Abstract
Although live-attenuated influenza vaccines (LAIV) are safe for use in protection against seasonal influenza strains, concerns regarding their potential to reassort with wild-type virus strains have been voiced. LAIVs have been demonstrated to induce enhanced mucosal and cell-mediated immunity better than inactivated vaccines while also requiring a smaller dose to achieve a protective immune response. To address the need for a reassortment-incompetent live influenza A virus vaccine, we have designed a chimeric virus that takes advantage of the fact that influenza A and B viruses do not reassort. Our novel vaccine prototype uses an attenuated influenza B virus that has been manipulated to express the ectodomain of the influenza A hemagglutinin protein, the major target for eliciting neutralizing antibodies. The hemagglutinin RNA segment is modified such that it contains influenza B packaging signals, and therefore it cannot be incorporated into a wild-type influenza A virus. We have applied our strategy to different influenza A virus subtypes and generated chimeric B/PR8 HA (H1), HK68 (H3), and VN (H5) viruses. All recombinant viruses were attenuated both in vitro and in vivo, and immunization with these recombinant viruses protected mice against lethal influenza A virus infection. Overall, our data indicate that the chimeric live-attenuated influenza B viruses expressing the modified influenza A hemagglutinin are effective LAIVs.
INTRODUCTION
Influenza A viruses (IAV), members of the Orthomyxoviridae family of negative-strand RNA viruses, are formidable pathogens, causing significant morbidity and mortality worldwide (18). Seasonal epidemics caused by slight permutations in the antigenic regions of the viral surface proteins kill thousands of people each year in the United States (18). Pandemic-scale influenza A virus disease occurs when the reassortment of genomic segments between different virus subtypes results in a virus that is virulent in humans and that is antigenically novel in the human population (18). During the last 100 years, there have been four global pandemics: the 1918 Spanish pandemic (H1N1 subtype), the 1957 Asian pandemic (H2N2 subtype), the 1968 Hong Kong pandemic (H3N2subtype), and the 2009 swine-origin flu (H1N1 subtype). Among those, the most devastating outbreak was the 1918 pandemic, which claimed the lives of an estimated 100 million people worldwide (1). Sixteen different influenza A virus hemagglutinin (HA) subtypes have been identified (7, 29); however, most modern humans have been exposed to only two to three of those subtypes (H1, H2, and H3); this limited immune exposure is concerning, since a virus from any of the other 13 to 14 subtypes that gained the ability to transmit and cause disease in humans would not be controlled by existing human immunity (11). The severe disease in humans caused by sporadic primary transmission events of H5 highly pathogenic influenza A viruses (HPAI) highlights the threat that novel subtypes pose to the human population (3, 23).
Vaccination is still the best means of protection from influenza virus infections (17, 19). There are presently two types of FDA-approved influenza vaccines: an inactivated vaccine and a cold-adapted live-attenuated vaccine. Although the inactivated form has long been the mainstay weapon in the fight against influenza, the live-attenuated influenza vaccine (LAIV) does have several advantages over the inactivated vaccine. The intranasal delivery of the LAIV can induce a broader immunologic response, engaging both enhanced mucosal and cellular immunity. While providing effective and possibly enhanced immunity against influenza disease, serious concerns exist regarding the administration of LAIVs in prepandemic circumstances. Although the risk is low, one cannot exclude the possibility that an attenuated vaccine strain, containing a novel hemagglutinin gene, will recombine with a wild-type virus to generate a fully virulent virus expressing the hemagglutinin from the vaccine strain. Consequently, even though multiple LAIV candidates have been generated and demonstrated to have protective efficacy (4, 13, 15, 22, 24, 28), these live vaccines are not likely to be used in prepandemic situations.
To address this safety concern, we have designed a system for generating reassortment-incapable virus strains that could be used as LAIVs in prepandemic circumstances. Our system is based on a chimeric influenza B/A approach in which a recombinant and attenuated influenza B virus (rIBV) is rescued that expresses a modified ectodomain of the influenza A hemagglutinin (see Fig. 1A and 6A). Previous studies have demonstrated the feasibility of generating recombinant influenza A viruses expressing the hemagglutinin of a B virus (5, 10); there is no known reassortment capability between influenza A and B viruses (8, 12, 14, 26). We have generated three reassortment-incapable viruses using this influenza A/B chimera strategy in an attenuated influenza B virus background. Our influenza B-based vaccine strains express the hemagglutinin from either the H1 or H3 hemagglutinin or the hemagglutinin from a prototypic H5 subtype influenza A virus.
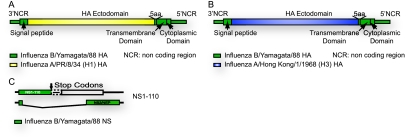
Fig. 1.
Schematic design of HA and nonstructural gene (NS) segments used to generate chimeric recombinant influenza B/Ya88 viruses (rIBVs). Diagram of the rIBV expressing the A/PR/8/34 (H1) HA ectodomain (A) and the A/Hong Kong/1/1968 (H3) HA ectodomain (B). The majority of the HA ectodomain was from the influenza A virus, whereas the rest of the HA protein was from the influenza B virus. (C) The truncated form of NS1-110 was generated by the introduction of two stop codons (indicated by the arrow) after the codons corresponding to amino acid 80 or 110 and a deletion of 100 nucleotides (indicated by a dotted box). NCR, noncoding region.
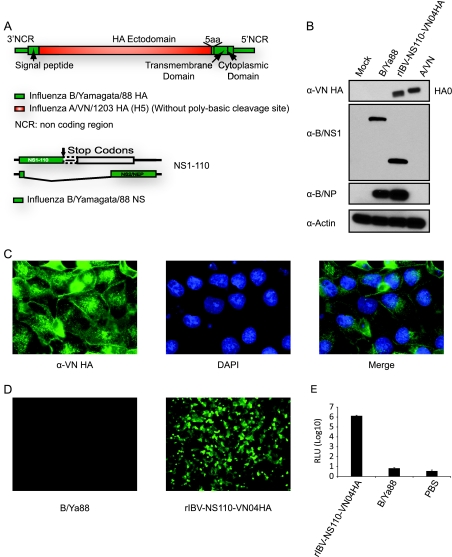
Fig. 6.
Characterization of recombinant influenza B/Yamagata/88 viruses expressing VN H5 HA ectodomain (rIBV-NS110-VN04HA). (A) Schematic design of HA and NS segments used to generate rIBV-NS110-VN04HA. The stable expression of chimeric hemagglutinin during infection was monitored by Western blotting (B) and immunofluorescence analysis (C). Extracts from MDCK cells mock infected or infected with the indicated viruses (15 hpi) were probed with specific antibodies against VN HA (M08) (α-VN HA), B/NS1 (polyclonal antibody) (α-B/NS1), and B/NP (B017) (α-B/NP) to monitor viral infection, and β-actin (α-Actin) was used as an internal loading control. (C) Immunofluorescence analysis was performed using a VN HA-specific monoclonal antibody. Nuclei were stained with DAPI. MDCK pIFNβ cells were infected with the recombinant B viruses or mock-treated with PBS to evaluate IFN-α/β induction by rIBV-NS110-VN04HA. Fifteen hours postinfection, the activation of the IFN-β promoter was determined by assessing the GFP expression level (D) and firefly luciferase activity (E).
