Abstract
The cytokine erythropoietin (Epo) promotes erythropoietic progenitor cell proliferation and is required for erythropoietic differentiation. We have found that the Epo gene is a direct transcriptional target gene of retinoic acid signaling during early erythropoiesis (prior to embryonic day E12.5) in the fetal liver. Mouse embryos lacking the retinoic acid receptor gene RXRα have a morphological and histological phenotype that is comparable with embryos in which the Epo gene itself has been mutated, and flow cytometric analysis indicates that RXRα-deficient embryos are deficient in erythroid differentiation. Epo mRNA levels are reduced substantially in the fetal livers of RXRα−/− embryos at E10.25 and E11.25, and genetic analysis shows that the RXRα and Epo genes are coupled in the same pathway. We furthermore show that the Epo gene is retinoic acid inducible in embryos, and that the Epo gene enhancer contains a DR2 sequence that represents a retinoic acid receptor-binding site and a retinoic acid receptor transcriptional response element. However, unlike Epo-deficient embryos that die from anemia, the erythropoietic deficiency in RXRα−/− embryos is transient; Epo mRNA is expressed at normal levels by E12.5, and erythropoiesis and liver morphology are normal by E14.5. We show that HNF4, like RXRα a member of the nuclear receptor family, is abundantly expressed in fetal liver hepatocytes, and is competitive with retinoic acid receptors for occupancy of the Epo gene enhancer DR2 element. We propose that Epo expression is regulated during the E9.5–E11.5 phase of fetal liver erythropoiesis by RXRα and retinoic acid, and that expression then becomes dominated by HNF4 activity from E11.5 onward. This transition may be responsible for switching regulation of Epo expression from retinoic acid control to hypoxic control, as is found throughout the remainder of life.
Keywords: Erythropoiesis, HNF4, retinoic acid regulation, RXRα, fetal liver, DR2
The generation of red blood cells is a necessary step in vertebrate embryogenesis to progress from diffusion-limited growth to circulatory system-mediated growth. Red blood cell production (erythropoiesis) involves the commitment of a pluripotent hematopoietic stem cell to the erythroid lineage, progression through erythroid BFU-E and CFU-E progenitor cell stages, proerythroblast and several types of erythroblast stages, and ultimately culminating in the terminally differentiated erythrocyte. During development, the location of erythropoiesis shifts from the yolk sac in early embryos, to the fetal liver in midgestation embryos, to the bone marrow and spleen in late embryos and in postnatal life. Yolk sac erythropoiesis is referred to as primitive erythropoiesis, and is contrasted with definitive erythropoiesis, which occurs in the fetal liver and throughout postnatal life. In mouse embryos, yolk sac erythropoiesis initiates around embryonic day 7.5 (E7.5) and ceases around E12.5, although primitive erythrocytes persist for another 2 or 3 d. Erythropoietic activity initiates in the fetal liver around E9.5 (Houssaint 1981; Palis et al. 1999), concurrent with the formation of the liver from the hepatic diverticulum of the foregut, and ends late in gestation when bone marrow erythropoiesis is initiated. Thus, there is a brief window during which primitive and definitive erythroid cells coexist. In a normal mouse embryo, 25% of the peripheral red blood cells at E12.5 are derived from the fetal liver, whereas at E14.5, 90% of these cells are of definitive origin (Mucenski et al. 1991; Trimborn et al. 1999).
A number of cytokines are established to control the erythroid differentiation process. Among these, the glycoprotein erythropoietin (Epo) has two critical roles in definitive erythropoiesis. First, Epo contributes to the survival and proliferation of definitive erythropoietic progenitor cells in the fetal liver. Thus, in Epo−/− embryos (Wu et al. 1995) the CFU-E and proerythroblast populations are reduced several-fold in number. Second, Epo is essential for these cells to reach the terminal steps of definitive erythropoiesis, as no fetal liver-derived erythroblasts or red blood cells form in Epo-deficient embryos. Epo is not required for primitive erythropoiesis, in that mouse embryos lacking Epo support yolk sac production of red blood cells. Epo-deficient embryos die of anemia at E13.5, as the yolk sac-derived red blood cells that sustained development from E7.5–E12.5 die out and are not replaced through definitive erythropoiesis. These embryos show a characteristic small and pale fetal liver at E12.5 and E13.5, a consequence of the absence of terminal erythropoiesis. In histological sections, Epo deficiency is manifest in a dramatic increase in cell death, indicative of the aforementioned role in erythroid cell survival. Mouse embryos lacking the Epo receptor (Wu et al. 1995) or the JAK2 (Neubauer et al. 1998; Parganas et al. 1998) component of the Epo receptor signal transduction cascade have a comparable midgestation anemic phenotype.
The primary physiological regulator of Epo expression in late embryos and in postnatal stages is oxygen tension (Bunn et al. 1998). A mostly unknown hypoxia sensing mechanism (Zhu and Bunn 1999) results in the activity of the transcription factor HIF1 (hypoxia-inducible factor 1), which binds to a defined sequence in the 3′ enhancer of the Epo gene (Semenza et al. 1991) and initiates Epo expression. In the fetal liver, Epo is expressed primarily by hepatocytes (Koury et al. 1991), a property which is conserved in hepatocellular carcinoma cell lines such as Hep3B and HepG2, in which Epo expression is induced in response to hypoxia (Goldberg et al. 1987). Adjacent to the HIF1-binding site in the mouse Epo 3′ enhancer is the sequence TGACCTCTTGACCC, which is known as a DR2 element because of the direct repeat of the hexameric sequence TGACC(C/T) spaced by two nucleotides. The Epo enhancer DR2 element substantially augments hypoxic induction of Epo gene reporter constructs in transfected Hep3B cells, but is itself not responsible for responding to hypoxia (Blanchard et al. 1992). HNF4 (hepatocyte nuclear factor 4) is currently believed to be the primary factor that is responsible for Epo gene regulation through the DR2 element (Bunn et al. 1998). HNF4 is expressed in the fetal liver and postnatal kidney, the two major sites of Epo expression, and introduction of an HNF4 expression construct in transfected HeLa cells (which do not normally express HNF4) confers hypoxic inducibility to an Epo reporter gene (Galson et al. 1995). HNF4 appears to function synergistically with HIF1 on the Epo enhancer by direct protein–protein interaction and through the recruitment of transcriptional coactivators (Bunn et al. 1998).
We have studied the biological function of the retinoic acid receptor, which is comprised of a heterodimer of RAR and RXR (Evans 1988). The RXR–RAR heterodimer is well established to bind to and to transactivate through generic DR2 elements; however, there has been no prior evidence that retinoic acid or retinoic acid receptors are involved in Epo gene expression. Thus, retinoic acid treatment does not activate an Epo reporter gene, nor alter hypoxic induction of the reporter gene, in transfected Hep3B cells (Blanchard et al. 1992). We have described previously a loss-of-function mutation of the RXRα gene (Sucov et al 1994) that results in embryonic lethality at E14.5–E15.5 as a consequence of cardiac failure and placental dysfunction. However, transiently at E11.5–E12.5 in RXRα−/− embryos, there is a deficiency in liver size and appearance that is strikingly similar to the appearance of Epo-deficient embryos. The RXRα fetal liver phenotype is corrected from E13.5, such that when these mutant embryos die of cardiac failure 1 or 2 d later, their livers appear normal, unlike Epo−/− embryos, which die at E13.5. In this study, we show that the initial expression of the Epo gene in the fetal liver is regulated by retinoic acid receptors, and that the transient liver phenotype in RXRα−/− embryos is a consequence of impaired erythropoiesis. Furthermore, we show that RXR–RAR and HNF4 are mutually competitive for activation through the Epo enhancer DR2 element, and propose a model in which regulation of Epo gene expression in the fetal liver transitions from a retinoic acid- and RXRα/RAR-dependent mechanism to a HNF4- and hypoxia-dependent mechanism.
