Abstract
Emerging technologies allow the high-throughput profiling of metabolic status from a blood specimen (metabolomics). We investigated whether metabolite profiles could predict the development of diabetes. Among 2,422 normoglycemic individuals followed for 12 years, 201 developed diabetes. Amino acids, amines, and other polar metabolites were profiled in baseline specimens using liquid chromatography-tandem mass spectrometry. Cases and controls were matched for age, body mass index and fasting glucose. Five branched-chain and aromatic amino acids had highly-significant associations with future diabetes: isoleucine, leucine, valine, tyrosine, and phenylalanine. A combination of three amino acids predicted future diabetes (>5-fold higher risk for individuals in top quartile). The results were replicated in an independent, prospective cohort. These findings underscore the potential importance of amino acid metabolism early in the pathogenesis of diabetes, and suggest that amino acid profiles could aid in diabetes risk assessment.
Metabolic diseases are often present for years before becoming clinically apparent. For instance, by the time relative insulin deficiency manifests itself as hyperglycemia and the diagnosis of type 2 diabetes is made, significant pancreatic α-cell insufficiency has already occurred1. Current clinical and laboratory predictors such as body mass index or fasting glucose can be helpful in gauging diabetes risk2, but they often reflect extant disease, are most useful when assayed in temporal proximity to the development of overt diabetes, and may provide little additional insight regarding pathophysiologic mechanisms. Given the availability of effective interventions for delaying or preventing the onset of type 2 diabetes and the increasing burden of the condition worldwide, earlier identification of individuals at risk is particularly important3-6.
Emerging technologies have enhanced the feasibility of acquiring high-throughput profiles of a whole organism’s metabolic status (metabolite profiling, or metabolomics)7-10. These techniques, which allow assessment of large numbers of metabolites that are substrates and products in metabolic pathways, are particularly relevant for studying metabolic diseases such as diabetes. Furthermore, in addition to serving as potential biomarkers of disease11, metabolites may have unanticipated roles as regulatory signals with hormone-like functions12,13 or effectors of the disease process itself14.
Recent cross-sectional studies have documented differences in blood metabolite profiles before and after glucose loading15-17 and in obese compared with lean individuals14. These studies have noted differences in levels of C3 and C5 acylcarnitines, glutamine/glutamate, additional amino acids, and other small molecules. These observations raise the possibility that alterations in plasma metabolite levels could presage the onset of overt diabetes and therefore aid in the identification of ‘at risk’ individuals by adding information over standard clinical markers. We performed metabolite profiling in participants from two large, longitudinal studies, with the goal of identifying early pathophysiological changes that might also serve as novel predictors of future diabetes.
RESULTS
Metabolite profiling in the Framingham Heart Study
We performed a nested case-control study in the Framingham Offspring Study. Among 2,422 eligible, non-diabetic attendees to a routine examination between 1991 and 1995, 201 individuals developed new-onset diabetes during a 12-year follow-up period. Metabolite profiling was performed on the baseline samples from 189 of these individuals, for whom 189 propensity-matched controls could be identified from the same baseline examination who did not develop diabetes. Cases and controls were closely matched with respect to age, sex, body mass index (BMI), and fasting glucose (Table 1). BMI and waist circumference were also available from four years (for both measures) to eight years (for BMI) prior to the index examination. There were no significant differences in BMI and waist circumference at these earlier time periods between cases and controls (data not shown).
Table 1.
Baseline characteristics (Framingham Heart Study)
| Cases (n=189) |
Matched Controls (n=189) |
Random cohort (n=400) |
|
|---|---|---|---|
| Clinical characteristics | |||
| Age, years | 56 ± 9 | 57 ± 8 | 55 ± 9 |
| Women, % | 42% | 42% | 58% |
| Body mass index, kg/m2 | 30.5 ± 5.0 | 30.0 ± 5.5 | 26.8 ± 4.6 |
| Waist circumference, cm | 102.3 ± 12.1 | 99.6 ± 13.5 | 90.8 ± 13.8 |
| Hypertension, % | 53% | 53% | 27% |
| Parental history of diabetes*, % | 31% | 18% | 21% |
| Physical activity index | 35 ± 6.2 | 35 ± 7.3 | 35 ± 6.0 |
| Total caloric intake, kcal | 1,982 ± 660 | 1,866 ± 600 | 1854 ± 581 |
| Total protein intake, g | 82 ± 28 | 78 ± 28 | 76 ± 26 |
| Phenylalanine intake, g | 3.6 ± 1.2 | 3.4 ± 1.3 | 3.4 ± 1.1 |
| Tyrosine intake, g | 3.0 ± 1.0 | 2.8 ± 1.1 | 2.8 ± 1.0 |
| Leucine intake, g | 6.5 ± 2.2 | 6.1 ± 2.3 | 6.0 ± 2.1 |
| Isoleucine intake, g | 3.9 ± 1.3 | 3.7 ± 1.4 | 3.6 ± 1.3 |
| Valine intake, g | 4.3 ± 1.5 | 4.1 ± 1.5 | 4.0 ± 1.4 |
| Other laboratory tests | |||
| Fasting glucose, mg/dl | 105 ± 9 | 105 ± 9 | 94 ± 9 |
| 2-hour glucose (OGTT), mg/dl | 126 ± 32 | 118 ± 30 | 103 ± 27 |
| Serum triglycerides, mg/dl | 192 ± 114 | 151 ± 90 | 138 ± 93 |
| Fasting insulin, uU/ml | 13.7 ± 9.9 | 11.9 ± 8.8 | 8.1 ± 7.2 |
| HOMA-IR | 3.5 ± 2.6 | 3.1 ± 2.3 | 1.9 ± 1.8 |
| HOMA-B | 2.7 ± 2.0 | 2.4 ± 1.7 | 1.8 ± 1.5 |
Values are mean ± SD, or percentage.
