Abstract
Human immunodeficiency virus type 1 (HIV-1) has the ability to adapt to the host environment by escaping from host immune responses. We previously observed that escape from humoral immunity, both at the individual and at a population level, coincided with longer variable loops and an increased number of potential N-linked glycosylation sites (PNGS) in the viral envelope glycoprotein (Env) and, in particular, in variable regions 1 and 2 (V1V2). Here, we provide several lines of evidence for the role of V1V2 in the resistance of HIV-1 to neutralizing antibodies. First, we determined that the increasing neutralization resistance of a reference panel of tier-categorized neutralization-sensitive and -resistant HIV-1 variants coincided with a longer V1V2 loop containing more PNGS. Second, an exchange of the different variable regions of Env from a neutralization-sensitive HIV-1 variant into a neutralization-resistant escape variant from the same individual revealed that the V1V2 loop is a strong determinant for sensitivity to autologous-serum neutralization. Third, exchange of the V1V2 loop of neutralization-sensitive HIV-1 variants from historical seroconverters with the V1V2 loop of neutralization-resistant HIV-1 variants from contemporary seroconverters decreased the neutralization sensitivity to CD4-binding site-directed antibodies. Overall, we demonstrate that an increase in the length of the V1V2 loop and/or the number of PNGS in that same region of the HIV-1 envelope glycoprotein is directly involved in the protection of HIV-1 against HIV-specific neutralizing antibodies, possibly by shielding underlying epitopes in the envelope glycoprotein from antibody recognition.
INTRODUCTION
The HIV-1 envelope glycoprotein (Env) is a major target of the humoral immune response in HIV-1-infected individuals. Antibodies directed against Env can be detected early in infection and are able to neutralize autologous virus variants with increasing titers over time in most patients (1, 35, 40, 51). HIV-1 Env has developed multiple mechanisms to evade neutralizing antibodies, including the inaccessibility of relevant epitopes due to the trimeric structure of Env, the density of glycosylation, and the presence of occluding variable loops on the outer domain of Env (11, 14, 17, 22, 51). Moreover, some epitopes for neutralizing antibodies only emerge after the conformational changes that occur upon the engagement of Env with the CD4 receptor, when spatial constraints between cell and viral membrane no longer allow binding of the relatively large immunoglobulins to Env (21, 22, 23, 29, 52).
The HIV-1 Env is synthesized as a gp160 precursor protein, which is subsequently cleaved into two subunits, surface protein gp120 and transmembrane protein gp41. Three subunits of gp120 bind noncovalently to three subunits of gp41 to form a trimeric complex on the surface of the virion. Gp120 is composed of five conserved regions (C1 to C5) that are interspersed with 5 variable regions (V1 to V5) (47). The conserved regions form a central core consisting of an inner domain, which interacts with gp41 and is important for trimer formation, and an outer domain, which interacts with the (co)receptors. The variable regions can be highly diverse, both between viruses from different patients and within the viral quasispecies of one patient, and form flexible loop structures on the outer domain of gp120 (54).
Neutralizing antibody pressure results in the rapid selection of escape variants with changes in their variable loops, such as large insertions, deletions, and changes in the number of potential N-linked glycosylation sites (PNGS). In particular, the length and glycosylation characteristics of the V1V2 loop seem to play a role in resistance against neutralizing antibodies (8, 9, 10, 33, 41, 42, 43, 46, 49, 52), possibly by shielding underlying regions of Env from antibody recognition (18, 37) and, especially, in the protection against anti-V3 and anti-CD4-binding site antibodies (12, 30, 37, 43).
We previously reported on the adaptation of the HIV-1 Env to humoral immunity at a population level, reflected in an increasing resistance of recently transmitted HIV-1 to neutralizing antibodies over a time course of 20 years (6). The increased neutralization resistance of recently transmitted HIV-1 from contemporary seroconverters, which is most obvious for CD4-binding site-directed antibodies, coincided with changes in the viral envelope, mainly a longer V1 loop with an increased number of PNGS (6, 17a).
In our present study, we investigated whether these changes in Env are indeed causally related to the differences in neutralization sensitivity of HIV-1 variants. For this reason, we first compared the molecular characteristics of Envs of reference viruses that are categorized from tier 1 to tier 3 based on their decreasing neutralization sensitivity (26, 27, 31, 45). We also examined whether changes in the V1V2 loop are directly responsible for increased neutralization resistance by generating chimeric viruses in which Env fragments were exchanged between neutralization-sensitive and neutralization-resistant HIV-1 variants that were isolated from a single individual early and late in infection, respectively, and between neutralization-sensitive and neutralization-resistant HIV-1 variants from historical and contemporary seroconverters, respectively. The results from these studies strongly suggest that the V1V2 loop of the envelope glycoprotein is directly involved in the protection of HIV-1 from CD4-binding-site-directed neutralizing antibodies, possibly by shielding the targeted epitopes.
MATERIALS AND METHODS
Viral variants.
The viral variants used to construct the chimeric NL4-3/Env viruses were isolated from HIV-1-infected men who have sex with men of the Amsterdam Cohort Studies (ACS) on HIV and AIDS and were all HIV-1 subtype B. None of the individuals received combination antiretroviral therapy during the sampling period used for this study. Clonal virus variants were obtained from peripheral blood mononuclear cells (PBMC) as previously described (44, 50). From patient ACH19642, one virus that was sensitive to autologous neutralization was isolated from PBMC obtained 29 months after seroconversion (SC) (GenBank accession number GU455427), and one virus that had escaped from autologous neutralization was isolated from PBMC obtained 144 months post-SC (GenBank accession number HQ902005). These virus variants were used to exchange different regions of Env in order to investigate the effects of these regions on autologous-neutralization sensitivity. In addition, 5 recently transmitted viruses from individuals who seroconverted between 1985 and 1989 (historical seroconverters) (GenBank accession numbers EU43976, EU44098, EU44014, HQ902003, and HQ902004) that were sensitive to neutralization by HIVIG (a pool of purified IgG obtained in 1995 from chronically HIV-1-infected individuals) and 5 recently transmitted viruses from individuals who seroconverted between 2003 and 2006 (contemporary seroconverters) (GenBank accession numbers HQ901998 to HQ902002) that were resistant to neutralization by HIVIG were selected from a previous study (6) to analyze the effects of the V1V2 loops on neutralization sensitivity.
