Abstract
Budding of retroviruses from cell membranes requires ubiquitination of Gag and recruitment of cellular proteins involved in endosome sorting, including endosome sorting complex required for transport III (ESCRT-III) protein complex and vacuolar protein sorting 4 (VPS4) and its ATPase. In response to infection, a cellular mechanism has evolved that blocks virus replication early and late in the budding process through expression of interferon-stimulated gene 15 (ISG15), a dimer homologue of ubiquitin. Interferon treatment of DF-1 cells blocks avian sarcoma/leukosis virus release, demonstrating that this mechanism is functional under physiological conditions. The late block to release is caused in part by a loss in interaction between VPS4 and its coactivator protein LIP5, which is required to promote the formation of the ESCRT III-VPS4 double-hexamer complex to activate its ATPase. ISG15 is conjugated to two different LIP5-ESCRT-III-binding charged multivesicular body proteins, CHMP2A and CHMP5. Upon ISGylation of each, interaction with LIP5 is no longer detected. Two other ESCRT-III proteins, CHMP4B and CHMP6, are also conjugated to ISG15. ISGylation of CHMP2A, CHMP4B, and CHMP6 weakens their binding directly to VPS4, thereby facilitating the release of this protein from the membrane into the cytosol. The remaining budding complex fails to release particles from the cell membrane. Introducing a mutant of ISG15 into cells that cannot be conjugated to proteins prevents the ISG15-dependent mechanism from blocking virus release. CHMP5 is the primary switch to initiate the antiviral mechanism, because removal of CHMP5 from cells prevents ISGylation of CHMP2A and CHMP6.
INTRODUCTION
Retroviruses such as human immunodeficiency virus type 1 (HIV-1) and avian sarcoma/leukosis virus (ASLV) release from cells using similar but different pathways that are mediated by small protein-binding domains (L domains) found in the structural proteins of the viruses (8, 41, 42). L domains recruit host cell proteins Nedd4 (11) and Tsg101 (38); the latter protein is involved in membrane vesicle biogenesis and forms part of the endosomal sorting complex required for transport (ESCRT). The ESCRT proteins are subdivided into several complexes, referred to as ESCRT-0, -I, -II, and -III (2). HIV-1 and ASLV utilize ESCRT-I and -II proteins, respectively, to assemble the same late budding complex (6, 23, 24) that contains 10 different charged multivesicular body proteins (CHMPs) that comprise the ESCRT-III complex. In addition, the viruses appear to passage through different membranes in the process (17). The ESCRT-III proteins in turn contain microtubule-interacting and transport (MIT) interaction motif (MIM) domains which recruit the vacuolar protein sorting protein 4 (VPS4) AAA ATPase to the site of budding through interaction with the VPS4 MIT domain. There are two types of MIM domains known, MIM1 (found in CHMP1A, -1B, -2A, and -2B) and MIM2 (found in CHMP4A to -C and CHMP6) (10, 34). Membrane scission, which releases particles from the cell membrane, is catalyzed by the ESCRT-III proteins (12). When VPS4 hydrolyzes ATP, the ESCRT-III complex is disassembled into the cytosol. VPS4 is normally found in the cytosol as an inactive dimer. It is activated on membranes when it forms a double hexamer ring structure in the presence of a coactivator protein, LIP5 (30). Point mutations that inactivate the ATPase block the release of virus from the cell membrane (6, 9, 16). The coactivator, LIP5, is delivered to membranes by binding to several ESCRT-III proteins, including CHMP1B, -2A, -3, and -5 (32). CHMP5 differs from the other ESCRT-III proteins in that its LIP5-binding site is unique and it does not directly interact with VPS4 (1).
A cellular innate immunity mechanism which targets retrovirus budding both early and late in the release process induces the expression of interferon-stimulated gene 15 (ISG15) and its specific ISG15 E1, E2, and E3 ligase complex (3, 4, 25, 27, 33, 43, 45). ISG15 is a dimer homologue of ubiquitin. Ubiquitination of viral Gag is required for normal release of retroviruses from cells, and the addition of ubiquitin is catalyzed by its E1, E2, and E3 ubiquitin ligase complex (5, 15, 21, 37). Neither HIV-1 Gag nor Tsg101 is ISGylated. Several reports show that ISG15 expression inhibits the E3 subunit of the ligase involved in Gag or Tsg101 ubiquitination, suggesting a possible mechanism for the observed inhibition (14, 19, 20, 44). Disruption of the late release process is a more general antiviral mechanism in which the VPS4 ATPase is removed from the budding complex (25). When ISG15 and its ligase are expressed in cells, CHMP5 becomes ISGylated and accumulates in the membrane fraction (25). This blocks the interaction of LIP5 with VPS4, thereby preventing the activation of the ATPase, and promotes the release of the protein from the membrane. If CHMP5 expression is suppressed in cells by specific targeting small interfering RNAs (siRNAs), ISG15 no longer inhibits virus budding, and the complex between VPS4 and LIP5 is restored (25, 40). The latter observation suggests that CHMP5 is not the sole delivery system of LIP5 to VPS4 on the membrane. The finding that expression of ISG15 in cells blocks virus release provides a mechanistic explanation for an old observation in the literature that murine leukemia virus is inhibited by interferon expression and results in virus trapped on the cell surface (26).
In the present paper, we further characterize the mechanism of inhibition late in the budding process. We present evidence that when CHMP5 is ISGylated, it loses the ability to interact with LIP5, thus failing to deliver LIP5 to the budding complex. Several other ESCRT-III proteins, including CHMP2A, CHMP3, CHMP4B, and CHMP6, are also ISGylated. Modification of CHMP2A, similar to that of CHMP5, causes the loss of this protein's ability to interact with LIP5. In addition, ISGylation of CHMP2A, CHMP4B, and CHMP6 weakens their binding directly to VPS4. The combination of both causes the release of VPS4 from the budding complex, resulting in the budding defect. If CHMP5 is removed from cells, CHMP2A and CHMP6 are not ISGylated. This indicates that CHMP5 is the key regulatory protein to initiate the antiviral mechanism. This mechanism functions under physiological conditions, because interferon treatment of DF-1 cells blocks the release of ASLV. Also, the introduction of a mutant of ISG15 that cannot be conjugated to protein prevents the inhibition, indicating that covalent linkage of ISG15 to protein is required.
MATERIALS AND METHODS
Reagents.
