Abstract
Histone deacetylation plays an important role in methylated DNA silencing. Recent studies indicated that the methyl-CpG-binding protein, MBD2, is a component of the MeCP1 histone deacetylase complex. Interestingly, MBD2 is able to recruit the nucleosome remodeling and histone deacetylase, NuRD, to methylated DNA in vitro. To understand the relationship between the MeCP1 complex and the NuRD complex, we purified the MeCP1 complex to homogeneity and found that it contains 10 major polypeptides including MBD2 and all of the known NuRD components. Functional analysis of the purified MeCP1 complex revealed that it preferentially binds, remodels, and deacetylates methylated nucleosomes. Thus, our study defines the MeCP1 complex, and provides biochemical evidence linking nucleosome remodeling and histone deacetylation to methylated gene silencing.
Keywords: Transcriptional repression, DNA methylation, nucleosome remodeling, histone deacetylation
ATP-dependent nucleosome remodeling and core histone tail acetylation play important roles in chromatin function (Kornberg and Lorch 1999). The purification and functional characterization of the nucleosome remodeling and histone deacetylase complex, NuRD/Mi-2 complex, suggests that the two chromatin modifying enzymatic activities could be coupled (Tong et al. 1998; Wade et al. 1998; Xue et al. 1998; Zhang et al. 1998a). NuRD has been purified from both HeLa cells and Xenopus eggs (Zhang et al. 1998a; Wade et al. 1999). The NuRD complex from HeLa cells contains seven major polypeptides, including Mi2, MTA2, MBD3 and the histone deacetylase core, HDAC1/2 and RbAp46/48 (Zhang et al. 1998a; Zhang et al. 1999). Mi2 is an SWI2/SNF2 type helicase/ATPase domain-containing protein likely to be responsible for the chromatin remodeling activity of the NuRD complex. MTA2 is a novel protein that is highly similar (65% identical) to the candidate metastasis-associated protein MTA1 (Toh et al. 1994; Zhang et al. 1999). Biochemical characterization of MTA2 indicates that it plays an important role in modulating the histone deacetylase activity of the NuRD complex (Zhang et al. 1999). MBD3 is a methyl-CpG-binding domain-containing protein, similar to MBD2 (Hendrich and Bird 1998).
The identification of the methyl-CpG-binding domain-containing protein MBD3 in the NuRD/Mi2 complex suggests that this complex may be recruited to methylated DNA for transcriptional silencing. Therefore, considerable efforts have been devoted to establishing a link between the NuRD/Mi-2 complex and DNA methylation (Wade et al. 1999; Zhang et al. 1999). Consistent with the finding that the bulk of mammalian MBD3 is not localized to methylated DNA foci in vivo (Hendrich and Bird 1998), mammalian MBD3, either by itself or in association with NuRD, does not show affinity binding to methylated DNA in gel shift assays (Hendrich and Bird 1998; Zhang et al. 1999). Interestingly, MBD2, although not an integral component of the NuRD complex, has the ability to mediate the interaction between NuRD and methylated DNA in vitro (Zhang et al. 1999). In contrast, recombinant Xenopus MBD3 (xMBD3) does show affinity for binding to methylated DNA (Wade et al. 1999). Thus, the Xenopus Mi-2 complex was proposed to couple DNA methylation to chromatin remodeling and histone deacetylation (Wade et al. 1999), although it remains to be determined whether the Xenopus Mi-2 complex has affinity to methylated DNA. Nevertheless, the above observations suggest that NuRD may be involved in methylated DNA silencing.
