and UL34-null/US9R58Stop mutant viruses resulted in a product of 395 bp, as expected (Fig. 8, lanes 2 and 4). The wild-type US9 product from the UL34-null virus was cut by HincII into two fragments of 216 and 179 bp, as expected, but no digestion products were detected from the UL34-null/US9R58Stop (Fig. 8, compare lanes 3 and 5). The UL34-null, UL34-null/US9R58Stop, and Y68ARev viruses were tested for suppression in a plaque assay on Y68A pUL34-expressing cells in which plaques were allowed to develop for 1 week (Fig. 9). UL34-null virus formed only minute plaques after 1 week, whereas the Y68ARev virus forms quite large plaques with this extended growth time (compare Fig. 9A and B). However, addition of the US9R58Stop mutation to the UL34-null virus does not improve its ability to form plaques on Y68A UL34-expressing cells (Fig. 9C). This suggests that the phenotypic improvement in plaque size in the Y68ARev strain is polygenic and that the UL34-null/US9R68Stop virus will be an important tool in mapping additional contributors to this phenotype.
Fig. 8.
Characterization of mutation at the UL34 and US9 loci. Digital images show electrophoretically separated PCR products. (A) Amplification products from the UL34 locus using genomes from rescued viruses as the template. The BAC used for rescue is indicated above each lane. The sizes of the PCR products are indicated to the right of the gel. Lambda BstEII digest size standards are shown in lane 1, and the sizes of standard bands are indicated to the left of the gel. (B) Amplification products from the US9 locus that are either undigested (lanes 2 and 4) or digested with restriction enzymes HincII (lanes 3 and 5). A 100-bp ladder is shown in lane 1.
Fig. 9.
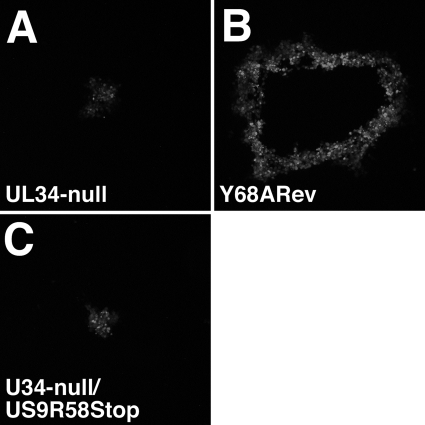
Growth of BAC-derived UL34-null and US9R58Stop virus on WT and mutant pUL34-expressing cells. Digital micrographs are shown of representative plaques formed for 1 week on Y68A pUL34-expressing cells. Plaques were immunofluorescently stained for gD. The infecting virus is indicated in each panel.
DISCUSSION
Mutation of conserved tyrosine 68 of pUL34 reveals new functions for pUL34.
It has been clear for some time that the nuclear envelopment complex (NEC) consisting of pUL34, pUL31, and other viral and cellular proteins is multifunctional. HSV-1 pUL34 and pUL31 have each been shown to have the ability to disrupt the nuclear lamina and to function at multiple steps in capsid envelopment, including capsid docking, curvature of the membrane around capsids, and deenvelopment at the outer nuclear membrane (3, 24, 27, 40, 45, 47, 50, 51, 58, 59). All of these functions are consistent with the concentration of pUL34 and pUL31 at the nuclear envelope in infected cells and consistent with a role as master regulators of the nuclear egress process. Tyrosine 68 is one of the most conserved residues in pUL34, and therefore it is unsurprising that it may be important for multiple pUL34 activities.
While most of the functions of NEC components have been identified using analysis of deletion mutants, functional analyses can be greatly enhanced by analysis of more subtle mutations. Analysis of small insertion mutations in the murine cytomegalovirus homologs of UL34 and UL31 (M50 and M53, respectively) has allowed identification of functional domains of these proteins required for their interaction with each other, nuclear envelope localization, capsid envelopment, and capsid maturation (6, 32, 46, 53). These studies showed that the residue homologous to HSV-1 Y68 (M50 residue Y57) is critical for function of the protein (6). Surprisingly, however, the effect of mutation at this site is to disrupt interaction with M53 and consequently prevent proper localization of M50 and M53 at the nuclear envelope (6). In HSV pUL34, the sequences required for interaction with pUL31 and proper localization to the nuclear envelope have been mapped to a different part of the protein (29), and we observed no defect in recruitment of pUL34 to the nuclear envelope (Fig. 5). This suggests that despite its conservation, tyrosine 68 has different functions in different herpesvirus families. Those functions may, however, overlap. Because M50 function is disrupted very early by mutation of Y57, downstream functions, including possible roles in capsid envelopment and virus spread, may be masked.
