Abstract
The quaking viable mice have myelination defects and develop a characteristic tremor 10 d after birth. The quaking gene encodes at least five alternatively spliced QUAKING (QKI) isoforms that differ in their C-terminal 8–30-amino-acid sequence. The reason for the existence of the different QKI isoforms and their function are unknown. Here we show that only one QKI isoform, QKI-7, can induce apoptosis in fibroblasts and primary rat oligodendrocytes. Heterodimerization of the QKI isoforms results in the nuclear translocation of QKI-7 and the suppression of apoptosis. The unique C-terminal 14 amino acids of QKI-7 confers the ability to induce apoptosis to heterologous proteins such as the green fluorescent protein and a QKI-related protein, Caenorhabditis elegans GLD-1. Thus, the unique C-terminal sequences of QKI-7 may function as a life-or-death ‘sensor’ that monitors the balance between the alternatively spliced QKI isoforms. Moreover, our findings suggest that nuclear translocation is a novel mechanism of inactivating apoptotic inducers.
Keywords: quaking, QKI, nuclear translocation, apoptosis, alternative splicing
Apoptosis, or programmed cell death, is a physiological process characterized by a cascade of events culminating in the destruction of the cell. There are numerous protein families that have been shown to induce apoptosis including the TNF/Fas ligands (Tartaglia and Goeddel 1992; Ashkenazi and Dixit 1998), oncogenes (Evan and Littlewood 1998), the Bcl-2 family (Adams and Cory 1998; Reed 1998; Gross et al. 1999; Porter 1999), and p53 (Livingstone et al. 1992; Ko and Prives 1996). Heterogeneous nuclear ribonucleoprotein particle K (KH) homology domain containing RNA-binding proteins (Gibson et al. 1993; Siomi et al. 1993) are an emerging class of apoptotic inducers. We have shown that mouse QKI-7 and Drosophila KEP1 and Sam50 are potent inducers of apoptosis (Chen and Richard 1998; Di Fruscio et al. 1998). Two other KH domain-containing proteins, Drosophila FMRP and MCG10, have also been shown to induce apoptosis (Wan et al. 2000; Zhu and Chen 2000). The mechanism by which this family of RNA-binding proteins induce cell death is unknown.
The quaking viable (qkv) mice have been studied for more than thirty years and represent an animal model for dysmyelination (Hogan and Greenfield 1984). Ten days after birth these animals develop a rapid tremor that is especially pronounced in the hind limbs (Hogan and Greenfield 1984). The gene responsible for the defect was cloned and termed the quaking (qk) gene (Ebersole et al. 1996). The mouse qk gene expresses at least five alternatively spliced mRNA including QKI-5, QKI-6, QKI-7, and QKI-G that differ in their C-terminal 30 amino acids (Ebersole et al. 1996; Cox et al. 1999; Kondo et al. 1999). The KH domain of the QKI proteins is embedded in a larger conserved domain of ∼200 amino acids called the GSG (GRP33, Sam68, GLD-1) domain (Jones and Schedl 1995; Di Fruscio et al. 1998) or the STAR (signal transduction activator of RNA metabolism; Vernet and Artzt 1997) domain. The GSG domain of the QKI proteins is required for RNA binding and dimerization (Chen et al. 1997; Zorn and Krieg 1997; Chen and Richard 1998; Wu et al. 1999).
The cellular localization of the QKI isoforms differ in oligodendrocytes (OLs), the myelinating cells of the central nervous system. QKI-5 is predominantly nuclear; QKI-6 and QKI-7 are localized in the perikaryal cytoplasm with lower levels in the nucleus (Hardy et al. 1996). In the qkv mice, part of the quaking enhancer/promoter is deleted (Ebersole et al. 1996) and, as a result, QKI-6 and QKI-7 isoforms are not expressed in OLs (Hardy et al. 1996). Several missense mutations have been generated in the qk gene by using ethylnitrosourea, and these mutations are known to be embryonic lethal in mice (Justice and Bode 1986, 1988; Shedlovsky et al. 1988). One allele, qkkt4, was found to alter QKI glutamic acid 48 to glycine (Ebersole et al. 1996). This amino acid substitution disrupts a predicted coiled-coil region, prevents dimerization, and may be the underlying mechanism for the embryonic lethality (Chen and Richard 1998).