Histone deacetylation is a major mechanism of methylated DNA silencing (Bird and Wolffe 1999). Recent studies have revealed that the methyl-CpG-binding protein MBD2 is a component of the MeCP1 histone deacetylase complex that also contains HDAC1/2 and RbAp46/48 (Ng et al. 1999). The findings that MeCP1 contains MBD2 and shares the same histone deacetylase core with the NuRD complex (Zhang et al. 1998a; Ng et al. 1999), and that MBD2 can interact with the NuRD complex in vitro (Zhang et al. 1999) prompted us to ask whether the two complexes are related. Here, we purified the MeCP1 complex and found that it is composed of 10 major polypeptides including MBD2 and all of the known NuRD components. In addition, we demonstrate that the purified MeCP1 complex preferentially binds, remodels, and deacetylates methylated nucleosomes. Importantly, expression of an Mi2 mutant that is defective in its ATPase activity relieved methylation-dependent transcriptional repression. Thus, our results define the molecular composition of the MeCP1 complex, and provide the first biochemical evidence linking nucleosome remodeling and histone deacetylation to methylated gene silencing.
Results and Discussion
MeCP1 copurifies with a methyl-CpG-binding activity as a 1 megadalton protein complex
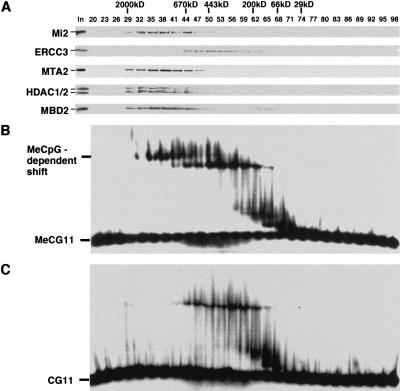
The methyl-CpG-binding protein MBD2 has been shown to be a component of the MeCP1 complex that also includes the histone deacetylase core complex, HDAC1/2 and RbAp46/48 (Ng et al. 1999). To determine whether the MeCP1 complex contains proteins other than the five polypeptides mentioned above, we determined the size of the native MeCP1 complex. Fractionation of HeLa nuclear extracts on a gel filtration column revealed that, similar to the NuRD complex (represented by Mi2 and MTA2), the bulk of MBD2 coeluted with the histone deacetylases HDAC1/2 as a large protein complex of about 1 MD (Fig. 1A). However, less than 10% of MBD2 eluted in a smaller complex of about 100 kD. The general transcription factor TFIIH (represented by ERCC3) eluted at its expected size (Drapkin and Reinberg 1994), confirming proper separation of proteins on this column. Thus, more than 90% of MBD2 exists in a 1 MD protein complex that likely represents the MeCP1 complex. Since the combined weights of HDAC1/2, RbAp46/48, and MBD2 cannot account for the estimated size of the MeCP1 complex, additional polypeptides are likely to be present in the MeCP1 complex.
Figure 1.
MBD2 and NuRD copurify with a methyl-CpG-binding activity as a 1MD protein complex. (A) Western blot analysis of fractions derived from a gel filtration Superose-6 column. The elution profiles of the protein size markers and the proteins analyzed are indicated. (B,C) Gel mobility shift assays using the same fractions as in panel A. The probes used in panels B and C are methylated and nonmethylated, respectively. Methylation-dependent shift and free probes are indicated.
MBD2 was found to be responsible for the methyl-CpG-binding activity of the MeCP1 complex (Ng et al. 1999). To ensure that the methylation-dependent gel shift assay can be used to monitor the MeCP1 complex purification, the fractions shown in Fig. 1A were analyzed by gel shift assays. The results shown in Fig. 1B revealed several DNA binding activities. However, only the largest shift is methylation-dependent (Fig. 1, cf. B and C). This methylation-dependent DNA binding activity coeluted with MBD2 and NuRD (Fig. 1A,B).