The pcDNA4HisMaxC vector encoding ISG15 was a generous gift from Deborah J. Lenschow (Washington University School of Medicine) and was previously described (7). All His-tagged vectors encode 6His residues. The His-ISG15 GG-to-AA mutant (13) was assembled with a 156 and 157 Gly-to-Ala substitution by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The pcDNA3 vectors encoding UbE1L and UbcH8 were kindly provided by Robert M. Krug at University of Texas (44). Herc5, ISG15 E3 ligase, was cloned into pCNA3 vector and generously provided by Jon M. Huibregtse (University of Texas). Herc5 is an interferon-induced homologous to the E6-AP carboxyl terminus (HECT) E3 enzyme required for conjugation of ISG15 to protein in human cells (4). DNA encoding human wild-type VPS4A (a gift from Wesley Sundquist, University of Utah) was as previously described (6). The cDNA sequences of CHMP1B, CHMP2A, CHMP3, CHMP4B, CHMP5, and CHMP6 were cloned into the pIRES-hrGFP-1a vector (FLAG-CHMPs) (also a generous gift from Wesley Sundquist, University of Utah). DNAs encoding hemagglutinin (HA)-CHMP5, HA-CHMP2A, and HA-CHMP3 were constructed as follows: p2036 (11) was doubly digested with KpnI and XbaI to remove the green fluorescent protein (GFP) coding region. By using PCR-based methods, a 5′ KpnI site was introduced upstream of the HA start codon. Additionally, a 3′ XbaI site was introduced downstream of the CHMP5, CHMP2A, and CHMP3 coding sequence. FLAG-CHMPs were used as a template. The PCR product was then ligated to the KpnI and XbaI doubly digested p2036 vector DNA to produce p2036HA-CHMPs. A 3× FLAG tag fused to the C terminus of CHMP proteins allows for detection with an anti-FLAG monoclonal serum (Sigma-Aldrich, Milwaukee, WI). The following probes were purchased. Antibodies recognizing CHMP3 (Santa Cruz Biotechnology, Santa Cruz, CA), influenza virus HA (Covance, Berkeley, CA), CHMP2A (Proteintech Group, Chicago, IL), and ISG15 (Cell Signaling Technology, Danvers, MA) were purchased from the sources indicated. Antiserum directed at CHMP5 and LIP5 were generous gifts from Jerry Kaplan and Wes Sundquist (University of Utah). The recombinant chicken alpha interferon (rChIFN-α) (31) was a generous gift from Philip I. Marcus (University of Connecticut).
Transfection of 293E cells.
293E cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (1,000 units/ml), and streptomycin (1,000 μg/ml) and grown to 60% confluence at 37°C. Expression of plasmids from p2036 was high in 293E cells, because these cells stably express the EBNA1 protein of Epstein-Barr virus (EBV) and the p2036 constructs contain the EBV family of repeats (FR) plasmid maintenance element that EBNA1 binds. In all experiments, six-well plates of 293E cells were transfected with the indicated DNA with FuGene 6 transfection reagent (Roche Diagnostics, Alameda, CA) according to the manufacturer's instructions. Three micrograms of His-ISG15-expressing plasmid was transfected into cells along with E1 (0.5 μg) and E2 (0.5 μg) ligases. When the E3 expression plasmid was included, 0.5 μg of His-ISG15, 0.25 μg of E1, 0.25 μg of E2, and 0.5 μg of E3 were transfected into cells.
Detection of proteins by Western blotting.
For detection of exogenous or endogenous proteins in 293E cells, cell lysate fractions were prepared by suspension in radioimmunoprecipitation assay (RIPA) buffer (phosphate-buffered saline [PBS] containing 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor mixture tablets) 48 h posttransfection. For coimmunoprecipitation experiments, 90% cell lysate fractions were incubated with the antibody and equilibrated protein A-agarose beads overnight at 4°C (37). The remaining 10% of the cell lysates was used to detect protein expression. The expressed or precipitated proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. After blocking the membrane with wash buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 0.1% Tween 20) containing 5% nonfat dry milk, proteins were detected with the primary antibody indicated in the legends to Fig. 1 to 6, and anti-mouse or anti-rabbit IgG-horseradish peroxidase (HRP) secondary antibody was used for enhanced chemiluminescence (ECL) (Denville Scientific; Metuchen, NJ). For the CHMP5 and CHMP2A ISGylation assay, the cell lysate fractions were prepared by suspension in cell lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% Triton X-100, and protease inhibitor mixture tablets). CHMP5 and CHMP2A conjugates were purified by suspension with Ni+-agarose beads. The beads were then washed three times with 20 mM imidazole-containing cell lysis buffer, and the ISG15-conjugated CHMP5 and CHMP2A proteins were then eluted with 300 mM imidazole-containing cell lysis buffer. The proteins were separated by SDS-PAGE and transferred to PVDF membranes. The ISG-conjugated bands were detected using a mouse anti-FLAG serum. For detection of the ISG conjugated HA-CHMPs, a mouse anti-HA serum was used.
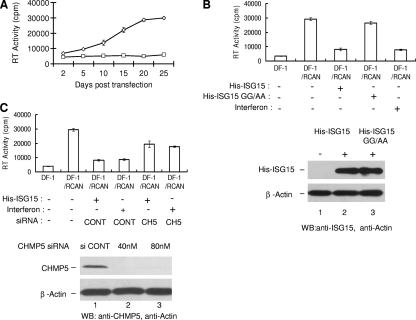
Fig. 1.
Expression of ISG15 and its ligase complex induced by transfection or interferon treatment of DF-1 cells blocked virus release. (A) The RCAN vector was transfected into DF-1 cells, and the appearance of ASLV in the culture supernatants was determined by RT activity in detergent-activated virions as described in Materials and Methods. ⋄, RCAN-transfected cells; ◊, untransfected cells. (B) DF-1 cells or DF-1 cells at 25 days posttransfection with RCAN vectors were transfected with pHis-ISG15 or the pHis-ISG15 mutant form containing a GG-to-AA substitution plus the E1, E2, and E3 ligase plasmids or treated with rChIFN-α (2,000 units) as described in Materials and Methods. At 48 h after transfection or treatment with rChIFN-α, ASLV in the medium fraction was analyzed as in panel A. His-ISG15 and His-ISG15 GG-to-AA mutant expression in lysates was detected by Western blotting (WB) with anti-ISG15 serum (lower panel). Anti-actin serum was used to detect β-actin as a loading control. (C) DF-1 cells or DF-1 cells at 25 days posttransfection with RCAN vectors were transfected with pHis-ISG15 or treated with chicken interferon along with an siRNA pool (40 nM or 80 nM concentration) targeting CHMP5 (CH5) or a nontargeting siRNA (80 nM) control (CONT). Forty-eight hours after transfection or treatment with interferon, ASLVs in the medium fraction were collected and analyzed as in panel A. To confirm knockdown of CHMP5 in DF-1 cells, endogenous CHMP5 expression in cells in the presence of control siRNA (80 nM; lower panel, lane 1) or the siRNA targeting CHMP5 (40 nM, lane 2; 80 nM, lane 3) was analyzed by Western blotting with anti-ISG15 serum. β-Actin again served as a loading control. The RT activity data in panels A, B, and C represent the averages from three independent experiments.
