Abstract
Methionine sulfoxide reductase A (MsrA), a member of the Msr gene family, can reduce methionine sulfoxide residues in proteins formed by oxidation of methionine by reactive oxygen species (ROS). Msr is an important protein repair system which can also function to scavenge ROS. Our studies have confirmed the expression of MsrA in mouse embryonic stem cells (ESCs) in culture conditions. A cytosol-located and mitochondria-enriched expression pattern has been observed in these cells. To confirm the protective function of MsrA in ESCs against oxidative stress, a siRNA approach has been used to knockdown MsrA expression in ES cells which showed less resistance than control cells to hydrogen peroxide treatment. Overexpression of MsrA gene products in ES cells showed improved survivability of these cells to hydrogen peroxide treatment. Our results indicate that MsrA plays an important role in cellular defenses against oxidative stress in ESCs. Msr genes may provide a new target in stem cells to increase their survivability during the therapeutic applications.
Keywords: MsrA, ANOXIA/REOXYGENATION, CELL DEATH, siRNA
Patients suffering from acute ischemia/reperfusion of any tissue (e.g., heart infarct) or chronic insufficient blood perfusion (e.g., vascular stenosis) are prone to cellular injuries induced by oxidative stress. During oxidative stress, the overproduced reactive oxygen species (ROS) induce the oxidative damage to many cellular components, including proteins (amino acids), lipids, and nucleic acids, which will finally contribute to the cell death [Honig and Rosenberg, 2000; Boldyrev et al., 2004; Onyango et al., 2005]. One novel treatment method for these diseases is to transplant normal stem cells into the patients to differentiate into appropriate mature cells to replace the function of the lost host cells. To protect against the oxidative insults induced by a variety of causes, including ischemia/reperfusion, cells themselves have multiple anti-oxidation mechanisms [Boldyrev et al., 2004]. For example, superoxide dismutase, catalase, and glutathione peroxidase scavenge the superoxide anion and H2O2 to prevent ROS-induced damages. Cells that are modified to overexpress these genes may achieve better protection against the oxidative stress [Blass, 2001]. However, a certain level of ROS is an essential signal in the regulation of normal cell metabolism and differentiation, such as angiogenesis by tumor cells and cardiomyocyte differentiation from embryonic stem cells (ESCs) [Sauer et al., 2000; Boldyrev et al., 2004; Thiruchelvam et al., 2005; Wo et al., 2008]. Thus, reducing ROS inside cells using direct anti-oxidants such as vitamin E could interfere with normal cell function. This might pose a problem for stem cell therapy since the ROS level must be balanced such that the cells undergo normal differentiation without resulting in damage or cell death in the perfused tissues. If this is the case, then an indirect mechanism to reduce ROS could have therapeutic advantages over a direct ROS mechanism, eliminating agents such as vitamin E, during the use of stem cells for tissue repair.
Interestingly, there exists an indirect defense system to ROS in normal cells, the methionine sulfoxide reductase A and B (MsrA/B) system whose function is not to eliminate ROS directly, but rather to reverse its damaging effects in the cells. Furthermore, the amino acid methionine in proteins is oxidized by ROS, leading to the formation of the R (Met-R-O) and S (Met-S-O) epimers of methionine sulfoxide (met-O) and possible loss of protein function. MsrA and MsrB can reduce protein bound Met-S-O and Met-R-O, respectively, and restore their function. MsrA has been studied in detail, and has been shown to reduce Met-S-O in a number of oxidized proteins. The Msr system may also play a role in scavenging ROS by permitting methionine residues in proteins to act as catalytic anti-oxidants. Evidence for the scavenger role of the Msr system has come from recent studies on the overexpression of the Msr system, or knocking out the Msr system in cultured cells. Overexpression of MsrA by adenovirus-mediated delivery significantly diminishes the hypoxia-induced increase in ROS and facilitates cell survival in PC12 cells [Huang et al., 1997]. It also has been found by siRNA knockdown of MsrA and MsrBs in human lens cells that the MsrA/B system plays a protective role against H2O2-induced cell injury [Kantorow et al., 2004; Yermolaieva et al., 2004].
MsrA, unlike the three MsrBs that all can reduce the R form methionine sulfoxide, is the only known Msr family gene whose protein reduces the S form methionine sulfoxide [Kim and Gladyshev, 2004, 2005]. In the present study we examine whether MsrA can protect ESCs from oxidative damage using both siRNA to knockdown MsrA expression, and a MsrA-GFP fusion protein overexpression system in mouse ESCs. Our results demonstrate that MsrA plays an important role in protecting stem cells against oxidative damage and thus has important implications in stem cell therapy.
MATERIALS AND METHODS
MOUSE EMBRYONIC STEM CELL CULTURE
The mouse embryonic stem (MES) cells (CCE-24; L. Robertson, Columbia University) were routinely grown on 0.1% gelatin-coated dishes in Dubecco’s Modified Eagle’s medium (DMEM) containing 15% heat-inactivated fetal bovine serum (catalog # 10100, Invitrogen, Carlsbad, CA), 10 ng/ml human leukemia inhibitory factor (LIF) (LIF2010, Millipore, Billerica, MA), and monothioglycerol (Sigma, St. Louis, MO) at 4.5 × 10−4 M, split every 2 days at 1:4 to 1:10 and immunostained for stem cell-specific markers (SSEA-1 (Mab4301) and SSEA-4 (Mab4304, Millipore). Only cells within passage 20 were used for assays.
ANOXIA/REOXYGENATION TREATMENT
The anoxic treatment of mouse ESCs was performed by incubation in an anaerobic chamber (Sheldon Manufacturing, Inc., Cornelius, OR) supplied with 90% nitrogen, 5% hydrogen, and 5% carbon dioxide at 37°C. Cells were removed after set periods and incubated in a regular cell culture incubator at 37°C for designated times.
siRNA TRANSFECTION
MsrA-specific siRNAs were synthesized by Qiagen (Valencia, CA). One of the MsrA-specific siRNAs, 168 after transfection, showed the reduction of MsrA mRNA levels in mouse ESCs. The sequences:
sense: CCAUGAAUCAUUUGCCAAAUCGCUU;
antisense: AAGCGAUUUGGCAAAUGAUUCAUGG
The negative control siRNA (numbered as 882c) sequences:
sense: CCAGCACUAACACCCAUCCCACAAA;
antisense: UUUGUGGGAUGGGUGUUAGUGCUGG
For each transfection sample, siRNA-Lipofectamine™ 2000 (Invitrogen) complexes were prepared as follows: (a) Dilute 2 µg siRNA in 250 µl of DMEM. (b) Dilute 5 µl Lipofectamine™ 2000 in 250 µl of DMEM (no serum). Incubate for 15 min at room temperature. (c) After the 15-min incubation, combine the diluted siRNA with the diluted Lipofectamine™ 2000 (total volume ~505 µl) and incubate for another 15 min at room temperature. (d) During the 15-min incubation, dissociate mouse ESCs by trypsinization and prepare a single cell suspension. Plate 5 × 105 cells per well in six-well plates in a DMEM complete medium (with serum) in suspension. (e) Add the ~505 µl of siRNA-Lipofectamine™ 2000 complexes to each well containing cells and medium. Incubate the cells at 37°C in a humidified CO2 incubator for 24 h, collect samples for RNA and protein extraction as well as H2O2 treatment. The medium was changed at 24 h and cultures were continued for 2 days. At each 24-h time point, samples were collected for RNA and protein extractions.