Parental history information missing in 57 participants. HOMA-IR: homeostasis model assessment of insulin resistance. HOMA-B: homeostasis model assessment of beta cell function. OGTT: oral glucose tolerance test.
We assessed the correlations between baseline concentrations of metabolites. Mean correlations within groups of related molecules were highest for urea cycle metabolites (age- and sex-adjusted r=0.49; Figure 1), metabolites involved in nucleotide metabolism (r=0.38), amino acids (r=0.34), and methyl transfer metabolites (r=0.34).
Figure 1. Correlation matrix for plasma metabolite levels.
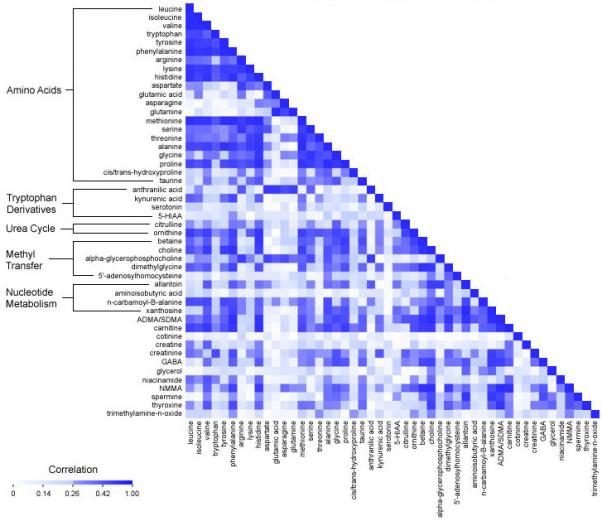
Age- and sex-adjusted Pearson correlation coefficients for baseline metabolite values in controls in the Framingham Offspring Study.
Selected metabolites predict future diabetes
In paired analyses, five metabolites had p-values of 0.001 or smaller for the baseline differences between cases and controls (Supplementary Table 1). Fasting concentrations were higher in the cases in all instances. Three of these metabolites were branched chain amino acids: leucine (p=0.0005), isoleucine (p=0.0001), and valine (p=0.001). The other two were aromatic amino acids: phenylalanine (p<0.0001) and tyrosine (p<0.0001). A third aromatic amino acid, tryptophan, had a p-value of 0.003.
The change in levels of the five branched chain or aromatic amino acids during the OGTT was not associated with incident diabetes, suggesting that levels after OGTT did not add predictive information to the baseline concentrations. For the other metabolites studied, only lysine exhibited a differential response to OGTT when cases were compared with controls (p=0.0005).
In additional analyses stratified by duration of follow-up, there was no evidence of an interaction between follow-up year and case-control difference for any of the amino acids (P >0.10 for all tests of interaction). Thus, the amino acids appeared to retain their predictive value for the development of new-onset diabetes up to 12 years after the baseline examination at which metabolite profiling was performed.
Predictive value adds to standard clinical or insulin resistance measures
Conditional logistic regression models were fitted to assess the association between baseline metabolite levels and future diabetes, adjusting for age, sex, BMI, and fasting glucose (Table 2). For the five amino acids of interest, each SD increment in log marker was associated with a 57% to 102% increased odds of future diabetes (p=0.0002 to 0.002). Individuals in the top quartile of individual plasma amino acid levels had a 2- to 3.5-fold higher odds of developing diabetes over the 12-year follow-up period, compared with those whose plasma amino acid levels were in the lowest quartile. Odds ratios for the metabolites remained strong when models were further adjusted for parental history of diabetes, and serum triglycerides, which were higher in cases than controls. Findings were also similar after adjustment for dietary intake of protein, amino acids, and total calories, and in the subgroup of individuals with propensity scores in the lowest tertile.
Table 2.