Results
A transient reduction in fetal liver erythropoiesis in RXRα-deficient embryos
The initial steps of liver morphogenesis occur normally in RXRα-deficient embryos. Thus, in both wild-type and RXRα−/− embryos at E10.5, the endodermal hepatic cords have become invaded by nonparenchymal mesoderm, and endothelial cells line all sinusoids (Fig. 1a–f). However, over the next 2 d, the liver tissue of RXRα−/− embryos fails to expand normally. At E12.5, the liver is a prominent and externally visible organ in the wild-type embryo, but in RXRα-deficient embryos is only marginally evident (Fig. 1g). Upon dissection, the mutant livers appeared to be organized properly, but contained approximately fourfold less tissue than wild-type littermates (estimated by counting cell numbers, by measurement of weight, and by reconstruction of liver tissue volume from histological sections). The penetrance of this phenotype at E12.5 was near 100% in inbred strains and in a mixed background of C57Bl/6 and 129/Sv, and more variable in other mixed strain backgrounds (data not shown). No wild-type embryo was ever deficient in the fetal liver in this regard. RXRα-deficient embryos were not growth retarded at E12.5, and with the exception of the liver deficiency, were otherwise mostly normal. The mutant embryos were also not obviously anemic, although this is almost certainly because of sufficient yolk sac-derived primitive erythropoiesis, which at E12.5 predominates peripheral circulation. Furthermore, mutant embryos at E12.5 are not yet altered in placental (Sapin et al. 1997) or cardiac (Dyson et al. 1995) function.
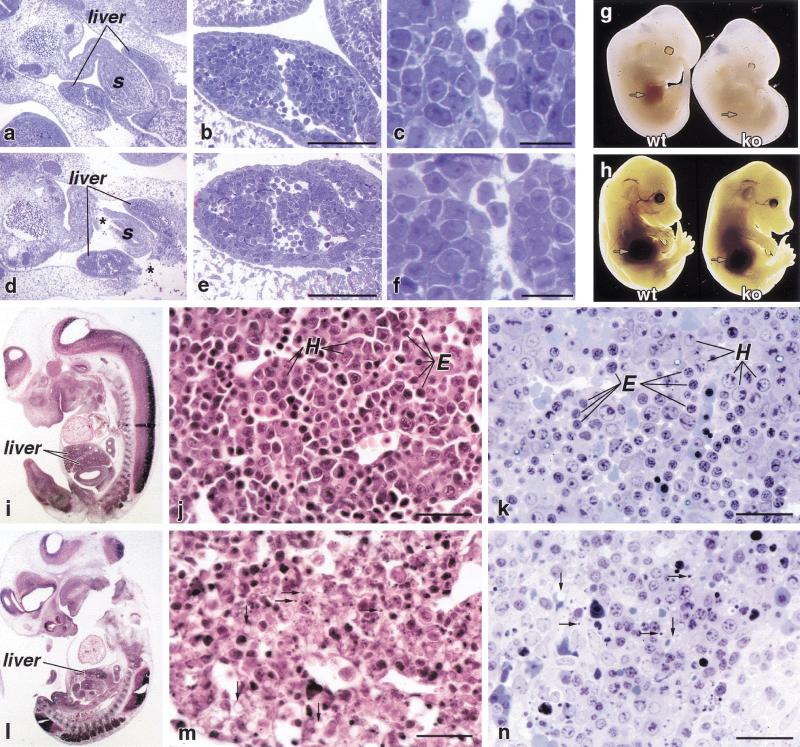
Figure 1.
Liver morphology in RXRα−/− embryos. (a–f) Morphology of liver tissue at E10.5. Richardson's stained Epon sections of a wild-type embryo (1 μm) (a) and littermate RXRα−/− (d) embryo indicate two lobes of the developing liver flanking the stomach(s); the overall morphology of the mutant tissue is normal. (b,c,e,f) Successively higher magnifications of a and d, respectively. (Asterisk) Regions of tissue damage caused by histology artifact. (g,h) External appearance of littermate embryos at E12.5 (g) and E14.5 (h). The arrows point to the externally apparent fetal liver in wild-type (wt) embryos at E12.5, and to the absence of evident tissue in RXRα−/− (ko) embryos at the same stage, whereas at E14.5, the fetal liver is prominent in embryos of both genotypes. An eye defect (Kastner et al. 1994) can be noted in mutant embryos at both stages. (i–n) Morphology of liver tissue at E12.5. Both 5 μ paraffin hematoxylin and eosin-stained sections (i,j,l,m) and 1 μ Richardson's stained Epon sections (k,n) of different embryos are shown. Erythropoietic cells (E) and hepatocytes (H) are morphologically distinguishable in wild-type tissue, although because of section angle or stage of cell cycle, it is not possible to identify all cells by morphology. Note the widespread presence of pycnotic nuclei and cellular debris (horizontal arrows) in RXRα−/− tissue. It is uncertain as to what extent the debris is from erythroblasts or hepatocytes, but material from both cell types may be represented. There is extensive vacuolization in the RXRα−/− liver tissue (vertical arrows) that is present, but to a much lesser extent, in wild-type tissue, and may be an indication of impending cell death, or aberrant hepatocyte glycogen or lipid storage. Cellular debris and vacuolization are both seen in Epo−/− livers as well. Scale bars, b,e,j,k,m, and n, 100 μm; c and f, 20 μm.
In wild-type E12.5 embryos, differentiating erythropoietic cells can be identified in liver tissue on the basis of their smaller size and highly condensed and deeply staining nuclei (Fig. 1j,k). These cells are likely to be erythroblasts, as they are still nucleated, but are not stem or progenitor cells, which are not strongly eosin positive. Furthermore, these cells can be distinguished from circulating erythrocytes, which at E12.5, are mostly yolk sac derived and thus nucleated (primitive erythropoiesis), as the latter are found within sinusoids and vessels enclosed by endothelium. Hepatocytes are recognizable as larger cells with larger nuclei that stain more diffusely. The overall appearance of the normal tissue was highly uniform and organized.
In contrast, in littermate embryos lacking RXRα, there was a less compact organization in the mutant tissue (Fig. 1m,n). Extended regions of cell death were prominent, appearing as a loss of cellular structure with fractured nuclei, with extensive cellular vacuolization and with extracellular and subcellular granular material apparent. Differentiating erythropoietic cells were less common in the mutant fetal liver, suggestive of a defect in erythropoietic survival and/or differentiation. Because erythropoiesis is a major aspect of the growth of the liver through midgestation, a deficiency in erythropoiesis is a plausible explanation for the observation that RXRα−/− embryos are deficient in liver tissue at E12.5.
The liver deficiency in RXRα−/− embryos is transient. At E13.5, mutant embryos had externally visible liver tissue, and at E14.5, the mass and volume of liver in RXRα−/− embryos was less than twofold reduced relative to that of wild-type littermates, and was substantially greater than what was present 2 or 3 d earlier (Fig. 1h). Liver tissue from E14.5 mutant embryos was histologically normal (data not shown; see also Sucov et al. 1994). The RXRα−/− background is lethal at this time because of cardiac defects (Sucov et al. 1994).
Flow cytometric analysis of fetal liver populations
To obtain a more precise description of the RXRα-deficient liver phenotype, we quantitatively characterized the fetal liver cell population by flow cytometric analysis (Fig. 2). For this analysis, we used the cell surface markers c-kit and TER119; c-kit expression is high in hematopoietic and erythroid progenitor cells and is reduced and ultimately eliminated as differentiation ensues, whereas TER119 is specifically expressed in erythroblasts and erythrocytes, but not in BFU-E and CFU-E populations (Ikuta et al. 1990; Kina et al. 2000). The population of cells that is positive for both markers is believed to be at the proerythroblast stage. The double-negative cell population includes hepatocytes and differentiated nonerythroid hematopoietic cells, although the latter population is insignificant at E12.5 as almost all hematopoiesis is devoted to erythropoiesis at this stage.
Figure 2.
Flow cytometric analysis of fetal liver cell populations. (a) Distribution profiles of cells expressing c-kit and TER119 are shown for littermate wild-type and RXRα−/− embryos isolated at E12.25. The scale for both axes is presented in log units. The designations of regions R1–R4 was made on the basis of the presence of troughs between peaks. R1 includes progenitor cells, R2 includes proerythroblasts, R3 represents erythroblasts and erythrocytes, and R4 includes hepatocytes and potentially other nonerythroid hematopoietic cells. Note the concentration of cells in the R3 region in the wild-type embryo, and the disproportionate number of cells in the R1 and R2 regions in the homozygous embryo. (b,c) Compiled representation of cellular distributions, expressed in terms of total cell number per fetal liver for each region. (b) Cellular distributions at E12.25. (c) Cellular distributions at E14.25. (Solid bars) Wild-type plus heterozygous embryos; (hatched bars) RXRα homozygous embryos; error bars, ±S.E.M.