The MeCP1 complex is composed of 10 polypeptides including all the NuRD components
Using the gel mobility shift assay and Western blot analysis described above, we purified the MeCP1 complex by a six-step chromatographic procedure (Fig. 2A). The NuRD complex, represented by Mi2, was also monitored during the purification process. As shown in Fig. 2B, MBD2 elutes in two protein complexes on DEAE-5PW column peaking in fractions 35 and 56, respectively. Although the elution profile of Mi2 overlaps the first peak of MBD2, the bulk of Mi2 does not completely coelute with the two MBD2 peaks, indicating that the majority of the NuRD complex does not associate with MBD2. To identify the proteins that associate with MBD2 at the first peak, the peak fractions were pooled and purified further as outlined in Fig. 2A. A gel mobility shift assay of the fractions derived from the last purification step indicated that fractions 30 to 57 could shift the methylated probe MeCG11 (Fig. 2C). However, the same fractions failed to shift the nonmethylated CG11 probe (data not shown). Silver staining revealed that about ten major polypeptides coeluted with the binding activity (Fig. 2D). The molecular weights (MW) of most of the coeluted polypeptides were strikingly similar to the components of the NuRD complex, suggesting that the coeluted polypeptides are likely to be NuRD components. This was confirmed by Western blot analysis (Fig. 2E). Thus, at least a portion of MBD2 copurifies with NuRD and the methyl-CpG-binding activity.
Figure 2.
The MeCP1 complex contains MBD2 and NuRD. (A) Schematic representation of the steps used to purify the MeCP1 complex. The range of salt concentrations where the MeCP1 complex eluted out of the columns is indicated. (B) Western blot analysis of the fractions derived from DEAE-5PW column. (C) Gel mobility shift assay of the fractions derived from the last purification step. Methylation-dependent shift and free probes are indicated. (D) A silver-stained gel showing that a group of 10 polypeptides copurify with the methyl-CpG-binding activity. The identities of these polypeptides are confirmed by Western blot analysis, shown in panel E.
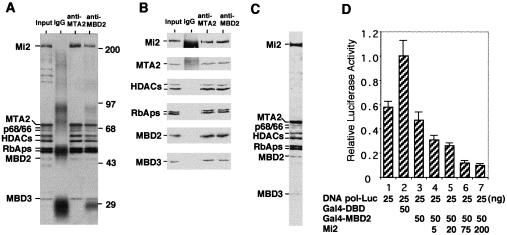
Extensive copurification through a variety of columns suggests that MBD2 and NuRD may exist in the same protein complex. To explore this possibility, the last column fractions containing methyl-CpG-binding activity were pooled and used as input for immunoprecipitation using antibodies against MTA2 and MBD2. To avoid the possibility that MBD2 antibodies immunoprecipitate the NuRD complex by recognizing MBD3, we used antibody S923, which only recognizes MBD2 (Ng et al. 1999). As a negative control for specificity, rabbit IgG was also used. Immunoprecipitated proteins were analyzed by silver staining and Western blotting. The results shown in Figure 3A and B indicated that each of the two antibodies immunoprecipitated the same set of proteins, including the seven characterized NuRD components, MBD2, and two polypeptides of 66 and 68 kD. Consistent with the result shown in Fig. 2D, where the polypeptides of MW between 100 and 200 kD do not copurify with the putative MeCP1 complex, neither MTA2 nor MBD2 antibodies immunoprecipitated these polypeptides, indicating that the immunoprecipitated polypeptides are specific. Therefore, we conclude that the MeCP1 complex contains ten polypeptides including MBD2, NuRD, and two uncharacterized polypeptides of 66 and 68 kD.
Figure 3.
MBD2 and NuRD exist in the same protein complex. (A) A silver-stained gel showing that the same 10 polypeptides are immunoprecipitated by both MTA2 and MBD2 antibodies. MBD2 has characteristically faint staining with silver (Figs. 2D,3A), but stains better with Coomassie (Fig. 3C). The identities of the proteins and size markers are indicated. (B) Western blot analysis of the same samples used for silver staining in panel A. Antibodies used for immunoprecipitation and Western blotting are indicated on the top and left of the panel, respectively. (C) Coomassie staining of the MeCP1 complex. (D) Mi2 enhances MBD2-mediated transcription repression. The amount of reporter and effector plasmids used in each transfection is indicated. Transfection efficiencies were normalized using β-galactosidase assays. The data shown represent the average of two independent experiments. Variations between experiments are depicted by the error bars.