Relation of baseline amino acid levels to risk of future diabetes (Framingham Heart Study)
| Model | Isoleucine | Leucine | Valine | Tyrosine | Phenylalanine | Isoleucine, tyrosine, and phenylalanine |
|---|---|---|---|---|---|---|
| Models adjusting for age, sex, BMI, and fasting glucose (n=378) | ||||||
| Metabolite as continuous variable | ||||||
| Per SD | 1.70 (1.27-2.28) | 1.62 (1.20-2.17) | 1.57 (1.17-2.09) | 1.85 (1.35-2.55) | 2.02 (1.40-2.92) | 2.42 (1.66-3.54) |
| P | 0.0004 | 0.001 | 0.002 | 0.0001 | 0.0002 | <0.0001 |
| Metabolite as categorical variable | ||||||
| 1st quartile | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) |
| 2nd quartile | 1.11 (0.58-2.10) | 2.40 (1.24-4.68) | 1.49 (0.75-2.94) | 1.89 (0.94-3.81) | 1.39 (0.74-2.59) | 3.48 (1.68-7.23) |
| 3rd quartile | 2.14 (1.07-4.27) | 3.15 (1.46-6.84) | 2.15 (1.05-4.42) | 3.26 (1.56-6.84) | 2.12 (1.04-4.32) | 2.82 (1.25-6.34) |
| 4th quartile | 3.14 (1.51-6.55) | 3.66 (1.61-8.29) | 3.14 (1.43-6.86) | 2.82 (1.25-6.34) | 2.28 (1.00-5.20) | 5.99 (2.34-15.34) |
| P for trend | 0.001 | 0.004 | 0.003 | 0.010 | 0.035 | 0.0009 |
| Models adjusting for age, sex, BMI, fasting glucose, and parental history (n=272) | ||||||
| Metabolite as continuous variable | ||||||
| Per SD | 1.70 (1.19-2.44) | 1.67 (1.16-2.40) | 1.54 (1.07-2.22) | 2.02 (1.34-3.05) | 2.01 (1.29-3.12) | 2.62 (1.61-4.28) |
| P | 0.004 | 0.006 | 0.02 | 0.0008 | 0.002 | 0.0001 |
| Metabolite as categorical variable | ||||||
| 1st quartile | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) |
| 2nd quartile | 0.92 (0.41-2.09) | 1.95 (0.84-4.52) | 0.94 (0.39-2.29) | 1.64 (0.71-3.79) | 1.39 (0.64-2.99) | 4.60 (1.72-12.32) |
| 3rd quartile | 1.62 (0.68-3.85) | 2.99 (1.13-7.90) | 1.89 (0.78-4.56) | 3.26 (1.29-8.24) | 2.07 (0.84-5.08) | 3.69 (1.16-11.78) |
| 4th quartile | 2.98 (1.21-7.38) | 4.12 (1.51-11.30) | 2.85 (1.06-7.65) | 2.91 (1.03-8.25) | 2.10 (0.76-5.78) | 7.60 (2.14-27.07) |
| P for trend | 0.009 | 0.006 | 0.012 | 0.034 | 0.134 | 0.007 |
Values are odds ratios (95% confidence intervals) for diabetes, from conditional logistic regressions. All models are adjusted for age, sex, BMI, and fasting glucose. The amino acid combination is modeled according to the formula β1X1 + β2X2 + β3X3, with Xj denoting the standardized value for the jth amino acid, and βj denoting the regression coefficient from the regression model containing the indicated metabolites.
Fasting levels of the five amino acids were moderately correlated with biochemical measures of insulin resistance and α-cell function, including HOMA-IR and HOMA-B (r = 0.24 to 0.37, p<0.001; Supplementary Table 2). Nonetheless, the association of the plasma amino acid levels and incident diabetes was unchanged even after adjusting for these measures (Table 3).
Table 3.
Relation of individual amino acid levels to risk of future diabetes, with adjustment for insulin measures
| Model | Isoleucine | Leucine | Valine | Tyrosine | Phenylalanine |
|---|---|---|---|---|---|
| Adjusted odds ratios, per SD increment in metabolite (95% confidence interval) | |||||
| Basic model | 1.70 (1.27-2.28) | 1.62 (1.20-2.17) | 1.57 (1.17-2.09) | 1.85 (1.35-2.55) | 2.02 (1.40-2.92) |
| + fasting insulin | 1.65 (1.23-2.23) | 1.58 (1.17-2.12) | 1.52 (1.13-2.04) | 1.81 (1.31-2.50) | 2.00 (1.38-2.89) |
| + HOMA-IR | 1.65 (1.23-2.23) | 1.58 (1.17-2.12) | 1.52 (1.13-2.04) | 1.81 (1.31-2.50) | 2.00 (1.39-2.89) |
| + HOMA-B | 1.65 (1.23-2.23) | 1.58 (1.17-2.12) | 1.52 (1.13-2.04) | 1.81 (1.31-2.50) | 2.00 (1.38-2.88) |
| + OGTT (2-hr glucose) | 1.60 (1.18-2.17) | 1.53 (1.13-2.06) | 1.49 (1.11-2.00) | 1.79 (1.30-2.47) | 1.98 (1.37-2.86) |
Basic model: age, sex, body mass index, fasting glucose. Subsequent models include the basic clinical variables plus the insulin resistance/sensitivity measure shown. HOMA-IR: homeostasis model assessment of insulin resistance. HOMA-IR: homeostasis model assessment of beta cell function. OGTT: oral glucose tolerance test.