A depiction of the distribution of fetal liver cell populations (expressed in terms of normalized frequency per 50,000 fetal liver cells) from a wild-type embryo at E12.25 and from a littermate RXRα−/− embryo with a prominent phenotype is shown in Figure 2a. In wild-type embryos, the majority of cells were found in the c-kit negative, TER119-positive category (region 3 [R3] in Fig. 2a), representing erythroblasts and erythrocytes. However, in mutant embryos, a disproportionate percentage of cells were found in regions 1 and 2, with a compensatory reduction in the percentage of cells in the R3 population. It should be noted that the R3 population in both wild-type and mutant livers is contaminated by yolk sac-derived red blood cells within hepatic blood vessels, which are also c-kit negative and TER119 positive. Consequently, the impairment of definitive erythropoietic differentiation in mutant liver is even more severe than suggested by the data shown in Figure 2.
We analyzed a total of 37 fetal liver samples from 5 different litters of embryos isolated at E12.25, representing 13 wild-type, 11 heterozygous, and 13 RXRα homozygous embryos. The compiled results, expressed in terms of the absolute number of cells in each category per fetal liver, are shown in Figure 2b. Because of the decrease in total cell number in mutant embryos (3.4-fold average decrease for the 5 litters of Fig. 2), there was a 2.4–2.6-fold decrease in the total number of cells in the R1, R2, and R4 populations, despite an increase in the normalized frequency of cells in these categories. However, the R3 population of differentiating erythroid cells was compromised to a much greater degree (5.2-fold). Of the 3.4-fold reduction in total fetal liver cell number in mutant embryos relative to wild-type and heterozygous embryos, 90% of this reduction occurs in the hematopoietic population (R1–R3), and 59% occurs specifically in the erythrocyte (R3) population. As noted above, this is a minimum estimate, because of the presence of contaminating primitive erythrocytes in both samples.
These results show a modest reduction in erythroid progenitor cell populations, and a more severe block in erythropoietic differentiation, in RXRα−/− embryos at E12.25. An independent assessment of erythroid progenitor cells at E12.5 in a colony outgrowth assay indicated a comparable frequency of BFU-Es in wild-type (75 ± 18 per 105 nucleated fetal liver cells) and mutant (67 ± 8) tissue, which is consistent with the flow cytometric analysis. The cellular phenotype and the histological appearance together indicate a defect in erythroid progenitor cell survival and differentiation. The reduction in the nonhematopoietic R4 cell population may be a secondary consequence of the reduced number of erythroid progenitor cells and/or of the increased cell death present in fetal liver tissue, or may represent some additional function of RXRα in hepatocytes.
At E14.25, the RXRα-deficient fetal liver was histologically normal (data not shown), and contained no more than a twofold reduction in total cell number. Flow cytometric analysis of E14.25 fetal liver samples indicated that all populations were fully or almost normalized at this developmental time, both in frequency and in absolute numbers (Fig. 2c), although there was still a small residual decrease in the R3 population at this time.
Epo expression is reduced in RXRα-deficient fetal liver
Because the fetal liver phenotype of RXRα−/− embryos at E12.25–E12.5 is very similar to that of Epo-deficient embryos, we addressed whether a deficiency in Epo expression might underlie the RXRα−/− phenotype. RNA was isolated from fetal livers of individual littermate embryos at developmental stages through midgestation. Because the amount of total RNA recovered per fetal liver is small, we used a commercially packaged quantitative RT–PCR assay in which 18S ribosomal RNA serves as an internal standard; the amount of 18S rRNA amplification product is attenuated by the inclusion of competimer oligonucleotides that are titrated to ensure that the level of amplification of the 18S rRNA and Epo products are in a linear range. We found (Fig. 3a) that Epo mRNA levels were reduced ninefold relative to wild-type embryos at E10.25 (the earliest time point sampled). However, at E12.25 there were equal levels of Epo message in knockout and wild-type liver samples (Fig. 3b). We anticipate that there is a lag of ∼2 or more d between the onset of Epo gene expression and the phenotypic consequences of that expression, representing the time necessary for proliferation and differentiation of erythroid progenitor cells (e.g., it takes 2–3 d for CFU-E erythroid progenitor cells to differentiate in vitro). Thus, the substantially reduced level of Epo mRNA expression in RXRα-deficient fetal liver at E10.25 is consistent with a failure in the survival, proliferation, and differentiation of erythroid progenitor cells as evaluated at E12.25, whereas normal Epo expression at E12.25 is consistent with the phenotypic recovery and normal appearance of the fetal liver in RXRα−/− embryos at E14.25, as observed (Figs. 1 and 2). Note that the substantial reduction in Epo expression at E10.25 occurs at a time when the overall morphology of the mutant liver tissue is normal, and that the normal expression at E12.25 occurs in tissue that is highly dysmorphic but soon to recover (Fig. 1). At times between E10.25 and E12.25, we observed an intermediate level of Epo expression in RXRα−/− embryos (i.e., a threefold reduction at E11.25) and an intermediate level of tissue dysmorphology (data not shown).
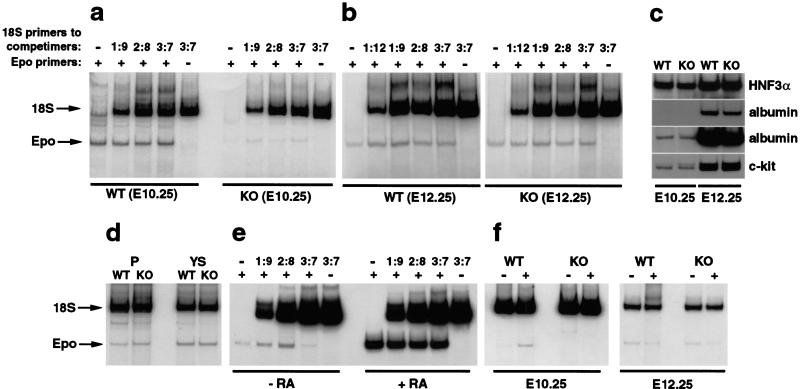
Figure 3.
Epo expression is specifically reduced in early RXRα−/− fetal liver and is retinoic acid inducible. Fetal liver samples were analyzed by relative RT–PCR, using 18S RNA as an internal standard. Varying the ratios of 18S primers ensures that a linear range is achieved for this product; for assays in which this titration is not shown, prior amplifications established the linear range of amplification. (a) Analysis of fetal liver samples from E10.25 littermate embryos. (b) Analysis of fetal liver samples from littermate E12.25 embryos. (c) Relative RT–PCR analysis of HNF3α, albumin, and c-kit expression in E10.25 and E12.25 fetal liver tissue. 18S RNA amplifications are not shown but were equal between corresponding lanes. Long and short exposures of the same gel for the albumin product are both shown to point out comparable amplification from wild-type and RXRα−/− samples at both time points. Quantitation of c-kit amplification relative to 18S RNA amplification indicated that at E12.25 there is a modest (1.6-fold) increase in the abundance of this message in mutant tissue relative to wild type, which is consistent with flow cytometry analysis. (d) Epo expression in placenta and yolk sac of E10.25 wild-type and RXRα−/− embryos. (e) Analysis of fetal liver samples from E10.0 wild-type embryos cultured in vitro for a further 9 h in the absence or presence of retinoic acid. (f) Epo expression in wild-type and RXRα−/− hepatocytes isolated from E10.25 and E12.25 embryos and cultured without and with retinoic acid.