To prevent a change in neutralization sensitivity of the virus variants during in vitro culture, the number of virus passages in PBMC was kept to a minimum (2).
The Amsterdam Cohort Studies are conducted in accordance with the ethical principles set out in the declaration of Helsinki, and written consent was obtained prior to data collection. The study was approved by the Academic Medical Center institutional medical ethics committee.
Preparation of chimeric viruses.
Chimeras of gp160 proteins were created using a PCR overlap strategy followed by recombination of the PCR product into an HIV-1 backbone. The variable region inserts of env and the flanking regions of env were amplified in separate reactions using an Expand high fidelity PCR system (Roche Applied Science). Separate PCR products were then combined by PCR overlap into a chimeric gp160 product that spanned from nucleotide (nt) 5658 to nt 9171 of reference strain HXB2. All primer combinations are listed in Table S1 in the supplemental material. Chimeric NL4-3/Env viruses were produced by homologous recombination of the env PCR products with a pNL4-3 vector (a kind gift from J. Alcami). In short, pNL4-3 was restricted with XbaI (HXB2 nt 6114) and XhoI (HXB2 nt 8898) and was subsequently cotransfected with an env PCR product into 293T cells in a 24-well plate using the calcium phosphate method. After 2 days, phytohemagglutinin (PHA)-stimulated PBMC from healthy seronegative blood donors were added to the culture, and the next day the PBMC were transferred to a culture flask. Supernatants were harvested when positive for p24, as determined using an in-house p24 antigen capture enzyme-linked immunosorbent assay (48). The presence of the correct env in NL4-3 was confirmed by sequencing.
PBMC-based antibody neutralization assay.
The chimeric NL4-3/Env viruses, in which different regions of Env were exchanged between a virus that was sensitive to autologous neutralization and a virus that had escaped autologous neutralization, were tested for their relative neutralization sensitivities against autologous serum obtained 77 months post-SC, the serum pool from HIV-negative individuals, HIVIG, and monoclonal antibodies (MAbs) 447-52D, b12, 2G12, 4E10, and 2F5. The chimeric NL4-3/Env viruses in which the V1V2 regions were exchanged between virus variants from historical and contemporary seroconverters were tested for their relative neutralization sensitivities against HIVIG and MAbs VRC01, b12, 447-52D, and 2F5.
PBMC were obtained from buffy coats from 10 healthy seronegative blood donors and pooled prior to use. Cells were isolated by Ficoll-Isopaque density gradient centrifugation and then stimulated for 3 days in Iscove's modified Dulbecco medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (100 U/ml), ciproxin (5 μg/ml), and PHA (5 μg/ml) at a cell concentration of 5 × 106/ml. After inoculation, the cells (1 × 106/ml) were grown in the absence of PHA in medium supplemented with recombinant interleukin-2 (20 U/ml; Chiron Benelux, Amsterdam, the Netherlands) and Polybrene (hexadimethrine bromide, 5 μg/ml; Sigma, Zwijndrecht, Netherlands). To prevent possible complement-mediated antibody inhibition of virus infection, complement in human sera and fetal bovine serum was inactivated by a 30-min incubation at 56°C.
From each virus isolate, an inoculum of 20 50% tissue culture infective doses, in a total volume of 50 μl for serum or HIVIG and a total volume of 100 μl for MAbs, was incubated for 1 h at 37°C with decreasing concentrations of serum (range, 1:50 to 1:3,200), HIVIG (range, 23 to 1,500 μg/ml), MAb VRC01 (range, 0.078 to 5 μg/ml), or MAb b12, 2G12, 4E10, 447-52D, or 2F5 (range, 0.03 to 25 μg/ml) in 96-well microtiter plates. Subsequently, 105 PHA-stimulated PBMC were added to the mixtures of virus with serum. After 4 h of incubation with serum or HIVIG, PBMC were washed once in 100 μl phosphate-buffered saline, after which fresh medium was added. On day 7, virus production in culture supernatants was analyzed in an in-house p24 antigen capture enzyme-linked immunosorbent assay (48). Experiments were performed in triplicate. For serum neutralization experiments, background measurements were performed using pooled sera from uninfected individuals. Neutralization sensitivities were expressed as the MAb concentration or reciprocal serum dilution that established 50% inhibition (IC50) of virus infection, as determined by linear regression.
Sequence analyses.
The gp120 sequences of 116 reference viruses, categorized into tiers 1 to 3 (26, 27, 31, 45), were obtained from the Los Alamos Sequence Database (http://www.hiv.lanl.gov/).
envs of clonal HIV-1 variants and chimeric NL4-3 viruses were amplified from DNA that was isolated from in vitro-infected healthy donor PBMC. env PCR products were subsequently sequenced as described previously (3, 5, 39). The nucleotide sequences of all virus clones were aligned using ClustalW in the software package BioEdit (19) and edited manually. The reference sequence HXB2 was included in the alignment to number each aligned residue according to the corresponding position in this reference sequence. Potential N-linked glycosylation sites (PNGS) were identified using the N-glycosite tool at the HIV database website (http://www.hiv.lanl.gov/content/sequence/GLYCOSITE/glycosite.html) (53).
Statistical analyses.
Statistical analyses were performed using the SPSS 16 software package. The differences in amino acid length and number of PNGS between viruses from the three tier groups were evaluated for statistical significance by a Jonckheere-Terpstra test, while differences between viruses from two of the tier groups were assessed by a Student's t test. Correlations between neutralization sensitivity and calendar year of isolation of the tier-categorized reference HIV-1 variants were evaluated for statistical significance by Spearman's correlation. Differences in neutralization sensitivities to HIVIG and MAbs of the V1V2 exchange chimeras compared to the neutralization sensitivities of the corresponding NL4-3/Env chimeras containing the wild-type Env were evaluated for statistical significance by a Wilcoxon signed rank test.