Amino acid combinations and prediction of diabetes
We assessed the predictive performance of clinical models with and without fasting plasma amino acids (Supplementary Table 3). The basic clinical model had a −2 log-likelihood ratio (LHR statistic) < 3, which was expected given the matched-pair design that included age, sex, BMI, and fasting glucose. Addition of any one of the five branched chain or aromatic amino acids improved model fit substantially, as indicated by large increases in the LHR statistic (+9 to 16, p<0.05). Combinations of three amino acids further improved the −2 log-likelihood ratio (+6 to 9, p<0.05), when compared with individual amino acids. There was only small additional increment in the LHR statistic when all five amino acids were included. Similar patterns were observed with changes in the c-statistic across different models. The top combination of three amino acids based on LHR statistic and c-statistic was comprised of isoleucine, phenylalanine, and tyrosine.
We performed conditional logistic regression models with the 3-amino acid combination (isoleucine, phenylalanine, tyrosine; Table 2). Individuals in the top quartile of the amino acid score had a 5- to 7-fold higher risk of developing diabetes, compared with individuals in the lowest quartile (p for trend, 0.007 to 0.0009).
Replication analyses (Malmö Diet and Cancer study)
We measured the five amino acids of interest in an independent replication sample comprising 163 cases and 163 controls (mean age 58 years, 55% women). Four of the five individual amino acids (leucine, valine, tyrosine, and phenylalanine) were significantly associated with incident diabetes (adjusted odds ratios per SD increment were similar to Framingham, 1.37 to 2.01; p=0.009 to 0.04; Table 4). The remaining amino acid, isoleucine, had a non-significant association (p=0.09).
Table 4.
Replication in the Malmo Diet and Cancer Study
| Model | Isoleucine | Leucine | Valine | Tyrosine | Phenylalanine | Isoleucine, tyrosine, and phenylalanine |
|---|---|---|---|---|---|---|
| Metabolite as continuous variable | ||||||
| Per SD | 1.37 (0.95-1.96) | 1.60 (1.13-2.27) | 2.01 (1.18-3.42) | 1.41 (1.05-1.91) | 1.37 (1.01-1.84) | 1.52 (1.10-2.11) |
| P | 0.09 | 0.009 | 0.01 | 0.02 | 0.04 | 0.01 |
| Metabolite as categorical variable | ||||||
| 1st quartile | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) | 1.0 (referent) |
| 2nd quartile | 1.02 (0.53-1.98) | 1.97 (0.86-4.54) | 2.11 (0.99-4.48) | 1.92 (0.92-4.01) | 2.37 (1.08-5.16) | 2.08 (0.97-4.46) |
| 3rd quartile | 1.62 (0.77-3.42) | 2.53 (1.09-5.87) | 1.82 (0.59-5.65) | 2.17 (1.03-4.57) | 4.30 (1.87-9.88) | 2.59 (1.09-6.15) |
| 4th quartile | 2.37 (0.97-5.81) | 3.81 (1.39-10.46) | 2.85 (0.77-10.51) | 3.16 (1.31-7.59) | 3.04 (1.20-7.70) | 3.93 (1.54-10.04) |
| P for trend | 0.05 | 0.01 | 0.09 | 0.01 | 0.007 | 0.006 |
Values are odds ratios (95% confidence intervals) for diabetes, from conditional logistic regressions. All models are adjusted for age, sex, BMI, and fasting glucose. The amino acid combination is modeled according to the formula β1X1 + β2X2 + β3X3, with Xj denoting the standardized value for the jth amino acid, and βj denoting the regression coefficient from the conditional logistic regression model containing the indicated metabolites.
We also tested the 3-amino acid combination (isoleucine, phenylalanine, tyrosine) derived in FHS. Individuals in the upper quartile of the 3-amino acid combination had a 4-fold higher risk of incident diabetes in MDC, compared with those in the lowest quartile (p for trend across quartiles, 0.006; Table 4).
Assessment of biomarkers in a random population
Because diabetes propensity was used to match individuals in the case-control studies, the study sample was enriched for individuals with “high risk” features, such as obesity and elevated fasting glucose. As a consequence, results in the case-control samples reflect what would be expected in a high-risk cohort, but not necessarily in a more heterogeneous sample. Thus, we performed metabolomic profiling on an additional 400 controls randomly selected from all individuals in the Framingham Offspring cohort who were free of diabetes or cardiovascular disease (n=2,422). As expected, the new sample (referred to as the “random cohort”) had a lower mean fasting glucose and BMI, compared with the original case-control sample (Table 1).
We repeated the analyses for the amino acid profiles identified in the case-control study. After adjustment for standard diabetes risk factors, including fasting glucose, body mass index, and parental history, the amino acid profile was still associated with future diabetes development (adjusted odds ratio, 1.36, per SD increment in the amino acid score, p=0.008; Table 5 and Supplementary Table 3). Individuals with the highest amino acid scores (top quartile), had an approximately 2-fold higher adjusted risk of developing diabetes over 12 years of follow up.
Table 5.