To demonstrate that the deficiency of Epo expression at E10.25 is a specific effect, the expression of other genes was investigated. HNF3α is a transcription factor that is expressed in definitive endoderm (plus other nonhepatic tissues), including hepatocytes from the onset of hepatogenesis. Albumin is a structural protein gene that is exclusively expressed in hepatocytes. As noted above, c-kit is expressed in hematopoietic stem and erythroid progenitor cells. As shown in Figure 3c, all three genes were comparably expressed in the E10.25 liver of RXRα−/− embryos. Normal expression of c-kit also shows that hematopoietic stem cells have colonized the liver of RXRα−/− embryos at E10.25 in an apparently normal manner, at least to the extent that c-kit is a reliable marker of this population. Thus, the reduction in Epo expression seen in RXRα−/− embryos is a specific effect and not an indirect consequence of globally altered fetal liver gene expression. Epo expression in the placenta and yolk sac, the two major non-hepatic sites of Epo expression, was unaltered in RXRα−/− embryos (Fig. 3d), indicating that the phenotype is specific to the liver.
RXRα and Epo are epistatically related
The results described above suggested that the Epo gene might be downstream of RXRα in a common genetic pathway. To further confirm this model, we evaluated embryos that were heterozygous at both the RXRα and Epo loci. Embryos that were heterozygous at either locus individually were normal (see below), which indicates that fetal liver hematopoiesis is insensitive to a 50% reduction in the amount of RXRα or Epo protein. If RXRα is a regulator of Epo expression prior to E12.5, then reduction of Epo gene dosage by half, coupled with a reduced transactivation of the remaining allele by the reduced level of RXRα, might bring the level of Epo expression to below a threshold level such that a phenotype might result. In genetic terms, the evaluation of such double heterozygotes (or trans-heterozygotes; Avery and Wasserman 1992) is a way of determining if the RXRα locus is a genetic enhancer of the Epo mutant phenotype.
Epo−/+ mice were mated to RXRα−/+ animals to obtain embryos at E12.0 that were either wild-type, single heterozygotes, or double heterozygotes, from which fetal liver samples were analyzed as above by flow cytometry with c-kit and TER119. These embryos were isolated at a slightly earlier stage relative to those described above (Fig. 2), on the basis of RNA analyses (Fig. 3a,b), which suggested the likelihood of a more significant response with younger embryos. In embryos singly heterozygous for either the RXRα or the Epo gene mutations, there was no significant decrease in total fetal liver cell number, and the distributions of the R1–R4 populations were comparable with wild-type littermates. In contrast, in double heterozygous embryos, there was a slightly less than twofold decrease in total fetal liver cell number, and about the same decrease in the normalized frequency of cells specifically in the R3 population, resulting in a decrease in the total number of cells in the R3 population of greater than threefold (Fig. 4). The R1, R2, and R4 populations were only marginally altered in the double heterozygous background. Double heterozygous embryos were visually apparent at the time of isolation as being compromised in their fetal livers (data not shown). Thus, this analysis shows that the Epo and RXRα genes are both components of a common genetic pathway, as there is phenotypic synergy when the heterozygous mutations are combined. The observation that Epo message levels are reduced in RXRα−/− embryos (Fig. 3) suggests that the Epo gene lies downstream of RXRα in this pathway. Furthermore, this analysis strongly suggests that the morphological deficiency of E12.5 RXRα−/− embryos is a direct effect of the decreased level of Epo expression seen earlier in development, rather than an indirect or secondary consequence of other manifestations elsewhere in the embryo.
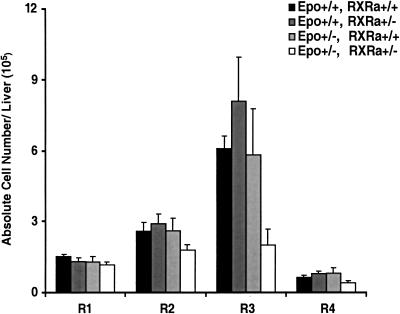
Figure 4.
Epistasis analysis of RXRα and Epo gene function. The compiled representation of cell distributions in E12.0 wild-type (n = 3), RXRα heterozygous (n = 6), Epo heterozygous (n = 4), and RXRα/Epo double heterozygous (n = 3) fetal liver samples from three litters is expressed in terms of total cell number per fetal liver for each region, similar to Fig. 2.
The Epo gene is retinoic acid responsive in embryos
RXRα is a heterodimeric partner for multiple nuclear receptors in complexes that are activated by a variety of ligands. In principle, the absence of RXRα could interfere with normal transcriptional activation mediated by any of these partner receptors and their ligands. Therefore, we asked whether the Epo gene is ligand inducible by any of the known compounds that activate RXR partner receptors. For this purpose, wild-type embryos were isolated at E10.0 intact within their yolk sacs and cultured in vitro (see Materials and Methods) in the presence of various ligands for 9 h. Of those tested, only retinoic acid treatment resulted in an induction of Epo gene expression (Fig. 3e); ligands for the thyroid hormone receptor (TR), bile acid receptor (FXR), fatty acid receptor (PPARα), and pregnane/steroid receptor (PXR) all failed to induce or only marginally induced Epo gene expression (data not shown).
To confirm the role of RXRα in this process, we isolated primary fetal liver cells (enriched for hepatocytes) from individual embryos at E10.25 and cultured in the presence or absence of retinoic acid. RA treatment induced Epo expression in wild-type cells but not in cells isolated from RXRα-deficient embryos (Fig. 3f). There was a higher basal level of expression in untreated wild-type cells relative to untreated RXRα−/− cells, which may be a consequence of the low level of retinoids present in the serum used to culture the fetal liver cells and consequent activation of Epo expression in wild-type cells. At E12.25, primary hepatocytes from embryos of both genotypes expressed Epo at equivalent levels, and in neither case was Epo expression induced by RA treatment. Thus, the Epo gene is RA inducible in E10.25 hepatocytes in a RXRα-dependent manner, whereas the gene is refractory to RA treatment at E12.25 regardless of RXRα genotype.
The Epo enhancer contains a retinoic acid response element
The results described above are consistent with the Epo gene being a direct target of RXRα transactivation during the early phase of liver development. We next asked whether a luciferase reporter gene utilizing the Epo gene promoter and enhancer was retinoic acid responsive in a transfection assay. The reporter gene we used, called wtEpo–LUC, contained the sequences from −411 to +10 of the mouse Epo gene promoter, plus a 96-bp fragment containing the Epo gene enhancer (including the HIF1-binding site and the DR2 element). This construct was only marginally active in transfected normoxic Hep3B cells, and was not retinoic acid inducible. However, under hypoxic (1% O2) conditions, this reporter gene was inducible by retinoic acid treatment (Fig. 5a). Because DR2 elements are known as retinoic acid response elements, we asked whether retinoic acid responsiveness was conferred through this sequence. When this site was mutated, the Epo promoter/enhancer reporter gene was no longer responsive to retinoic acid (mutEpo–LUC construct; Fig. 5a).
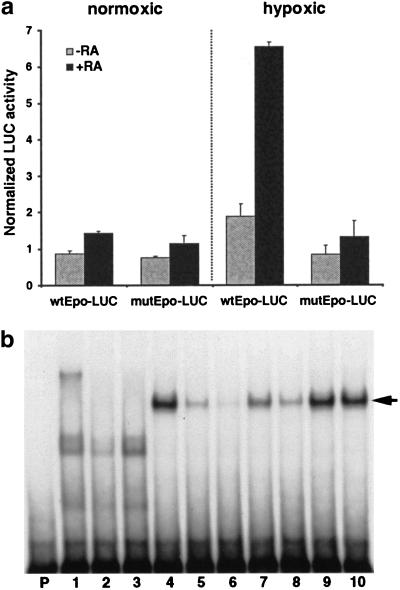
Figure 5.