The recombinant viruses generated demonstrate attenuated growth in vitro and are not pathogenic in mice. Further, the immunization of mice with the chimeric A/B viruses provided protective immunity against disease caused by the homologous influenza A virus. We suggest that a robust human vaccine against H5 or other potential pandemic subtypes (16) can be generated using this novel replication-incompetent live-attenuated vaccine system.
MATERIALS AND METHODS
Cells and viruses.
293T and Madin-Darby canine kidney (MDCK) cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (DMEM) and minimal essential medium (both from Gibco, Carlsbad, CA), respectively, supplemented with 10% fetal calf serum (HyClone, Logan, UT) and 1% penicillin-streptomycin (Gibco). The MDCK cell line, expressing both green fluorescent protein-chloramphenicol acetyltransferase (GFP-CAT) and firefly luciferase under the control of the beta interferon (IFN-β) promoter (MDCK pIFNβ), was maintained in the medium containing 1 mg of hygromycin B/ml and 2 mg of Geneticin/ml (9).
All recombinant influenza B viruses used in these experiments were propagated in 8-day-old embryonated chicken eggs for 3 days at 33°C (9). All recombinant influenza A viruses were propagated in 8-day-old embryonated chicken eggs for 2 days at 37°C.
Construction of plasmids.
The reverse-genetic plasmids used for generating influenza B viruses were constructed in our previous study (9). The plasmids encoding the chimeric A/B HAs were derivatives of the corresponding wild-type (WT) A/HA segment. Briefly, the chimeric pDZ-B/VN HA was constructed by swapping the A/VN/1203 hemagglutinin (without a polybasic cleavage site) into the B/HA sequence using QuikChange XL site-directed mutagenesis (see Fig. 6A). A similar strategy was applied to construct the plasmids for pDZ-B/PR8 HA and pDZ-B/HK68 HA (Fig. 1A and B).
Rescue of recombinant chimeric IBVs.
Rescue of influenza B viruses from plasmid DNA was performed as previously described (6, 9). Briefly, for the generation of recombinant B viruses, 293T-MDCK cell cocultures were cotransfected with 1 μg of each of the eight plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). At 12 h posttransfection, the medium was replaced with Dulbecco's modified essential medium (DMEM) containing 0.3% bovine serum albumin (BSA), 10 mM HEPES, and 1.5 μg/ml TPCK (l-1-tosylamide-2-phenylethyl chloromethyl ketone)-treated trypsin. At 3 days posttransfection, virus-containing supernatant was inoculated into 8-day-old embryonated chicken eggs. Allantoic fluid was harvested after 3 days of incubation at 33°C and assayed for the presence of virus by the hemagglutination of chicken red blood cells and by plaque formation in MDCK cells.
Growth kinetics of recombinant viruses in MDCK cells.
To analyze viral replication, confluent IFN-competent MDCK cells were infected at a multiplicity of infection (MOI) of 0.05 and incubated at 33°C in minimal essential medium containing 0.3% BSA and 1.5-μg/ml TPCK-treated trypsin. Viral titers in supernatants were determined by plaque assay on MDCK cells.
Western blotting and indirect immunofluorescence analysis.
One well of a six-well dish of confluent MDCK cells was infected (MOI of 2) with the indicated recombinant influenza viruses or mock infected with phosphate-buffered saline (PBS) for 1 h at 33°C. At 15 h postinfection (hpi), cells were lysed in 1× protein loading buffer as described previously (9). The reduced cell lysates were subjected to Western blot analysis by using monoclonal antibody against A/PR8/HA (PY102), A/HK68/HA (12D1), A/VN04/HA (M08), and B/NP (B017), as well as polyclonal antibody against NS1 or monoclonal anti-actin (Sigma, St. Louis, MO). The final Western blotting bands were visualized using an enhanced chemiluminescence protein detection system (PerkinElmer Life Sciences, Boston, MA).
For immunofluorescence analysis, confluent monolayers of MDCK cells on 15-mm coverslips were infected with recombinant viruses at an MOI of 2. At 15 hpi, cells were fixed and permeabilized by treatment with methanol-acetone (ratio, 1:1) at −20°C for 20 min. After being blocked with 1% bovine serum albumin in PBS containing 0.1% Tween 20, cells were incubated for 1 h with a monoclonal antibody directed against A/PR8/HA (PY102), A/HK68/HA (12D1), and A/VN04/HA (M08) as mentioned above. After three washes with PBS containing 0.1% Tween 20, cells were incubated for 1 h with Alexa Fluor 594-conjugated anti-mouse immunoglobulin G (IgG; Invitrogen, Carlsbad, CA). After one additional wash, cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI; Invitrogen, Carlsbad, CA). Following the final two washes, infected cells were analyzed by fluorescence microscopy with an Olympus IX70 microscope.
Bioassay to measure IFN production.
We used the MDCK pIFNβ cell line to evaluate the levels of IFN produced in cells infected with influenza B viruses. The experimental procedure was adapted from our former study (9). Briefly, six-well dishes of confluent MDCK pIFNβ cells were infected at an MOI of 2 with different recombinant B viruses for 12 h, with PBS as the mock infection control. The expression of GFP was visualized by fluorescence microscopy with an Olympus IX70 microscope. Cell lysates were prepared for luciferase assay.
Mouse immunization and challenge.
Eight-week-old female BALB/c mice (Jackson Laboratory, Bar Harbor, ME) were anesthetized with a mixture of ketamine and xylazine administered intraperitoneally and infected intranasally with 10-fold serial dilutions of different rIBVs (in 50-μl volumes) as indicated below. To determine lung virus titers, mice were euthanized at day 3 or 6 postinfection. Lungs were homogenized and resuspended in 1 ml sterile PBS containing 0.3% BSA, and the titers were evaluated on MDCK cells.
Three weeks after immunization, mice were challenged by intranasal infection with the influenza A/PR8 H1N1 virus at 1 × 103 PFU (pathology) or 1 × 102 PFU (virus lung replication), the influenza A/PR8 HK68:HA/NA H3N2 virus at 1 × 106 PFU (pathology) or 5 × 104 PFU (virus lung replication), or the influenza A/PR8 VN:HA/NA H5N1 virus at 5 × 103 PFU (pathology) or 1 × 103 PFU (virus lung titer).
Eight-week-old female C57BL/6 PKR−/− mice were anesthetized and infected intranasally with 10-fold serial dilutions of the recombinant B/VN HA NS1-110 chimeric viruses. Three weeks postimmunization, mice were challenged by intranasal infection with influenza B/Ya88 virus at 1 × 106 PFU.
All animal procedures performed in this study were in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines and have been approved by the IACUC of the Mount Sinai School of Medicine.
Passive immunization and challenge.
Eight-week-old BALB/c mice were passively immunized with 200 μl of polyclonal sera from mice previously vaccinated with different recombinant viruses and then lethally challenged with influenza A viruses 24 h later. Survival and body weight loss were monitored for 14 days postchallenge.