QKI homologs have been identified in many species including Xenopus (Zorn et al. 1997), chicken (Mezquita et al. 1998), zebrafish (Tanaka et al. 1997), and Drosophila (Baehrecke 1997; Zaffran et al. 1997). Caenorhabditis elegans does not have a QKI homolog but has a closely related protein called GLD-1 (germ-line defective; Francis et al. 1995) that has a high degree of sequence identity with the QKI GSG domain (Jones and Schedl 1995; Vernet and Artzt 1997). GLD-1 is required for germ cell differentiation and has been shown to function as a tumor suppressor (Jones and Schedl 1995) and as a translational repressor (Jan et al. 1999). The relatedness between the GSG domains of QKI and GLD-1 suggest that these proteins may recognize similar RNA targets as dimers. We have shown that C. elegans GLD-1 is able to homodimerize and that QKI and GLD-1 associate with one another when ectopically expressed in HeLa cells (Chen et al. 1997). It was also shown that QKI-6 can replace GLD-1 in repressing the translation of a GLD-1-specific RNA target, tra-2 (Saccomanno et al. 1999). These observations demonstrate the functional similarity between the QKI and GLD-1 GSG domains. The overexpression of GLD-1 in NIH 3T3 cells did not induce apoptosis as was observed with QKI-7 (Chen and Richard 1998). These findings suggested that the QKI-7 GSG domain may not be required for the induction of apoptosis.
In this study, we identified a short region of 14 amino acids in QKI-7 that confers the ability to induce apoptosis to heterologous proteins. The other QKI isoforms were unable to induce apoptosis, but induced cell survival when expressed with QKI-7. Here we describe the possible mechanism by which the QKI-7 apoptotic inducer is suppressed by the QKI isoforms. Our data suggest that a balance between the alternatively spliced QKI isoforms is required for cell survival.
Results
Characterization of the apoptosis induced by QKI-7
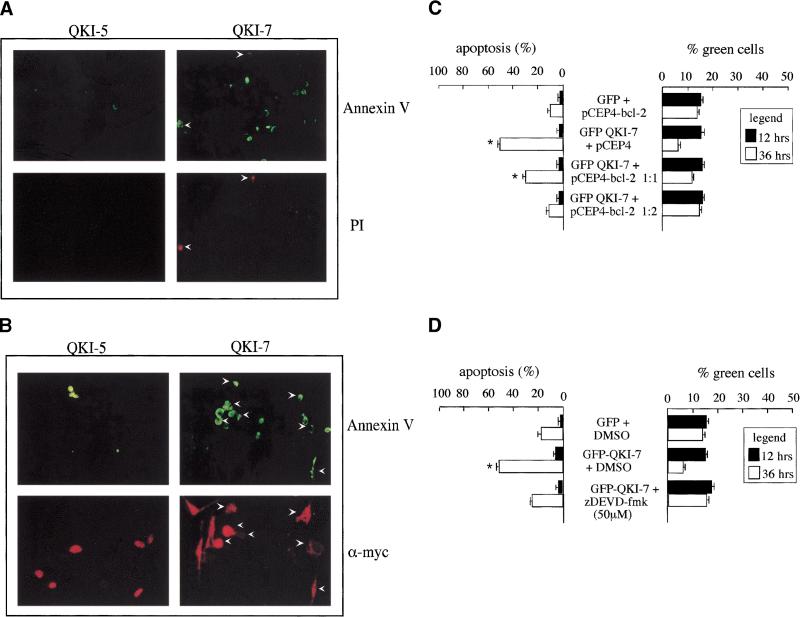
The expression of QKI-7 in NIH 3T3 cells induces cell death (Chen and Richard 1998). To further assess the apoptosis and determine whether early signs of apoptosis were observed, we stained QKI-7-expressing cells with annexin V conjugated with fluorescein isothiocyanate (FITC) and propidium iodide. Abundant staining with annexin V–FITC was observed in NIH 3T3 cells transfected with QKI-7 compared with the control cells transfected with QKI-5 (Fig. 1A, top panels). Some of the QKI-7 transfected cells that stained with annexin V also stained with propidium iodide (Fig. 1A, bottom panels), implying that they represent later stages of cell death. To confirm that the transfected cells were staining with annexin V-FITC, we performed double staining by using annexin V–FITC and indirect immunofluorescence using anti-myc antibodies followed by a rhodamine-conjugated secondary antibody. Most of the myc epitope-tagged QKI-7 transfected cells that stained with annexin V–FITC also stained with anti-myc antibodies (Fig. 1B, right panels), demonstrating that the QKI-7 transfected cells were dying. In comparison, the myc-QKI-5 transfected cells that stained with annexin V–FITC did not stain with anti-myc antibodies (Fig. 1B, left panels), demonstrating that the cell death observed in these cells corresponds to background cell death and not from the presence of QKI-5.
Figure 1.