The Epo gene is retinoic acid responsive through the enhancer DR2 element. (a) Transfection analysis. The wild-type Epo promoter/enhancer reporter gene construct (wtEpo-LUC) or a variant in which the DR2 element was mutated (mutEpo-LUC) were cotransfected with RXR and RAR expression constructs into Hep3B cells under normoxic conditions or under hypoxic conditions (1% oxygen) in the absence or presence of 10−6 M all trans RA, and cultured for 24 h. Luciferase activity was normalized to β-galactosidase activity. (b) Gel shift analysis. In vitro-translated human RXR and RAR protein was used under standard conditions with labeled probe representing the human Epo gene DR2 sequence. (Lane P) is probe alone; (lane 1) mock in vitro translation only; (lane 2) RXRα only; (lane 3) RARα only; (lanes 4–10) RXRα plus RARα. Cold competitor oligonucleotides included were a 10× and 50× molar excess of the mouse Epo enhancer DR2 (lanes 5,6), 10× and 50× excess of the human Epo enhancer DR2 (lanes 7,8), or 10× and 50× excess of an irrelevant sequence (a loxP sequence) of the same length (lanes 9,10). The arrow indicates the bound RXR–RAR complex.
We next addressed whether the enhancer DR2 element functions as a RXR–RAR-binding site and as a retinoic acid response element. Gel shift assays (Fig. 5b) using in vitro-translated RXRα and RARα protein showed that neither protein alone bound to the Epo DR2, but together the RXR–RAR heterodimer formed a specific complex on this element that was competed by unlabeled DR2 sequences from the human and mouse Epo genes, but not by unlabeled nonspecific competitor sequence. The human sequence contains a single base change relative to the mouse (the mouse sequence is TGACCTCTT GACCC; the human sequence changes the underlined T to a C), and the human sequence is approximately fourfold lower in affinity for RXR–RAR binding, consistent with the known sequence requirements for DNA recognition by retinoic acid receptors.
A reporter gene (4xDR2-TK-LUC) was prepared by insertion of four copies of the mouse Epo DR2 element into a minimal thymidine kinase promoter–luciferase construct. The element was multimerized to maximize responsiveness, a well-known requirement when the minimal TK promoter is used in transfection studies; we also assayed a reporter construct containing a single DR2 element with comparable, although less dramatic, results (data not shown). In CV-1 cells, which express a very low level of endogenous nuclear receptors, this reporter gene was not basally active and was not retinoic acid inducible. However, when RXR and RAR expression vectors were cotransfected into these cells, the reporter gene was dramatically induced by retinoic acid treatment (Fig. 6a). Thus, as shown by gel shift and transfection assays, the DR2 element in the Epo gene 3′ enhancer is a bonafide retinoic acid response element.
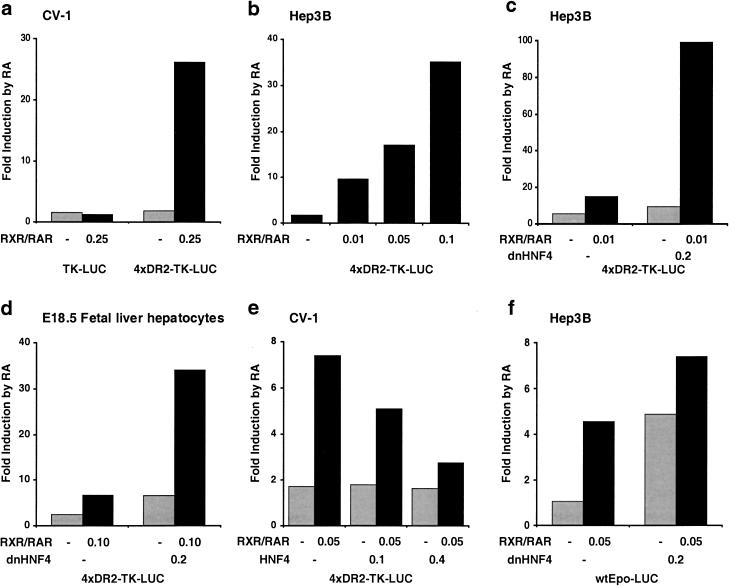
Figure 6.
HNF4 is competitive with RXR–RAR for occupancy of the Epo enhancer DR2 element. The indicated reporter genes were transfected into CV-1 cells (a,e), Hep3B cells (b,c,f), or primary E18.5 fetal hepatocytes (d) under normoxic conditions (a–e) or hypoxic conditions (f) in the presence or absence of 10−6 M all trans RA, with cotransfected receptor expression plasmids as indicated. Numbers indicate micrograms of transfected DNA.
Competitive regulation of the Epo enhancer DR2 element by RXR–RAR and HNF4
In Hep3B cells, the chromosomal Epo gene is inducible by hypoxia (unlike in CV-1 cells), and therefore represents a more authentic context for examination of Epo gene expression. Hep3B cells express endogenous RXR and RAR (see below). In our initial transfection assays, the 4xDR2-TK-LUC reporter gene was only modestly induced by RA treatment, although we found that we could increase responsiveness by increasing the amount of cotransfected RXR and RAR expression vector (Fig. 6b). We suspected that the known high level in Hep3B cells of HNF4, which is already established to bind as a homodimer to the Epo enhancer DR2 element (Galson et al. 1995), might interfere with transactivation by the retinoic acid receptor RXR–RAR heterodimer. To confirm this model, we made use of a dominant-negative HNF4 construct (dnHNF4), in which a mutation in the DNA binding domain renders the encoded protein fully able to dimerize with wild-type HNF4, but the dimeric complex is unable to bind DNA (Taylor et al. 1996). Introduction of dnHNF4 alone allowed significant (ninefold) RA-inducible transactivation of the reporter gene through endogenous RA receptors, and cotransfection of dnHNF4 with RXR and RAR expression vectors resulted in synergistic activation of reporter gene activity (Fig. 6c). Thus, in Hep3B cells, the Epo gene enhancer DR2 element is transactivated by retinoic acid receptors, in a manner that is inhibited by HNF4. We obtained similar results in primary E18.5 fetal hepatocytes (Fig. 6d).
In CV-1 cells, introduction of dnHNF4 did not alter expression of the reporter gene (data not shown), consistent with the absence of endogenous HNF4 expression in these cells. However, by cotransfecting wild-type HNF4 expression vector into CV-1 cells, we were able to decrease the ability of RXR–RAR to transactivate through the Epo enhancer DR2 element (Fig. 6e). Thus, the activity of this reporter gene is titratable by adjusting the relative levels of HNF4 and RXR–RAR.
Finally, we asked whether retinoic acid responsiveness of the Epo gene promoter/enhancer was also inhibited by HNF4. In transfected hypoxic Hep3B cells, the presence of dnHNF4 allowed a fivefold responsiveness to RA of the wtEpo–LUC reporter via endogenous receptors, and cotransfection of RXR–RAR and dnHNF4 together resulted in an even greater response (Fig. 6f). Collectively, these results indicate that HNF4 and RXR–RAR are mutually competitive for recognition and occupancy of the Epo enhancer DR2 element.
Discussion
The period between E9.5 and E12.5 in the mouse embryo is a particularly critical time in hematopoiesis, as this is when the embryo transitions from yolk sac (primitive) to fetal liver (definitive) hematopoiesis. We have found that the initial phase of erythropoiesis in the fetal liver is acutely dependent upon RXRα function. In RXRα−/− embryos prior to E12.5, erythroid differentiation in the fetal liver is severely compromised, although this deficiency does not cause any apparent consequence in other parts of the embryo as there is adequate provision of primitive red blood cells from the yolk sac (this process appears to be unaffected by RXRα gene mutation). As shown in this study by genetic and molecular approaches, the fetal liver phenotype prior to E12.5 is accounted for by a decrease in the level of Epo expression. Thereafter, control of hematopoiesis becomes independent of RXRα function; Epo expression is normal in these later embryos, and erythropoiesis is restored to normal as well.
Based on results described in this study, we propose that the initial expression of Epo in the fetal liver, from E9.5 through E11.5, is under retinoic acid control. The transduction of this retinoic acid signal requires RXRα, and occurs by occupancy of and transactivation through the DR2 element in the Epo gene 3′ enhancer. Most probably, the active complex is a heterodimer of RXRα with any of the RARs, as all three RAR genes are expressed in the early fetal liver (Dolle et al. 1990), and there is no known erythropoietic phenotype in any single or double RAR gene mutations (Mendelsohn et al. 1994), although it is possible that the phenotype described in this study could have been overlooked. In the absence of RXRα, Epo gene expression is reduced substantially, resulting in a deficiency in the expansion and differentiation of erythroid progenitors within the fetal liver. Starting around E11.5 and continuing for the remainder of fetal liver erythropoiesis, HNF4 activity supplants retinoic acid receptor function in controlling expression of the Epo gene, thereby restoring normal levels of Epo gene expression and promoting the recovery of the transient liver defect in RXRα−/− embryos.