Assessment of pathogenicity and infectivity for chickens.
The ability of the recombinant wild-type (rWT) B/Ya88 and rIBV-VN04HA to infect or cause disease in chickens was assessed by intranasal and intravenous inoculation studies, respectively, in 4-week-old specific-pathogen-free White Plymouth Rock chickens as previously described (24).
ELISAs and enzyme-linked immunospot (ELISpot) assays.
To assess the levels of virus-specific antibodies present in immunized mice, enzyme-linked immunosorbent assays (ELISAs) were performed on diluted serum samples and nasal and lung washes, as described earlier (9). In brief, serum was obtained from mice right before viral challenge and stored at −80°C. We coated 96-well ELISA plates (Immulon4; Dynex, Chantilly, VA) with 50 μl (10 μg/ml) of the appropriate influenza A viruses. After being washed with PBS, coated wells were blocked with PBS containing 1% BSA and then incubated with diluted serum. After 1 h of incubation at room temperature, wells were rinsed with PBS and incubated with a secondary anti-mouse IgG conjugated to peroxidase (Invitrogen, Carlsbad, CA). Rinsed wells were incubated with colorimetric substrate (4-nitrophenyl phosphate; Invitrogen, Carlsbad, CA) for 30 min and read with a plate reader that measured the optical density at 405 nm (OD405; DTX880 multimode detector; Beckman Coulter).
To detect antigen-specific cytotoxic T lymphocytes, we used the ELISpot assay to measure the number of IFN-γ-producing spleen cells in response to stimulation with influenza B/Ya88 viral peptide presented on major histocompatibility complex (MHC) class I, as described previously (25). The assay was performed using an ELISpot kit (R&D Systems). Pooled splenocytes of immunized mice were counted and incubated with anti-CD8 beads (Miltenyi Biotec) in buffer (PBS, pH 7.2, 0.5% BSA, and 2 mM EDTA; degassed for 15 min). CD8+ T cells were purified by adherence and elution from LS MACS columns (Miltenyi Biotec) and then counted. Splenocytes from naïve mice (including antigen-presenting cells) were infected with the rWT at an MOI of 5 for 2 h in a volume of 100 μl, treated with 6 μl of 0.5-mg/ml mitomycin C (Sigma) to stop cell growth, and used to stimulate CD8+ T cells from vaccinated mice. Infected splenocytes (2 × 105) from naïve mice were added to each well. Purified CD8+ T cells (1 × 106, 5 × 105, and 2.5 × 105) from vaccinated mice were added to wells in triplicate. The plate was read using an ELISpot plate reader (Cellular Technology, Ltd.).
RESULTS
Generation of an attenuated recombinant influenza B virus expressing either the PR8 or HK68 hemagglutinin.
The truncation of the NS1 protein in both influenza A and B viruses has been shown to significantly attenuate virus growth; NS1-truncated influenza viruses have been developed as candidate live-attenuated influenza vaccines (9, 21, 22). The recombinant influenza B virus with a truncated NS1 protein with a 110-amino-acid final length is currently a promising influenza B vaccine candidate, since it can grow to high titers in vitro and has proven safe and protective (9). Using this recombinant influenza B virus possessing a truncated NS1 (rIBV-NS110), we constructed two novel vaccine strains comprising the rIBV-NS110 virus expressing the hemagglutinin ectodomain of two influenza A viruses, A/Puerto Rico/8/34 (rIBV-NS110-PR8HA, H1) and A/Hong Kong/68 (rIBV-NS110-HK68HA, H3).
The segments coding for hemagglutinin in these chimeric viruses retain the signal peptide sequence and the transmembrane and cytoplasmic domains of B/Yamagata/16/88. Of note, we also replaced the last five amino acids of the A virus hemagglutinin ectodomain with those of the sequence from the B virus hemagglutinin. The resulting recombinant viruses are composed of seven genomic segments from B/Yamagata/88 (with a truncated NS1) plus a chimeric eighth segment coding for the influenza A hemagglutinin while retaining regions of the influenza B hemagglutinin segment required for packaging (Fig. 1).
Recombinant influenza B viruses express both truncated NS1 and influenza A hemagglutinin.
To determine whether the recombinant viruses express the corresponding influenza A hemagglutinin and the truncated NS1-110 protein, we infected MDCK cells with rIBV-NS110-PR8HA or rIBV-NS110-HK68HA. Mock-infected cells were used as a negative control. At 15 hpi, cells were lysed, the lysates were resolved on an SDS-PAGE gel, and viral proteins were detected by Western blot analysis (Fig. 2A and B). As shown in Fig. 2A, both rIBV-NS110 viruses express the truncated NS1 protein at the expected molecular weight using a polyclonal antibody against the aminoterminal portion of NS1, whereas the rWT B virus expresses the full-length NS1. In addition, both recombinant chimeric viruses express their corresponding influenza A hemagglutinin, with rIBV-NS110-PR8 and rIBV-NS110-HK68 expressing the PR8 hemagglutinin and the HK68 hemagglutinin, respectively. The expression of PR8 hemagglutinin and HK68 hemagglutinin in infected MDCK cells also was analyzed and detected by indirect immunofluorescence, further indicating that the influenza A HAs are indeed expressed in rIBV-NS110-AHA-infected cells (Fig. 2C).
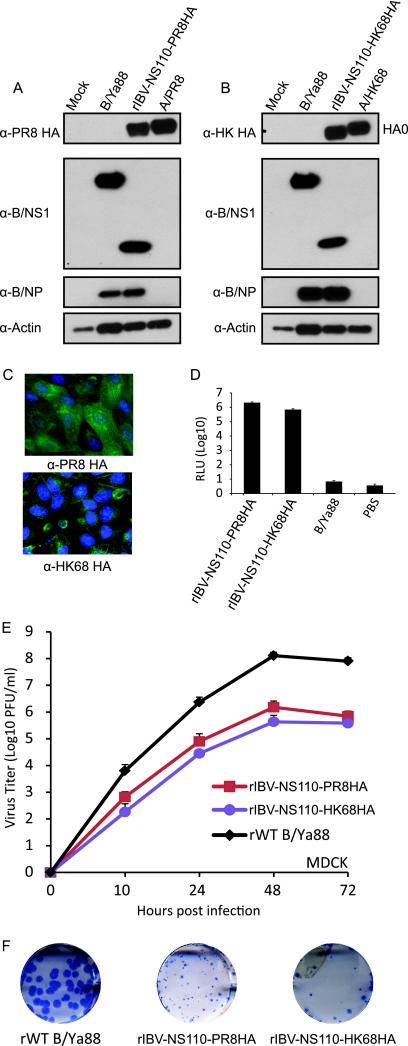
Fig. 2.