Results for the amino acid combination in the random cohort sample
| Model | 3 amino acids (Isoleucine, Phenylalanine, Tyrosine) |
5 amino acids (Isoleucine, Phenylalanine, Tyrosine, Leucine, Valine) |
|---|---|---|
| Models adjusting for age, sex, BMI, and fasting glucose (n=601) | ||
| Per SD (score) | 1.33 (1.08-1.63) | 1.33 (1.08-1.63) |
| P | 0.008 | 0.007 |
| 1st quartile | 1.0 (referent) | 1.0 (referent) |
| 2nd quartile | 1.12 (0.64-1.98) | 1.07 (0.60-1.90) |
| 3rd quartile | 1.43 (0.81-2.53) | 1.67 (0.95-2.95) |
| 4th quartile | 1.73 (0.96-3.10) | 1.94 (1.08-3.49) |
| P for trend | 0.03 | 0.005 |
| Models adjusting for age, sex, BMI, fasting glucose, and parental history (n=516) | ||
| Per SD (score) | 1.36 (1.08-1.70) | 1.38 (1.09-1.74) |
| P | 0.008 | 0.007 |
| 1st quartile | 1.0 (referent) | 1.0 (referent) |
| 2nd quartile | 1.42 (0.73-2.73) | 1.48 (0.76-2.89) |
| 3rd quartile | 1.93 (1.00-3.71) | 2.16 (1.11-4.20) |
| 4th quartile | 2.01 (1.02-3.99) | 2.23 (1.12-4.42) |
| P for trend | 0.03 | 0.01 |
Values are odds ratios (95% confidence intervals) for diabetes, from conditional logistic regressions. The amino acid combination is modeled according to the formula β1X1 + β2X2 + β3X3, with Xj denoting the standardized value for the jth amino acid, and βj denoting the regression coefficient from the conditional logistic regression model containing the indicated metabolites.
DISCUSSION
Using a mass spectrometry-based metabolite profiling platform, we identified a panel of amino acids whose fasting levels at a routine examination predicted the future development of diabetes in otherwise healthy, normoglycemic individuals. Indeed, fasting concentrations of these amino acids were elevated up to 12 years prior to the onset of diabetes. The risk of future diabetes was elevated at least 4-fold in those with high plasma amino acid concentrations in both the discovery and replication samples.
A growing number of studies have used mass spectrometry as a tool for biomarker discovery18,19, but these studies have been largely cross-sectional, providing limited information regarding the relation of metabolomic (or proteomic) biomarkers to the future development of disease. Thus, an important strength of the current investigation is the use of two well-characterized prospective cohorts, one for derivation and one for replication, each with more than 3,000 participants who have been followed longitudinally for decades. All individuals in our study were free of diabetes at the time the blood samples were collected, and matching for BMI and fasting blood glucose in our study design minimized confounding from existing glucose intolerance. The long period of observation is a distinctive feature of our study, because it enabled us to demonstrate that circulating amino acid elevations can occur well before any alteration in insulin action is detectable using standard biochemical measures.
Our findings, which highlight five branched chain and aromatic amino acids from 61 metabolites profiled, are particularly noteworthy in the context of experimental and clinical data suggesting that certain amino acids may be both markers and effectors of insulin resistance14,15,18,20,21. Several decades ago, Felig and colleagues studied 20 non-obese and obese individuals, and found that fasting concentrations of branched chain and aromatic amino acids correlated with obesity and serum insulin20. Additionally, glucose loading lowered amino acid concentrations in insulin-sensitive, but not insulin-resistant individuals. Both sets of findings have been corroborated by more recent studies using LC-MS-based metabolomics platforms14-16,18. Studies of branched chain amino acid supplementation in both animals14 and humans22 indicate that circulating amino acids may directly promote insulin resistance, possibly via disruption of insulin signaling in skeletal muscle. The underlying cellular mechanisms may include activation of the mTOR, JUN and IRS1 signaling pathways in skeletal muscle14,21. By contrast, others have demonstrated improved glucose homeostasis in animals fed a diet specifically enriched in leucine23.
In addition to insulin resistance, impaired insulin secretion plays a critical role in the pathogenesis of type 2 diabetes. In this regard, it is noteworthy that multiple amino acids, particularly the branched chain amino acids, are modulators of insulin secretion24-26. Thus, another possible mechanism by which hyperaminoacidemia could promote diabetes is via hyperinsulinemia leading to pancreatic α-cell exhaustion.
While circulating amino acids were correlated with standard biochemical measures of insulin resistance and α-cell function, amino acid concentrations were predictive even among individuals with similar fasting insulin and glucose levels. Furthermore, stimulation of the insulin axis with OGTT did not elicit differential amino acid changes between cases and controls. All of these findings support the notion that hyperaminoacidemia could be a very early manifestation of insulin resistance—one that presages the clinical onset of diabetes by years.
The ability to identify individuals prior to the onset of disease is particularly important for conditions such as diabetes, because proven, preventive therapies exist and end-organ complications accrue over time. Although traditional risk factors such as body mass index and fasting glucose provide important information about future diabetes risk, not all individuals who are obese develop diabetes. It is important to understand which “at-risk” individuals are most likely to progress to overt disease. There has been interest in genetic risk prediction, but the known diabetes polymorphisms add modestly to risk assessment27,28. For instance, known polymorphisms are only associated with 5% to 37% increases in the relative risk of diabetes, compared with the 60% to 100% increases in risk that we observed with elevation in amino acids. Indeed, the relative risks associated with elevated amino acids were comparable to, or higher than, those associated with higher age, fasting glucose, or body mass index in prior population-based studies28.