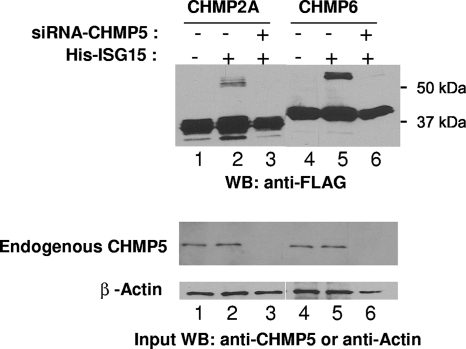
Fig. 6.
Removal of CHMP5 from cells prevents ISGylation of CHMP2A and CHMP6. CHMP2A-FLAG or CHMP6-FLAG was coexpressed in 293E cells with ISG15, E1, and E2 as described in the legend to Fig. 1. Where indicated, an siRNA targeting CHMP5 was introduced into the cells as previously described (21). CHMP2A-FLAG and CHMP6-FLAG and their ISGylated forms were detected by Western blotting using an anti-FLAG serum (upper panel). Endogenous CHMP5 was detected by Western blotting using an anti-CHMP5 serum (middle panel). β-Actin was detected (lower panel) as a sample loading control as described in the legend to Fig. 2.
Sedimentation assay.
293E cells in 6-well plates were transfected with the plasmid(s) using FuGene 6 transfection reagent. At 48 h posttransfection, cells were washed with PBS and solubilized in 350 μl lysis buffer (10 mM Tris-HCl [pH 7.5], 10% sucrose, 1 mM EDTA, 1% Triton X-100, complete protease inhibitor [Roche Diagnostics, Alameda, CA]) at 4°C for 60 min. Supernatant and pellet fractions were separated by centrifugation at 10,000 × g for 15 min at 4°C. Pellets were suspended into the same volume as the supernatant in lysis buffer and then sonicated 10 s to shear DNA. Equal volumes of fractions were analyzed by immunoblotting.
Detecting reverse transcriptase (RT) in detergent-activated virus particles released from DF-1 cells.
DF-1 cells were cultured in DMEM supplemented with 10% fetal bovine serum, 1% penicillin (1,000 units/ml), and 1% streptomycin (1,000 μg/ml). After cells were grown to 60% confluence at 37°C, they were transfected with 3 μg replication-competent ASLV LTR, no splice acceptor (RCAN) virus plasmid DNAs encoding ASLV using FuGene 6 transfection reagent. Uninfected and infected cells were then transfected with the DNAs or treated with recombinant chicken alpha interferon (rChIFN-α) at a concentration of 2,000 U/ml. At 48 h after transfection or treatment with rChIFN-α, the medium was collected, cell debris was removed by centrifugation at 4,000 rpm for 10 min at 4°C, and virus particles were collected by centrifugation through a 20% sucrose solution at 29,000 rpm for 45 min at 4°C. Pelleted virus particles were suspended in 50 μl of 5× TE buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA). The amount of reverse transcriptase was detected in NP-40-treated virions by analyzing serial dilutions for RT activity using an exogenous template and primer as previously described with some modifications (18). A 5-μl virus particle sample was mixed in 30 μl of reaction buffer containing 0.5 optical density unit at 260 nm (OD260)/ml poly(A), 0.25 OD260 units/ml oligo(dT), and 0.5 μCi [α-33P]dTTP (3,000 Ci/mmol; PerkinElmer, Waltham, MA). After being incubating at 37°C for 25 min, 10-μl reaction mixtures were spotted on Whatman DE81 disks, washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), dried, and counted for radioactivity in 3 ml of scintillation fluid (Ready Safe; Beckman).
siRNA depletion of CHMP5 in 293E cells.
siRNA pools directed against the coding sequence of human CHMP5 and a random, nontargeting siRNA sequence were purchased from Dharmacon (Lafayette, CO). The CHMP5 siRNA pool consists of a mixture of four siRNA duplexes and was previously described (25). Knockdown of CHMP5-FLAG expression was achieved by transfecting 80 nM CHMP5 siRNA into 293E cells in six-well plates with RNAiMax reagent (Invitrogen, Carlsbad, CA). Forty-eight hours posttransfection, cells were transfected with plasmid DNA encoding CHMP2A-FLAG or CHMP6-FLAG with FuGene 6 transfection reagent as described above. Lysate fractions were harvested 24 h later, and the levels of protein expression and ISGylation were analyzed by Western blotting as described above. Depletion of CHMP5 expression in DF-1/RCAN cells was obtained by transfecting the same amount of CHMP5 targeting siRNA (80 nM) as in the 293E cells. The same amount of nontargeting control siRNA (80 nM) was used in the control experiment.
RESULTS
ISG15 expression blocks ASLV release from DF-1 cells under physiological conditions.
The RCAN expression vector for ASLV was transfected into DF-1 cells, and release of virus over time was monitored by reverse transcriptase activity detected by incorporation of [33P]TTP in response to exogenously added poly(A)/oligo(dT) in detergent-activated virions recovered from the cell medium fraction (Fig. 1 A) as described in Materials and Methods. DNAs encoding His-tagged ISG15 and its E1, E2, and E3 ligases were transfected into the DF-1 cells 25 days posttransfection of the RCAN vector, and the level of virus released into the medium fraction 48 h later was monitored (Fig. 1B). The expression of ISG15 significantly blocked the release of virus from the cells under these conditions. If a mutant form of His-ISG15 containing a GG-to-AA substitution, which prevents ISG15 from being conjugated to protein (13), was substituted for the His-ISG15, little effect on virus release was detected (Fig. 1B). If the cells were treated with chicken interferon rather than transfection by the His-ISG15, E1, E2, and E3 expression DNAs, a similar degree of inhibition to virus release was observed (Fig. 1B). Equivalent amounts of His-ISG15 and His-ISG15 GG-to-AA mutant were expressed in the cell lysate fraction detected by Western blotting (Fig. 1B, lower panels).
We repeated the DF-1/ASLV experiment but introduced into the cells siRNAs directed at CHMP5 or nontargeting control siRNAs (Fig. 1C). The presence of the control siRNAs did not have any detectable effect on the amount of virus released from the DF-1 cells treated with the expression vectors for ISG15 and its ligase complex or with chicken interferon. In contrast, the siRNAs targeting CHMP5 partially rescued virus release (Fig. 1C) under either condition. We conclude from these experiments that ISG15 inhibits ASLV release from cells under physiological conditions.
ISG15 disrupts the interaction of LIP5 with CHMP5.