Characterization of recombinant chimeric influenza B/Yamagata/88 viruses. Western blot analysis of the rIBV-NS110-PR8HA (A) and rIBV-NS110-HK68HA (B). Extracts from MDCK cells mock infected or infected with the indicated viruses (15 hpi) were probed with specific antibodies against PR8 HA (PY102) (α-PR8 HA), HK68 HA (12D1) (α-HK68 HA), B/NS1 (polyclonal antibody) (α-B/NS1), and B/NP (B017) (α-B/NP) to monitor viral infection, and β-actin (α-Actin) was used as an internal loading control. (C) The stable expression of the chimeric hemagglutinin during infection was further assessed by immunofluorescence analysis using either a PR8 HA-specific or an HK68 HA-specific monoclonal antibody. Nuclei were stained with DAPI. IFN-α/β induction by B viruses was evaluated using the MDCK pIFNβ cell line, which expresses the firefly luciferase (FF-Lucif) gene under the control of the IFN-β promoter (9). (D) The cells were infected with the recombinant influenza B viruses. Fifteen hours postinfection, the activation of the IFN-β promoter was determined by assessing firefly luciferase activity. (E) Multicycle growth curves of the recombinant viruses in MDCK cells infected at an MOI of 0.05 and titrated by plaque assay on MDCK cells. (F) Plaque size phenotypes of the chimeric B NS1-110 viruses in MDCK cells at 3 days p.i.
The efficient induction of interferons (IFNs) during viral replication is a hallmark of NS1 truncation mutant viruses of both influenza A and B origins (9, 21). To characterize the IFN induction by rIBV-NS110-AHA viruses, we used the MDCK pIFNβ cell line. MDCK pIFNβ cells were infected with rWT B virus or the two rIBV-NS110-AHA viruses at an MOI of 2, and PBS was used as a negative control. At 15 hpi, we observed significantly higher GFP signals in rIBV-NS110-AHA-infected cells than in either the rWT B virus- or mock-infected cells (data not shown). In addition, rIBV-NS110-AHA infection induced 5-log-higher levels of luciferase activity than those induced by the wild-type B virus and mock infection (Fig. 2D).
rIBV-NS110-AHA viruses are attenuated in vitro and in vivo.
Safety is the primary concern in determining whether chimeric rIBV-NS110 viruses can be used as live-attenuated vaccines. To address this, we first investigated the multistep replication kinetics of rIBV-NS110-AHA in MDCK cells. MDCK cells were infected at an MOI of 0.05, and the virus present in supernatants from infected cells was measured at different time points postinfection by plaque assay on MDCK cells. The growth characteristics of the rIBV-NS110-AHA viruses were different from those of the rWT B virus (Fig. 2E). The peak titer of the rIBV-NS110-PR8HA virus was close to 2 logs lower than that of the rWT B virus, while the peak titer of the rIBV-NS110-HK68HA virus was even lower (Fig. 2E). These observations are consistent with those in our previous study of the influenza B NS1-110 mutant virus (9). In addition, the plaque sizes of the two rIBV-NS110-AHA viruses also were smaller than that of the rWT B virus (Fig. 2F). These results demonstrate that the two recombinant viruses are attenuated in vitro.
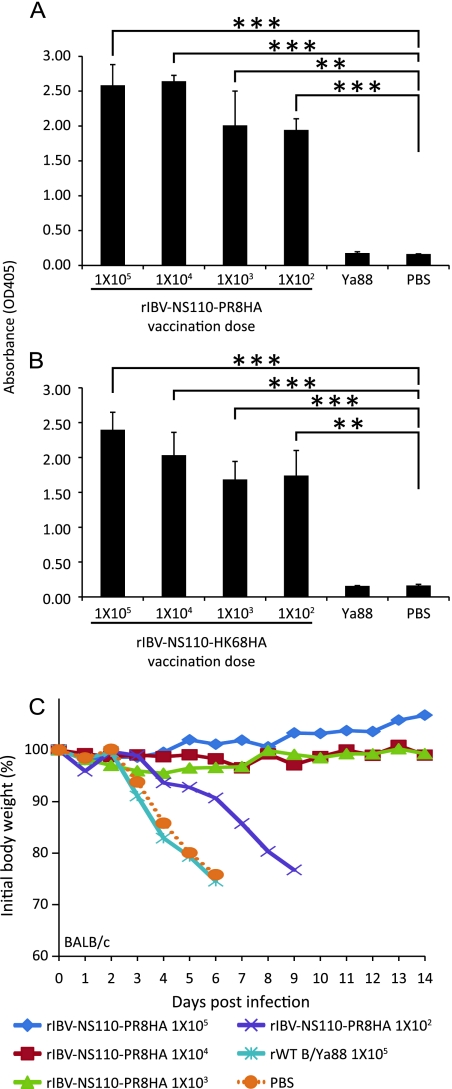
We next investigated the attenuation of rIBV-NS110-AHA in vivo by measuring viral replication in lungs, along with weight loss and survival rates in mice. To evaluate pulmonary viral replication, BALB/c mice were infected with 1 × 105 PFU of recombinant B viruses. As shown in Fig. 3, rIBV-NS110-AHA virus replication in the lungs was 2 to 3 logs lower than that of the rWT B virus at 3 days postinfection. In addition, rIBV-NS110-AHA viruses were cleared more rapidly and were undetectable in the lungs 6 days postinfection. Weight loss is an important index for evaluating influenza virus pathogenicity in vivo. We infected BALB/c mice (n = 4) intranasally with 10-fold serial dilutions of each recombinant virus (1 × 105 to 1 × 102 PFU) and measured body weight daily for 14 days postinfection. Only mice infected with 1 × 105 PFU of rIBV-NS110-PR8HA lost about 15% of their initial weight at 7 days postinfection, but they eventually regained the weight. The weight loss likely is due to the specific mouse-adapted character of the PR8 hemagglutinin. In summary, both rIBV-NS110-AHAs have an attenuated phenotype in vitro and in vivo.
Fig. 3.
Pathogenicity of chimeric rIBV-NS110-PR8HA or rIBV-NS110-HK68HA in BALB/c mice. (A) Pulmonary replication of chimeric virus in mice. Average lung titers ± standard deviations are depicted. The limit of detection was 5 PFU. Also shown is the percent change in weight following intranasal (i.n.) infection with 10-fold serial dilutions of rIBV-NS110-PR8HA (B) or rIBV-NS110-HK68HA (C), from 1 × 105 PFU to 1 × 102 PFU. Control mice were intranasally infected with 1 × 105 PFU of rWT B/Ya88 virus or mock treated with PBS.
Vaccination with rIBV-NS110-AHA elicits an antibody response in BALB/c mice.
Antibodies that target the influenza virus hemagglutinin protein are the main protective mechanism against influenza infection and therefore are a critical component of a successful vaccine. To analyze the efficiency of rIBV-NS110-AHAs in inducing a protective humoral response, we evaluated the antibody response following vaccination. Mice were immunized with rIBV-NS110-PR8HA, rIBV-NS110-HK68HA, or rWT B virus, and antibodies in the sera were analyzed by ELISA 21 days postvaccination (Fig. 4). Immunization with rIBV-NS110-PR8HA or rIBV-NS110-HK68HA induced virus HA-specific IgG antibodies in the sera of vaccinated mice (Fig. 4A and B). To further evaluate whether the antibodies could confer protection in vivo, we immunized mice with sera from previously vaccinated animals and found that sera from the three highest vaccine dose groups conferred full protection upon challenge with the same dose (1 × 103 PFU) of PR8 virus (Fig. 4C). Sera from the lowest vaccine dose (1 × 102 PFU) could not protect mice from death but did prolong their survival by 3 days compared to that of the control polyclonal sera treatment groups (Fig. 4C).