Additionally, our findings may provide insight regarding subgroups in which amino profiles could yield the most incremental information. Most of our analyses were based on “high risk” study samples, as a result of the matching scheme which paired cases with controls who had a high predicted risk of diabetes. In this setting, amino acid elevations were associated with very high relative risks for developing diabetes, and the amino acid profiles led to large improvements in model fit and discrimination (c-statistics). This result was noted in both the discovery and replication cohorts, attenuating concern for over-fitting the data. In a more heterogeneous study sample, obtained by looking at a random set of controls from the Framingham cohort, the relative risks associated with elevated amino acids were attenuated (though still significant, in the 2-fold range) and changes in c-statistics were modest. Baseline BMI and glucoses in the random cohort analysis were lower, on average, compared with the case-control analyses, and the distributions much broader. Most of the variation in diabetes risk in such a sample is attributable to variation in BMI and other standard diabetes risk factors. Overall, these findings suggest that amino acid profiling might have greater value in high-risk individuals, but confirmation in additional studies is needed.
Several limitations of the study deserve comment. We used a “targeted” approach that coupled liquid chromatography with a triple quadrupole tandem mass spectrometer. Although alternate LC-MS techniques or nuclear magnetic resonance spectroscopy can be used to acquire spectral data in a less “biased” manner, targeted LC-MS/MS provides much greater sensitivity, highly-specific identification of analytes, and the ability to quantify absolute analyte concentrations when appropriate standards are added. The platform used for the present study was geared toward small molecules such as amino acids, as well as urea cycle and nucleotide metabolites. This choice was informed by prior studies suggesting a cross-sectional association between insulin resistance and several metabolites18,20, yet the absence of prospective data linking metabolite concentrations to future risk of diabetes. That we identified a set of five amino acids whose fasting levels strongly predicted the future development of diabetes does not preclude that other metabolites may also predict disease. Identification of novel biomarkers will no doubt accelerate as platforms expand their metabolite coverage.
In the Framingham Heart Study, close surveillance of the participants over serial examinations ensured reliable ascertainment of the development of diabetes over time. In the replication cohort (MDC), incident diabetes cases were identified through the use of three registries. Although this introduces the possibility of misclassification of diabetes status in MDC, such misclassification would be expected to bias the results toward the null. Indeed, the robustness of the findings in two longitudinal cohorts with widely different methods for ascertaining diabetes further increases confidence in the validity of the results. Lastly, individuals in both cohorts were predominantly white and of European descent. Further studies are needed to determine whether the findings extend to other racial/ethnic groups.
In summary, from a panel of >60 metabolites, branched chain and aromatic amino acids emerged as predictors of the future development of diabetes. A single, fasting measurement of these amino acids provided information incrementally over standard risk factors (such as BMI, dietary patterns, and fasting glucose). Further investigation is warranted to test whether plasma amino acid measurements might help identify candidates for interventions to reduce diabetes risk, and to elucidate the biological mechanisms by which selected amino acids might promote type 2 diabetes.
METHODS
Study samples
The Framingham Offspring Study was initiated in 1971, when 5,124 individuals enrolled into a longitudinal cohort study29. The 5th quadrennial examination took place between 1991 and 1995. Of 3,799 attendees to the 5th examination, 2,422 were eligible for the present investigation because they were free of diabetes and cardiovascular disease, underwent routine oral glucose tolerance testing (OGTT), and were 35 years or older.
We tested findings for replication in the Malmö Diet and Cancer (MDC) study, a Swedish population-based cohort of 28,449 persons enrolled between 1991 and 1996. From this cohort, 6,103 persons were randomly selected to participate in the MDC Cardiovascular Cohort30. We obtained fasting plasma samples in 5,305 subjects in the MDC Cardiovascular Cohort, of whom 564 had prevalent diabetes or cardiovascular disease prior to baseline, and an additional 456 subjects had missing covariate data, leaving 4,285 subjects eligible for analysis.
The study protocols were approved by the Institutional Review Boards of Boston University Medical Center, Massachusetts General Hospital, and Lund University, Sweden, and all participants provided written informed consent. Detailed descriptions of the clinical assessment, diabetes definition, and subject selection are provided in the Supplementary Methods.
Metabolite profiling
We profiled amino acids, biogenic amines, and other polar plasma metabolites using liquid chromatography-tandem mass spectrometry (LC-MS). Formic acid, ammonium acetate, LC-MS grade solvents, and valine-d8 were purchased from Sigma-Aldrich. We purchased the remainder of the isotopically-labeled analytical standards from Cambridge Isotope Labs, Inc. We prepared calibration curves for a subset of the profiled analytes by serial dilution in stock pooled plasma using stable isotope-labeled reference compounds (leucine-13C, 15N, isoleucine-13C6, 15N, alanine-13C, glutamic acid-13C5, 15N, taurine-13C2, trimethylamine-N-oxide-d9). We ran samples with isotope standards for calibration curves at the beginning, middle, and end of each analytical queue. We prepared plasma samples for LC-MS analyses via protein precipitation with the addition of nine volumes of 74.9:24.9:0.2 v/v/v acetonitrile/methanol/formic acid containing two additional stable isotope-labeled internal standards for valine-d8 and phenylalanine-d8. The samples were centrifuged (10 min, 10,000 rpm, 4°C) and the supernatants were injected directly. Detailed methods are provided in the Supplementary Methods.