RESULTS
Env characteristics that coincide with neutralization sensitivity in a reference panel of tier-categorized HIV-1 variants with various neutralization sensitivities.
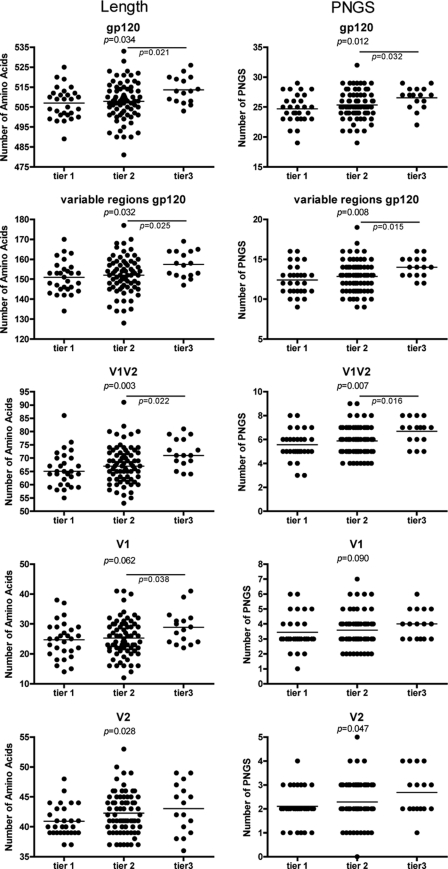
We have previously reported that escape from humoral immunity, both at the individual and at a population level, coincides with an increase in envelope glycoprotein length, mainly of the V1V2 loop, and an increase in the number of potential N-linked glycosylation sites (PNGS) within that same region. Here, we first analyzed whether these molecular differences coincide with HIV-1 neutralization sensitivity in general. To this end, we analyzed the length of Env and number of PNGS in Env in a reference panel of HIV-1 variants that were classified as tier 1 to tier 3 based on their neutralization sensitivities. These reference panels are being used for the assessment of the neutralizing ability of sera and monoclonal antibodies and are supposed to represent genetically and geographically diverse subsets of viruses with neutralization phenotypes that are representative of primary isolates. To better evaluate the neutralizing activity of a serum or monoclonal antibody, a tiered algorithm has been designed in which tier 1 viruses are the most sensitive and tier 3 viruses are the most resistant to antibody neutralization (26, 27, 31, 45). A total of 116 reference viruses that were categorized into tier 1 to tier 3 by Seaman et al. and by Li et al. and Mascola et al. (29 tier 1 viruses, 71 tier 2 viruses, and 16 tier 3 viruses) (26, 27, 31, 45) were analyzed. We found that the length of Env and the number of PNGS in Env were significantly increased in viruses that were more resistant to antibody neutralization. This increase was most obvious between the tier 2 and tier 3 viruses (Fig. 1). These differences in length and number of PNGS could be attributed to the variable but not the constant regions of gp120. Of the variable regions, we only observed a significant correlation between neutralization sensitivity and the length and number of PNGS of the Env V1V2 loops (Fig. 1).
Fig. 1.
Envelope glycoprotein characteristics of tier-categorized reference HIV-1 variants. The numbers of amino acids in the different regions of Env (from top to bottom: gp120, all variable regions, variable regions 1 and 2, variable region 1, and variable region 2) are depicted in the left panels, while the numbers of potential N-linked glycosylation sites (PNGS) are depicted in the right panels. The tier-categorized reference viruses are grouped according to their neutralization sensitivity, with tier 1 being the most neutralization sensitive and tier 3 the most neutralization resistant. Horizontal bars represent the mean values. The associations between numbers of amino acids or PNGS and neutralization resistance between viruses from the three tier groups were evaluated for statistical significance by the Jonckheere-Terpstra test and between two tier groups by Student's t test.
In line with our previous observation that HIV-1 is evolving toward a more neutralization-resistant phenotype over calendar time, we here observed a correlation between the neutralization resistance of the tier-categorized reference viruses and the year in which these viruses were isolated, albeit the correlation was not very strong (Spearman r = 0.369, P = 0.001; data not shown).
Regions in the envelope glycoprotein that are involved in escape from neutralizing activity in autologous serum.
Within a patient, HIV-1 has the ability to adapt to the humoral immune response and to escape from the neutralizing antibodies in serum. Here, we examined the effects of changes in different regions of Env on the sensitivity to neutralization by autologous serum through the introduction of gp41 or the variable region(s) V1, V1V2, V3, or V4 from a neutralization-sensitive virus that was isolated early in infection into the envelope background of a virus variant that was isolated late in infection and that had escaped from the neutralizing activity in serum.
Overall, amino acid sequence variation in Env between these two viruses was 14.5%, with much higher variations of 76.7%, 44.1%, and 38.5% in variable loops V1, V4, and V5, respectively. The V2 loop, V3 loop, and gp41 were more conserved, with 17.9%, 5.4%, and 8.4% difference in amino acid sequence, respectively. The variation in V1 was due to both single amino acid substitutions and an insertion of 7 amino acids in the autologous-neutralization-resistant virus, and these sequence changes also resulted in the introduction of two additional PNGS. Beside these changes, the autologous-neutralization-resistant virus had six additional PNGS in other regions of Env: one in the V2 loop, two in the C2 region, two in the V4 loop, and one in the C4 region. Moreover, the V4 loop in the autologous-neutralization-resistant virus was 2 amino acids longer than that same loop in the autologous-neutralization-sensitive virus.