Our data show three phenotypes for the Y68A substitution mutation in pUL34. (i) There is an exaggerated disruption of nuclear envelope structure, resulting in the formation of blebs that protrude from the nucleus into the cytoplasm. These appear from EM analysis to be extensions of both INM and ONM, still properly spaced with respect to one another, into the cytoplasm. Blebs like this are seen during normal infections with the wild-type virus and are likely the result of disconnection of nuclear envelope from the underlying lamina (44). Their much more extensive formation in infection with Y68A pUL34 suggests that this mutation may result in exaggerated disruption of the structure of the lamina. The lamina disruption activity of the NEC is tightly controlled and limited, presumably to maintain nuclear functions necessary for virus replication (3, 39). Lamina disruption is limited in part by the pUS3 protein kinase activity (3), but it is unsurprising that intrinsic sequence features of pUL34 might also play a role. (ii) There is a major defect in virus replication, and EM analysis suggests that this defect is caused by inhibition of nuclear egress, since capsids were only rarely observed outside the nucleus. Y68A mutant pUL34 accumulates normally at the nuclear envelope, suggesting that it makes a normal targeting interaction with pUL31. Since nuclear envelope disruption was observed, but no nuclear egress intermediates (e.g., docked capsids, perinuclear virions, etc.) accumulated, the Y68A nuclear egress defect likely follows lamina disruption but precedes docking of DNA-containing capsids at the inner nuclear membrane. (iii) There is a major defect in cell-to-cell spread of virus infection. This cannot be accounted for by the Y68A replication defect, since a similar growth defect that is associated with low-level wild-type pUL34 expression is not accompanied by inhibition of plaque formation. The spread defect also cannot be accounted for by a general inhibition of virus release from the cell, since the efficiency of release in cells that express Y68A pUL34 is just the same as that seen in cells that express WT pUL34 and that form robust plaques. The defect in cell-to-cell spread also cannot be accounted for by some UL34-independent peculiarity of the Y68A UL34-expressing cell line. These cells support completely normal replication and plaque formation by WT HSV-1, indicating that they have no general defect in the ability to support growth and cell-to-cell spread. The spread phenotype is evident only upon infection with UL34-null virus, clearly demonstrating that the spread phenotype is tied to the function of the mutant pUL34.
pUL34 in cell-to-cell spread.
pUL34 is the first viral protein other than those required for viral entry that is required for efficient cell-to-cell spread in Vero cells, and its effect on spread is far more dramatic than that seen for gE/gI in epithelial or fibroblast cells. The pUL34 mutant cell-to-cell spread defect seems not to be mediated by a pathway that requires either gE or US9, since deletion of either of these genes has no apparent effect on growth or spread in Vero cells (10, 31). A role for pUL34 in cell-to-cell spread is unexpected, since its localization at the nuclear envelope makes it unlikely to participate directly in trafficking of virions or viral components to the cell surface. It is more likely that the phenotype is indirect and reflects defective synthesis or trafficking of other essential viral or cellular components in the mutant-infected cell.
pUL34 expression is also required for proper localization of gE (Fig. 7). gE is required for efficient cell-to-cell spread in some cells (not including Vero cells), and this activity requires gE sorting to junctional surfaces, where it colocalizes with β-catenin (12, 16, 37, 64). The mechanism of sorting is not characterized but requires sequences in the cytoplasmic tail of gE (64). The requirement for gE in cell-to-cell spread is not absolute; gE-null mutants have only moderately diminished plaque sizes in all cells tested. Since gE is not essential for efficient spread in Vero cells, the pUL34 effect on gE localization is not likely the root of the spread defect. However, it suggests that localization of proteins at the cell periphery can be influenced by pUL34. The target(s) of pUL34 function that affects cell-to-cell spread in Vero cells remains to be discovered.
Mapping of extragenic suppressors.