Statistical analyses
We examined the association between plasma metabolite levels (pre- and post-OGTT) and incident diabetes in Framingham. We log transformed metabolite levels because the case-control differences did not exhibit a constant variance. We compared cases (those developing new-onset diabetes during the 12-year follow-up) versus propensity-matched controls, using paired t-tests for the 48 metabolites with <5% missing data. For the 13 metabolites with undetectable levels in ≥5% of samples, we used McNemar’s test to compare the proportion of detectable values. We also examined whether the change in metabolite levels during OGTT was associated with diabetes, by regressing the 2-hour metabolite level on the baseline level, case status, and an interaction term, using generalized estimating equations. We used a corrected p-value threshold of 0.001 to account for the 48 metabolites analyzed as continuous variables (Supplementary Methods). For metabolites meeting the p-value threshold, we performed conditional (matched-pairs) logistic regression analyses to estimate the relative risk of diabetes at different metabolite values, adjusting for age, sex, BMI, and fasting glucose. In additional analyses, we also adjusted for parental history, serum triglycerides, HDL cholesterol, hypertension, intake of dietary protein, amino acids, and total calories. We also assessed whether plasma metabolites predicted diabetes risk incrementally over biochemical measures of insulin resistance and α-cell function: fasting insulin, homeostasis model assessment of insulin resistance (HOMA-IR) and α-cell function (HOMA-B), and 2-hour post-OGTT glucose31. These models were adjusted for age, sex, BMI, fasting glucose, and the log-transformed insulin resistance or secretion measure. We analyzed metabolites as both continuous and categorical (using the quartile values in controls as cutpoints) variables. We also performed secondary analyses restricted to case-control pairs in the lowest tertile of propensity score.
To identify the most predictive biomarker combination, we calculated log-likelihood ratios and evaluated model discrimination using the c-statistic. The c-statistic was calculated by assessing the proportion of case-control pairs for which the biomarker value in cases exceeded that in controls. We assessed biomarker combinations by calculating the sum of standardized biomarker values weighted according to their corresponding beta coefficients from the regression analyses, and entering the weighted value into a separate logistic regression model. The values were grouped into quartiles, and tested using a class variable to estimate the odds ratio for each quartile.
We attempted to replicate our findings in the MDC cohort, by testing the most significant metabolite predictors identified in Framingham. A replication p-value <0.05 was considered significant. We also performed analyses in a random cohort sample from Framingham, to assess the association between amino acids and diabetes risk in a lower-risk, more heterogeneous group. A total of 201 cases were available for this analysis (overall N=601). We used Cox proportional hazards models to account for time to diabetes onset (discrete time based on inter-examination interval), with adjustment for age, sex, BMI, fasting glucose, parental diabetes history, and metabolite scores as described above.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH contract NO1-HC-25195, R01-DK-HL081572, the Donald W. Reynolds Foundation, the Leducq Foundation, and the American Heart Association. Dr. Florez is also supported by the Massachusetts General Hospital and a Clinical Scientist Development Award from the Doris Duke Charitable Foundation.
Footnotes
AUTHOR CONTRIBUTIONS T.J.W conceived the study, designed the experiments, analyzed and interpreted the data, and wrote the manuscript. A.A. and E.P.R., under the direction of C.B.C., developed the metabolic profiling platform, performed mass spectrometry experiments, and analyzed the data. S.A.C. and V.K.M. helped in the establishment of the metabolite profiling platform and manuscript revision. G.D.L. contributed to data analysis and manuscript generation. M.G.L., R.S.V., S.C. and E.M. helped in experimental design, performed statistical analyses, and assisted in manuscript generation. C.O.D. and C.S.F. helped in experimental design and manuscript revision. P.F.J. directed the dietary analyses in the Framingham Heart Study and contributed to manuscript revision. J.F. assisted in the interpretation of the data, and contributed to manuscript revision. O.M. and C.F. performed the replication analyses in the M.D.C. cohort and contributed to manuscript revision. R.E.G. conceived the study, designed the experiments, analyzed and interpreted the data, and wrote the manuscript.
CONFLICT OF INTEREST DISCLOSURE Drs. Wang, Vasan, Larson, Mootha, and Gerszten are named as co-inventors on a patent application pertaining to metabolite predictors of diabetes. Dr. Florez has received consulting honoraria from Publicis Healthcare, Merck, bioStrategies, XOMA and Daiichi-Sankyo, and has been a paid invited speaker at internal scientific seminars hosted by Pfizer and Alnylam Pharmaceuticals.