Our model is bolstered by genetic analysis of HNF4 function. Conventional HNF4−/− embryos die at gastrulation with a deficiency in extraembryonic endoderm (Chen et al. 1994). Very recently (Li et al. 2000), this phenotype has been rescued through E12.0 by use of tetraploid aggregation procedures, in which extraembryonic tissues are wild type in function and the embryo itself is HNF4-deficient. HNF4-deficient embryos at E12.0 are substantially compromised in fetal liver Epo expression, demonstrating that HNF4 is a required regulator of Epo gene expression at least from E12.0 onward. However, the morphology and histology of HNF4−/− fetal liver at E12.0 is normal, with apparently normal erythropoietic differentiation. Epo expression in earlier stage HNF4−/− embryos was not examined, but we predict that Epo expression must be normal in earlier embryos (i.e., at E10.5), on the basis of the lack of histological liver deficiencies in HNF4−/− embryos at E12.0 and the demonstrated necessity (Wu et al. 1995) of normal Epo function for normal liver histogenesis through E12.5. Thus, these observations indicate that HNF4 is not required for erythropoiesis during the early phase of liver development, but is required for normal Epo expression in later embryos, and support our model that RXR–RAR is the critical regulator of erythropoiesis during the initial period of erythropoiesis in the fetal liver.
We have presented evidence in this study that HNF4 and RXR–RAR can compete directly for occupancy of the Epo enhancer DR2 element. These results resolve a long-standing puzzling observation that the Epo enhancer DR2 element, although being nearly canonical in sequence to other retinoic acid response elements, is not retinoic-acid responsive in Hep3B cells (Blanchard et al. 1992). Thus, in Hep3B cells and in late-gestation hepatocytes, HNF4 is present in stochiometric excess relative to RXR–RAR, essentially preventing transactivation by retinoic acid. These results also explain why the chromosomal Epo gene itself is not RA inducible in the livers of embryos older than E12.0 or in postnatal animals (data not shown), although it is clearly so in younger embryos (Fig. 3).
To explain the observation that expression of the Epo gene transitions from RXR–RAR control to HNF4 control around E11.5, we favor a model in which Epo expression occurs in fetal hepatocytes. Accordingly, but for reasons we have not yet defined, RXR–RAR is dominant in activity over HNF4 prior to E11.5, but with HNF4 being dominant over RXR–RAR at times thereafter. HNF4 protein may be less abundant in the earlier period, its activity may be suppressed by modification or by ligand, or the context of the Epo promoter/enhancer prior to E11.5 may favor the activity of RXR–RAR even in the presence of excess HNF4 (possibly through some type of interaction with HIF1, which binds to the hypoxic response element immediately adjacent to the DR2 element, or through differential interaction with transcriptional coactivators).
There are two alternative models that bear consideration. First, RXRα function may occur outside of the fetal liver, and in a secondary or indirect manner affect Epo expression in the liver. Two feasible outside candidate tissues might be the heart and placenta, as there are defects in both tissues in RXRα−/− embryos, and the placenta in particular has been recently suggested to influence definitive erythropoiesis in the fetal liver (Ihle 2000). However, the placental (Sapin et al. 1997) and cardiac (Dyson et al. 1995) defects of RXRα−/− embryos are only first apparent at E12.5 and progressively worsen with advancing age, unlike the erythropoietic phenotype and the reduction in Epo expression which are most severe prior to E12.5 and resolve thereafter. Furthermore, there are no known defects of any type in RXRα heterozygous embryos that would account in an indirect manner for the erythropoietic phenotype of RXRα/Epo double heterozygous embryos (Fig. 4). As a second alternative, Epo expression prior to E11.5 may occur in a nonhepatocyte cell population of the fetal liver under RXRα control, and after E11.5 in hepatocytes under HNF4 control. We can exclude the hematopoietic and endothelial lineages, as chimeric embryos comprised of RXRα−/− cells and Flk1−/− cells (the latter are unable to contribute to these lineages; Shalaby et al. 1997) have no erythropoietic phenotype (C. Tran and H.M. Sucov, unpubl.), although we cannot yet formally rule out the possibility that RXRα might function in another nonhepatocyte lineage.
It is of interest to speculate as to the developmental logic that causes the Epo gene to be regulated transiently by retinoic acid receptors before becoming dominated by HNF4 activity. Because genetic analysis (Li et al. 2000) shows that HNF4 does not control Epo expression prior to E11.5, the availability of an alternative mechanism (i.e., retinoic acid receptors) through which to activate Epo gene expression from E9.5–E11.5 has two potential adaptive benefits. First, this mechanism allows the initiation of erythropoiesis in the fetal liver ∼2 d (in the mouse embryo; possibly longer in human embryos) earlier than otherwise, which provides an earlier contribution of mature red blood cells in peripheral circulation as the yolk sac-derived primitive erythrocytes are dying. Second, in addition to promoting erythropoietic differentiation, Epo also stimulates erythropoietic progenitor cell proliferation, and this may be of even greater benefit in establishing definitive hematopoiesis within the fetal liver prior to the time (around E12.5–E13.5) when definitive erythropoiesis is required for embryo viability.
Interestingly, occupancy of the DR2 element by HNF4 in the context of either the Epo promoter or the TK promoter does not lead to a substantial transcriptional response. That is, there was a fairly low basal level of activity of the DR2-containing reporter genes in Hep3B cells; this was also true in CV-1 cells transfected with HNF4 expression plasmids. HNF4 is constitutively active but is a relatively weak transcriptional activator, particularly in certain promoter and cellular contexts (Jiang et al. 1995; Sladek et al. 1999). This may explain why Epo gene regulation during the E9.5–E11.5 period has evolved to depend on the transcriptionally more potent RXR–RAR heterodimer, assuming that a high level of Epo expression is beneficial (see above). One clear consequence of the relatively weak transcriptional activity of HNF4 is that Epo expression in the later stage liver is not constitutive, as would occur if HNF4 was able to basally activate expression, but is hypoxic-dependent via the activity of HIF1. The transition from RXR–RAR control to HNF4 control may therefore serve to switch regulation of Epo expression from a paracrine mode controlled by local retinoic acid production to an environmentally responsive mode controlled solely by oxygen tension, as is found throughout the remainder of fetal and postnatal life.
Materials and methods
Histology
Embryos were isolated at appropriate gestational stages and genotyped retrospectively by analysis of yolk sac DNA. For hematoxylin/eosin staining, embryos were fixed in 10% phosphate-buffered formalin, paraffin embedded, sectioned at 5-μ thickness, and stained and processed by standard procedures. For Richardson's stained samples, whole embryos or isolated liver tissue were fixed in phosphate buffer containing 2% paraformaldehyde and 2.5% glutaraldehyde, followed by 1% osmium tetroxide in 0.1 M cacodylate buffer, and then embedded in Epon-812. The 1-μ sections were cut with a glass knife and stained in a solution containing 0.5% methylene blue, 0.5% azure II, and 0.5% borax.
Flow cytometry analysis
Fetal liver tissue was isolated by dissection from individual embryos and dissociated by trituration in Hank's balanced salt solution containing 0.02% EDTA, 0.2% BSA, and 0.03% sodium azide on ice. A total of 0.05–0.2 million fetal liver cells from each sample were stained with monoclonal antibodies for surface expression of c-Kit (2B8) and TER-119 (Ly-76) (both from Pharmingen), after blocking FcγII/III receptors by use of supernatant from the 2.4G2 hybridoma. Two-color analyses were performed on individual fetal liver samples by flow cytometry in a Becton Dickinson FACScalibur. A total of 50,000 events were collected per sample, and analyses performed after excluding dead cells. Because the actual developmental stage varied by up to half a day between litters at the same nominal stage, we calculated within each litter the fold increase or decrease in each category for mutant embryos relative to wild-type plus heterozygote embryos, and then combined these fold changes for all litters to generate a composite depiction of the normal and mutant phenotypes.