Fig. 4.
Mice immunized with chimeric viruses generate influenza A virus-specific antibodies. Eight-week-old female BALB/c mice (n = 3) were immunized with the indicated viruses or with rIBV and PBS as negative controls. Serum samples were collected 21 days postvaccination. Specific IgG antibodies against A/PR8 virus (A) or A/X31 virus (B) were detected by ELISA as described in Materials and Methods. Eight-week-old female BALB/c mice (n = 4) were passively immunized with a total 200 μl of the indicated polyclonal sera (intraperitoneal route) 24 h prior to viral challenge. After antibody administration, the mice were challenged with 1,000 PFU PR8 virus. (C) Body weight of passively immunized mice challenged with PR8 virus. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 versus PBS control).
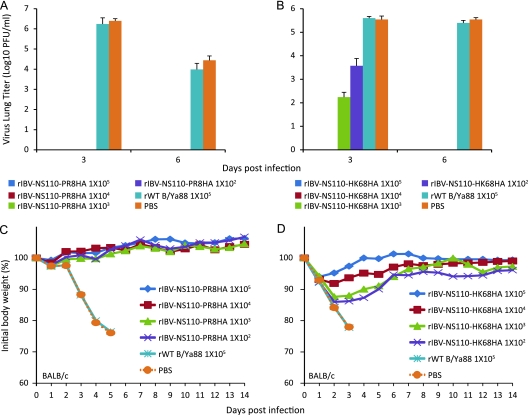
rIBV-NS110-AHA protects against lethal influenza A virus challenge in BALB/c mice.
To determine whether robust antibody response to two influenza A subtypes, H1 and H3, correlated with in vivo protection, mice were immunized with rIBV-NS110-PR8HA or rIBV-NS110-HK68HA and challenged with rPR8 or X31 (rPR8 expressing the HA and neuraminidase [NA] of HK68), and lungs were harvested at 3 and 6 days postchallenge to determine viral burden. Mice immunized with rIBV-NS110-PR8HA and challenged with rPR8 had no detectable virus in the lungs at both 3 and 6 days postinfection (Fig. 5A), whereas control mice (rWT B virus infected or mock treated with PBS) had significant amounts of virus in their lungs. Similarly, mice immunized with rIBV-NS110-HK68HA and challenged with X31 (Fig. 5B) had significantly smaller amounts of virus at day 3 postinfection than control groups and had no detectable virus at 6 days postinfection. These observations indicated that immunization with recombinant chimeric B viruses conferred protection against PR8 and X31 lung infection.
Fig. 5.
Vaccination with the chimeric rIBV-NS110-AHA protects BALB/c mice against lethal infection with influenza A virus. Eight-week-old female BALB/c mice (n = 10) were vaccinated with 10-fold serial dilutions of rIBV-NS110-PR8HA (A and C) or rIBV-NS110-HK68HA (B and D) (from 1 × 105 PFU to 1 × 102 PFU). Control groups were immunized with 1 × 105 PFU rWT B/Ya88 virus or mock treated with PBS. Three weeks postvaccination, for viral lung replication study the mice were infected with either 1 ×102 PFU of A/PR8 virus (n = 6) or 5 × 104 PFU of A/X31 virus (n = 6). For stringent viral survival studies, mice were infected with a higher dose of 1 × 103 PFU of A/PR8 virus (n = 4) or 1 ×106 PFU of A/X31 virus (n = 4). (A and B) The pulmonary replication of influenza A viruses in vaccinated mice on days 3 and 6 p.i. Average lung titers ± standard deviations are depicted. The limit of detection was 5 PFU. (C and D) Animals were monitored for body weight changes and survival for a total of 14 days postchallenge.
To further evaluate whether the vaccination could deliver full protection to animals against lethal viral infection, mice then were challenged with a lethal dose of either rPR8 or X31. Mice immunized with either rIBV-NS110-PR8HA (Fig. 5C) or rIBV-NS110-HK68HA (Fig. 5D) survived lethal viral challenge without exhibiting any symptoms during the course of 14 days postinfection. In contrast, both the rWT B virus-immunized and mock-immunized mice showed significant weight loss, and all animals succumbed to infection by day 5. Of note, mice immunized with 1 × 102 and 1 × 103 PFU rIBV-NS110-HK68HA lost about 15% of their initial weight in the first 3 days postchallenge but quickly regained their weight after day 4 postinfection. Ultimately, all vaccinated mice survived lethal challenge, whereas all control mice succumbed to lethal challenge with PR8 or X31 around 3 days postinfection.
Generation of rIBV-NS110 expressing the HA of VN04 (H5).
Thus far, we have demonstrated the feasibility of using the recombinant influenza B virus possessing a truncated NS1 and an influenza A hemagglutinin as candidate vaccines against two representative influenza A virus subtypes, namely, H1 and H3, which currently are cocirculating in the human population. As shown above, both rIBV-NS110-AHAs are attenuated in growth in vitro and apathogenic in mice, and more importantly, they induced an immune response that protects against lethal influenza A virus (H1 and H3) challenge. Because the influenza A virus of the H5 subtype has the potential to cause the next pandemic, we used a similar approach to construct an rIBV-NS110 expressing the ectodomain of A/Vietnam/1203/04 (rIBV-NS110-VN04HA), which is of the H5 subtype (Fig. 6A). We then carried out similar experiments to evaluate rIBV-NS110-VN04HA as a candidate for a prepandemic LAIV.
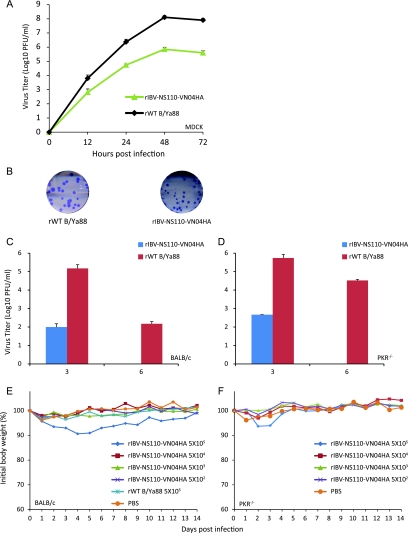
We first examined the proper expression of the chimeric VN04 hemagglutinin in rIBV-NS110-VN04HA-infected cells. As expected, the VN04 hemagglutinin protein was detected in rIBV-NS110-VN04HA-infected MDCK cells by Western blot analysis and immunostaining assays (Fig. 6B and C), indicating that the recombinant virus properly expresses VN04 hemagglutinin protein. We next examined the ability of rIBV-NS110-VN04HA to induce an IFN response. We observed an increase in GFP expression and an enhancement of firefly luciferase activity in MDCK pIFNβ cells after rIBV-NS110-VN04HA infection compared to those of the controls at 15 hpi (Fig. 6D and E). Thus, our data demonstrate that rIBV-NS110-VN04HA induces an appropriate IFN-β response which is known to be associated with attenuation in vivo of NS1-truncated influenza viruses (9, 21, 22).