REFERENCES
- 1.Tabak AG, et al. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet. 2009;373:2215–2221. doi: 10.1016/S0140-6736(09)60619-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson PW, et al. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch.Intern Med. 2007;167:1068–1074. doi: 10.1001/archinte.167.10.1068. [DOI] [PubMed] [Google Scholar]
- 3.Pan XR, et al. Effects of diet and exercise in preventing NIDDM in people with impaired glucose tolerance. The Da Qing IGT and Diabetes Study. Diabetes Care. 1997;20:537–544. doi: 10.2337/diacare.20.4.537. [DOI] [PubMed] [Google Scholar]
- 4.Tuomilehto J, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N.Engl.J.Med. 2001;344:1343–1350. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 5.The Diabetes Prevention Program Research Group Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerstein HC, et al. Effect of rosiglitazone on the frequency of diabetes in patients with impaired glucose tolerance or impaired fasting glucose: a randomised controlled trial. Lancet. 2006;368:1096–1105. doi: 10.1016/S0140-6736(06)69420-8. [DOI] [PubMed] [Google Scholar]
- 7.Nicholson JK, Wilson ID. Opinion: understanding ‘global’ systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discov. 2003;2:668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- 8.Raamsdonk LM, et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19:45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- 9.Allen J, et al. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol. 2003;21:692–696. doi: 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- 10.An J, et al. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat Med. 2004;10:268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- 11.Sabatine MS, et al. Metabolomic identification of novel biomarkers of myocardial ischemia. Circulation. 2005;112:3868–3875. doi: 10.1161/CIRCULATIONAHA.105.569137. [DOI] [PubMed] [Google Scholar]
- 12.Sapieha P, et al. The succinate receptor GPR91 in neurons has a major role in retinal angiogenesis. Nat Med. 2008;14:1067–1076. doi: 10.1038/nm.1873. [DOI] [PubMed] [Google Scholar]
- 13.He W, et al. Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature. 2004;429:188–193. doi: 10.1038/nature02488. [DOI] [PubMed] [Google Scholar]
- 14.Newgard CB, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaham O, et al. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol. 2008;4:214. doi: 10.1038/msb.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wopereis S, et al. Metabolic profiling of the response to an oral glucose tolerance test detects subtle metabolic changes. PLoS One. 2009;4:e4525. doi: 10.1371/journal.pone.0004525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X, et al. Changes of the plasma metabolome during an oral glucose tolerance test: is there more than glucose to look at? Am J Physiol Endocrinol Metab. 2009;296:E384–393. doi: 10.1152/ajpendo.90748.2008. [DOI] [PubMed] [Google Scholar]
- 18.Huffman KM, et al. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes Care. 2009 doi: 10.2337/dc08-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis GD, Asnani A, Gerszten RE. Application of metabolomics to cardiovascular biomarker and pathway discovery. J.Am.Coll.Cardiol. 2008;52:117–123. doi: 10.1016/j.jacc.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felig P, Marliss E, Cahill GF., Jr. Plasma amino acid levels and insulin secretion in obesity. N Engl J Med. 1969;281:811–816. doi: 10.1056/NEJM196910092811503. [DOI] [PubMed] [Google Scholar]
- 21.Patti ME, Brambilla E, Luzi L, Landaker EJ, Kahn CR. Bidirectional modulation of insulin action by amino acids. J Clin Invest. 1998;101:1519–1529. doi: 10.1172/JCI1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krebs M, et al. Mechanism of amino acid-induced skeletal muscle insulin resistance in humans. Diabetes. 2002;51:599–605. doi: 10.2337/diabetes.51.3.599. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. Increasing dietary leucine intake reduces diet-induced obesity and improves glucose and cholesterol metabolism in mice via multimechanisms. Diabetes. 2007;56:1647–1654. doi: 10.2337/db07-0123. [DOI] [PubMed] [Google Scholar]
- 24.Floyd JC, Jr., Fajans SS, Conn JW, Knopf RF, Rull J. Stimulation of insulin secretion by amino acids. J Clin Invest. 1966;45:1487–1502. doi: 10.1172/JCI105456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson M, Holst JJ, Bjorck IM. Metabolic effects of amino acid mixtures and whey protein in healthy subjects: studies using glucose-equivalent drinks. Am J Clin Nutr. 2007;85:996–1004. doi: 10.1093/ajcn/85.4.996. [DOI] [PubMed] [Google Scholar]
- 26.van Loon LJ, Saris WH, Verhagen H, Wagenmakers AJ. Plasma insulin responses after ingestion of different amino acid or protein mixtures with carbohydrate. Am J Clin Nutr. 2000;72:96–105. doi: 10.1093/ajcn/72.1.96. [DOI] [PubMed] [Google Scholar]
- 27.Meigs JB, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359:2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyssenko V, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 29.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families: the Framingham Offspring Study. Am J Epidemiol. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 30.Persson M, Hedblad B, Nelson JJ, Berglund G. Elevated Lp-PLA2 levels add prognostic information to the metabolic syndrome on incidence of cardiovascular events among middle-aged nondiabetic subjects. Arteriosclerosis, thrombosis, and vascular biology. 2007;27:1411–1416. doi: 10.1161/ATVBAHA.107.142679. [DOI] [PubMed] [Google Scholar]
- 31.Matthews DR, et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