Colony outgrowth assay
E12.5 fetal liver tissue was dissociated by trituration, and an aliquot counted to determine nucleated cell number. Cells were mixed with Methocult 3430 (Stem Cell Technologies, Vancouver, Canada) and incubated 7 d, then stained with benzidine and scored by the criteria of containing 20 or more cells, of which at least 15% were benzidine positive.
Quantitative RT–PCR
Total RNA was extracted from individual fetal liver tissue by guanidinium isothiocyanate extraction. A total of 1 μg of total RNA was reverse transcribed in a 20-μL reaction containing 2 μL (100–500ng) of Random Decamer Primer (Ambion), 100 ng of Epo gene-specific primer (5′-GAGCAAGTTCGTCGGTCCA-3′), 0.5 mM dNTPs, and 200 units of M-MuLV Reverse Transcriptase (GIBCO BRL) in First Strand buffer (50 mM Tris-HCl at pH 8.3, 75 mM KCl, 3 mM MgCl2, and 5 mM DTT). The RT reaction was diluted with water to 80 μL, and 1 μL was used for PCR. To initially determine the linear range of Epo amplification, PCR was performed with multiple samples in a 30-μL reaction containing 100 ng of each Epo primer (Zimmermann and Rich 1997), 5 μCi of [α-32P]dCTP, and 2.5 units of Taq DNA Polymerase (GIBCO) in PCR buffer [20% DMSO, 134 mM Tris at pH 8.8, 33 mM (NH4)2SO4, 20 mM β-mercaptoethanol, 6 mM MgCl2, and 1 mM dNTPs] with the following parameters: 1 min at 94°C, 2 min at 57°C, and 2 min at 72°C. The amplification profile was examined at 3-cycle intervals from 14 to 44 cycles. PCR products were separated by nondenaturing 4% polyacrylamide gel electrophoresis in 1 × TBE at 4°C, and signal intensity was analyzed by PhosphorImaging (Molecular Dynamics). For quantitative analysis of Epo transcripts level, PCR was performed with the addition of 0.4 μM of 18S ribosomal RNA primers and varying amounts of competimer mixture (Ambion) as an internal control. The varying ratios of 18S primers ensures that a linear range is achieved for this product as well, and allows comparison of Epo mRNA abundance between samples by measurement of the intensity of the Epo and 18S products at any 18S primer/competimer ratio in which linear amplification of both products occurs. Amplification of other genes was done exactly as above, except for substitution of primers for HNF3α (GGCCTACTCCTCTGTCCCTGTCAG and ATGCCAGCCA CAGCACCGGGACTC), albumin (CCCCACTTAGCCTCTG GCAAAAT and AGACTCATCGGCAACACACGTCT), or c-kit (ACAGGAGCAGAGCAAAGGTG and CGACCACAAAG CCAATGAGC). For in vitro whole embryo culture, wild-type embryos were isolated at E10.0 intact within their yolk sacs and cultured in DMEM with 50% FCS in the presence of 10−6 M all trans retinoic acid or solvent alone with gentle agitation on a rocker platform in a tissue culture incubator. Other compounds tested and their final concentrations included thyroid hormone (T3; 10−8 M), Wy14,643 (5 × 10−6 M), chenodeoxycholic acid (10−4 M), and pregnenalone 16α-carbonitrile (PCN; 10−5 M). After 9 h, fetal liver tissue was isolated from these embryos and processed as above. For primary cell culture, fetal liver tissue was dissociated from individual embryos by trituration, and plated in the presence of solvent only or 10−6 M all trans retinoic acid in replicate wells of rat tail collagen-coated dishes. After 4 h of incubation, nonadherent cells were washed off and adherent cells collected for analysis. These conditions enrich for hepatocytes (HNF3α-expressing) and decrease representation of hematopoietic cells (GATA1-expressing).
Electrophoretic mobility shift assay
Double-stranded oligonucleotide probe containing the human or mouse Epo DR2 element was labeled with [α-32P]dCTP by Klenow fill-in reaction. Preincubation was performed for 20 min on ice in a 19-μL reaction containing 150 ng of poly [d(I-C)] plus 60 ng of nonspecific single-strand oligomer in a binding buffer (75 mM KCl, 7.5% glycerol, 20 mM HEPES, 0.1% NP40, 2 mM DTT) in the presence or absence of 3 μL each of in vitro-translated hRXRα and hRARα, followed by addition of 0.8 ng of labeled probe and 20 min of further incubation on ice. DNA–protein complexes (20 μL) were separated by 5% polyacrylamide gel electrophoresis in 0.5× TBE at room temperature. Competition assays were performed in the presence of 8 ng (10×) and 40 ng (50×) of unlabeled human or mouse Epo DR2 sequences or a nonspecific (loxP) sequence.
DNA constructs
The wild-type Epo promoter enhancer reporter construct (wtEpo-LUC) was made by insertion of a 420-bp XbaI–SmaI fragment (positions 1–420 of GenBank M12482) representing the sequences between −410 and +10 relative to the transcription start site of the mouse Epo gene (Shoemaker and Mitsock 1986) into a luciferase vector lacking other promoter sequence. The original plasmid from which this fragment was obtained was described in Wu et al. (1995). A 96-bp fragment (positions 399–494 of GenBank L13456) containing the complete enhancer element as functionally defined previously (Pugh et al. 1991) was prepared by PCR amplification, subcloned and validated by sequencing, and inserted as a single copy 5′ of the promoter fragment. To specifically mutate the DR2 element, an enhancer fragment of the same length was prepared by PCR amplification but using a primer with an altered DR2 sequence, which was sequenced to confirm correct amplification and inserted 5′ of the Epo promoter fragment as above. The wild-type mouse DR2 element is TGACCTCTTGACCT; the mutant sequence is TTCATTCTGGCTAA. To make 4xDR2-TK-LUC, a 39-bp fragment containing the wild-type DR2 sequence was inserted as a four- copy head-to-tail multimer into the HindIII site of a parental TK-LUC plasmid.
Transient transfection assays
CV-1 cells were cultured in DMEM with 10% FBS and plated on 35-mm tissue culture dishes at 60% confluency. A total of 3 μg of reporter plasmid plus 0.25 μg of CMX–β-gal were cotransfected by calcium phosphate precipitation in the absence or presence of cotransfected CMX–hRXRα, CMX–hRARα, or pSG5-rHNF4α1 expression vectors. The total amount of CMX promoter and total plasmid (5 μg) was adjusted by addition of CMX–EGFP and pGEM4 plasmids, respectively. After a 16-h incubation, the transfection medium was replaced with fresh medium with or without 10−6 M all-trans retinoic acid (Sigma), and cells were incubated for 24 h before harvest. Hep3B cells were cultured in DMEM containing 10% FBS and plated into 35-mm tissue culture dishes at 80% confluency, and were transfected using Lipofectin (GIBCO BRL). Lipofection was performed for 14 h, followed by replacement with medium with or without 10−6 M of all-trans RA, and cells were cultured another 24 h before harvest. For normoxic conditions, cells were cultured in standard incubators. For hypoxic conditions, following transfection and replacement of medium, cells were transferred to a chamber that was flushed with 1%O2/5%CO2/94%N2, then sealed and incubated for 24 h. Hypoxic medium was first sparged before use, and CoCl2 was added at a concentration of 10−4 M. For all reporter gene assays, cells were rinsed with 1× PBS and scraped with 250 μL of 1× PBS. Cell suspensions were centrifuged and pellets were lysed in 50–100 μL of lysis buffer (TROPIX) by freeze-thaw. A total of 10 μL of cell lysate was assayed with the Luciferase Assay System (Promega), and with the Galacto-Light Plus (TROPIX) assay system. Primary hepatocytes were prepared by trituration of fetal liver tissue, plated in rat tail type IV collagen-coated dishes in DMEM with 10% FCS, allowed to adhere for 4 h, washed to remove nonadherent hematopoietic cells, and then transfected by calcium phosphate as above.