Mi2 enhances MBD2-mediated transcriptional repression
Since MBD2 physically associates with NuRD in the MeCP1 complex, we expect that overexpression of NuRD components, such as Mi2, should affect MBD2-mediated transcriptional repression. Similar to previous observations (Ng et al. 1999), tethering MBD2 to the DNA polymerase β promoter through Gal4 DNA binding domain resulted in transcriptional repression (Fig. 3D, cf. columns 2 and 3). Importantly, overexpression of Mi2 resulted in a dose-dependent enhancement of the MBD2-mediated transcription repression which is consistent with MBD2/NuRD association (Fig. 3D, cf. columns 4–7 with 3).
The MeCP1 complex preferentially binds, remodels and deacetylates methylated nucleosomes
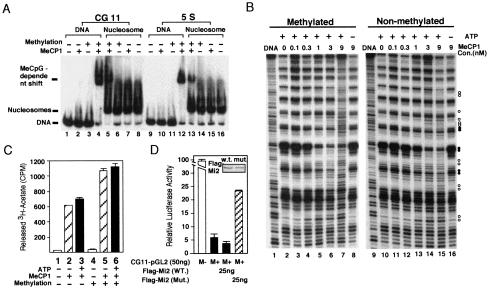
Having defined the molecular composition of the MeCP1 complex, we sought to address the function of the MeCP1 complex in methylated DNA silencing. We first asked whether MeCP1 could bind specifically to methylated nucleosomal DNA. Thus, nonmethylated and methylated CG11 probes were assembled into mononucleosomes (Steger et al. 1998). A gel mobility shift assay using the assembled nucleosomes indicated that MeCP1 can form a stable complex with nucleosomes as long as the DNA is methylated (Fig. 4A, cf. lanes 5 and 7). However, the affinity of MeCP1 for methylated nucleosomes is much less than that for methylated naked DNA (Fig. 4A, cf. lanes 4 and 5). About 10-fold more MeCP1 is required to completely shift a probe that is assembled into a nucleosome as compared to naked DNA (data not shown). Since the affinity of the MeCP1 complex for methylated DNA depends on the number of available methyl-CpGs (Meehan et al. 1989), the reduced binding activity of MeCP1 for nucleosomes is expected, because about half of the methyl-CpGs would be facing in towards the histone octamer and therefore would not be accessible when the DNA is assembled into nucleosomes.
Figure 4.
MeCP1 is able to preferentially bind, remodel, deacetylate and repress transcription from methylated nucleosomal templates. (A) Gel mobility shift assay comparing the binding of MeCP1 to methylated and nonmethylated DNA and nucleosomes. The four probes used are indicated on top of the panel. (B) Mononucleosome disruption assay comparing the efficiency of MeCP1 to disrupt methylated and nonmethylated nucleosomes. End-labeled mononucleosomes with a concentration of 10 nM were incubated with increasing concentrations of MeCP1, followed by DNase I digestion. The presence or absence of ATP in the reactions is indicated. Open circles indicate enhanced DNase I digestion; filled circles indicate reduced DNase I digestion. (C) Histone deacetylase assays comparing the efficiency of MeCP1 to deacetylate methylated or nonmethylated nucleosomal histones. Approximately 300 ng of 3H-labeled acetylated core histones was assembled into mononucleosomes with methylated or nonmethylated DNA (final concentration of 80 nM) and were used in a 30 μL reaction with or without the presence of 5 μL MeCP1 (final concentration of 20 nM) and 2 mM of ATP. The bar graph represents an average of two independent experiments. Variations between the two experiments are depicted by the error bars. (D) Expression of an Mi2 mutant, crippled in ATP-binding, partially relieved methylation-dependent transcriptional repression. Shown are relative luciferase activities of different transfections. M− and M + indicate that the reporter plasmid CG11–pGL12 is mock- or HhaI-methylated, respectively. The data shown represent the average of two independent experiments. Variations between experiments are depicted by the error bars. The insert is a Western blot probed with antibody against Flag.