REAL-TIME RT-PCR
Total RNA was extracted from mouse ESCs after various combinations of anoxia/reoxygenation treatments or MsrA siRNA transfection. After pretreatment by RNase-free DNase I, 2 µg of total RNA was used for cDNA syntheses with a ThermoScript RT-PCR System (Invitrogen). Real-time RT-PCR experiments were performed on a Capillary Lightcycler (Roche Applied Science, Indianapolis, IN) machine using a Roche Fast Start SYBR Green I Kit. Amplification of genes was confirmed by calculating melting temperatures (Tm) for the products from the melting peak curve (−dF/dT vs. temperature). All the amplicons were collected and confirmed by agarose gel electrophoresis and sequencing. A standard curve of cross-point versus Log concentration was created using one of the cDNA samples with serial dilutions or with known concentrations of plasmid DNA with a MsrA gene insert. Negative controls were included using cDNAs synthesized the same way as above but with no reverse transcriptase. Each cDNA sample was run in triplicate. The data were averaged and standard deviations calculated. The β-actin gene was used as a standard control. The primer sequences designed for real-time PCR from two consecutive exons flanking a piece of intron were as follows:
MsrA-for: 5′-TCTGGGTCTTGAAAGGAGTGTA;
MsrA-rev: 5′-AGGTATTGCTGGTGGTAGTCTTC; amplicon size: 395 bp.
β-actin-for: 5′-CCACTGCCGCATCCTCTTCCTC;
β-actin-for: 5′-CAGCAATGCCTGGGTACATGGTG; amplicon size: 249 bp.
CONSTRUCTION OF MsrA-eGFP FUSION EXPRESSION PLASMID AND TRANSFECTION INTO MOUSE EMBRYONIC STEM CELLS
MsrA full-length cDNA was cloned by RT-PCR from total RNA extracted from stem cells with a primer pair: MsrA-full-for: 5′-cctggctgcggaggtggagaaac and MsrA-full-rev: 5′-atggccatcgggcaggaaactcc. cDNA was purified and ligated to pGEM-T-easy vector (Promega, WI). The insert was cut by the SacII enzyme and ligated into pEGFP-N1 to generate the fusion expression vector, pEGFP-N1-MsrA. The orientation and integrity of the inserted MsrA cDNA sequence and the continuation of the open reading frame of the MsrA-eGFP fusion peptide were confirmed by restrictive enzyme digestion (EcoRI) and by sequencing.
Mouse ESCs were plated on day 1 at 5,000 cells/well in a 96-well plate (~15% confluency). On day 2, the cells were changed with fresh culture medium (no antibiotics) and continued in culture for 2–4 h. Plasmid transfection was performed by mixing 5 µl DMEM medium (Invitrogen) with 0.9 µl Fugene6 (Roche Applied Science) for 5 min, then adding 0.25–0.5 µg DNA of each sample to the Fugene6-DMEM mix, incubating for 15 min and transferring the mix to each well with 100 µl complete medium (no antibiotics). On day 4, after 36–48 h of culture and confirmation of the expression of green fluorescence, cells were treated with H2O2 for 16–24 h before the MTT assay or flow cytometry analysis.
TREATMENT OF THE STEM CELLS WITH H2O2
After transfection with MsrA-specific siRNA and negative control siRNA for 24 h, wells were changed with fresh 100 µl DMEM complete medium without phenol red and continued in culture at 37°C for 1 h. Different concentrations of H2O2 were prepared by diluting with the DMEM complete medium without phenol red. The final concentrations of H2O2 were 0, 25, 50, 100, 150, 200, 250, 300 µM. The cells then were changed into this new medium at different concentrations of H2O2 and cultured for 24 h prior to the MTT assay to test cell survival using the Vybrant Cell Proliferation Assay kit (Invitrogen).
WESTERN BLOTTING
Protein samples were heat denatured at 95°C for 5 min before loading onto an SDS–polyacrylamide gel (reducing or nonreducing) (4–12%) (Invitrogen) and run under standard conditions. Proteins were transferred to a nitrocellulose membrane and blocked with a 3% (w/v) BSA in TBST (0.05% Tween-20) for 1 h at room temperature. The anti-MsrA polyclonal antibody (catalog #: ab16803, Abcam, Cambridge, MA) was diluted in 3% BSA in TBST at 1:4,000 and incubated with the blot for 1.5 h at room temperature. The horseradish peroxidase (HRP)-labeled secondary antibody diluted 1/5,000 in blocking buffer was incubated with the blot for 30 min. The blot was rinsed in TBST solution with 0.5% Tween-20 before the ECL detection with RPN 2108 kit (Amersham, Buckinghamshire, UK). After exposure on X-ray film for MsrA protein, the blot was stripped in 1 M NaOH solution for 10 min at room temperature and hybridized using antibody to β-actin (Abcam) for normalization. The protein band densitometry was analyzed using a gel imaging system (Alpha Innotech Corp., San Leandro, CA).
CONFOCAL MICROSCOPY ON MsrA-eGFP FUSION PLASMID TRANSFECTED EMBRYONIC STEM CELLS
After the MsrA-eGFP expression plasmid was transfected into cells for 3 days and confirmation of the GFP fluorescence signal by epi-fluorescent microscopy, 500 nM Mitotracker (Molecular Probes, Carlsbad, CA) diluted in complete culture medium was added to the cells. Mitotracker was first diluted in DMSO (anhydrous) to 1 mM and stored in the dark at −20°C. The stock was diluted to 0.1 mM by DMSO and diluted again in DMEM complete cell culture medium (with serum) to a final concentration of 500 nM. Cells were incubated for 30 min, washed with PBS, fixed using 1% formaldhyde in PBS for 15 min, rinsed with PBS twice and incubated in a 1/10,000 dilution of DAPI (10 mg/ml) in PBS for 15–20 min, at room temperature. The cells were then mounted in Pro-Long (Molecular Probes, Carlsbad, CA), air-dried for 2 days in the dark, and sealed with nail polish. Confocal microscopy was carried out using a Carl Zeiss confocal microscope at the University of Miami Diabetes Research Institute. Five micron Z-series were scanned for each sample.
FLOW CYTOMETRY
Cells pretransfected with different plasmids or negative controls (DMEM culture only) were trypsinized, passed through a 50 µM filter (BD Biosciences, San Jose, CA), resuspended in filtered PBS solution and analyzed in a Becton-Dickinson FACSCalibur flow cytometer. Dual laser wavelengths (488 and 536 nm) were setup for two-dimensional plotting and data were collected/analyzed by CellQuest Pro software. For each sample, 20,000 cells were collected by the cytometer with three parallel samples analyzed for statistical analyses.
RESULTS
CONFIRMATION OF MsrA EXPRESSION AND STUDIES ON ITS EXPRESSION PATTERN UNDER OXIDATIVE STRESS INDUCED BY ANOXIA/REOXYGENATION IN MOUSE EMBRYONIC STEM CELLS
We have confirmed MsrA gene expression in mouse ESCs and have performed studies on MsrA gene expression during hypoxia/reoxygenation using real-time RT-PCR for mRNA transcription, as well as Western blotting with anti-MsrA antibody for protein synthesis.