Acknowledgments
We thank Todd Leff for provision of CMV-dnHNF4, Ron Evans for provision of CMX–hRXRα and CMX–hRARα, and Francie Sladek for provision of pSG5–rHNF4α1. This work was supported in its early phase by a pilot/feasibility project grant (to H.M.S.) as a component of the University of Southern California Research Center for Liver Diseases (PHS grant no. DK48522). G.H.-H. was supported by the Stowers Institute for Medical Research. H.W. is an Assistant Investigator of the Howard Hughes Medical Institute. T.C. was supported by a predoctoral fellowship from the American Heart Association.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
A recent report has described retinoic acid induction of chromosomal Epo gene expression in P19 cells (Kambe, T., Tada-Kambe, J., Kuge, Y., Yamaguchi-Iwai, Y., Nagao, M., and Sasaki, R. 2000. Blood 96: 3265–3271).
Footnotes
E-MAIL sucov@hsc.usc.edu; FAX (323) 442-2764.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.871601.
References
- Avery L, Wasserman S. Ordering gene function: The interpretation of epistasis in regulatory hierarchies. Trends Genet. 1992;8:312–316. doi: 10.1016/0168-9525(92)90263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard KL, Acquaviva AM, Galson DL, Bunn HF. Hypoxic induction of the human erythropoietin gene: Cooperation between the promoter and enhancer, each of which contains steroid receptor response elements. Mol Cell Biol. 1992;12:5373–5385. doi: 10.1128/mcb.12.12.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunn HF, Gu J, Huang LE, Park JW, Zhu H. Erythropoietin: A model system for studying oxygen-dependent gene regulation. J Exp Biol. 1998;201:1197–1201. doi: 10.1242/jeb.201.8.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell JE. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes & Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- Dolle P, Ruberte E, Leroy P, Morriss KG, Chambon P. Retinoic acid receptors and cellular retinoid binding proteins. I. A systematic study of their differential pattern of transcription during mouse organogenesis. Development. 1990;110:1133–1151. doi: 10.1242/dev.110.4.1133. [DOI] [PubMed] [Google Scholar]
- Dyson E, Sucov HM, Kubalak SW, Schmid-Schonbein GW, DeLano FA, Evans RM, Ross J, Chien KR. Atrial-like phenotype is associated with embryonic ventricular failure in RXRα−/− mice. Proc Natl Acad Sci. 1995;92:7386–7390. doi: 10.1073/pnas.92.16.7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galson DL, Tsuchiya T, Tendler DS, Huang LE, Ren Y, Ogura T, Bunn HF. The orphan receptor hepatic nuclear factor 4 functions as a transcriptional activator for tissue-specific and hypoxia-specific erythropoietin gene expression and is antagonized by EAR3/COUP-TF1. Mol Cell Biol. 1995;15:2135–2144. doi: 10.1128/mcb.15.4.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MA, Glass GA, Cunningham JM, Bunn HF. The regulated expression of erythropoietin by two human hepatoma cell lines. Proc Natl Acad Sci. 1987;84:7972–7976. doi: 10.1073/pnas.84.22.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houssaint E. Differentiation of the mouse hepatic primordium. II. Extrinsic origin of the haemopoietic cell line. Cell Differ. 1981;10:243–252. doi: 10.1016/0045-6039(81)90007-5. [DOI] [PubMed] [Google Scholar]
- Ihle JN. The challenge of translating knockout phenotypes into gene function. Cell. 2000;102:131–134. doi: 10.1016/s0092-8674(00)00017-9. [DOI] [PubMed] [Google Scholar]
- Ikuta K, Kina T, MacNeil I, Uchida N, Peault B, Chien YH, Weissman IL. A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell. 1990;62:863–874. doi: 10.1016/0092-8674(90)90262-d. [DOI] [PubMed] [Google Scholar]
- Jiang G, Nepomuceno L, Hopkins K, Sladek FM. Exclusive homodimerization of the orphan receptor hepatocyte nuclear factor 4 defines a new subclass of nuclear receptors. Mol Cell Biol. 1995;15:5131–5143. doi: 10.1128/mcb.15.9.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner P, Grondona JM, Mark M, Gansmuller A, LeMeur M, Decimo D, Vonesch JL, Dolle P, Chambon P. Genetic analysis of RXRα developmental function: Convergence of RXR and RAR signaling pathways in heart and eye morphogenesis. Cell. 1994;78:987–1003. doi: 10.1016/0092-8674(94)90274-7. [DOI] [PubMed] [Google Scholar]
- Kina T, Ikuta K, Takayama E, Wada K, Majumdar AS, Weissman IL, Katsura Y. The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br J Haematol. 2000;109:280–287. doi: 10.1046/j.1365-2141.2000.02037.x. [DOI] [PubMed] [Google Scholar]
- Koury ST, Bondurant MC, Koury MJ, Semenza GL. Localization of cells producing erythropoietin in murine liver by in situ hybridization. Blood. 1991;77:2497–2503. [PubMed] [Google Scholar]
- Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4α. Genes & Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- Mendelsohn C, Lohnes D, Decimo D, Lufkin T, LeMur M, Chambon P, Mark M. Function of the retinoic acid receptors (RARs) during development. (II) Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development. 1994;120:2749–2771. doi: 10.1242/dev.120.10.2749. [DOI] [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Potter SS. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, et al. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Pugh CW, Tan CC, Jones RW, Ratcliffe PJ. Functional analysis of an oxygen-regulated transcriptional enhancer lying 3′ to the mouse erythropoietin gene. Proc Natl Acad Sci. 1991;88:10553–10557. doi: 10.1073/pnas.88.23.10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapin V, Dolle P, Hindelang C, Kastner P, Chambon P. Defects of the chorioallantoic placenta in mouse RXRα null fetuses. Dev Biol. 1997;191:29–41. doi: 10.1006/dbio.1997.8687. [DOI] [PubMed] [Google Scholar]
- Semenza GL, Nejfelt MK, Chi SM, Antonarakis SE. Hypoxia-inducible nuclear factors bind to an enhancer element located 3′ to the human erythropoietin gene. Proc Natl Acad Sci. 1991;88:5680–5684. doi: 10.1073/pnas.88.13.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby F, Ho J, Stanford WL, Fischer KD, Schuh AC, Schwartz L, Bernstein A, Rossant J. A requirement for Flk1 in primitive and definitive hematopoiesis and vasculogenesis. Cell. 1997;89:981–990. doi: 10.1016/s0092-8674(00)80283-4. [DOI] [PubMed] [Google Scholar]
- Shoemaker CB, Mitsock LD. Murine erythropoietin gene: Cloning, expression, and human gene homology. Mol Cell Biol. 1986;6:849–858. doi: 10.1128/mcb.6.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek FM, Ruse MD, Nepomuceno L, Huang SM, Stallcup MR. Modulation of transcriptional activation and coactivator interaction by a splicing variation in the F domain of nuclear receptor hepatocyte nuclear factor 4α1. Mol Cell Biol. 1999;19:6509–6522. doi: 10.1128/mcb.19.10.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucov HM, Dyson E, Gumeringer CL, Price J, Chien KR, Evans RM. RXRα mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes & Dev. 1994;8:1007–1018. doi: 10.1101/gad.8.9.1007. [DOI] [PubMed] [Google Scholar]
- Taylor DG, Haubenwallner S, Leff T. Characterization of a dominant negative mutant form of the HNF-4 orphan receptor. Nucleic Acids Res. 1996;24:2930–2935. doi: 10.1093/nar/24.15.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimborn T, Gribnau J, Grosveld F, Fraser P. Mechanisms of developmental control of transcription in the murine α- and β-globin loci. Genes & Dev. 1999;13:112–124. doi: 10.1101/gad.13.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Liu X, Jaenisch R, Lodish HF. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 1995;83:59–67. doi: 10.1016/0092-8674(95)90234-1. [DOI] [PubMed] [Google Scholar]
- Zhu H, Bunn HF. Oxygen sensing and signaling: Impact on the regulation of physiologically important genes. Respiration Physiol. 1999;115:239–247. doi: 10.1016/s0034-5687(99)00024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann F, Rich IN. Mammalian homeobox B6 expression can be correlated with erythropoietin production sites and erythropoiesis during development, but not with hematopoietic or nonhematopoietic stem cell populations. Blood. 1997;89:2723–2735. [PubMed] [Google Scholar]