The CG11 probe is a synthetic sequence that contains 27 CpG pairs. Based upon the original report (Meehan et al. 1989), it was not clear whether the MeCP1 complex could bind in a DNA methylation-dependent manner to genes that do not contain such a high frequency of methyl-CpGs. To address this question, we used a 152 bp Xenopus 5S rRNA gene sequence containing 10 CpG pairs in the gel mobility shift assay. The results shown in Fig. 4A demonstrate that MeCP1 is able to bind specifically to methylated 5S DNA (lane 12). More importantly, it also binds specifically to methylated 5S nucleosomes (lane 13). The demonstration of specific binding of the MeCP1 complex to a methylated, naturally occurring gene packaged into nucleosomes strongly argues that the recruitment of the MeCP1 complex to methylated genes is likely to be biologically relevant.
The presence of NuRD in the MeCP1 complex suggests that this complex may be able to remodel nucleosome structure. An important question is whether the presence of methyl-CpG-binding protein MBD2 in the complex facilitates remodeling of methylated nucleosomes. To address this question, we compared the ability of MeCP1 to disrupt methylated and nonmethylated nucleosomes. As shown in Fig. 4B, in the absence of MeCP1, DNase I digestion of end-labeled nucleosomal DNA produces a periodic pattern of enhanced cutting every 10 base pairs, which is in contrast with the digestion pattern of naked DNA (Fig. 4B, cf. lanes 1 and 2, lanes 9 and 10). In the presence of sufficient amounts of MeCP1, however, the DNase I digestion patterns were altered (Fig. 4B, cf. lane 7 with 2, lane 15 with 10), indicating that the MeCP1 complex is capable of disrupting nucleosome structure. As expected, the ability of MeCP1 to disrupt nucleosome structure is ATP-dependent (Fig. 4B, cf. lanes 7 and 8, lanes 15 and 16). Although MeCP1 is able to disrupt both methylated and nonmethylated nucleosomes, it appears to be more efficient in disrupting methylated nucleosomes. For example, disruption of methylated nucleosomes is observed in the presence of 1 nM MeCP1 (Fig 4B, lane 5), while a similar level of disruption of nonmethylated nucleosomes requires the presence of 3 nM MeCP1 (Fig 4B, lane 14). This result indicates that recruitment of the MeCP1 complex to methylated nucleosomes facilitates nucleosome remodeling.
Having established that the MeCP1 complex preferentially disrupts methylated nucleosome structure in the presence of ATP (Fig. 4B), we next asked whether MeCP1 preferentially deacetylates histones when the nucleosomal DNA is methylated and whether nucleosomal histone deacetylation by MeCP1 is stimulated by ATP. 3H-labeled acetylated core histone octamers and unlabeled methylated and nonmethylated 5S rDNA were assembled into mononucleosomes. To ensure successful nucleosome assembly, parallel assembly reactions, in which 10% of the DNA was end-labeled with 32P, were also performed. To avoid potential contamination of nucleosomes by non-assembled 3H-labeled core histones, a 15% excess of DNA relative to core histones was used in these assembly reactions. The results shown in Fig. 4C indicate that MeCP1 preferentially deacetylates methylated nucleosomal histones (Fig. 4B, cf. columns 2 and 5, columns 3 and 6). The presence of ATP did not significantly increase deacetylation of nucleosomal histones (Fig. 4B, cf. columns 2 and 3, 5 and 6), suggesting that nucleosome disruption is not required for mononucleosomal histone deacetylation in vitro.