Growth characteristics of rIBV-NS110-VN04HA virus in cell culture, mice, and chickens.
Comparably to the H1 and H3 chimeric viruses, rIBV-NS110-VN04HA grew to a level of about 2 logs less than that of the rWT B virus (Fig. 7A), which translated to smaller plaque sizes (Fig. 7B). We next investigated rIBV-NS110-VN04HA replication characteristics in both BALB/c and C57BL/6 PKR−/− mice. C57BL/6 PKR−/− mice have compromised immunity due to a deficiency in protein kinase R (PKR) and previously have been shown to be an alternative animal model for studying the pathogenesis and growth kinetics of non-mouse-adapted B virus strains (9). Only about 100 PFU per ml of rIBV-NS110-VN04HA was detected in lung tissues of BALB/c mice 3 days postinfection, which was 3 logs lower than that in rWT B virus-infected mice (Fig. 7C). Only the infection of BALB/c mice with the highest dose (5 × 105 PFU) of rIBV-NS110-VN04HA led to a 10% weight loss in the first 5 days postinfection (Fig. 7E). Similar observations were obtained with C57BL/6 PKR−/− mice. rIBV-NS110-VN04HA viruses replicated at low levels early in the infection and caused no weight loss after 14 days postinfection. In summary, both mouse strains showed no mortality or morbidity after infection with rIBV-NS110-VN04HA, which indicates that this virus is attenuated in mice.
Fig. 7.
Chimeric rIBV-NS110-VN04HA is attenuated both in vitro and in vivo. (A) Multicycle growth curves of the recombinant viruses in MDCK cells infected at an MOI of 0.05 and titrated by plaque assay on MDCK cells. (B) Plaque size phenotypes of the rIBVs in MDCK cells at 3 days p.i. Eight-week-old BALB/c (C) and C57BL/6 PKR−/− (D) mice (n = 6) were infected intranasally with 1 × 105 PFU of the indicated viruses. Average lung titers ± standard deviations are depicted. The limit of detection was 5 PFU. Following viral infection, BALB/c (E) and C57BL/6 PKR−/− (F) mice were weighed daily, and the average body weights of surviving animals in each group up to day 14 p.i. are indicated as percentages of the original body weights.
As this is the first time that a hemagglutinin of avian origin is expressed on the B virus background, we tested whether such genetic manipulation of the B virus enabled it to infect chickens. To abolish the potential negative interference from the attenuation brought about by the truncated NS1 protein, we generated an rIBV-VN04HA virus with the wild-type NS1 gene. As shown in Table 1, no morbidity or mortality was observed in chickens infected with either the rWT B virus or the rIBV-VN04HA virus. No viruses were recovered from any intranasally inoculated chicken. Of note, we did detect seroconversion in the chickens intravenously inoculated with the chimeric B virus (Table 1). Seroconversion also was observed in animals receiving the rWT B virus (Table 1). Thus, we conclude that the rIBV-VN04HA virus is unlikely to cross the species barrier to infect chickens.
Table 1.
Direct assessment of infection of recombinant B viruses in chickensa
| Virus strain | No. of birds | Route | Morbidity | Mortality | HI serology (GMT) |
|---|---|---|---|---|---|
| rWT B/Ya88 | 5 | i.n. | 0/5 | 0/5 | 1/5 (<8) |
| rIBV-VN04HA | 5 | i.n. | 0/5 | 0/5 | 0/5 (<8) |
| rWT B/Ya88 | 10 | i.v. | 0/10 | 0/10 | 10/10 119 |
| rIBV-VN04HA | 10 | i.v. | 0/10 | 0/10 | 3/10 (<8) |
HI, hemagglutination inhibition assay; GMT, geometric mean titer; i.n., intranasal(i.n. dose, 105.7 50% egg infective doses [EID50]); i.v., intravenous.(i.v. dose, 106.9 EID50).
Vaccination with rIBV-NS110-VN04HA elicits antibody responses in BALB/c mice.
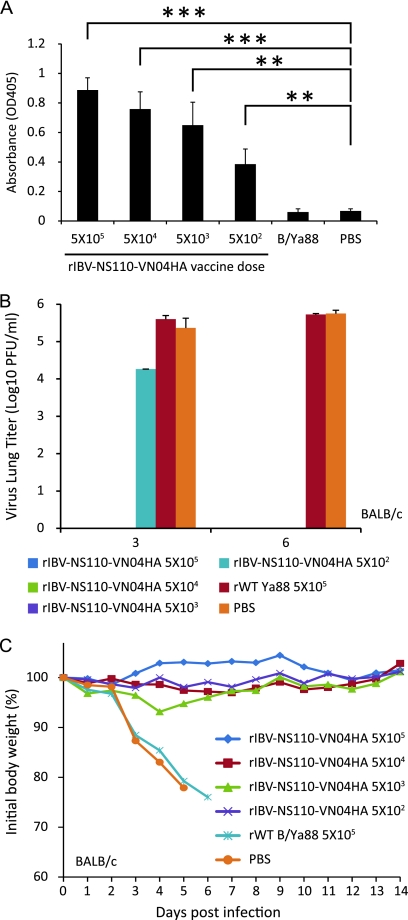
We next wished to evaluate the production of VN HA-specific IgGs in the sera of BALB/c mice immunized with rIBV-NS110-VN04HA. As shown in Fig. 8A, vaccination elicited a significant increase in levels of antibodies against VN hemagglutinin, and antibody titers increased in a dose-dependent manner with the amount of rIBV-NS110-VN04HA administered.
Fig. 8.
Vaccination with chimeric A/B viruses protects BALB/c mice from lethal infection with influenza A virus. Eight-week-old female BALB/c mice (n = 13) were immunized with the indicated viruses or rWT B/Ya88 virus, and PBS was used as a negative control. Serum samples were collected 21 days postvaccination (n = 3). A/PR8 VN HA/NA virus-specific antibodies (A) were detected by ELISA as described in Materials and Methods. The rest of the immunized mice were infected with either 1 ×103 PFU of rPR8-VN04HANA virus for viral lung replication study (n = 6) or 5 × 103 PFU of rPR8-VN04HANA virus for pathology study (n = 4). (B) Pulmonary viral replication in vaccinated BALB/c mice on days 3 and 6 p.i. Average lung titers ± standard deviations are depicted. The limit of detection was 5 PFU. (C) After viral challenge, animals (n = 4) were monitored for body weight changes and survival for a total of 14 days postchallenge. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 versus PBS control.
Immunization with rIBV-NS110-VN04HA protects against lethal H5 challenge in BALB/c mice.