Chimeric NL4-3/Env viruses that harbored the Env of the autologous-neutralization-resistant virus in which gp41 or one of the variable loop(s) V1, V1V2, V3, or V4 was exchanged with the same region from the autologous-neutralization-sensitive virus were all replication competent. The reciprocal chimeric NL4-3/Env viruses in which the different regions of Env of the autologous-neutralization-resistant virus were placed in the background of Env of the autologous-neutralization-sensitive virus were replication incompetent, despite the correct construction of the chimeric env PCR products as verified by sequencing.
Replacement of the V1 loop or the V1V2 loop of the neutralization escape variant by the shorter, potentially less densely glycosylated loops of the neutralization-sensitive virus resulted in a reversion toward a neutralization-sensitive phenotype to levels comparable with those of the original neutralization-sensitive virus variant (Table 1). Introduction of the V4 loop from the neutralization-sensitive virus into the Env background of the neutralization-resistant virus resulted in only a small increase in sensitivity to neutralization by autologous serum, while the exchange of V3 or gp41 had no effect on neutralization sensitivity. A serum pool from HIV-1-negative individuals was used as a negative control for the neutralization assay and did not show any neutralization of the chimeric NL4-3/Env viruses.
Table 1.
Neutralization sensitivities of original and chimeric NL4-3/Env viruses in which gp41 or gp120 variable regions have been exchanged
| Env in chimeric NL4-3/Env virus | IC50 (1/serum dilution) a |
IC50 (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|---|
| Autologous serum 77 mo post-SC | HIV-negative serum | MAb b12 | MAb 2G12 | MAb 2F5 | MAb 4E10 | MAb 447-52D | HIVIG | |
| Env of sensitive virus | 510 | <50 | 3.4 | 0.6 | 1.9 | 11.0 | >25 | >1,500 |
| Env of resistant virus | <50 | <50 | >25.0 | 1.2 | >25.0 | >25.0 | >25 | >1,500 |
| Env of resistant virus with V1 of sensitive virus | 837 | <50 | >25.0 | 0.1 | >25.0 | 11.3 | >25 | >1,500 |
| Env of resistant virus with V1V2 of sensitive virus | 680 | <50 | 1.7 | 1.0 | >25.0 | >25.0 | >25 | >1,500 |
| Env of resistant virus with V3 of sensitive virus | 71 | <50 | >25.0 | 1.2 | >25.0 | 14.7 | >25 | >1,500 |
| Env of resistant virus with V4 of sensitive virus | 153 | <50 | >25.0 | 2.4 | >25.0 | >25.0 | >25 | >1,500 |
| Env of resistant virus with gp41 of sensitive virus | <50 | <50 | >25.0 | 0.5 | 6.5 | >25.0 | >25 | >1,500 |
IC50, 50% inhibitory concentration; SC, seroconversion.
We also tested the neutralization sensitivities of the chimeric NL4-3/Env viruses to HIVIG (a pool of purified IgG obtained in 1995 from chronically HIV-1-infected individuals) (25) and MAbs b12, 2G12, 2F5, and 4E10. In addition, sensitivity to neutralization by MAb 447-52D was tested, since a change in sensitivity to this V3-directed antibody may point to unintended conformational changes in the chimeric NL4-3/Env viruses. The virus that was sensitive to neutralization by autologous serum was also sensitive to neutralization by MAbs b12, 2G12, 2F5, and 4E10, while the virus that was resistant to neutralization by autologous serum was also resistant to neutralization by all of these antibodies except MAb 2G12. Replacement of the V1V2 loop in the neutralization-resistant escape variant by the V1V2 loop of the neutralization-sensitive virus resulted in a chimeric virus that was sensitive to neutralization by MAb b12. This change in phenotype was not observed when only the V1 loop was replaced, indicating that the V2 loop is involved in the neutralization resistance to MAb b12 of the neutralization-resistant escape variant (Table 1). The virus that was sensitive to neutralization by autologous serum was also sensitive to neutralization by the gp41-directed MAbs 2F5 and 4E10. This phenotype could be transferred to the envelope of the virus that resisted neutralization by autologous serum by exchange of the gp41 region. All original and chimeric NL4-3/Env viruses were resistant to neutralization by MAb 447-52D and HIVIG (Table 1), suggesting that the exchange of gp41 or variable regions did not change the overall Env conformation of these viruses or result in an overall better accessibility of epitopes.
Effect of V1V2 loops on the neutralization sensitivity of HIV-1 over calendar time.
We previously observed that recently transmitted HIV-1 variants from historical seroconverters were more sensitive to neutralizing antibodies than recently transmitted viruses from contemporary seroconverters and that the viruses from contemporary seroconverters had a longer V1V2 loop and more PNGS in this region than recently transmitted viruses from historical seroconverters (6) (Table 2). To determine if these Env characteristics are indeed involved in the neutralization sensitivity of these viruses, we constructed a total of 36 replication-competent chimeric NL4-3/Env viruses in which the V1V2 loop was reciprocally exchanged between 5 recently transmitted viruses from historical seroconverters and 5 recently transmitted viruses from contemporary seroconverters. Our attempt to generate 14 of these chimeric NL4-3/Env viruses failed. This was due either to an inability to generate chimeric env PCR products in which the V1V2 was properly exchanged (n = 4) or to our inability to clone the chimeric env PCR products into the NL4-3 background (n = 10). For accurate evaluation of the viral phenotype, all wild-type (wt) env genes (n = 10) were also cloned into the background of NL4-3.
Table 2.