To begin to understand the mechanism of inhibition of late budding caused by ISG15 expression in cells, we examined the formation of the complex of LIP5 and CHMP5. When HA-CHMP5 and FLAG-LIP5 were coexpressed in 293E cells, LIP5 was immunoprecipitated from the lysate fraction with an anti-FLAG serum. Following SDS-PAGE, the coimmunoprecipitation with CHMP5 could be demonstrated by Western blotting of the gel using an anti-HA serum (Fig. 2 A, upper panel, lane 2). In contrast, in the presence of ISG15 expression, coimmunoprecipitation of the two proteins was not detected (lane 3). FLAG-LIP5 and HA-CHMP5 were expressed in the cell lysate fraction under these conditions (Fig. 2A, lower panels). When HA-CHMP5 was not expressed, a band of protein was not detected by Western blotting with the HA serum (lane 1). The gel was also probed for β-actin as a sample loading control. We conclude from this experiment that ISGylated CHMP5 no longer interacts with LIP5.
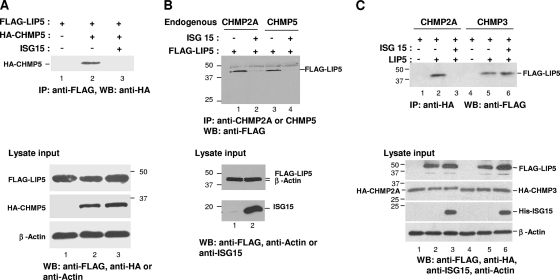
Fig. 2.
ISG15 expression blocks the interaction of LIP5 with CHMP5 and CHMP2A but not CHMP3. (A) 293E cells were transfected with plasmids expressing FLAG-LIP5 and/or HA-CHMP5 (lanes 1 and 2) and, where indicated, pHis-ISG15, E1, and E2 (lane 3) as described in Materials and Methods. Forty-eight hours posttransfection, 10% of total cell lysate input was resolved by 10% SDS-PAGE to verify expression of FLAG-LIP5 and HA-CHMP5 by Western blotting with anti-HA or anti-FLAG serum (bottom panels, as indicated). The lysate was probed with an anti-actin serum as a loading control. FLAG-LIP5 was immunoprecipitated (IP) from the remainder of the cell lysate fraction with an anti-FLAG serum and resolved by 10% SDS-PAGE. HA-CHMP5 was detected by Western blotting with an anti-HA serum (top panel). (B) 293E cells were transfected with plasmids expressing FLAG-LIP5 and ISG15, E1, and E2 where indicated. Cell lysates were prepared, endogenous CHMP5 and CHMP2A were immunoprecipitated with the respective antisera, and samples were analyzed as in panel A. (C) 293E cells were transfected with plasmids expressing FLAG-LIP5 and HA-CHMP2A (lane 1 to 3) or HA-CHMP3 (lanes 4 to 6). Expression plasmids for pHis-ISG15, E1, E2, and E3 were cotransfected into cells where indicated (lane 3 and lane 6). Samples were analyzed as in panel A except that HA-CHMP2A and HA-CHMP3 were immunoprecipitated with an antiserum directed at the respective proteins and an anti-HA or -FLAG serum was used in Western blotting to detect the three proteins (top panel). Molecular size markers (in kilodaltons) are indicated by the numbers at the sides of the panels.
Because of the possibility of experimental artifacts caused by overexpression of exogenous proteins, we repeated the above experiment but measured formation of the complex of FLAG-LIP with endogenous CHMP5 (Fig. 2B). To do this experiment, we immunoprecipitated CHMP5 from the cell lysate with a specific CHMP5-targeted antiserum and detected the presence of FLAG-LIP5 by Western blotting using the FLAG serum. As shown in Fig. 2B, we detected the coimmunoprecipitation of endogenous CHMP5 with FLAG-LIP5 (lane 3). When ISG15, E1, and E2 were coexpressed, coimmunoprecipitation was barely or not detected (lane 4). These results are consistent with those obtained by expressing exogenous CHMP5 in cells (Fig. 2A).
ISG15 expression disrupts the interaction of CHMP2A with LIP5.
Other ESCRT-III proteins besides CHMP5, including CHMP2A and CHMP3, bind to LIP5. We therefore determined if ISG15 expression affected interaction of these proteins with LIP5. As shown in Fig. 2C, we detected coimmunoprecipitation of FLAG-LIP5 with exogenous HA-CHMP2A or HA-CHMP3 in the absence of ISG15, E1, E2, and E3 expression (Fig. 2C, upper panel, lane 2 or 5, respectively). When ISG15 was coexpressed, the coimmunoprecipitation of FLAG-LIP5 with HA-CHMP2A (Fig. 2C, lane 3) was not detected. In contrast, the interaction of FLAG-LIP5 with HA-CHMP3 was unaffected (lane 6). Thus, the binding of LIP5 to CHMP2A but not CHMP3 was prevented by coexpression of ISG15, E1, E2, and E3. We repeated the above experiment but monitored endogenous rather than exogenous CHMP2A. As shown in Fig. 2B (upper panel, lanes 1 and 2), we detected the coimmunoprecipitation of endogenous CHMP2A and FLAG-LIP5 in the absence of ISG15 expression and found it to be significantly reduced in the presence of coexpression of ISG15, E1, and E2. The lower panels are an analysis of the same extracts, showing expression of FLAG-LIP5 and ISG15 under these conditions. Thus, the results monitoring endogenous protein mirror those obtained with the exogenous proteins, as shown in Fig. 2C.
ISG15 expression results in LIP5 released from the budding complex into the cytosol.
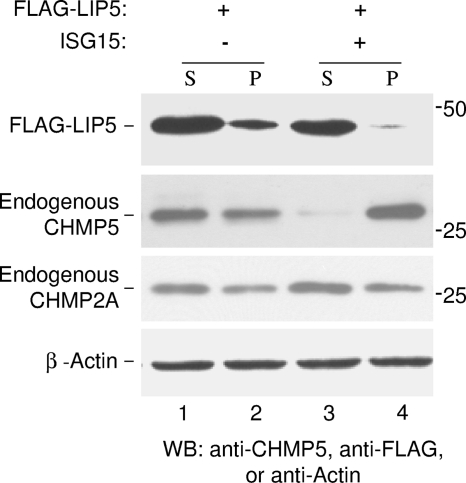
The finding that LIP5 still binds to CHMP3 in the presence of ISG15 expression might suggest that not all of the LIP5 would be released into the cytosol. We therefore examined the distribution of FLAG-LIP5 between the cytosol and membrane pellet fraction in the presence and absence of ISG15, E1, and E2 coexpression as described in Materials and Methods. Proteins were separated by SDS-PAGE, and endogenous CHMP5 or CHMP2A and exogenous FLAG-LIP5 were detected by Western blotting using antiserum directed at CHMP5, CHMP2A, and the FLAG tag, respectively. As shown in Fig. 3 (FLAG-LIP5 panel, lanes 1 and 2), FLAG-LIP5 was detected in both the soluble and membranes fractions in the absence of ISG15 expression. In contrast, most but not all LIP5 was found in the cytosol when ISG15, E1 and E2 were coexpressed (FLAG-LIP5 panel, lanes 3 and 4). As previously reported (25), endogenous CHMP5 accumulated in the membrane pellet fraction when ISG15, E1 and E2 were coexpressed (Endogenous CHMP5 panel, lanes 3 and 4). The distribution between the soluble and membrane fractions of endogenous CHMP2A was mostly unaffected (Endogenous CHMP2A panel, lanes 1 to 4). Again, β-actin was probed as a sample loading control (β-Actin panel, lanes 1 to 4).