To our knowledge, this is the first demonstration of the use of high-throughput sequencing (HTS) to facilitate mapping of extragenic suppressor mutations in HSV-1. As anticipated, this method revealed potential interactors with the UL34 pathway that would not have been considered in a candidate-based approach. The availability of sequences from several different HSV-1 strains allowed us to filter down the list of potential candidate mutations further, and this approach will be further aided as additional HSV-1 genomes are added to the databases. As sequencing technology and analysis improve, it may become practical to apply this technique further, for example, to an HTS screen in parallel with the parental strain and the additional five Y68A suppressors described here. Although it is beyond the scope of this paper, this approach might reveal a polygenic basis to the observed phenotype, if for example the additional suppressors shared not only the US9 R58Stop mutation tested for by PCR but also an untested SNP from Table 2 or even a noncoding mutation. The UL34-null/US9R58Stop strain generated here provides an important substrate for further testing of such mutations, since conserved candidates from further HTS screening can be added sequentially to the BAC and tested phenotypically. Since polygenic mutations are typically lost from mutant and suppressor screens due to failure to complement, future application of HTS may greatly improve this aspect of mutational mapping.
The appearance of a US9 nonsense mutation in five out of six pUL34 Y68A suppressor mutants is consistent with the observed cell-to-cell spread phenotype for the Y68A mutant. It was unexpected, however, because pUS9 is not thought to influence cell-to-cell spread in cells other than neurons, and there is no other evidence for a physical or functional interaction between pUS9 and pUL34. pUS9 is a type II membrane protein found on cytoplasmic membranes, including Golgi membranes, whereas pUL34 is highly concentrated at the nuclear membrane (4, 34, 48).
Phenotypic suppression of missense mutations can occur by several mechanisms. True reversion (restoration of the original UL34 sequence) and intragenic suppression (restoration of function by other mutations in UL34) were eliminated by our selection strategy. Extragenic suppressors are mutations in a different gene, and the US9 mutation is a candidate for this type of suppressor. In some cases of extragenic suppression, an initial mutation in one protein disrupts a critical functional interaction with a second protein, and the suppressor mutation changes the sequence of a second protein, restoring the interaction or its functional consequence. In this scenario, however, deletion of either gene would be expected to result in the same defective phenotype. Deletion of US9 has no cell-to-cell spread phenotype in Vero cells, making it unlikely that pUS9 and pUL34 interact in some way. A more likely possibility is that the US9 nonsense mutation is a bypass suppressor. This suppressor class opens an alternative pathway for achievement of the same biological result (35).
How US9 mutation might open an alternative spread pathway depends on the effect of the R58Stop mutation. The exact R58Stop mutation described here as correlated with the Y68ARev phenotype has been previously found by Negatsch and colleagues in the supposedly wild-type strain HSV-1 KOS (42). They showed that the truncated protein is not detectable in HSV-1 KOS-infected cells and suggested that the mutation creates a null allele. If the US9R58Stop mutant is indeed a null allele of US9, it may be that US9 expression normally inhibits the function of an alternative cell-to-cell spread pathway that is exposed only in a US9-null infection. However, there are many single nucleotide substitutions that would result in even more severely truncated proteins and abrogate US9 expression. Truncation of US9 in the same position in these disparate strains suggests that this specific mutation is advantageous for viral growth in some circumstances, that the viral DNA has features facilitating mutation at this position, or both. This truncation preserves US9's endocytosis motif and key phosphorylation sites but removes its transmembrane domain and the preceding cluster of basic, positively charged amino acids. It is possible that the truncated protein, even if expressed at very low levels, might interact in interesting ways with virion trafficking pathways.
ACKNOWLEDGMENTS
We thank the staff of the Central Microscopy Research Facility of the University of Iowa and especially Jean Ross for expertise and help with TEM analysis.
These studies were supported by the University of Iowa and Public Health Service award AI 41478. We acknowledge funding from a Center Grant (NIH/NIGMS P50 GM071508), the New Jersey Commission on Spinal Cord Research (M.L.S.), NIH P40 RR018604 (L.W.E. and M.L.S.), and a supplement to NIH R01 AI 033063 (M.L.S.).
Footnotes
Published ahead of print on 11 May 2011.