Neutralization sensitivities and Env characteristics of the HIV-1 variants that were isolated from historical and contemporary seroconvertersa
| Virus and period of isolation | Yr of SC | IC50 (μg/ml) |
Length (aa) |
No. of PNGS |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIVIG | VRC01 | b12 | 2F5 | 447-52D | V1 | V2 | V1V2 | V1 | V2 | V1V2 | ||
| Historical | ||||||||||||
| H19999 | 1985 | 831.3 | 0.121 | 1.12 | >25.00 | >25 | 23 | 39 | 62 | 4 | 2 | 6 |
| H18766 | 1988 | >1,500.0 | 0.094 | >25.00 | 7.29 | >25 | 25 | 38 | 63 | 3 | 2 | 5 |
| H19768 | 1986 | 805.4 | 0.099 | 1.46 | 1.51 | >25 | 25 | 39 | 64 | 3 | 2 | 5 |
| H19342 | 1986 | 846.3 | 0.090 | 1.17 | 2.17 | >25 | 25 | 39 | 64 | 3 | 2 | 5 |
| H19542 | 1985 | 632.9 | 0.524 | 1.41 | 0.69 | >25 | 25 | 42 | 67 | 3 | 3 | 6 |
| Contemporary | ||||||||||||
| P197 | 2005 | 1,009.0 | 0.482 | >25.00 | 0.53 | >25 | 29 | 39 | 68 | 4 | 2 | 6 |
| P180 | 2004 | >1,500.0 | 0.753 | >25.00 | >25.00 | >25 | 27 | 41 | 68 | 5 | 2 | 7 |
| P004 | 2003 | >1,500.0 | 0.345 | >25.00 | 1.76 | >25 | 33 | 39 | 72 | 5 | 1 | 6 |
| P002 | 2003 | 1,316.3 | >5.000 | 5.30 | 0.11 | >25 | 30 | 46 | 76 | 6 | 5 | 11 |
| P127 | 2005 | >1,500.0 | <0.078 | 0.21 | 4.29 | >25 | 38 | 68 | 106 | 4 | 5 | 9 |
| P valueb | 0.042 | 0.283 | 0.204 | 0.883 | 1.000 | 0.008 | 0.230 | 0.088 | 0.006 | 0.380 | 0.043 | |
SC, seroconversion; IC50, 50% inhibitory concentration; aa, amino acid; PNGS, potential N-linked glycosylation site(s).
Differences in neutralization sensitivity and Env characteristics between the two groups of HIV-1 variants from historical and contemporary seroconverters were evaluated for statistical significance using Student's t test.
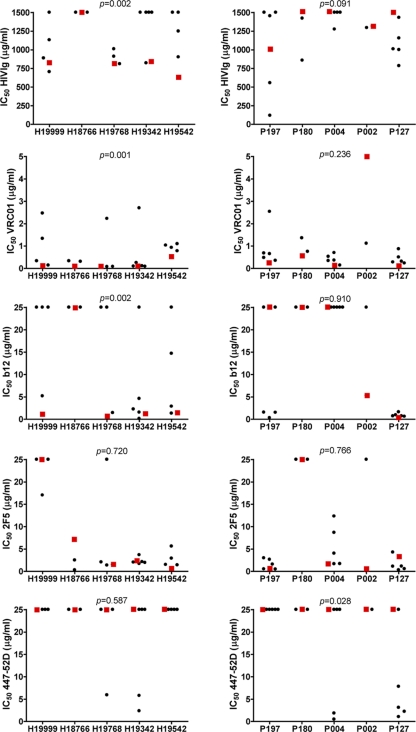
The 36 chimeric NL4-3/Env viruses that were successfully generated, now harboring envelope genes that were chimeric for the V1V2 region, were tested for their sensitivities to neutralization by HIVIG and MAbs b12, VRC01, 2F5, and 447-52D, which were compared to the neutralization sensitivities of the chimeric NL4-3/Env viruses with the Env of the original wt viruses.
With the exception of chimeric NL4-3/Env-wtH18766, all chimeric NL4-3/Env viruses containing wt Envs of HIV-1 from historical seroconverters were sensitive to neutralization by HIVIG, VRC01, and b12 (Table 2). The introduction of a longer V1V2 loop with more PNGS of HIV-1 from contemporary seroconverters into the background of Env of HIV-1 from historical seroconverters increased the levels of resistance to HIVIG neutralization to >1,500 μg/ml (highest antibody concentration tested) for 7 out of 18 chimeric NL4-3/Env viruses and raised the levels of neutralization resistance to a lesser extent for an additional 6 variants (P = 0.002) (Fig. 2). For the remaining 5 chimeric NL4-3/Env viruses, the levels of neutralization sensitivity to HIVIG did not change compared to those of the chimeric NL4-3/Env wt viruses. The introduction of the V1V2 region of HIV-1 from contemporary seroconverters into the background of Env of HIV-1 from historical seroconverters resulted in a >2-fold increase in neutralization resistance to MAb VRC01 for 10 out of 18 viruses (P = 0.001) (Fig. 2) and to MAb b12 for 11 out of 18 viruses (P = 0.002) (Fig. 2). For 6 viruses, increased levels of neutralization resistance were observed for both VRC01 and b12. In line with our previous observation that the neutralization sensitivity of HIV-1 for 2F5 did not change over the course of the epidemic (6), the levels of sensitivity to neutralization by MAb 2F5 were similar for viruses with either wt or chimeric envs. The comparable sensitivities to neutralization by MAb 447-52D of viruses with wt Env or chimeric Env implied that the overall conformation of Env, which may result in enhanced exposure and accessibility of the V3 loop, was intact (Fig. 2).
Fig. 2.
Neutralization sensitivities of the chimeric NL4-3/Env viruses in which the V1V2 loop was exchanged between viruses from historical and contemporary seroconverters. The chimeric NL4-3/Env viruses consisting of Env of an HIV-1 variant from a historical seroconverter with the V1V2 region of an HIV variant from a contemporary seroconverter are depicted in the left panels, and the chimeric NL4-3/Env viruses consisting of Env of an HIV-1 variant from a contemporary seroconverter with the V1V2 region of an HIV variant from a historical seroconverter are depicted in the right panels. IC50s, determined by linear regression, are indicated for HIVIG and MAbs VRC01, b12, 2F5, and 447-52D (from top to bottom), with a red square representing the original wild-type NL4-3/Env virus and each black circle representing a corresponding chimeric NL4-3/Env virus in which the V1V2 loop has been exchanged. Differences in neutralization sensitivities to HIVIG and MAbs of the V1V2 exchange chimeras compared to the neutralization sensitivities of the corresponding NL4-3 chimera containing the wild-type Env were evaluated for statistical significance by a Wilcoxon signed rank test, and P values are shown at the top of each box.