Fig. 3.
ISG15 expression results in release of LIP5 from the budding complex in membranes into the cytosol. 293E cells were transfected with plasmids expressing FLAG-LIP5 and, where indicated, pHis-ISG15, E1, and E2. Forty-eight hours posttransfection, cell lysates were prepared and fractionated into soluble and pellet fractions as described in Materials and Methods. Distribution of FLAG-LIP5 and endogenous CHMP2A or CHMP5 in the resulting soluble (S) (lanes 1 and 3) and membrane pellet (P) (lanes 2 and 4) fractions were visualized by Western blotting with anti-FLAG serum to detect LIP5 or anti-CHMP2A and anti-CHMP5 sera to detect endogenous CHMP2A and CHMP5, respectively. β-Actin served as a loading control.
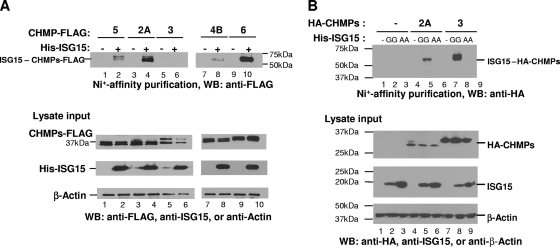
ISG15 conjugates to C-terminal FLAG tagged-CHMP2A, CHMP4B, and CHMP6.
The above results suggested that CHMP2A might be targeted for ISGylation. To test this hypothesis, we coexpressed CHMP5-FLAG, CHMP6-FLAG, CHMP4B-FLAG, CHMP3-FLAG, and CHMP2A-FLAG separately in cells with His-ISG15, E1, and E2. Cell lysates were prepared, and proteins containing the His-ISG15 were affinity purified using a Ni+ resin as described in Materials and Methods. After the beads were washed, the His-ISG15-containing proteins were eluted with 300 mM imidazole and analyzed by SDS-PAGE (Fig. 4 A). The presence of the respective CHMP protein was detected by Western blotting with an anti-FLAG serum. When ISG15 was coexpressed with CHMP5-FLAG, we detected modified CHMP5 migrating at a molecular mass that would correspond to His-ISG15 conjugated to CHMP5 with a size indicative of a one-to-one molar ratio (upper panel, lane 2) as previously reported (25). Detection of this protein was dependent upon coexpression of His-ISG15 in cells (lane 1). When CHMP2A-FLAG was substituted for CHMP5-FLAG, we detected protein of an estimated molecular mass that would correspond to that of His-ISG15-CHMP2A-FLAG (lane 4). Its detection was again dependent upon coexpression of His-ISG15 (lane 3). These results indicate that CHMP2A was conjugated to ISG15. When we substituted CHMP3-FLAG for CHMP5-FLAG, we did not detect an ISGylated form of this protein (lane 6). When CHMP4B-FLAG and CHMP6-FLAG were expressed in the cells in the presence but not the absence of His-ISG15, ISGylated forms of these proteins were also detected (upper panels, lanes 7 to 10). Each of the CHMP-FLAG proteins and His-ISG15 were expressed under these conditions (lower panels).
Fig. 4.
ISG15 conjugates to ESCRT-III proteins. (A) ISG15 conjugates to CHMP2A, CHMP4B, and CHMP6. 293E cells were transfected with plasmids expressing C-terminal FLAG-tagged CHMP2A, -3, -4B, -5, or -6 in the presence or absence of a plasmid expressing CHMP-ISG15, E1, and E2 ligases as described in Materials and Methods. Forty-eight hours posttransfection, cells were lysed and ISGylated proteins were affinity purified using Ni+-agarose beads as described in Materials and Methods. Proteins were resolved by 12% SDS-PAGE. ISGylated CHMP5 (lane 2), CHMP2A (lane 4), CHMP4B (lane 8), and CHMP6 (lane 10) were visualized by Western blot analysis with a monoclonal anti-FLAG serum (top panel). No CHMP3-ISG15 conjugate was detected (lane 6). The bottom panel shows a Western blot of the 10% input of total cell lysates used in the affinity purification step, to verify expression of CHMP5-FLAG (lanes 1 and 2), CHMP2A-FLAG (lanes 3 and 4), CHMP3-FLAG (lanes 5 and 6), CHMP4B-FLAG (lanes 7 and 8), and CHMP6-FLAG (lanes 9 and 10), respectively. β-Actin served as a loading control. (B) ISG15 also conjugates to CHMP3 and is blocked by the ISG15 GG-to-AA mutant. 293E cells were transfected with plasmids expressing HA-CHMP2A or -3 in the presence or absence of a plasmid expressing His-ISG15 or the His-ISG15 GG-to-AA mutant and E1, E2, and E3 as described in Materials and Methods. Samples were analyzed as in panel A except that HA-CHMP2A and HA-CHMP3 were detected by using an anti-HA serum. GG denotes His-tagged wild-type ISG15; AA denotes the His-tagged ISG15 mutant.
ISG15 conjugates to N-terminal HA-tagged CHMP2A and CHMP3.
The reason for the multiple bands detected for the ESCRT-III-FLAG-tagged proteins in Fig. 4A is not known but may be due to degradation. These multiple bands are not observed when N-terminal HA-tagged CHMP2A or CHMP3 was substituted for C-terminal FLAG-tagged CHMP2A or CHMP3, respectively (Fig. 4B). In agreement with the results shown in Fig. 4A, we again detect ISGylation of CHMP2A. However, in contrast to the results shown in Fig. 4A, lane 6, we detected ISGylation of HA-CHMP3 (lane 8). When the His-ISG15 GG-to-AA mutant, which cannot be conjugated to protein, was substituted for His-ISG15 expression in the 293E cells, we did not detect the ISGylation of HA-CHMP2A (lane 6) or HA-CHMP3 (lane 9).
VPS4 interaction with ESCRT-III proteins is weakened by ISG15 expression.