REFERENCES
- 1. Balan P., et al. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J. Gen. Virol. 75:1245–1258 [DOI] [PubMed] [Google Scholar]
- 2. Bjerke S. L., et al. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J. Virol. 77:7601–7610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bjerke S. L., Roller R. 2006. Roles for herpes simplex type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347(2):261–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brideau A. D., Banfield B. W., Enquist L. W. 1998. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J. Virol. 72:4560–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brideau A. D., Card J. P., Enquist L. W. 2000. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. J. Virol. 74:834–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bubeck A., et al. 2004. Comprehensive mutational analysis of a herpesvirus gene in the viral genome context reveals a region essential for virus replication. J. Virol. 78:8026–8035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cai J. S., et al. 1999. Identification and structure of the Marek's disease virus serotype 2 glycoprotein M gene: comparison with glycoprotein M genes of Herpesviridae family. J. Vet. Med. Sci. 61:503–511 [DOI] [PubMed] [Google Scholar]
- 8. Darlington R. W., Moss L. H. 1968. Herpesvirus envelopment. J. Virol. 2:49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Desai P., Person S. 1999. Second site mutations in the N terminus of the major capsid protein (VP5) overcome a block at the maturation cleavage site of the capsid scaffold proteins of herpes simplex virus type 1. Virology 261:357–366 [DOI] [PubMed] [Google Scholar]
- 10. Dingwell K. S., et al. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dingwell K. S., Doering L. C., Johnson D. C. 1995. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J. Virol. 69:7087–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dingwell K. S., Johnson D. C. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 72:8933–8942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ejercito P. M., Kieff E. D., Roizman B. 1968. Characteristics of herpes simplex virus strains differing in their effect on social behavior of infected cells. J. Gen. Virol. 2:357–364 [DOI] [PubMed] [Google Scholar]
- 15. Farina A., et al. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 79:3703–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farnsworth A., Johnson D. C. 2006. Herpes simplex virus gE/gI must accumulate in the trans-Golgi network at early times and then redistribute to cell junctions to promote cell-cell spread. J. Virol. 80:3167–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Forrester A., et al. 1992. Construction and properties of a mutant herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fuchs W., Klupp B. G., Granzow H., Osterrieder N., Mettenleiter T. C. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonnella R., et al. 2005. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: interactions with BFRF1 and with the nuclear lamina. J. Virol. 79:3713–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jacobson J. G., Yang K., Baines J. D., Homa F. L. 2006. Linker insertion mutations in the herpes simplex virus type 1 UL28 gene: effects on UL28 interaction with UL15 and UL33 and identification of a second-site mutation in the UL15 gene that suppresses a lethal UL28 mutation. J. Virol. 80:12312–12323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson D. C., Feenstra V. 1987. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J. Virol. 61:2208–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson D. C., Frame M. C., Ligas M. W., Cross A. M., Stow N. D. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J. Virol. 62:1347–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson D. C., Spear P. G. 1982. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J. Virol. 43:1102–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klupp B. G., et al. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. U. S. A. 104:7241–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klupp B. G., Granzow H., Mettenleiter T. C. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063–10073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lake C. M., Hutt-Fletcher L. M. 2004. The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 320:99–106 [DOI] [PubMed] [Google Scholar]
- 27. Leach N., et al. 2007. Emerin is hyperphosphorylated and redistributed in herpes simplex virus type 1-infected cells in a manner dependent on both UL34 and US3. J. Virol. 81:10792–10803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H., Ruan J., Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang L., Baines J. D. 2005. Identification of an essential domain in the herpes simplex virus 1 UL34 protein that is necessary and sufficient to interact with UL31 protein. J. Virol. 79:3797–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ligas M. W., Johnson D. C. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 62:1486–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Longnecker R., Roizman B. 1987. Clustering of genes dispensable for growth in culture in the S component of the HSV-1 genome. Science 236:573–576 [DOI] [PubMed] [Google Scholar]
- 32. Lötzerich M., Ruzsics Z., Koszinowski U. H. 2006. Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J. Virol. 80:73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lyman M. G., Feierbach B., Curanovic D., Bisher M., Enquist L. W. 2007. Pseudorabies virus Us9 directs axonal sorting of viral capsids. J. Virol. 81:11363–11371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lyman M. G., Kemp C. D., Taylor M. P., Enquist L. W. 2009. Comparison of the pseudorabies virus Us9 protein with homologs from other veterinary and human alphaherpesviruses. J. Virol. 83:6978–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Manson M. 2000. Allele-specific suppression as a tool to study protein-protein interactions in bacteria. Methods 20:18–34 [DOI] [PubMed] [Google Scholar]