Replacement of V1V2 in HIV-1 of contemporary seroconverters by the V1V2 loop of HIV-1 from historical seroconverters resulted in a trend toward increased neutralization sensitivity to HIVIG (P = 0.091) (Fig. 2) but did not consistently change the neutralization phenotypes for MAbs b12, VRC01, and 2F5 compared to the NL4-3 with the corresponding wt Env from viruses of contemporary seroconverters. We did, however, observe a significant increase in neutralization sensitivity to MAb 447-52D for these viruses (P = 0.028) (Fig. 2), mainly caused by the chimeric NL4-3/Env viruses constructed from wt virus P127.
DISCUSSION
HIV-1 has developed multiple mechanisms to evade neutralizing antibodies. The viral envelope glycoprotein can change dramatically during the course of infection due to the positive selection of escape mutations under the immune pressure of neutralizing antibodies. In particular, the length and glycosylation characteristics of the V1V2 loop seem to play a role in resistance against neutralizing antibodies, possibly by shielding underlying regions of the envelope glycoprotein from antibody recognition (8, 9, 10, 18, 33, 37, 41, 42, 43, 46, 49, 52).
We recently observed that HIV-1 has become more resistant to neutralizing antibodies at a population level over a time course of 20 years (6), which also coincided with an increased length of the V1 loop of the HIV-1 envelope glycoprotein and an increased number of PNGS in that same region. The outcome of that study was seemingly in contrast with a previous report that did not observe a change in the number of PNGS in HIV-1 envelope over the calendar time of the epidemic (53). The facts that we studied HIV-1 evolution over a longer period, that we only compared HIV-1 variants from historical and contemporary seroconverters rather than including HIV-1 variants from individuals who seroconverted during the in-between time period, and that we excluded CXCR4-using HIV-1 variants from the analysis may have contributed to the discrepancy between these results.
Our observations imply a major role of the V1V2 loop of Env in the resistance to antibodies, both at an individual and at a population level (6, 37, 43). This idea is further strengthened by our present analysis of envelope gp120 sequences of reference viruses that are categorized from tier 1 to tier 3 based on their susceptibility to neutralization by antibodies and sera (26, 27, 31, 45). We found that both the length of Env and the number of PNGS in Env were significantly increased in reference viruses that were more resistant to antibody neutralization (tier 3 viruses) compared to those in reference viruses that were more sensitive to neutralization (tier 1 and tier 2 viruses). This association was particularly evident for the V1V2 region of Env. Although the association between neutralization sensitivity and both Env length and the number of PNGS in Env was significant for the tier 1- to 3-categorized reference viruses, the variation in envelope characteristics among viruses from each tier was quite large, suggesting that other regions are likely to play a role in neutralization sensitivity as well. In line with our previous observation that HIV-1 is evolving toward a more neutralization-resistant phenotype over calendar time (6), the neutralization resistance of the tier-categorized reference viruses was also correlated with the year of virus isolation, albeit the correlation was not very strong.
The association between V1V2 loop characteristics and neutralization sensitivity of tier-categorized reference viruses does not provide direct proof for a causal relationship. Results from earlier studies do, however, imply a direct role for the V1V2 loop in the resistance to neutralizing antibodies. Replacement of the V1V2 loop of neutralization-sensitive T cell line-adapted HIV-1 strains or highly neutralization-sensitive tier 1 viruses, such as SF162 (12, 13, 34, 37), with the V1V2 loop of a more neutralization-resistant virus resulted in less neutralization-sensitive chimeric viruses. In our present study, we used primary HIV-1 variants that are representative of the viruses that occur in human infection (16). However, our results with primary viruses are in full agreement with the results of previous studies (20, 24, 37, 41, 43), confirming that the V1V2 loop plays an important role in the escape of HIV-1 from neutralizing antibodies over the course of infection. In the patient we studied longitudinally here, the large increases in the length of the V1V2 loop and in the number of PNGS in this same region are likely to reflect the large selective pressure on this part of the viral envelope glycoprotein. As we did not map the antibody specificities in the serum of this patient, we cannot conclude whether the selection of these changes is driven by direct escape from V1V2-directed antibodies or whether the increased size of the V1V2 loops may protect part of the outer domain of gp120 against antibody recognition. In addition, other regions of Env, such as V4, which have not been investigated in other studies (20, 24, 37, 41, 43), may also contribute to the escape of HIV-1 from neutralizing antibodies, as we observed here that exchange of the V4 loop also influenced neutralization sensitivity.
Our data illustrate that the longer V1V2 loops with more PNGS on viruses from contemporary seroconverters compared to those from historical seroconverters are directly related to a decreased sensitivity to neutralization by HIVIG and CD4-binding-site-directed antibodies b12 and VRC01. This implies that longer, more heavily glycosylated V1V2 loops may occlude the CD4-binding site and prevent the binding of CD4-binding-site antibodies. Given the relatively large proportion of HIV-1-infected individuals that develop CD4-binding-site-directed antibodies (4, 15, 28), it seems likely that the adaptation of HIV-1 toward increased neutralization resistance to this antibody specificity is a direct consequence of selective antibody pressure. However, we cannot exclude other mechanisms that may have influenced this process, such as positive selection for HIV-1 variants that have an increased binding affinity to the CD4 receptor. Indeed, changes in the CD4-binding site resulting from competition for binding to the CD4 receptor may at the same time increase the sensitivity of HIV-1 to neutralization by CD4-binding-site-directed antibodies. In this scenario, the changes we observed in the V1V2 loops may compensate for the increased exposure of the CD4-binding site to neutralizing antibodies. The observation that neutralization sensitivity to gp41-directed antibodies has not changed over the course of the epidemic (6) indicates that gp41-directed antibody specificities are rare or that they do not provide strong selection pressure on the viral envelope glycosylation. Our data do not discriminate whether a particular glycan or region of the V1V2 loop is involved in the protection against CD4-binding-site-directed neutralizing antibodies, nor do they exclude the involvement of other regions of the viral envelope.