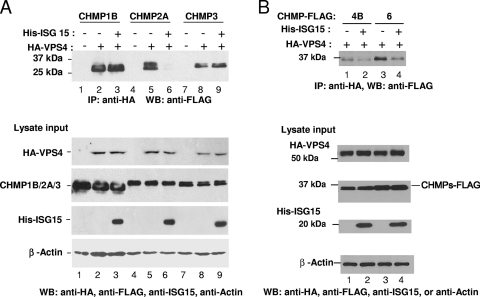
The interaction of LIP5 with VPS4 is required to form the double-hexamer structure that activates its ATPase. As shown above, ISG15 expression in cells interferes with the association of LIP5 with VPS4 by preventing LIP5 interaction with CHMP5 and CHMP2A. However, because not all of the LIP5 was released from the membrane into the cytosol cell fraction when ISG15 was expressed in cells (Fig. 3), we considered the possibility that other factors, such as a weakening in the binding of ESCRT-III proteins for VPS4, contribute to the release of VPS4 from the membrane budding complex. As shown in Fig. 5 A, we detected coimmunoprecipitation of CHMP1B-FLAG, CHMP2A-FLAG, and CHMP3-FLAG (upper panel, lanes 2, 5, and 8, respectively) with HA-VPS4 in the cell lysate fraction. When ISG15, E1, E2, and E3 were coexpressed, the coimmunoprecipitation of CHMP2A-FLAG with HA-VPS4 was significantly decreased (lane 6). In contrast, there was little or no detectable change in the coimmunoprecipitation of the complex of CHMP1B-FLAG or CHMP3-FLAG with HA-VPS4 (Fig. 5A, lane 3 or 9, respectively). Each ESCRT-III protein and VPS4 was expressed in cells under these conditions (lower panels). As a control, when HA-VPS4 was not expressed in cells, we did not detect coimmunoprecipitation with the anti-FLAG serum (lanes 1, 4, and 7).
Fig. 5.
ISG15 expression weakens the interaction of VPS4 with ESCRT-III proteins. (A) 293E cells were cotransfected with plasmids expressing HA-VPS4 and FLAG-tagged CHMP1B, CHMP2A, or CHMP3 and 3 μg of pHis-ISG15 where indicated. At 48 h posttransfection, HA-VPS4 was immunoprecipitated from cell extracts with an anti-HA serum, and samples were resolved by 10% SDS-PAGE. For the top panel, precipitation of CHMP1B-FLAG (lanes 1 to 3), CHMP2A-FLAG (lanes 4 to 6), and CHMP3-FLAG (lanes 7 to 9) was determined by Western blotting with anti-FLAG antibody. The bottom panel shows a Western blot of the 10% input of total cell lysates used in the coimmunoprecipitation assay to verify even expression of HA-VPS4 (lanes 2 and 3, 5 and 6, and 8 and 9) and CHMP1B-FLAG (lanes 1 to 3), CHMP2A-FLAG (lanes 4 to 6), and CHMP3-FLAG (lanes 7 to 9), respectively. β-Actin served as a loading control. (B) 293E cells were transfected as described in the legend to panel A except that expression vectors of CHMP4B-FLAG (lanes 1 and 2) and CHMP6-FLAG (lanes 3 and 4) were substituted for the other ESCRT-III proteins. These experiments were repeated three different times with similar results.
We repeated the above analysis to examine the interaction of HA-VPS4 with CHMP4B or CHMP6 in the presence and absence of ISG15 expression. As shown in Fig. 5B, we detected coimmunoprecipitation of CHMP4B or CHMP6 with HA-VPS4 in the absence of ISG15 expression (lane 1 or 3, respectively). When ISG15 was coexpressed, the amount of coimmunoprecipitation of HA-VPS4 with either ESCRT-III protein was decreased (lanes 2 and 4). All proteins were expressed under these conditions (lower panels).
ISGylation of CHMP2A and CHMP6 are dependent upon CHMP5.
To determine if ISGylation of CHMP5 is the key event initiating the late inhibition mechanism, we asked if ISGylation of CHMP2A-FLAG or CHMP6-FLAG were dependent upon CHMP5. We coexpressed CHMP2A-FLAG or CHMP6-FLAG in 293E cells in the presence of ISG15, E1, and E2 and analyzed the cell lysate fractions for CHMP2A-FLAG or CHMP6-FLAG and their ISG15-modified forms by SDS-PAGE and Western blotting using an antiserum against the FLAG tag. We detected CHMP2A-FLAG and CHMP6-FLAG along with their ISGylated forms in the presence of ISG15 as shown in Fig. 6 (lanes 2 and 5, respectively). When an siRNA targeting CHMP5 was introduced in cells under the latter conditions, the ISGylated forms of both proteins were no longer detected (lanes 3 and 6, respectively). These results indicated that ISGylation of CHMP2A and CHMP6 was dependent upon CHMP5. The expression level of endogenous CHMP5 is shown in the middle panel and demonstrates that the siRNA targeting CHMP5 was effective in lowering the level of protein to below detectable levels (lower panel, lanes 3 and 6). β-Actin was included as a sample loading control.
DISCUSSION
The interferon-induced gene ISG15 inhibited budding late in the release process by preventing the formation of the VPS4-LIP5 complex required to activate its ATPase (25). At the same time, there was a significant decrease in VPS4 cosedimentation with insoluble CHMP5 polymers on cell membranes. Because CHMP5 was known to bind to LIP5, we asked if the interaction between these two proteins was altered. This appears to be the case (Fig. 2), highlighting an important role for CHMP5 in delivering LIP5 to the membrane. However, when CHMP5 expression in cells was suppressed, the ability of ISG15 to inhibit virus release was blocked but the VPS4/LIP5 interaction was restored (25). This indicated that there must be an alternative pathway to deliver LIP5 to the budding complex in membranes. Three other ESCRT-III proteins, CHMP1B, -2A, and -3, are known to bind to LIP5 (32). CHMP2A and -3 are required for virus infection, while CHMP1B is not (39). We therefore asked if the LIP5 binding to any of these proteins was changed by ISG15 coexpression in cells. We found that the binding of endogenous or exogenous CHMP2A to LIP5 was not detected when ISG15 was coexpressed, similar to results for CHMP5 (Fig. 2). In contrast, the binding of LIP5 to CHMP3 was not affected under the same conditions (Fig. 2). Because coimmunoprecipitation of CHMP2A with LIP5 was adversely affected by ISG15 coexpression, we examined whether this ESCRT-III protein was conjugated to ISG15. Using a His-tagged ISG15, we purified and then detected CHMP2A covalently linked to ISG15 (Fig. 4), similar to the case with CHMP5. If we substituted an ISG15 GG-to-AA mutant that cannot be conjugated to protein, we did not detect ISGylation of CHMP2A. Thus, CHMP2A is a second ESCRT-III protein targeted for ISGylation that when modified results in the loss of its binding to LIP5. These observations strongly suggest that there are two parallel LIP5 membrane delivery pathways; one via CHMP5, which does not bind to VPS4 (25), and one via CHMP2A, which does bind to VPS4 (Fig. 5). We did not detect ISGylation of CHMP3-FLAG. However, when we substituted an HA-CHMP3 for the CHMP3-FLAG, we did detect ISG15 conjugation to this protein even though we did not detect a concomitant change in its binding to LIP5. In this regard, it is different from CHMP5 and CHMP2A. Because modification of this protein influences the degree of its ISGylation, it is difficult to estimate the amount of total protein that is ISGylated in cells. Using the same selection protocol, we detected ISGylation of the non-LIP5-binding proteins, CHMP4B and CHMP6. Thus, many of the ESCRT-III proteins found in the budding complex are conjugated to ISG15 under these conditions.