An ATPase-deficient Mi2 mutant partially relieved methylation-dependent transcriptional repression
Preferential binding, remodeling, and deacetylating of methylated nucleosomes by the MeCP1 complex (Fig. 4A–C) indicate that this complex is likely to play a role in methylation-dependent transcriptional repression, and that this repression can be specifically relieved by overexpression of a dominant negative component of the MeCP1 complex. Since nucleosome remodeling requires ATP hydrolysis, mutation in the ATP-binding pocket of Mi2 should inactivate its ATPase activity, and therefore cripple the remodeling activity of the MeCP1 complex. Previous studies of SWI2/SNF2 and ISWI have demonstrated that mutations on the lysine residue of the conserved ATP-binding pocket, GXGK, abolished the ATPase activities of these proteins (Richmond and Peterson 1996; Corona et al. 1999). Therefore, we analyzed the effect of a similar mutation in the Mi2 ATP-binding pocket (K757R) on transcription activity of a previously described reporter, CG11-pGL2 (Ng et al. 1999). The results shown in Fig. 4D indicate that methylation of the reporter plasmids by HhaI significantly reduced its transcriptional activity when compared with the same reporter that was mock methylated (cf. the first two columns). Importantly, while cotransfection of a plasmid encoding wild-type Mi2 slightly reduced the methylated reporter activity, cotransfection of a plasmid encoding the ATPase-deficient Mi2 mutant partially relieved methylation-dependent transcriptional repression (cf. the last three columns). This differential effect on methylated reporter is not a result of differential expression of the effector plasmids, since Western blot analysis using anti-Flag antibodies indicated that both proteins expressed at a similar level (Fig. 4D, insert). This result strongly suggests that the MeCP1 complex contributes, at least partially, to the methylation-dependent transcriptional repression.
The finding that MBD2 exists together with NuRD in the MeCP1 complex seems contrary to the initial characterization of the NuRD complex, which does not include MBD2 (Zhang et al. 1998a). This may be due to the different purification strategies employed. It is possible that there is a tightly associated NuRD core complex that does not always associate with DNA-binding proteins (Fig. 5). Depending on the physiological state of the cells, the NuRD core complex can associate with different DNA-binding proteins, such as the sequence-specific DNA binding proteins Hunchback (Kehle et al. 1998), Ikarose (Kim et al. 1999), the tumor suppressor p53 (Luo et al. 2000), and the methyl-CpG-binding protein MBD2 (Fig. 5). In this scenario, different purification strategies would result in purification of slightly different protein complexes. In the original NuRD purification, histone deacetylase activity and Mi2 protein were followed. As a result, the bulk of the tightly associated NuRD core complex, which is devoid of DNA binding protein, was purified. In the present study, however, the MeCP1 complex was purified by following its methyl-CpG-binding activity. Consequently, only the population of NuRD that associates with MBD2 was selected during purification. Several pieces of evidence support the existence of a core NuRD complex that does not always associate with MBD2. First, NuRD only partially overlaps with the first MBD2 peak in DEAE-5PW column (Fig. 2B). Second, the bulk of MBD2 is reported to be concentrated in the methylated DNA foci, while the bulk of MBD3, a NuRD core component, is not (Hendrich and Bird 1998). Third, with the use of crude nuclear extracts as an input, MTA2 antibody immunoprecipitated the core NuRD complex, which does not contain detectable MBD2 (Zhang et al. 1999). However, the same antibody immunoprecipitated the MBD2-containing MeCP1 complex when a partially purified MeCP1 complex was used as input (Fig. 3A,B). It is interesting to note that several components of the MeCP1 complex, including MBD2, MBD3, and p66/68, appear to be substoichiometric with respect to the rest of the MeCP1 components (Fig. 3C). This is surprising given that the methyl-CpG-binding protein MBD2 was selected during purification. How the stoichiometry of the different MeCP1 components affects the stability of the complex remains to be determined. However, the important role of MBD2 in methyl-CpG-binding is clear. Consistent with the lack of MBD2 in the originally purified NuRD complex and the finding that MBD2 was responsible for the methyl-CpG-binding activity of the MeCP1 complex, the originally purified NuRD core complex lacks methyl-CpG-binding activity (Zhang et al. 1999). The existence of a NuRD core complex which can be recruited by either the methyl-CpG-binding protein MBD2 or other sequence-specific DNA binding proteins provides cells with an efficient way of using the NuRD complex to regulate gene activity (Fig. 5).