The Envs of recently transmitted viruses from five historical seroconverters and the Envs of recently transmitted viruses from five contemporary seroconverters showed substantial sequence differences within the V1V2 region but also in other parts of the envelope glycoprotein. Indeed, sequence diversity in the V4 loop of recently transmitted viruses from historical and contemporary seroconverters was observed at 44% of the amino acid positions. Although we did not observe specific variations between recently transmitted viruses from historical and contemporary seroconverters in this region, the possibility that the mutations in the V4 loop might also have contributed to the increase in neutralization sensitivity cannot be excluded. The observation that replacement of the V1V2 loop in Env of HIV-1 from contemporary seroconverters by the V1V2 loop of viruses from historical seroconverters did not result in a significant difference in neutralization sensitivity, while the replication capacity was unchanged (data not shown), might suggest that other regions or characteristics of Env could also play a role in the increase in neutralization resistance in viruses of contemporary seroconverters.
Despite these unknowns, the results from our study seem to suggest that escape from neutralizing antibodies might be mediated by changes in the variable regions of Env and not necessarily by mutations in the epitopes of the antibodies themselves. This is in line with previous observations that escape from neutralizing antibodies and even cross-reactive neutralizing antibodies, which are most likely directed against highly conserved regions, does not have a major impact on HIV-1's replication capacity (7, 38, 49).
Changing a part of Env, such as the V1V2 loop, might be incompatible with the original conformation of the envelope glycoprotein, which may explain why certain chimeric NL4-3/Env viruses lacked replication competence. Conformational changes of Env may influence the neutralization sensitivity of the virus. It has been shown that deletion of the V1V2 loop can result in a more open conformation of the envelope, with better exposure of the V3 loop, and that even relatively small changes in the V1V2 loop may result in a more open envelope conformation, making the virus more susceptible to anti-V3 antibodies (20, 32, 36). Despite the fact that all of the original viruses had the epitope for V3 antibody 447-52D (55), they were all resistant to its neutralizing activity, indicating that the relevant epitope on the V3 loop was not exposed in these viruses. However, some of the chimeric NL4-3/Env viruses were more sensitive to MAb 447-52D, suggesting that the envelope conformation of these viruses was more open and no longer occluding the V3 loop. This was most obvious for the chimeric NL4-3/Env variants of the original P127 virus, which were much more sensitive to MAb 447-52D than the original virus. The V1V2 region in virus P127 was at least 30 amino acids longer than the V1V2 loop from the other viruses included in this study, which may explain the effect of V1V2 exchange on envelope conformation.
In conclusion, we here demonstrate that the increased length of the HIV-1 envelope V1V2 loop with an increased number of PNGS is directly responsible for the protection of HIV-1 against CD4-binding-site-directed neutralizing antibodies, possibly by shielding underlying epitopes in the envelope glycoprotein from antibody recognition. However, our findings do not exclude the possibility that changes in other regions of the HIV-1 envelope may cause a neutralization-resistant phenotype as well. For vaccine immunogen design, the properties of the V1V2 loop should be taken into account to achieve optimal exposure of certain conserved epitopes. Moreover, as HIV-1 may continue to evolve, it remains to be established whether the epitopes that will be included in a vaccine immunogen will indeed be accessible on HIV-1 variants that will be circulating once a vaccine becomes available.
Supplementary Material
Acknowledgements
This work is financially supported by the Netherlands Organization for Scientific research (NWO), grant number 918.66.628, by the European Community's Sixth Framework Programme Europrise (FP6/2007-2012), grant number 037611, and by the European Community's Seventh Framework Programme NGIN (P7/2007-2013), grant number 201433. The Amsterdam Cohort Studies on HIV Infection and AIDS, a collaboration between the Amsterdam Health Service, the Academic Medical Center of the University of Amsterdam, Sanquin Blood Supply Foundation, the University Medical Center Utrecht, and the Jan van Goyen Clinic, are part of the Netherlands HIV Monitoring Foundation and financially supported by the Center for Infectious Disease Control of the Netherlands National Institute for Public Health and the Environment.