When CHMP5 is ISGylated, VPS4 and LIP5 are released from and/or not recruited to the budding complex in the membrane (see reference 24) (Fig. 3 and 7). Because CHMP5 does not directly interact with VPS4, we examined whether the interaction between VPS4 and other ESCRT-III proteins with MIM1 domains was changed when ISG15 was coexpressed in cells. CHMP2A interaction with VPS4 detectably decreased in the presence of overexpression of ISG15 (Fig. 5A). In contrast, CHMP1B-VPS4 and CHMP3-VPS4 interactions were barely or not affected. CHMP4B and CHMP6 are also known to bind to VPS4 via the MIM2 domain (10). We detected decreases in the binding of both proteins to VPS4 when ISG15 was coexpressed in cells. Thus, the release of VPS4 into the cytosol is caused by the sum of ISGylations of several ESCRT-III proteins.
Fig. 7.
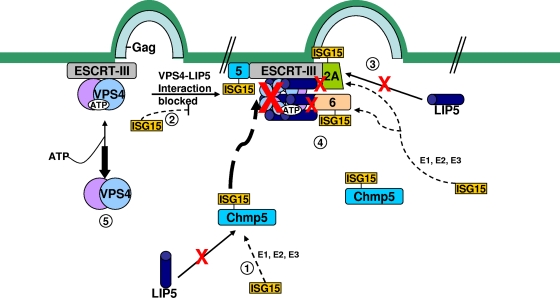
Mechanism of inhibition of late budding caused by ISG15 expression in cells. (Step 1) The ESCRT-III protein, CHMP5, binds to LIP5 in the cytosol. However, in the presence of ISG15-specific E1, E2, and E3 enzymes, ISG15 conjugates to CHMP5 and the binding to LIP5 is lost. (Step 2) The CHMP5-ISG15 conjugate accumulates in the budding complex on the membrane without LIP5. The interaction on the membrane between VPS4 and its coactivator LIP5 is thereby blocked, preventing activation of the ATPase through formation of the VPS4/LIP5 double-hexamer structure. (Step 3) CHMP2A is ISGylated in the presence of ISG15-CHMP5. This results in the loss of its binding to LIP5 and the weakening of its direct interaction to VPS4. (Step 4) CHMP6 is ISGylated in the presence of ISG15-CHMP5. This results in weakening of its binding to VPS4. (Step 5) VPS4 is released into the cytosol, and virus budding is arrested.
The fact that removal of CHMP5 from cells blocks the action of ISG15 suggests that ISGylation of CHMP5 is the key to initiate the antirelease mechanism. This hypothesis is supported by the findings that removal of CHMP5 from cells blocks the effects of ISG15 expression. Moreover, ISGylation of CHMP2A and CHMP6 were dependent upon CHMP5 (Fig. 6). This probably reflects a requirement for CHMP5-ISG15, because it is this form of the protein that appears to accumulate in the budding complex on the membrane (25). As yet, we do not know the reason ISGylated CHMP5 accumulates on the membrane. It is likely that ISGylation of CHMP4B was also dependent upon CHMP5, but we did not get sufficient expression of this protein in the lysate fraction to determine if this was the case. Independently, we examined whether there was a change in distribution of CHMP2A when ISG15 was coexpressed in cells. In contrast to that of CHMP5 (25), we detected little change in distribution of CHMP2A between the membrane and cytosol fractions (Fig. 3). A similar result was obtained for endogenous CHMP4B and CHMP6 (data not shown). This was not unexpected, because in the absence of the VPS4/LIP5 ATPase, ESCRT-III proteins would not be released from the membrane. Taken together, these results indicate that CHMP5 is the key regulatory molecule for initiation of disruption of the late budding complex, and this is summarized in the model presented in Fig. 7.
While many of these experiments employed expression vectors for viral Gag, ISG15, and its ligase complex, and ESCRT proteins to follow the release of virus-like particles (VLPs), we have now shown that ISG15 inhibition occurred under more physiological conditions, under which release of ASLV from DF-1 cells was blocked by chicken interferon treatment. The degree of inhibition was the same as that achieved by introducing expression vectors for ISG15 and its ligase complex (Fig. 1). The inhibition to virus release was also blocked by substituting the GG-to-AA mutant of ISG15 for the wild-type protein, indicating that conjugation of ISG15 to protein was required. Moreover, if CHMP5 expression was significantly lowered in cells, the inhibition of virus release caused by either interferon treatment or expression of ISG15 and its ligase complex was partially restored. As described above, CHMP5 is the key to initiation of the inhibition of the late budding step by controlling the delivery of the LIP5 coactivator to VPS4 on the membrane and promoting ISGylation of many of the other ESRT-III proteins which, in turn, weakens the binding of VPS4 for these ESCRT-III proteins, resulting in its release into the cytosol. Because ISG15 expression in cells also inhibits early steps in the budding process not dependent upon CHMP5, such as inhibiting the E3 ubiquitin ligase, we would not expect full restoration of the budding of ASLV by removal of CHMP5 from the cytosol. We estimate from the experiments shown in Fig. 1 that about half of the inhibition to release of ASLV was caused by blocking early steps in the process and half by blocking the CHMP-dependent steps late in the budding process.
Finally, the mechanism for blocking release of retroviruses from cell membranes described in this study has broader implications for many enveloped viruses, including filo-, paramyxo-, and rhabdoviruses (22, 28, 29, 35, 36). This is because these viruses depend on the ESCRT-III/VPS4 ATPase complex to release particles from cell membranes. Also, the findings that many different ESCRT-III proteins are ISGylated and that this results in changes in the binding properties of each protein to LIP5 or VPS4 strongly suggest that interferon-induced expression of ISG15 will shut down normal cellular endocytosis and multivesicular body (MVB) biogenesis.
ACKNOWLEDGMENTS
This work was supported in part by U.S. Public Health Service grant AI079025 (to J.L.).