Studies at mRNA level
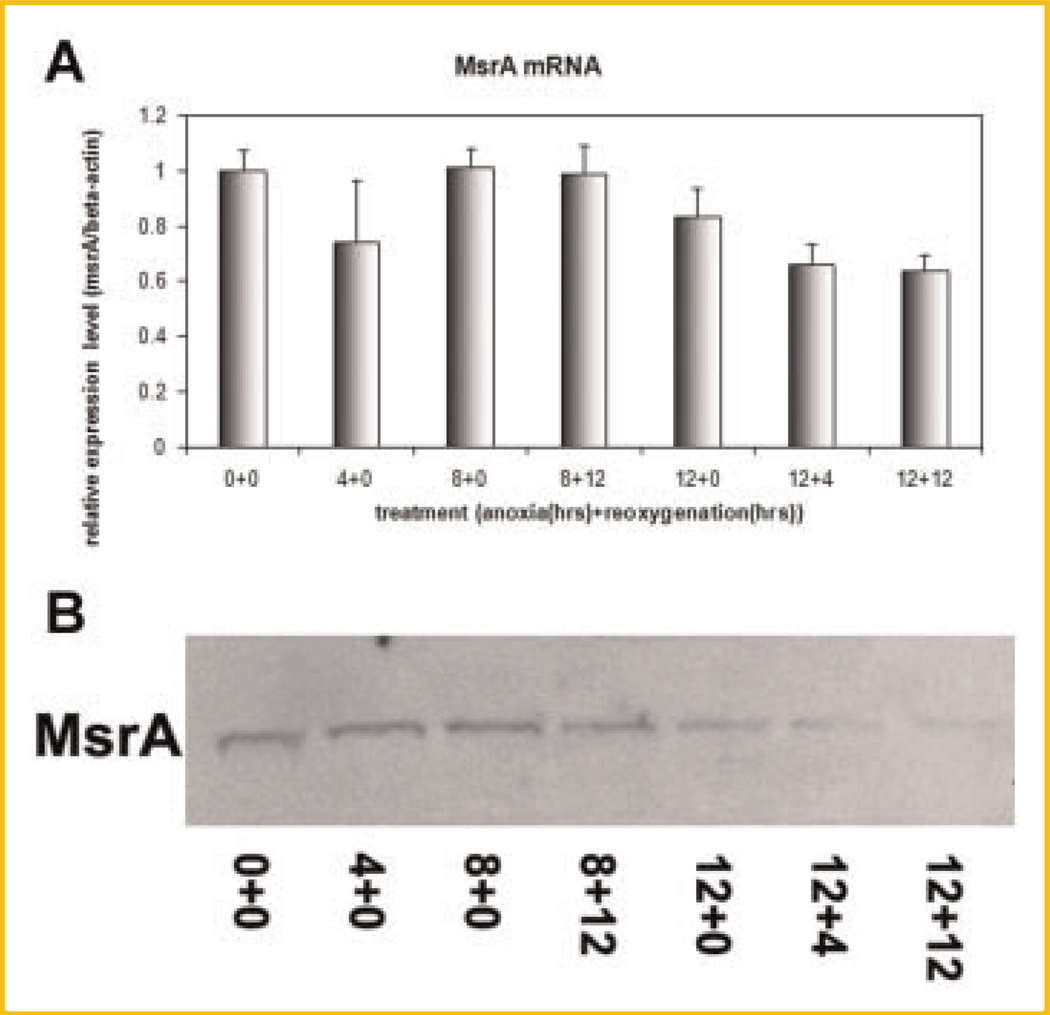
Using real-time RT-PCR, we have compared the mRNA transcription level regulation of the MsrA gene under different levels of oxidative stress. The oxidative stress levels were induced by treating cells with increasing time periods of anoxia (in 90% N2, 5% H2, and 5% CO2) and reoxygenation combinations in a hypoxia chamber. Results indicated that MsrA mRNA levels gradually decreased with prolonged hypoxic and reoxygenation treatments (Fig. 1A). Decreases of MsrA mRNA could be observed after 4 h of oxygen depletion but regained its expression level at 8 h (Fig. 1A). This could be explained by a transient, instant response of stem cells to oxygen depletion since mRNA transcription decreases can be observed for MsrA, MsrBs, and Matrix Metalloproteinase-9 (MMP9).
Fig. 1.
MsrA expression decreases at both mRNA and protein levels in response to the increasing levels of oxidative stress generated by anoxia/reoxygenation treatments. A: Real-time RT-PCR of MsrA mRNA on total RNA samples from cells treated by anoxia and reoxygenation (e.g., 12 h of anoxia followed by 4 h of reoxygenation treatment is labeled as 12 + 4 on the X-axis). B: Western blotting of MsrA protein using total protein extracts from cells pretreated by anoxia and reoxygenation.
Studies at protein expression level
After the above treatments, mouse ESC protein samples were collected. Equal amounts of protein from each sample were loaded onto 4–12% gradient NuPAGE polyacrylamide gels (Invitrogen). Western blotting using anti-MsrA antibody (Abcam) have shown gradually decreased expression of MsrA protein (Fig. 1B), with prolonged treatments of anoxia/reoxygenation, consistent with results from mRNA level studies.
DISCOVERY OF THE CYTOSOL AND MITOCHONDRIAL LOCALIZATIONS OF MsrA PROTEINS BY CONFOCAL MICROSCOPY
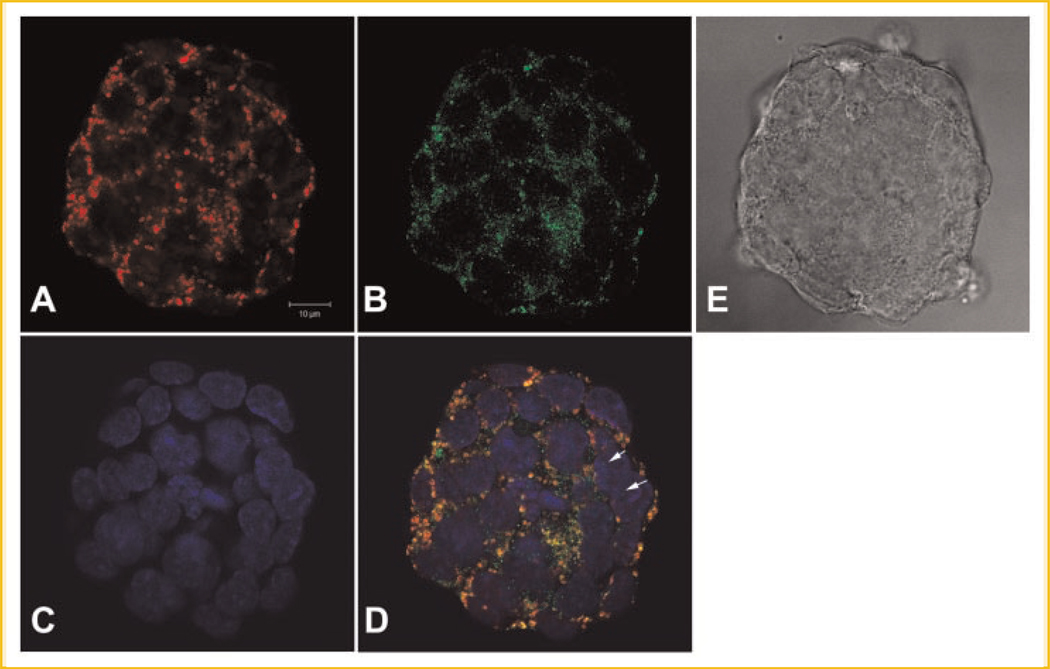
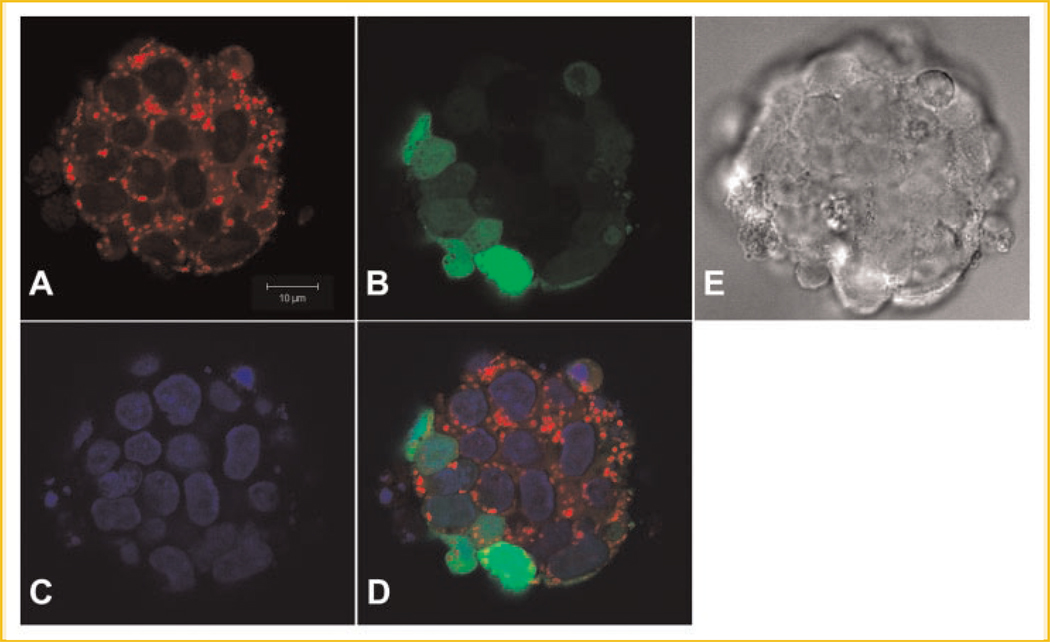
To study the subcellular localizations of MsrA in mouse ESCs, we have constructed a MsrA-eGFP fusion plasmid in the pEGFP-N1 vector (Clontech, CA) and transfected the plasmids into the ES cells. Fourty-eight to 72 h after transfection, green fluorescence could be observed in these cells indicating the expression of MsrA-eGFP fusion proteins. Cells were incubated with MitoTracker (Molecular Probes, WI) for mitochondrial staining before fixation and later mounted in DAPI for nuclear staining. Confocal microscopy (Fig. 2) showed strong colocalization of green fluorescence signals (MsrA-eGFP) with red MitoTracker signals (mitochondria). We also noted a more general cytosol distribution pattern of MsrA-eGFP signals outside of the nuclei and between mitochondria. This general cytosol localization with the more concentrated mitochondrial localization pattern of MsrA-eGFP was not observed in stem cells with only pEGFP-N1 vector transfection. The enhanced GFP (eGFP) expression inside the stem cells showed a nonspecific localization pattern with signals detected in both nuclei and cytosol (Fig. 3).