Figure 5.
Model depicting the relationship between the NuRD and the MeCP1 complexes and how each might be recruited to distinct gene promoters by either methylation-specific or sequence-specific DNA-binding proteins.
Since the initial report linking histone deacetylation to methylated gene silencing (Jones et al. 1998; Nan et al. 1998), accumulating evidence suggests that histone deacetylation is one of the major mechanisms in methylated gene silencing (Bird and Wolffe 1999; Li 1999). Our finding that MBD2 associates with NuRD in the MeCP1 complex in vivo leaves little doubt about the function of MBD2 in targeting the NuRD complex to methylated DNA. In addition, we have shown that the purified MeCP1 complex is able to preferentially bind, remodel, and deacetylate methylated nucleosomes (Fig. 4A–C). Importantly, a mutant Mi2 protein crippled in its ATPase activity is able to partially relieve methylation-dependent transcriptional repression (Fig. 4D). Given that multiple histone deacetylase complexes are involved in methylated DNA silencing (Bird and Wolffe 1999) and that methylation-dependent transcriptional silencing cannot be completely reactivated with trichostatin A (TSA) treatment (Cameron et al. 1999), it is likely that MeCP1 complex only accounts for part of the methylation-dependent transcriptional repression. Therefore, the Mi2 mutant did not completely relieve methylation-dependent transcriptional repression (Fig. 4D). We note that this same mutant failed to relieve MBD2-mediated transcription repression when MBD2 is tethered to promoter through the Gal4 DNA binding domain (data not shown). Whether the differential effects of the mutant Mi2 on the two reporters are the result of different promoters or different MBD2 recruiting methods remains to be determined. However, the demonstration that MBD2 and NuRD associate in vivo provides a platform for further studies of the roles of NuRD in methylated DNA silencing.
Materials and methods
MeCP1 purification and gel mobility shift assay
The MeCP1 complex was purified by following the procedure described in Figure 2A. Fractionation of HeLa nuclear extracts through the first two columns were performed as previously described (Zhang et al. 1998a). The DEAE52-bound materials were dialyzed into buffer D (40 mM HEPES at pH 7.9, 0.2 mM EDTA, 1 mM DTT, 0.2 mM PMSF, and 10% glycerol) containing 50 mM ammonium sulfate (BD50) and loaded onto an HPLC-DEAE-5PW column (TosoHaas, 45 mL). Proteins bound to the column were eluted with a 10-column-volume (cv) linear gradient from BD50 to BD400. The first MBD2 peak fractions were pooled and dialyzed into BD400, and loaded onto a 22 mL FPLC Phenyl Sepharose column (Pharmacia). Bound proteins were eluted with a linear gradient (15 cv) from BD400 to BD0. The fractions containing the MeCP1 complex were pooled, concentrated, and separated on a Superose-6 column (Pharmacia). The MeCP1 complex pool was dialyzed into BC50 and loaded onto a 1 mL Mono S column (Pharmacia) and eluted with a 20 cv linear gradient from BC50 to BC400. The gel mobility shift assay was performed as described (Zhang et al. 1999), with the following modifications. The 186 bp CG11 probe was generated from plasmid pCG11 (Meehan et al. 1989) by digestion with EcoRI and end labeled with [α-32P]dATP and Klenow enzyme, followed by digestion with HindIII. The 152 bp 5S probe was generated from plasmid pXP10 in a similar fashion except that RsaI was used after labeling. Purified probes were methylated with SssI (New England Biolabs). We used 2–10 μL of column fractions in 20 μL of binding reactions containing 0.1 ng of probe, 100 ng of poly-[d(GC)], 20 mM HEPES (pH 7.9), 5 mM MgCl2, 100 mM NaCl, 1 mM DTT, 2 μg BSA, 0.1% Triton X-100, and 3.5% glycerol. Binding reactions were allowed to proceed for 30 min at room temperature before loading onto a 1.5% agarose gel and resolved in 0.5× TBE buffer.