To evaluate whether vaccination with rIBV-NS110-VN04HA can protect against infection with a potentially pandemic H5 virus, we challenged rIBV-NS110-VN04HA-vaccinated mice with 5 × 103 PFU of an A virus expressing the HA (without the polybasic cleavage site) and NA of VN04 on a PR8 backbone (rPR8-VN04HANA; 50% lethal dose, 316 PFU), which previously has been used in our laboratory as a substitute challenge virus for WT HPAI H5 virus (27).
BALB/c mice were immunized with different doses of rIBV-NS110-VN04HA, rWT B virus, or PBS. At days 3 and 6 postchallenge, three mice per group were euthanized, and their lungs were harvested for the analysis of viral burden. We detected around 104 PFU viruses in animals receiving the lowest vaccine dose (5 × 102 PFU) at day 3 postchallenge, which still was 1 log lower than the level in control animals receiving the rWT B virus or PBS (Fig. 8B). At day 6 postchallenge, we did not detect any residual virus from any of the rIBV-NS110-VN04HA-vaccinated animals, whereas 105 PFU viruses were detected in the lungs of the two control groups. These results indicate that vaccination with rIBV-NS110-VN04HA can significantly decrease viral burden after challenge with rPR8-VN04HANA. To further evaluate the protection of animals from disease or death, we challenged the vaccinated animals with 5 × 103 PFU of rPR8-VN04HANA. We observed a 5% weight loss in mice immunized with 5 × 103 PFU rIBV-NS110-VN04HA (Fig. 8C). In contrast, all control mice immunized with either rWT B virus or PBS succumbed to viral challenge (Fig. 8C).
Vaccination with rIBV-NS110-VN04HA elicits antibody responses and increases influenza-specific IFN-γ-secreting CD8+ T cells in PKR−/− mice.
We measured the humoral response against the rWT B virus in rIBV-NS110-VN04HA-immunized C57BL/6 PKR−/− mice. Similarly to what we observed regarding the humoral response against influenza A viruses in BALB/c mice, the vaccination of C57BL/6 PKR−/− mice induced a potent increase in anti-B virus-specific antibodies (Fig. 9A). In addition, to determine whether rIBV-NS110-VN04HA vaccination induced a CD8+ T-cell response, we also examined the production of IBV-specific CD8+ T cells in immunized C57BL/6 PKR−/− mice. Purified CD8+ T cells were pooled from vaccinated mice (n = 3) at 21 days postvaccination and stimulated with splenocytes from naïve mice that were infected in vitro with rWT B virus for 2 h (Fig. 9B). We quantified the number of IBV-specific CD8+ T cells using an ELISpot reader and found that the vaccination of C57BL/6 PKR−/− mice with rIBV-NS110-VN04HA enhanced the number of CD8+ T cells that recognized B virus peptides (Fig. 9B).
Fig. 9.
Vaccination with chimeric A/B viruses protects C57BL/6 PKR−/− mice from lethal infection with the parental influenza B/Ya88 virus. Eight-week-old female C57BL/6 PKR−/− mice (n = 10) were immunized with 10-fold serial dilutions of rIBV-NS110-VN04HA, from 1 × 105 PFU to 1 × 102 PFU, and PBS was used as a negative control. At 21 days postvaccination, serum samples were collected (n = 3). (A) B/Ya88 virus-specific antibodies were detected by ELISA as described in Materials and Methods. CD8+ T cells were isolated from splenocytes (n = 3). (B) Splenocytes from naïve mice were infected with rWT B/Ya88 viruses and used as antigen-presenting cells in an ELISpot assay with CD8+ T cells from vaccinated mice. The experiment was performed in triplicate, and the average number of spots per well ± standard deviations is graphed. In each group, the other four immunized mice per group were infected with 1 × 106 PFU of rWT B virus for pathology study (n = 4). (C) After viral challenge, C57BL/6 PKR−/− mice were monitored for body weight changes and survival for a total of 14 days postchallenge. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 versus PBS control.
Immunization with rIBV-NS110-VN04HA protects against lethal B virus challenge in PKR−/− mice.
Given that influenza B viruses contribute to a large number of influenza cases annually, it would be of practical benefit to have a vaccine that protects against both A and B viruses. To this end, we wished to determine whether our recombinant B virus-based vaccine expressing the ectodomain of an A virus hemagglutinin would have protective efficacy against lethal challenge with IBV. We immunized C57BL/6 PKR−/− mice with different doses of rIBV-NS110-VN04HA and then challenged the mice with 1 × 106 PFU rWT B/Ya88. No weight loss was observed in mice immunized with rIBV-NS110-VN04HA in the 14 days postchallenge, whereas all control animals immunized with PBS succumbed to virus infection before day 8 postinfection (Fig. 9C).
In conclusion, we have shown that mice immunized once with a recombinant B virus expressing an A virus hemagglutinin is able to induce a robust immune response and protect against both influenza A and B virus challenge in mice.
DISCUSSION
Preexposure immunization, either through the use of inactivated vaccines or LAIVs, has been proven to be the most cost-effective method to prevent influenza virus infection (2). With regard to LAIVs, however, the possibility that the hemagglutinin segment from a vaccine strain could reassort with a wild-type circulating IAV would prevent the use of live vaccines in prepandemic scenarios. To address this issue, we propose a different approach in vaccine design by using a recombinant B virus-based LAIV expressing the hemagglutinin of an A virus, which is reassortment incompetent with circulating A viruses. Our vaccine model takes advantage of the fact that there is no known reassortment between influenza A and B viruses; we have incorporated the portion of the influenza A hemagglutinin that is required for the induction of protective immunity into an influenza B virus, and in doing so we have removed the influenza A packaging signals that would enable the incorporation of the gene segment into a wild-type influenza A virus. We show that our design strategy could be applied to multiple influenza A virus subtypes, including two seasonal subtypes (H1 and H3) and one potential pandemic virus subtype (H5).
In light of what has been described, the generation of this recombinant B virus-based LAIV against A viruses has several innovations over the previous generation of LAIVs. The chimeric influenza A/B virus that initially was rescued by Flandorfer at al. grew poorly in vitro (5). However, after several passages, it grew to titers comparable to those of wild-type IAV strains. Flandorfer at al. identified one mutation, the most C-terminal amino acid of the IBV HA ectodomain, which was mutated from an IBV-specific histidine back to an IAV-specific tyrosine; that single change appeared to be responsible for the observed enhancement in replication (5). Based on their observations, we designed our chimeric HA segment to include the last five amino acids of the IBV HA ectodomain (Fig. 1 and 6A).
When the chimeric HA segment was inserted into a WT B virus background, the recombinant influenza B virus containing the chimeric hemagglutinin segment replicated as well as the rWT B virus (data not shown). Although we are not sure why those membrane-proximal amino acids are required for the enhanced replication, there are two possible explanations for this observation: (i) the membrane-proximal amino acids are important for maintaining the stability of the chimeric protein, or (ii) the membrane-proximal amino acids are important for the efficient incorporation of the protein into the viral membrane. Of note, we also attempted to rescue a virus that maintained the membrane-proximal 10 amino acids of the B virus sequence. However, we were not able to rescue that virus. This indicated that the extra five amino acids from the B virus are genetically incompatible with the A hemagglutinin ectodomain. The interaction between these hemagglutinin elements may play a vital role in the expression or stability of the hemagglutinin protein.