Fig. 2.
Confocal microscopy of the subcellular localizations of MsrA-eGFP fusion protein in mouse embryonic stem cells after pEGFP-N1-MsrA fusion expression plasmid transfection for 3 days. A: Stem cells scanned for Mitotracker staining for mitochondria. B: Green fluorescence shows the MsrA-eGFP fusion protein. C: DAPI staining shows nuclei. D: Merged image of A–C showing that MsrA-eGFP localizes in both mitochondria and cytosol, occasionally in nuclei (arrows). E: Phase contrast image of a colony of stem cells. The bar is 10 µm.
Fig. 3.
Confocal microscopy of the subcellular localizations of eGFP protein in mouse embryonic stem cells after pEGFP-N1 vector transfection for 3 days. A: Stem cells scanned for Mitotracker staining for mitochondria. B: Green fluorescence shows eGFP protein. C: DAPI staining shows nuclei. D: Merged image of A–C showing that eGFP localizes randomly in the cells present in both cytosol and nuclei. E: Phase contrast image of a colony of stem cells. The bar is 10 µm.
KNOCKDOWN OF MsrA EXPRESSION IN MOUSE EMBRYONIC STEM CELLS USING siRNAs REDUCED RESISTANCE OF THE CELLS TO H2O2-MEDIATED OXIDATIVE STRESS
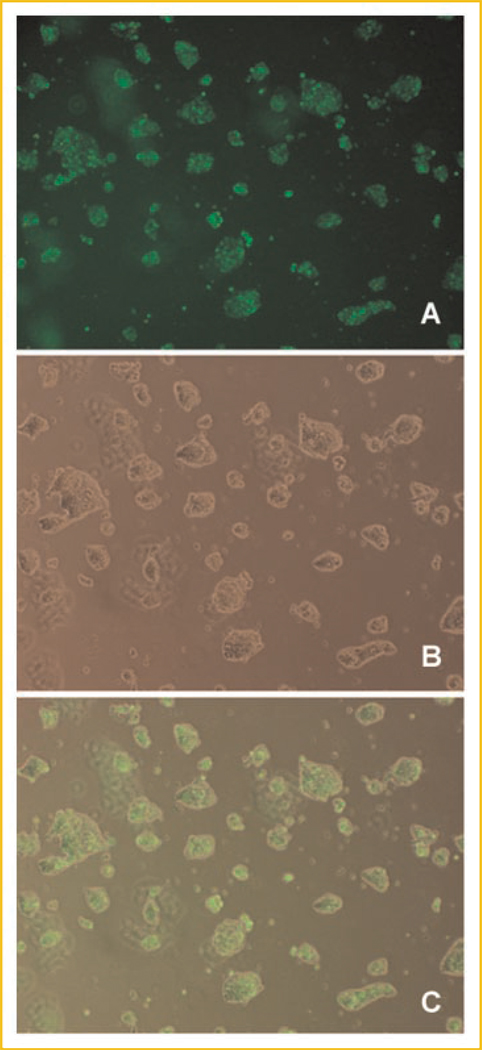
To find out whether MsrA can protect stem cells from oxidative stress, we designed and commercially synthesized three siRNAs for a MsrA knockdown experiment (Invitrogen). A negative control siRNA and a FITC-labeled siRNA also were synthesized. Using FITC-labeled siRNA, we optimized the transfection condition so that nearly 100% stem cells could be transfected with the siRNA (Fig. 4).
Fig. 4.
Epi-fluorescent microscopy shows nearly 100% transfection efficiency of siRNA in mouse embryonic stem cells. A: The green fluorescence inside the cells shows successful transfection of FITC-labeled siRNA. B: A phase contrast image of the stem cells. C: Merged image of panel A and B.
Real-time RT-PCR confirms the knockdown of MsrA at the mRNA level
Based on the optimized transfection method, we performed siRNA transfection on the stem cells and collected stem cell total RNA samples after 1, 2, and 3 days post-transfectionally. Both a siRNA designed specifically for the MsrA gene (168) and a negative control siRNA with a randomized sequence (882c) were transfected into the stem cells under the same conditions. Real-time RT-PCR experiments were conducted using MsrA-specific primers designed against its coding sequence near the c-terminal end, thus avoiding the alternative spliced 5′ end [Kim and Gladyshev, 2006]. When comparing the MsrA mRNA levels after MsrA-specific siRNA transfections with the negative control siRNA, we found that one siRNA, out of the three we designed successfully reduced the mRNA level of MsrA in the stem cells to only 20% of the nontransfected control cells (Fig. 5A). The reduced levels of MsrA mRNA could be maintained for 72 h post-transfection, with a slight increase returning after the 72-h period.