The funding organizations had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank J. Alcami (Instituto de Salude Carlos III, Spain) for his kind gift of the pNL4-3 vector, D. Burton and A. Hessell (The Scripps Research Institute, La Jolla, CA) for their generous supply of IgG1b12, D. Katinger (Polymun, Austria; as a partner in EUROPRISE) for his supply of MAbs 2G12, 2F5, and 4E10, J. Mascola (Vaccine Research Center, NIH, Bethesda, MD) for his supply of VRC01, and S. Zolla-Pazner (New York University Medical Center, New York, NY) for MAb 447-52D. The HIVIG was obtained from North American Biologicals and the National Heart, Lung, and Blood Institute through the U.S. National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Albert J., et al. 1990. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS 4:107–112 [DOI] [PubMed] [Google Scholar]
- 2. Beaumont T., Quakkelaar E., van Nuenen A., Pantophlet R., Schuitemaker H. 2004. Increased sensitivity to CD4 binding site-directed neutralization following in vitro propagation on primary lymphocytes of a neutralization-resistant human immunodeficiency virus IIIB strain isolated from an accidentally infected laboratory worker. J. Virol. 78:5651–5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beaumont T., et al. 2001. Reversal of HIV-1 IIIB towards a neutralization resistant phenotype in an accidentally infected laboratory worker with a progressive clinical course. J. Virol. 75:2246–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Binley J. M., et al. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boom R., et al. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bunnik E. M., et al. 2010. Adaptation of HIV-1 envelope gp120 to humoral immunity at a population level. Nat. Med. 16:995–997 [DOI] [PubMed] [Google Scholar]
- 7. Bunnik E. M., Lobbrecht M. S. D., van Nuenen A. C., Schuitemaker H. 2010. Escape from autologous humoral immunity of HIV-1 is not associated with a decrease in replicative capacity. Virology 397:224–230 [DOI] [PubMed] [Google Scholar]
- 8. Bunnik E. M., Pisas L., van Nuenen A. C., Schuitemaker H. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J. Virol. 82:7932–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cao J., et al. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808–9812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chackerian B., Rudensey L. M., Overbaugh J. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719–7727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen B., et al. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834–841 [DOI] [PubMed] [Google Scholar]
- 12. Ching L., Stamatatos L. 2010. Alterations in the immunogenic properties of soluble trimeric human immunodeficiency virus type 1 envelope proteins induced by deletion or heterologous substitutions of the V1 loop. J. Virol. 84:9932–9946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ching L. K., Vlachogiannis G., Bosch K. A., Stamatatos L. 2008. The first hypervariable region of the gp120 Env glycoprotein defines the neutralizing susceptibility of heterologous human immunodeficiency virus type 1 isolates to neutralizing antibodies elicited by the SF162gp140 immunogen. J. Virol. 82:949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Decker J. M., et al. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dhillon A. K., et al. 2007. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J. Virol. 81:6548–6562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Edo-Matas D., et al. 2010. Genetic composition of replication competent clonal HIV-1 variants isolated from peripheral blood mononuclear cells (PBMC), HIV-1 proviral DNA from PBMC and HIV-1 RNA in serum in the course of HIV-1 infection. Virology 405:492–504 [DOI] [PubMed] [Google Scholar]
- 17. Edwards T. G., et al. 2001. Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J. Virol. 75:5230–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a. Euler Z., et al. 2011. Activity of broadly neutralizing antibodies, including PG9, PG16, and VRC01, against recently transmitted subtype B HIV-1 variants from early and late in the epidemic. J. Virol. 85: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gray E. S., et al. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81:6187–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 20. Krachmarov C. P., et al. 2006. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J. Virol. 80:7127–7135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kwong P. D., et al. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682 [DOI] [PubMed] [Google Scholar]
- 22. Kwong P. D., et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Labrijn A. F., et al. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laird M. E., Igarashi T., Martin M. A., Desrosiers R. C. 2008. Importance of the V1/V2 loop region of simian-human immunodeficiency virus envelope glycoprotein gp120 in determining the strain specificity of the neutralizing antibody response. J. Virol. 82:11054–11065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lambert J. S., et al. 1997. Safety and pharmacokinetics of hyperimmune anti-human immunodeficiency virus (HIV) immunoglobulin administered to HIV-infected pregnant women and their newborns. Pediatric AIDS Clinical Trials Group Protocol 185 Pharmacokinetic Study Group. J. Infect. Dis. 175:283–291 [DOI] [PubMed] [Google Scholar]
- 26. Li M., et al. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li M., et al. 2006. Genetic and neutralization properties of acute and early subtype C human immunodeficiency virus type 1 molecular env clones from heterosexually acquired infections in southern Africa. J. Virol. 80:11776–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Y., et al. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13:1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu J., Bartesaghi A., Borgnia M. J., Sapiro G., Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ly A., Stamatatos L. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mascola J. R., et al. 2005. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J. Virol. 79:10103–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moore P. L., et al. 2008. The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J. Virol. 82:1860–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moore P. L., et al. 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morikita T., et al. 1997. The V1/V2 region of human immunodeficiency virus type 1 modulates the sensitivity to neutralization by soluble CD4 and cellular tropism. AIDS Res. Hum. Retrovir. 13:1291–1299 [DOI] [PubMed] [Google Scholar]
- 35. Pilgrim A. K., et al. 1997. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term-nonprogressive infection. J. Infect. Dis. 176:924–932 [DOI] [PubMed] [Google Scholar]
- 36. Pinter A. 2007. Roles of HIV-1 Env variable regions in viral neutralization and vaccine development. Curr. HIV Res. 5:542–553 [DOI] [PubMed] [Google Scholar]
- 37. Pinter A., et al. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Quakkelaar E. D., et al. 2007. Escape of human immunodeficiency virus type 1 from broadly neutralizing antibodies is not associated with a reduction of viral replicative capacity in vitro. Virology 363:447–453 [DOI] [PubMed] [Google Scholar]
- 39. Quakkelaar E. D., et al. 2007. Susceptibility of recently transmitted subtype B human immunodeficiency virus type 1 variants to broadly neutralizing antibodies. J. Virol. 81:8533–8542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Richman D. D., Wrin T., Little S. J., Petropoulos C. J. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rong R., et al. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J. Virol. 81:1350–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rong R., et al. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 5:e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sagar M., Wu X., Lee S., Overbaugh J. 2006. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J. Virol. 80:9586–9598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schuitemaker H., et al. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J. Virol. 66:1354–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Seaman M. S., et al. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stamatatos L., Cheng-Mayer C. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72:7840–7845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Starcich B. R., et al. 1986. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45:637–648 [DOI] [PubMed] [Google Scholar]
- 48. Tersmette M., et al. 1989. Detection and subtyping of HIV-1 isolates with a panel of characterized monoclonal antibodies to HIV-p24 gag. Virology 171:149–155 [DOI] [PubMed] [Google Scholar]
- 49. van Gils M. J., et al. 2010. Rapid escape from preserved cross-reactive neutralizing humoral immunity without loss of viral fitness in HIV-1-infected progressors and long-term nonprogressors. J. Virol. 84:3576–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Van 't Wout A. B., et al. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral and vertical transmission. J. Clin. Invest. 94:2060–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wei X., et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 52. Wyatt R., et al. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang M., et al. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246 [DOI] [PubMed] [Google Scholar]
- 54. Zhou T., et al. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zolla-Pazner S., et al. 2004. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res. Hum. Retrovir. 20:1254–1258 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.