The present recombinant viruses are unable to donate their A virus hemagglutinins to influenza A viruses because of the restrictions imposed by the packaging sequences. However, these viruses could accept influenza B virus hemagglutinin genes from circulating wild-type strains. Such reassortant viruses would be comparable to the wild-type strains in circulation.
Even though vaccination with different chimeric B viruses at different doses protected A virus-infected mice from death (Fig. 5 and 8), the data indicated that there were differences in the immunogenicity of the different chimeric B virus vaccines. Immunization with PR8 HA chimeric B virus induced higher IgG antibody responses (Fig. 4A and B) and lower viral replication of the challenge virus in the lung (Fig. 5A and B) than vaccination with HK68 HA chimeric B virus. The difference likely is due to the higher replication capacity of the PR8 HA chimeric virus compared to that of the HK68 HA chimeric virus (Fig. 2E and 3A).
It is interesting that recombinant B viruses expressing an H5 hemagglutinin do not infect chickens productively, indicating that the expression of a single surface protein of an avian A virus on an otherwise B virus backbone does not yield a recombinant virus that has the means to transmit to avian species. This observation underscores the importance of other B virus factors which block a successful infection of species other than humans.
Our serologic and cellular studies indicate that both humoral and cellular immune responses specific for non-hemagglutinin viral proteins, such as the neuraminidase and nucleoprotein, were sufficient to protect mice from lethal rWT B virus infection. A similar observation has been made in an earlier study of influenza A virus (20). It is not clear if this phenomenon would translate to human immunity, since people typically need to be immunized regularly with novel circulating influenza B viruses despite the high conservation of internal genes between seasonal B strains.
In summary, we have designed a live-attenuated vaccine model that might be safely used during prepandemic disease scenarios. Importantly, we also have demonstrated that our reassortment-incapable live vaccine strains can elicit a protective immune response in mice against a potential pandemic influenza virus subtype.
ACKNOWLEDGEMENTS
We are grateful to Janice Chen, Gene S. Tan, and Taia T. Wang for helpful discussions and critical reviews. We also thank Lily Ngai for excellent technical assistance. We express our appreciation to John Steel for supplying the construct containing the VN04 hemagglutinin gene without a polybasic cleavage site.
This work was supported by CRIP (Center for Research on Influenza Pathogenesis; NIAID contract HHSN266200700010C) and by the following grants from the NIH: UO1 AI070469, 1RC1 AI086061, P01 AI 058113, and U54 AI057158.
Footnotes
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Basler C. F., et al. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. U. S. A. 98:2746–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beyer W. E., Palache A. M., de Jong J. C., Osterhaus A. D. 2002. Cold-adapted live influenza vaccine versus inactivated vaccine: systemic vaccine reactions, local and systemic antibody response, and vaccine efficacy. A meta-analysis. Vaccine 20:1340–1353 [DOI] [PubMed] [Google Scholar]
- 3. Claas E. C., et al. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472–477 [DOI] [PubMed] [Google Scholar]
- 4. Fan S., et al. 2009. Immunogenicity and protective efficacy of a live attenuated H5N1 vaccine in nonhuman primates. PLoS Pathog. 5:e1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Flandorfer A., Garcia-Sastre A., Basler C. F., Palese P. 2003. Chimeric influenza A viruses with a functional influenza B virus neuraminidase or hemagglutinin. J. Virol. 77:9116–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fodor E., et al. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fouchier R. A., et al. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. U. S. A. 101:1356–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghate A. A., Air G. M. 1999. Influenza type B neuraminidase can replace the function of type A neuraminidase. Virology 264:265–277 [DOI] [PubMed] [Google Scholar]
- 9. Hai R., et al. 2008. Influenza B virus NS1-truncated mutants: live-attenuated vaccine approach. J. Virol. 82:10580–10590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horimoto T., et al. 2003. Generation of influenza A viruses with chimeric (type A/B) hemagglutinins. J. Virol. 77:8031–8038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson N. P., Mueller J. 2002. Updating the accounts: global mortality of the 1918-1920 “Spanish” influenza pandemic. Bull. Hist. Med. 76:105–115 [DOI] [PubMed] [Google Scholar]
- 12. Kaverin N. V., et al. 1983. Studies on heterotypic interference between influenza A and B viruses: a differential inhibition of the synthesis of viral proteins and RNAs. J. Gen. Virol. 64:2139–2146 [DOI] [PubMed] [Google Scholar]
- 13. Li S., et al. 1999. Recombinant influenza A virus vaccines for the pathogenic human A/Hong Kong/97 (H5N1) viruses. J. Infect. Dis. 179:1132–1138 [DOI] [PubMed] [Google Scholar]
- 14. Mikheeva A., Ghendon Y. Z. 1982. Intrinsic interference between influenza A and B viruses. Arch. Virol. 73:287–294 [DOI] [PubMed] [Google Scholar]
- 15. Min J. Y., et al. 2010. A live attenuated H7N7 candidate vaccine virus induces neutralizing antibody that confers protection from challenge in mice, ferrets, and monkeys. J. Virol. 84:11950–11960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nabel G. J., Wei C. J., Ledgerwood J. E. 2011. Vaccinate for the next H2N2 pandemic now. Nature 471:157–158 [DOI] [PubMed] [Google Scholar]
- 17. Palese P. 2006. Making better influenza virus vaccines? Emerg. Infect. Dis. 12:61–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Palese P., Shaw M. L. 2007. Orthomyxoviridae: the viruses and their replication, p. 1647–1689 In Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 19. Palese P., Garcia-Sastre A. 2002. Influenza vaccines: present and future. J. Clin. Investig. 110:9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rangel-Moreno J., et al. 2008. B cells promote resistance to heterosubtypic strains of influenza via multiple mechanisms. J. Immunol. 180:454–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Solórzano A., et al. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 79:7535–7543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Steel J., et al. 2009. Live attenuated influenza viruses containing NS1 truncations as vaccine candidates against H5N1 highly pathogenic avian influenza. J. Virol. 83:1742–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Subbarao K., et al. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393–396 [DOI] [PubMed] [Google Scholar]
- 24. Suguitan A. L., Jr., et al. 2006. Live, attenuated influenza A H5N1 candidate vaccines provide broad cross-protection in mice and ferrets. PLoS Med. 3:e360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Talon J., et al. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. U. S. A. 97:4309–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tanaka T., Urabe M., Goto H., Tobita K. 1984. Isolation and preliminary characterization of a highly cytolytic influenza B virus variant with an aberrant NS gene. Virology 135:515–523 [DOI] [PubMed] [Google Scholar]
- 27. Wang T. T., et al. 2010. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. U. S. A. 107:18979–18984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Watanabe T., Watanabe S., Kim J. H., Hatta M., Kawaoka Y. 2007. Novel approach to the development of effective H5N1 influenza A virus vaccines: use of M2 cytoplasmic tail mutants. J. Virol. 82:2486–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Webster R. G., Bean W. J., Gorman O. T., Chambers T. M., Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]