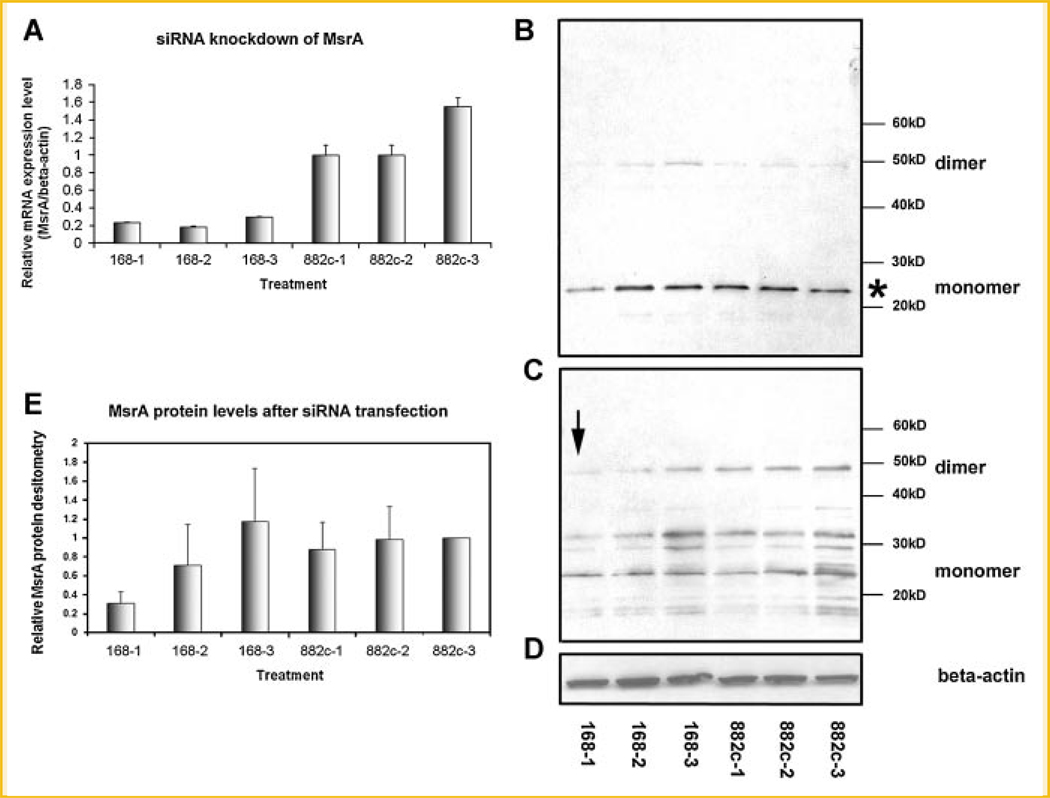
Fig. 5.
A: Confirmation of the knockdown of MsrA mRNA by MsrA-specific siRNA transfection using real-time RT-PCR. MsrA-specific siRNA (168) successfully reduces MsrA mRNA levels to only ~20% compared to the negative control siRNA (882c) transfected cells 1 day post-transfectionally. The mRNA levels remain low even 3 days after transfection. 168-1 stands for MsrA-specific siRNA (168) transfection for 1 day. B: Confirmation of the knockdown of MsrA protein by MsrA-specific siRNA tranfection using Western blots. Western blot was done with the protein samples from stem cells transfected by MsrA-specific siRNA (168) or negative control siRNA (882c) for various days (days 1, 2, and 3) in a reducing PAGE gel. C: Western blot after running the same protein samples as in panel B in a nonreducing PAGE gel. The arrow points to the significantly reduced MsrA dimers after 1 day transfection of MsrA-specific siRNA (168). D: The same blot as used in B was reprobed for β-actin protein showing equal loading of protein samples in each lane. E: Densitometry studies after scanning the monomer bands shown in panel B (shown with an asterisk) confirmed the significantly decreased expression level of MsrA after 1 day transfection of MsrA-specific siRNA compared to the negative control siRNA transfected cells (168-1 vs. 882c-1). Data are pooled from four separate experiments. 168-1 and 882c-1 stand for transfection of MsrA-specific siRNA (168) or a negative control siRNA (882c) for 1 day.
Western blots confirm the knockdown of MsrA at the protein level
Protein samples from stem cells were also collected after siRNA transfection at 24, 48, and 72 h post-transfectional time points. Western blotting experiments were performed using anti-MsrA antibody which recognizes a 24 kDa MsrA protein band (Fig. 5B). The membranes were stripped and rehybridized with β-actin antibody to normalize the MsrA concentrations in each sample (Fig. 5D). From Western blotting data as well as the densitometry of the blots (Fig. 5E) scanned by a gel imaging system (Alpha Innotech Corp.), we have clearly demonstrated a significant reduction of MsrA proteins 24 h after transfection of MsrA-specific siRNA in the stem cells as compared to the negative control siRNA transfections. It is interesting to note that the MsrA protein levels quickly returned to normal levels on the second day after transfection (Fig. 5B,E), even though the mRNA levels still remained low (see Fig. 5A). We have detected a faint band corresponding to MsrA protein dimers (~50 kDa) from Western blotting after protein samples were run in a reducing PAGE gel (Fig. 5B). To confirm the existence of the dimer form of MsrA protein in mouse ESCs, we performed the same Western blotting but used a nonreducing PAGE gel for electrophoresis. It clearly shows that the dimer form is present in significant amounts in the stem cells (Fig. 5C). The data suggest that it is the dimer form instead of a monomer form that is significantly reduced after siRNA knockdown of MsrA mRNA (arrow, Fig. 5C). Densitometry in Figure 5E shows the mean values from data pooled from four separate experiments using reducing PAGE gel electrophoreses (i.e., from four blots as shown in Fig. 5B). Only densitometry of the monomer forms (indicated by * in Fig. 5B) after reducing PAGE gel electrophoreses were performed and shown in Figure 5E.