Nucleosome assembly, mononucleosome disruption, and histone deacetylase assays
Nucleosome assembly was performed with the salt dilution method (Steger et al. 1998). Each assembly reaction contained 1 μg of DNA and an equimolar amount of HeLa core histone octamers. To generate the nucleosomes used in Fig. 4A and 4B, we used a 9 to 1 mass ratio of sonicated herring sperm DNA (Boehringer Mannheim) to labeled CG11 or 5S DNA. To generate the nucleosomes used in Fig. 4C, 2 μg of Hat1 acetylated 3H-labeled core histone octamers (Zhang et al. 1999) were assembled with a 15% molar excess of unlabeled 5S DNA. For the mononucleosome disruption assay, 5 μL of assembled nucleosomes and various amounts of MeCP1 were mixed in 20 μL of reaction containing 10 mM HEPES (pH 7.9), 100 mM KCl, 3 mM MgCl2, 2 mM ATP, 1mM DTT, 0.5 mM EDTA and 10% glycerol. The reactions were incubated at 30°C for 1 h before the addition of CaCl2 to a final concentration of 10 mM for DNase I digestion. After the removal of proteins, the digested DNA fragments were resolved on a 7% sequencing gel. Histone deacetylase assays were performed as described (Zhang et al. 1998b) except that 3 mM MgCl2 and 2 mM ATP were included to allow nucleosome remodeling.
Plasmids, mutagenesis, ATPase, transfection and reporter assays
Reporters CG11-pGL2 and DNA pol-β-Luc have been previously described (Ng et al. 1999). Plasmid encoding Flag-tagged Mi2 was made by subcloning the human Mi2 cDNA into the NotI and XbaI sites of a modified pCDNA3 vector. PCR-based mutagenesis was used in generating the Flag-Mi2(K757R) mutant. Transfection was performed using the Effectene transfection reagent (QIAGEN). Twenty-four hours after transfection, samples were collected and luciferase and β-gal assays were performed using the Promega kit.
Antibodies, Western blots, and immunoprecipitation
All antibodies used have been described previously (Ng et al. 1999; Zhang et al. 1999). Methods for immunoprecipitation, Western blots, and silver staining have also been described (Zhang et al. 1998a, 1999).
Acknowledgments
We thank Adrian Bird, Alan Wolffe, and Jeffrey Milbrandt for antibody and reporter constructs; Li Xia for help with the mutagenesis; and Danny Reinberg for continued support. Y.Z. is a V-foundation scholar and is supported by a startup fund from the Lineberger Comprehensive Cancer Center.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL yi_zhang@med.unc.edu; FAX (919) 966-9673.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.876201.
References
- Bird AP, Wolffe AP. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- Corona DF, Langst G, Clapier CR, Bonte EJ, Ferrari S, Tamkun JW, Becker PB. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/s1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- Drapkin R, Reinberg D. The multifunctional TFIIH complex and transcriptional control. Trends Biochem Sci. 1994;19:504–508. doi: 10.1016/0968-0004(94)90139-2. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, Muller J. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Li E. The mojo of methylation. Nat Genet. 1999;23:5–6. doi: 10.1038/12595. [DOI] [PubMed] [Google Scholar]
- Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Richmond E, Peterson CL. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996;24:3685–3692. doi: 10.1093/nar/24.19.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Eberharter A, John S, Grant PA, Workman JL. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc Natl Acad Sci. 1998;95:12924–12929. doi: 10.1073/pnas.95.22.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh Y, Pencil SD, Nicolson GL. A novel candidate metastasis-associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. cDNA cloning, expression, and protein analyses. J Biol Chem. 1994;269:22958–22963. [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998a;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sun ZW, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998b;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes & Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]