We thank Carol Carter and Tom Hope for critically reading the manuscript.
Footnotes
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Azmi I. F., et al. 2008. ESCRT-III family members stimulate VPS4 ATPase activity directly or via Vta1. Dev. Cell 14:50–61 [DOI] [PubMed] [Google Scholar]
- 2. Babst M. 2005. A protein's final ESCRT. Traffic 6:2–9 [DOI] [PubMed] [Google Scholar]
- 3. Bednarik D. P., Mosca J. D., Raaj N. B., Pitha P. M. 1989. Inhibition of human immunodeficiency virus (HIV) replication by HIV-trans-activated alpha 2-interferon. Proc. Natl. Acad. Sci. U. S. A. 86:4958–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dastur A., Beaudenon S., Kelley M., Klug R., Huibregtse J. 2006. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J. Biol. Chem. 281:4334–4338 [DOI] [PubMed] [Google Scholar]
- 5. Demirov D. G., Freed E. O. 2004. Retrovirus budding. Virus Res. 106:87–102 [DOI] [PubMed] [Google Scholar]
- 6. Garrus J. E., et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65 [DOI] [PubMed] [Google Scholar]
- 7. Giannakopoulos N. V., et al. 2009. ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus. J. Virol. 83:1602–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Göttlinger H. G., Dorfman T., Sodriski J. G., Haseltine W. A. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. U. S. A. 88:3195–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gottwein E., et al. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474–9485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kieffer C., et al. 2008. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell 15:62–73 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 11. Kikonyogo A., et al. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for Gag budding from cells. Proc. Natl. Acad. Sci. U. S. A. 98:11199–11204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lata S., et al. 2009. Structure and function of ESCTR-III. Biochem. Soc. Trans. 37:156–160 [DOI] [PubMed] [Google Scholar]
- 13. Lenschow D., et al. 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 79:13974–13983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malakhova O., Zhang D. 2008. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J. Biol. Chem. 283:8783–8787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin-Serrano J. 2007. The role of ubiquitin in retroviral egress. Traffic 8:1297–1303 [DOI] [PubMed] [Google Scholar]
- 16. Medina G., et al. 2005. The functionally exchangeable L domains in RSV and HIV-1 Gag direct particle release through pathways linked by Tsg101. Traffic 6:880–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Medina G., et al. 2008. Tsg101 can replace Nedd4 function in ASV Gag release but not membrane targeting. Virology 377:30–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller J., Ge Z., Morris S., Das K., Leis J. 1997. Multiple biological roles associated with the Rous sarcoma virus 5′ untranslated RNA U5-IR stem and loop. J. Virol. 71:7648–7656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okumura A., Lu G., Pitha-Rowe I., Pitha P. M. 2006. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc. Natl. Acad. Sci. U. S. A. 103:1440–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Okumura A., Pitha P. M., Harty R. N. 2008. ISG15 inhibits Ebola VP40 budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc. Natl. Acad. Sci. U. S. A. 105:3974–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patnaik A., Chau V., Wills J. W. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. U. S. A. 97:13069–13074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pawliczek T., Crump C. 2009. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of Tsg101 and Alix expression. J. Virol. 83:11254–11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pincetic A., Medina G., Carter C., Leis J. 2008. Avian sarcoma virus and human immunodeficiency virus, type 1 use different subsets of ESCRT proteins to facilitate the budding process. J. Biol. Chem. 283:29822–29830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pincetic A., Leis J. 2009. The mechanism of budding of retroviruses from cell membranes. Adv. Virol. 2009:6239691–6239699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pincetic A., Kuang Z., Seo E. J., Leis J. 2010. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J. Virol. 84:4725–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pitha P. M., Fernie B., Maldarelli F., Hattman T., Wivel N. A. 1980. Effect of interferon on mouse leukaemia virus (MuLV). V. Abnormal proteins on virions of Rauscher MuLV produced in the presence of interferon. J. Gen. Virol. 46:97–110 [DOI] [PubMed] [Google Scholar]
- 27. Poli G., Orenstein J. M., Kinter A., Folks T. M., Fauci A. S. 1989. Interferon-alpha but not AZT suppresses HIV expression in chronically infected cell lines. Science 244:575–577 [DOI] [PubMed] [Google Scholar]
- 28. Schmitt A., Leser G., Waning D., Lamb R. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J. Virol. 76:3952–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmitt A., Leser G., Morita E., Sunquist W., Lamb R. 2005. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J. Virol. 79:2988–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scott A., et al. 2005. Structural and mechanistic studies of VPS4 proteins. EMBO J. 24:3658–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sekellick M., Ferrandino A., Hopkins D., Marcus P. 1994. Chicken interferon gene: cloning, expression, and analysis. J. Interferon Res. 14:71–79 [DOI] [PubMed] [Google Scholar]
- 32. Shim S., Merrill S. A., Hanson P. I. 2008. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol. Biol. Cell 19:2661–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shirazi Y., Pitha P. M. 1992. Alpha interferon inhibits early stages of the human immunodeficiency virus type 1 replication cycle. J. Virol. 66:1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stuchell-Brereton M. D., et al. 2007. ESCRT-III recognition by VPS4 ATPases. Nature 449:740–744 [DOI] [PubMed] [Google Scholar]
- 35. Taylor G., Hanson P., Kielian M. 2007. Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting alphavirus budding. J. Virol. 81:13631–13639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Timmins J., et al. 2003. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326:493–502 [DOI] [PubMed] [Google Scholar]
- 37. Vana M. L., et al. 2004. The role of Nedd4 and ubiquitination of RSV Gag in budding of virus-like particles from cells. J. Virol. 78:13943–13953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. VerPlank L., et al. 2001. Tsg101, the prototype of a class of dominant-negative ubiquitin regulators, binds human immunodeficiency virus type 1 Pr55Gag: the L domain is a determining of binding. Proc. Natl. Acad. Sci. U. S. A. 98:7724–772911427703 [Google Scholar]
- 39. von Schwedler U., et al. 2003. The protein network of HIV budding. Cell 114:701–713 [DOI] [PubMed] [Google Scholar]
- 40. Ward D. M., et al. 2005. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem. 280:10548–10555 [DOI] [PubMed] [Google Scholar]
- 41. Wills J., et al. 1994. An assembly domain of Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605–6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xiang Y., Cameron C., Wills J., Leis J. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yuan W., Krug R. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao C., Denison C., Huibregtse J. M., Gygi S., Krug R. M. 2005. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 102:10200–10205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zou W., et al. 2005. ISG15 modification of ubiquitin E2 Ubc13 disrupts its ability to form thioester bond with ubiquitin. BBRC 336:61–68 [DOI] [PubMed] [Google Scholar]