Reduced resistance of stem cells to H2O2 treatments after MsrA siRNA transfection
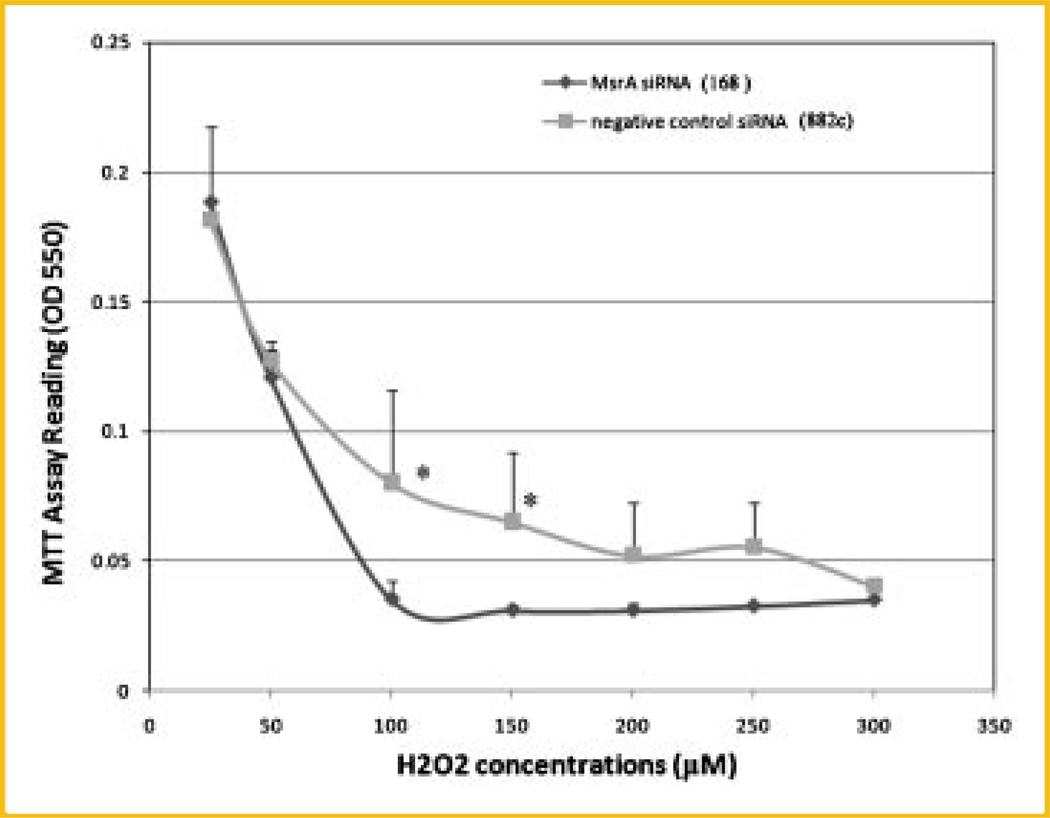
After testing the effects of siRNA we designed for the MsrA knockdown, we transfected the siRNA into mouse ES cells again and then applied H2O2 to the cells to evaluate the resistance of the cells to H2O2 after the MsrA protein was downregulated maximally (at the 24-h post-transfectional time point). H2O2 was diluted into the culture medium at various concentrations and incubated with the cells overnight before the MTT assays to compare the cell numbers in each group that survived. Results clearly showed a significantly reduced resistance of stem cells to H2O2 treatment with less cell survival after MsrA being knocked down (Fig. 6). Importantly, the difference between MsrA-specific siRNA and the negative control siRNA transfected cells were the most significant at the higher H2O2 concentrations used, that is, between 100 and 300 µM. No differences were observed at the lower concentrations when cells begin to die, for example, at concentrations of 25–50 µM.
Fig. 6.
MTT assays on stem cells treated by MsrA siRNA (168) versus a negative control siRNA (882c). After MsrA-specific siRNA (168) transfection for 24 h, the cells are significantly more prone to oxidative damage mediated by high concentrations of H2O2 treatment compared to a negative control siRNA (882c) transfected cells. The asterisks indicate P < 0.05.
OVEREXPRESSION OF MsrA-eGFP PROVIDES ADDITIONAL PROTECTION TO STEM CELLS AGAINST H2O2-INDUCED OXIDATIVE STRESS
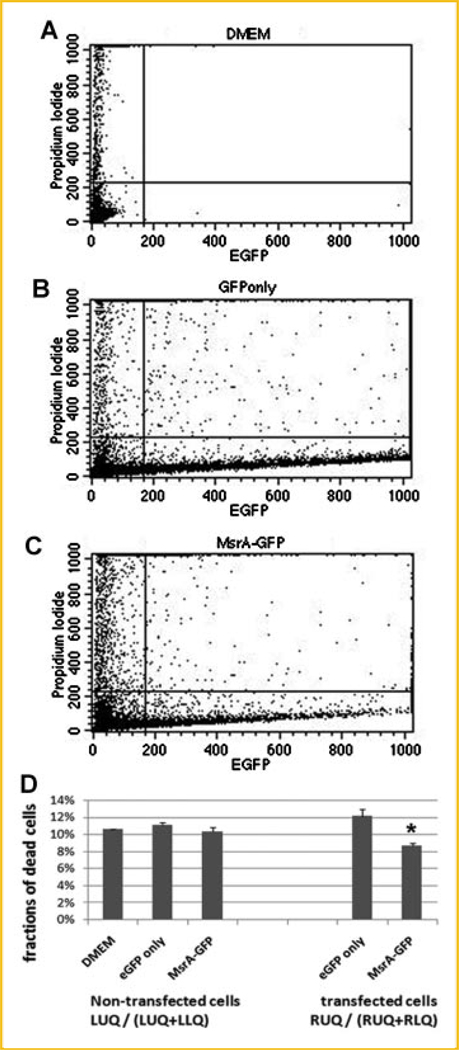
The MsrA-eGFP fusion constructs have been transfected to the cultured stem cells for 2 days. With confirmation of the expression of fusion proteins by the presence of green fluorescence using a fluorescence microscope, H2O2 at a concentration of 75 µM was added to the medium for 2 h before the cells were incubated with propidium iodide to stain for dead cells. The reason for using 75 µM is that at this concentration, around 50% cells survive from previous serial dilution assays (Fig. 6). Cells were then trypsinized and flow cytometry was performed to compare the differences of the ratios of cell death in cells with or without MsrA-eGFP fusion protein expression. Results from flow cytometry (Fig. 7) confirm expression of the MsrA-eGFP fusion protein, when comparing the cell numbers of EGFP channels (excitation wavelength at 488 nm) from MsrA-eGFP fusion plasmid transfected cells with the nontransfected cells. Based on cell flow cytometry, the cells were divided into four groups: ULQ: upper left quadrant (dead cells with no eGFP expression); URQ: upper right quadrant (dead cells with eGFP or MsrA-GFP expression); LLQ: lower left quadrant (live cells with no eGFP expression); LRQ: lower right quadrant (dead cells with eGFP or MsrA-GFP expression). Our results show that of those cells expressing the MsrA-eGFP fusion protein (RUQ plus RLQ), only 8.67% of cells are dead (stained by propidium iodide) (RUQ/(RUQ + RLQ)). However, in cells that are expressing eGFP only, 12.14% of the cells are showing propidium iodide positive staining (dead). Clearly, the MsrA-eGFP significantly protected the cells from H2O2-mediated oxidative damage, decreasing cell death by 28.6% compared to the eGFP only transfection (Fig. 7D). There are no significant differences in cell death ratios among nontransfected cells within DMEM medium cultured cells (no DNA transfection), eGFP transfected cells, and MsrA-eGFP transfected cells.
Fig. 7.
Flow cytometry studies confirming protective functions of MsrA when overexpressed in mouse ES cells. Cells are analyzed by eGFP fluorescent signals (indicating eGFP or MsrA-eGFP fusion protein expression) and propidium iodide (PI) staining (positive stain indicates dead cells). A: Cells incubated in DMEM medium only. B: Cells transfected with eGFP. C: Cells transfected with MsrA-eGFP fusion plasmid. D: Fractions of dead cells were calculated and compared within the positive green fluorescence (RUQ/(RUQ + RLQ) cells or those without fluorescent signals (LUQ/(LUQ + LLQ)). Asterisk indicates statistical significance compared to cells cultured in DMEM medium only without transfection as well as compared to cells with eGFP only transfection.
DISCUSSION
There have been several previous studies on methionine sulfoxide reductase (Msr) in mammalian cell lines such as lens cells, neuronal cells, fibroblasts, etc. [Huang et al., 1997; Petropoulos et al., 2001; Kantorow et al., 2004; Yermolaieva et al., 2004] as well as in animal models [Ruan et al., 2002; Moskovitz and Stadtman, 2003; Wodarz, 2007]. However, regulation of the Msr system as an antioxidant mechanism for stem cells was totally unknown before our studies. It is extremely important to understand more about stem cell anti-oxidative stress capabilities to provide guidelines for future therapeutic applications of stem cells placed in high oxidative stress environments. Our in vitro studies using a cell culture system have demonstrated the vital importance of the MsrA gene in mouse ESCs for protecting the cells from H2O2-mediated oxidative damage. It is our belief that the same protection may exist also in vivo which could be a promising approach to improve the cell survivability for stem cell transplantation in patients having ischemia/reperfusion oxidative damage in such tissues as an infarcted myocardial area. In addition, Msr genes might also play a significant role in protecting adult stem cells or stored ESCs in adult tissues from oxidative damage, since such cells appear to be maintained in the tissues in a quiescent state for long periods, perhaps even throughout life [Ding et al., 2003; Ryu et al., 2006].