- 36. McGraw H. M., Awasthi S., Wojcechowskyj J. A., Friedman H. M. 2009. Anterograde spread of herpes simplex virus type 1 requires glycoprotein E and glycoprotein I but not Us9. J. Virol. 83:8315–8326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McMillan T. N., Johnson D. C. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 75:1928–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morris J. B., Hofemeister H., O'Hare P. 2007. Herpes simplex virus infection induces phosphorylation and delocalization of emerin, a key inner nuclear membrane protein. J. Virol. 81:4429–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mou F., Forest T., Baines J. D. 2007. Us3 of herpes simplex type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 81:6459–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mou F., Wills E., Baines J. D. 2009. Phosphorylation of the U(L)31 protein of herpes simplex virus 1 by the U(S)3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids. J. Virol. 83:5181–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Muranyi W., Haas J., Wagner M., Krohne G., Koszinowski U. H. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854–857 [DOI] [PubMed] [Google Scholar]
- 42. Negatsch A., Mettenleiter T. C., Fuchs W. 2011. Herpes simplex virus type 1 strain KOS carries a defective US9 and a mutated US8A gene. J. Gen. Virol. 92:167–172 [DOI] [PubMed] [Google Scholar]
- 43. Neubauer A., Rudolph J., Brandmuller C., Just F. T., Osterrieder N. 2002. The equine herpesvirus 1 UL34 gene product is involved in an early step in virus egress and can be efficiently replaced by a UL34-GFP fusion protein. Virology 300:189–204 [DOI] [PubMed] [Google Scholar]
- 44. Nii S., Morgan C., Rose H. M. 1968. Electron microscopy of herpes simplex virus. II. Sequence of development. J. Virol. 2:517–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park R., Baines J. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80:494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Popa M., et al. 2010. Dominant negative mutants of the murine cytomegalovirus M53 gene block nuclear egress and inhibit capsid maturation. J. Virol. 84:9035–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reynolds A. E., Liang L., Baines J. D. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes UL31 and UL34. J. Virol. 78:5564–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reynolds A. E., et al. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reynolds A. E., Wills E. G., Roller R. J., Ryckman B. J., Baines J. D. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roller R. J., Bjerke S. L., Haugo A. C., Hanson S. 2010. Analysis of a charge cluster mutation of herpes simplex virus type 1 UL34 and its extragenic suppressor suggests a novel interaction between pUL34 and pUL31 that is necessary for membrane curvature around capsids. J. Virol. 84:3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roller R. J., Zhou Y., Schnetzer R., Ferguson J., DeSalvo D. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roop C., Hutchinson L., Johnson D. C. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells and its particles lack glycoprotein H. J. Virol. 67:2285–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rupp B., et al. 2007. Random screening for dominant-negative mutants of the cytomegalovirus nuclear egress protein M50. J. Virol. 81:5508–5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ryckman B. J., Roller R. J. 2004. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78:399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Santarelli R., et al. 2008. Identification and characterization of the product encoded by ORF69 of Kaposi's sarcoma-associated herpesvirus. J. Virol. 82:4562–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schnee M., Ruzsics Z., Bubeck A., Koszinowski U. H. 2006. Common and specific properties of herpesvirus UL34/UL31 protein family members revealed by protein complementation assay. J. Virol. 80:11658–11666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Scott E. S., O'Hare P. 2001. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 75:1818–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Simpson-Holley M., Colgrove R. C., Nalepa G., Harper J. W., Knipe D. M. 2005. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 79:12840–12851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Simpson-Holley M., Baines J., Roller R., Knipe D. 2004. Herpes simplex virus 1 UL31 and UL34 promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 78:5591–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Snyder A., Polcicova K., Johnson D. C. 2008. Herpes simplex virus gE/gI and US9 proteins promote transport of both capsids and virion glycoproteins in neuronal axons. J. Virol. 82:10613–10624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Szpara M. L., Parsons L., Enquist L. W. 2010. Sequence variability in clinical and laboratory isolates of herpes simplex virus 1 reveals new mutations. J. Virol. 84:5303–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tischer B. K., von Einem J., Kaufer B., Osterrieder N. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]
- 63. Tomishima M. J., Enquist L. W. 2001. A conserved alpha-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. Cell Biol. 154:741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wisner T., Brunetti C., Dingwell K., Johnson D. C. 2000. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J. Virol. 74:2278–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yamauchi Y., et al. 2001. Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J. Gen. Virol. 82:1423–1428 [DOI] [PubMed] [Google Scholar]