In mammals, three MsrBs have been cloned (MsrB1, MsrB2, and MsrB3), which are targeted to various subcellular compartments [Hansel et al., 2003; Moskovitz, 2005] and show differential expression patterns among tissues [Hamada et al., 2005]. In contrast, only a single mammalian MsrA gene is known and its protein products are located both in the cytosol and in mitochondria [Vougier et al., 2003]. Our studies using the eGFP fusion protein to track MsrA expression show that the majority of MsrA proteins are localized in mitochondria and the cytosol, the same subcellular localization pattern as shown by the study on rat liver cells from Vougier et al. [2003]. In our study, only very limited fluorescent signals have been seen in the cell nuclei, indicating low concentrations of MsrA-eGFP; however, we cannot rule out the possibility that the low staining levels for MsrA-eGFP fusion protein was due to the disturbance of eGFP being conjugated to MsrA [Hansel et al., 2002]. Studies from Hansel et al. [2002] on human MsrA demonstrated the exclusive mitochondrial localization of its protein. Studies from Kim and Gladyshev [2005] on mouse MsrA gene using the eGFP fusion protein approach in monkey kidney CV-1 cells revealed the subcellular localization of MsrA in mitochondria, cytosol, and nuclei. It was proposed by Kim and Gladyshev [2005] that the N-terminal mitochondrial signal peptide is responsible for directing the MsrA protein to localize in mitochondria. There also exist specific structures in the middle and c-terminal sequences of mouse MsrA protein that are required in targeting a fraction of the mature proteins back to cytosol that were once localized in mitochondria [Kim and Gladyshev, 2005]. We believe it is highly possible that MsrA subcellular localization has species- and cell type-variations.
It has been long suspected that in mammalian cells, MsrA is a constantly expressed gene without many exogenous influences except for a recent report that ecdysone can induce MsrA expression in fruit flies (Drosophila) during development [Roesijadi et al., 2007]. Our studies using combinations of anoxia and reoxygenation to treat mouse ESCs have shown a unique result that the levels of MsrA mRNA and protein expression decreases with increased periods of anoxia or reoxygenation treatment. Increasing periods of anoxia or reoxygenation result in increasing intracellular levels of ROS [Huang et al., 1997]. Results indicate that with increasing levels of oxidative stress, MsrA mRNA, and proteins in stem cells are downregulated. These results are consistent with the findings reported by Petropoulos et al. [2001] that rat MsrA gene expression is decreasing along with aging and the report that MsrA gene expression is downregulated during replicative senescence of human WI-38 fibroblasts [Choi et al., 2005]. In both cases, aging is concurrent with higher levels of oxidative stress. It is interesting to note that all of these studies show that the MsrA expression levels decrease instead of increase. Increasing MsrA expression would hypothetically provide more protection for cells when exposed to higher levels of oxidative damage. It remains unknown whether this is a mechanism that exists in nature as a protective mechanism to prevent stem cells in the body from turning into cancer by allowing cells to die by ROS damage once the hypoxic environment is established in a growing tumor. It is also possible that by decreasing MsrA via oxidative stress, ESCs are allowing higher and longer periods of ROS spikes to signal for differentiation initiation in which case overexpression of MsrA in stem cells will be detrimental to their plasticity. It will be interesting to further investigate this phenomenon for future guidance in manipulating MsrA expression in stem cells in response to oxidative damage to increased cell survivability and differentiation.
We have found that after siRNA knockdown of MsrA, its mRNA levels remain low for at least 72 h. However, MsrA protein levels bounce back quickly, after only 48 h. MsrA protein levels after normalization with β-actin at 48 h vary significantly at 48 h after siRNA transfection among the repeated experiments indicating it is within the transitional period when MsrA protein expression is recovering. The translational level regulation of MsrA protein in stem cells has never been reported before this current study and is still unknown as to its mechanism. Nonetheless, the tight control in mouse ESCs to keep the MsrA protein level high, even with low levels of mRNA present, suggests that this protein is essential to cellular function and survivability.
At 24 h after MsrA siRNA transfection, we have confirmed successful knockdown of MsrA mRNA levels in the stem cells. Nearly 80% reduction at mRNA levels and 70% reduction at protein levels have been detected in the cells at this time point. Therefore, we chose this time point to apply H2O2 to the cells to determine if there is a loss of protective effect after MsrA knockdown at high H2O2 concentrations. Our results showed that the loss of protection after MsrA knockdown is not significant when H2O2 concentrations are low (<50 µM), suggesting that the protective effects from other methionine sulfoxide reductases, such as MsrBs, are intact in the stem cells to fight against oxidative damage. To confirm this hypothesis and also to prove that the loss of protective effects is due to MsrA alone rather than simultaneous loss of MsrBs expression after MsrA siRNA transfection, we conducted real-time RT-PCR on all three reported MsrB genes (MsrB1, MsrB2, and MsrB3) using samples with MsrA siRNA transfection. Our results confirm that all three MsrBs are expressed in the stem cells at 24 h siRNA posttransfection, with no MsrBs mRNA expression levels being altered. Results strongly suggest that there are no direct interactions between MsrA and MsrBs gene expression in mouse ESCs.
CONCLUSION
Our results have demonstrated additional protective effects with MsrA overexpression in mouse ESCs. Based on these studies, we believe that by overexpressing MsrA in stem cells, we can provide significant protection to stem cells against harsh environments such as ischemia/reperfusion during strokes or heart attacks. Currently new approaches to treat diseases, such as neurodegenerative diseases or ischemia/reperfusion-induced brain or heart damage, are being developed using adult or ESC transplantation. One severe limitation of using these stem cells is the low survivability of the cells after surgery partly due to the oxidative environment into which the cells are implanted. Without getting stem cells properly prepared and protected to resist high oxidative stress prior to transplanting them into damaged tissue is destined to certain failure and death of the implanted cells. Results of our current study show a very promising approach to solving this problem by enhancing the resistance of stem cells to oxidative damage once they are transplanted into an oxidatively stressing environment. It will be most interesting to further investigate the differentiation capability of stem cells overexpressing MsrA. Due to the Msr genes’ direct function of reversing oxidized proteins back to their functional forms, thus indirectly controlling ROS levels in the cells, we believe that overexpressing Msr genes in stem cells could function to reduce ROS damage to the cells without losing the cells’ sensitivity to ROS as a differentiation signal. Thus, the Msr gene family could potentially serve a key role in engineering stem cells to obtain higher resistance to oxidative damages, while retaining the cells’ potential for differentiation into adult cell types.
ACKNOWLEDGMENTS
This study is supported by NIH grants HL-58435 and HL-061246 to L.F.L., a grant from American Heart Association to X. H. and a Research Fellowship from NIH to C. Z. We are grateful to Mrs. Amy Patrick for outstanding secretarial and administrative assistance in the preparation and submission of this manuscript. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung and Blood Institute.
Footnotes
Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Provision of study materials or patients, collection and/or assembly of data, data analysis and interpretation, manuscript writing.
Conception and design, administrative support, data analysis and interpretation, manuscript writing, final approval of manuscript.
Conception and design, financial support, administrative support, provision of study materials or patients, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Conception and design, financial support, administrative support, provision of study materials or patients, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript, principal investigator of laboratory/NIH Grants.
REFERENCES
- Blass JP. Brain metabolism and brain disease: Is metabolic deficiency the proximate cause of Alzheimer dementia? J Neurosci Res. 2001;66:851–856. doi: 10.1002/jnr.10087. [DOI] [PubMed] [Google Scholar]
- Boldyrev A, Bulygina E, Makhro A. Glutamate receptors modulate oxidative stress in neuronal cells. A mini-review. Neurotox Res. 2004;6:581–587. doi: 10.1007/BF03033454. [DOI] [PubMed] [Google Scholar]
- Choi D, Lee HJ, Jee S, Jin S, Koo SK, Paik SS, Jung SC, Hwang SY, Lee KS, Oh B. In vitro differentiation of mouse embryonic stem cells: Enrichment of endodermal cells in the embryoid body. Stem Cells. 2005;23:817–827. doi: 10.1634/stemcells.2004-0262. [DOI] [PubMed] [Google Scholar]
- Ding S, Wu TY, Brinker A, Peters EC, Hur W, Gray NS, Schultz PG. Synthetic small molecules that control stem cell fate. Proc Natl Acad Sci USA. 2003;100:7632–7637. doi: 10.1073/pnas.0732087100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H, Kobune M, Nakamura K, Kawano Y, Kato K, Honmou O, Houkin K, Matsunaga T, Niitsu Y. Mesenchymal stem cells (MSC) as therapeutic cytoreagents for gene therapy. Cancer Sci. 2005;96:149–156. doi: 10.1111/j.1349-7006.2005.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel A, Kuschel L, Hehl S, Lemke C, Agricola HJ, Hoshi T, Heinemann SH. Mitochondrial targeting of the human peptide methionine sulfoxide reductase (MSRA), an enzyme involved in the repair of oxidized proteins. FASEB J. 2002;16:911–913. doi: 10.1096/fj.01-0737fje. [DOI] [PubMed] [Google Scholar]
- Hansel A, Jung S, Hoshi T, Heinemann SH. A second human methionine sulfoxide reductase (hMSRB2) reducing methionine-R-sulfoxide displays a tissue expression pattern distinct from hMSRB1. Redox Rep. 2003;8:384–388. doi: 10.1179/135100003225003429. [DOI] [PubMed] [Google Scholar]
- Honig LS, Rosenberg RN. Apoptosis and neurologic disease. Am J Med. 2000;108:317–330. doi: 10.1016/s0002-9343(00)00291-6. [DOI] [PubMed] [Google Scholar]
- Huang JQ, Trasler JM, Igdoura S, Michaud J, Hanal N, Gravel RA. Apoptotic cell death in mouse models of GM2 gangliosidosis and observations on human Tay-Sachs and Sandhoff diseases. Hum Mol Genet. 1997;6:1879–1885. doi: 10.1093/hmg/6.11.1879. [DOI] [PubMed] [Google Scholar]
- Kantorow M, Hawse JR, Cowell TL, Benhamed S, Pizarro GO, Reddy VN, Hejtmancik JF. Methionine sulfoxide reductase A is important for lens cell viability and resistance to oxidative stress. Proc Natl Acad Sci USA. 2004;101:9654–9659. doi: 10.1073/pnas.0403532101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Methionine sulfoxide reduction in mammals: Characterization of methionine-R-sulfoxide reductases. Mol Biol Cell. 2004;15:1055–1064. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Role of structural and functional elements of mouse methionine-S-sulfoxide reductase in its subcellular distribution. Biochemistry. 2005;44:8059–8067. doi: 10.1021/bi0501131. [DOI] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Alternative first exon splicing regulates subcellular distribution of methionine sulfoxide reductases. BMC Mol Biol. 2006;7:11. doi: 10.1186/1471-2199-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J. Roles of methionine suldfoxide reductases in antioxidant defense, protein regulation and survival. Curr Pharm Des. 2005;11:1451–1457. doi: 10.2174/1381612053507846. [DOI] [PubMed] [Google Scholar]
- Moskovitz J, Stadtman ER. Selenium-deficient diet enhances protein oxidation and affects methionine sulfoxide reductase (MsrB) protein level in certain mouse tissues. Proc Natl Acad Sci USA. 2003;100:7486–7490. doi: 10.1073/pnas.1332607100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyango IG, Bennett JP, Jr, Tuttle JB. Endogenous oxidative stress in sporadic Alzheimer’s disease neuronal cybrids reduces viability by increasing apoptosis through pro-death signaling pathways and is mimicked by oxidant exposure of control cybrids. Neurobiol Dis. 2005;19:312–322. doi: 10.1016/j.nbd.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Petropoulos I, Mary J, Perichon M, Friguet B. Rat peptide methionine sulphoxide reductase: Cloning of the cDNA, and down-regulation of gene expression and enzyme activity during aging. Biochem J. 2001;355:819–825. doi: 10.1042/bj3550819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesijadi G, Rezvankhah S, Binninger DM, Weissbach H. Ecdysone induction of MsrA protects against oxidative stress in Drosophila. Biochem Biophys Res Commun. 2007;354:511–516. doi: 10.1016/j.bbrc.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Ruan H, Tang XD, Chen ML, Joiner ML, Sun G, Brot N, Weissbach H, Heinemann SH, Iverson L, Wu CF, Hoshi T. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc Natl Acad Sci USA. 2002;99:2748–2753. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu BY, Orwig KE, Oatley JM, Avarbock MR, Brinster RL. Effects of aging and niche microenvironment on spermatogonial stem cell self-renewal. Stem Cells. 2006;24:1505–1511. doi: 10.1634/stemcells.2005-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer H, Rahimi G, Hescheler J, Wartenberg M. Role of reactive oxygen species and phosphatidylinositol 3-kinase in cardiomyocyte differentiation of embryonic stem cells. FEBS Lett. 2000;476:218–223. doi: 10.1016/s0014-5793(00)01747-6. [DOI] [PubMed] [Google Scholar]
- Thiruchelvam M, Prokopenko O, Cory-Slechta DA, Buckley B, Mirochnitchenko O. Overexpression of superoxide dismutase or glutathione peroxidase protects against the paraquat+maneb-induced Parkinson disease phenotype. J Biol Chem. 2005;280:22530–22539. doi: 10.1074/jbc.M500417200. [DOI] [PubMed] [Google Scholar]
- Vougier S, Mary J, Friguet B. Subcellular localization of methionine sulphoxide reductase A (MsrA): Evidence for mitochondrial and cytosolic isoforms in rat liver cells. Biochem J. 2003;373:531–537. doi: 10.1042/BJ20030443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wo YB, Zhu DY, Hu Y, Wang ZQ, Liu J, Lou YJ. Reactive oxygen species involved in prenylflavonoids, icariin and icaritin, initiating cardiac differentiation of mouse embryonic stem cells. J Cell Biochem. 2008;103:1536–1550. doi: 10.1002/jcb.21541. [DOI] [PubMed] [Google Scholar]
- Wodarz D. Effect of stem cell turnover rates on protection against cancer and aging. J Theor Biol. 2007;245:449–458. doi: 10.1016/j.jtbi.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Yermolaieva O, Xu R, Schinstock C, Brot N, Weissbach H, Heinemann SH, Hoshi T. Methionine sulfoxide reductase A protects neuronal cells against brief hypoxia/reoxygenation. Proc Natl Acad Sci USA. 2004;101:1159–1164. doi: 10.1073/pnas.0308215100. [DOI] [PMC free article] [PubMed] [Google Scholar]