QKI-7 is a potent apoptotic inducer. (A) Detection of early apoptosis by annexin V. Myc–QKI-5 or myc–QKI-7 were transfected into NIH 3T3 cells and stained with annexin V–FITC and propidium iodide (PI). The cells were visualized by fluorescence microscopy. The arrowheads align the cells in top and bottom panels. (B) Myc–QKI-5 or myc–QKI-7 were transfected into NIH 3T3 cells stained live with annexin V–FITC and fixed, and the myc-tagged QKI-7 was visualized by indirect immunofluorescence with anti-myc antibody followed by a rhodamine-conjugated secondary antibody. (C) Suppression of apoptosis by Bcl-2 overexpression. GFP alone or GFP–QKI-7 were cotransfected into NIH 3T3 cells with increasing amounts of a Bcl-2 expression vector (pCEP4–Bcl-2) or empty plasmid (pCEP4). After 12 h (solid bars) and 36 h (open bars) the cells were fixed and stained with DAPI to visualize the apoptotic nuclei. (D) The caspase inhibitor Z-DEVD.fmk suppresses QKI-7-induced apoptosis. GFP or GFP–QKI-7 were transfected into NIH 3T3 cells and treated with either DMSO (control) or 50 μM Z-DEVD.fmk as indicated. After 12 h (solid bars) and 36 h (open bars) the cells were fixed and stained with DAPI to visualize the apoptotic nuclei. (C,D) The presence of apoptotic nuclei was scored as cells undergoing apoptosis and expressed as % green cells. Each bar represents the mean + S.E. of 3 experiments (n > 450, n = number of cells counted). Statistical evaluation was calculated by paired Student's t-test. (*) Values that differ significantly from GFP at 36 h (P < 0.01).
Two additional experiments were performed to assess whether classical apoptotic/survival pathways were being utilized. We examined the effect of overexpressing a known survival protein such as Bcl-2 (Adams and Cory 1998; Gross et al. 1999). NIH 3T3 cells were cotransfected with expression vectors encoding GFP alone and Bcl-2, GFP–QKI-7 with pCEP4 empty vector, and GFP–QKI-7 and Bcl-2. The transfected green cells were counted and scored as apoptotic if they displayed irregular, condensed, or fragmented nuclei with the nuclear stain DAPI. The expression of GFP–QKI-7 with pCEP4 empty vector induced >50% apoptosis at 36 h (Fig. 1C), consistent with our previous observations (Chen and Richard 1998). The expression of GFP alone and Bcl-2 had background levels of cell death (∼15%) at 36 h (Fig. 1C). The cotransfection of Bcl-2 suppressed the apoptosis mediated by GFP–QKI-7 in a dose-dependent manner (Fig. 1C, left panel). The percentage of green cells or the transfection efficiency ranged from 15%–20% 12 h after transfection and decreased by 36 h, in some cases to ∼5%, indicating that some green cells had died and did not remain attached to the dish (Fig. 1C, right panel). To examine whether the apoptosis was caspase dependent, GFP–QKI-7 transfected cells were treated with 50 μM Z-DEVD.fmk, a known caspase 3 inhibitor (Nicholson et al. 1995), or dimethylsulfoxide as a control. The addition of Z-DEVD.fmk suppressed the apoptosis induced by QKI-7 to near background levels (Fig. 1D, left panel). In adition, the number of green cells was maintained at 15%, 36 h post-transfection in the presence of Z-DEVD.fmk, consistent with cell survival. Taken together, our results suggest that QKI-7-expressing NIH 3T3 cells exhibit the hallmarks of apoptosis. The QKI-7 transfected cells stain by using the TUNEL assay (Chen and Richard 1998) and annexin V–FITC. The apoptotic process is caspase dependent and suppressed by the overexpression of the survival protein Bcl-2.
The C terminus of QKI-7 is necessary and sufficient to signal apoptosis
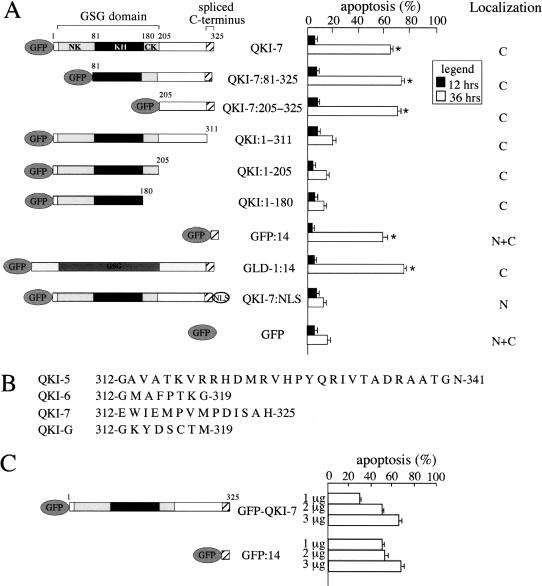
A structure analysis of QKI-7 was performed to determine the regions or domains required for the induction of apoptosis. Truncations and chimeric proteins were engineered as GFP fusion proteins, transfected in NIH 3T3 cells, and analyzed for their ability to induce apoptosis. The QKI GSG domain is a tripartite protein module with a central KH domain flanked by the N terminus of KH region (NK) and the C terminus of KH region (CK). The NK and CK regions are also referred to as the QUA1 and the QUA2 regions (Vernet and Artzt 1997). To investigate the role of the NK region in QKI, known to mediate dimerization (Chen and Richard 1998), or the entire GSG domain in QKI-7-mediated apoptosis, deletion analyses were performed. The truncated proteins deleted for the NK region (QKI-7:81–325) or entire GSG domain (QKI-7:205–325) induced apoptosis in NIH 3T3 cells to the same extent as QKI-7 (Fig. 2A). These findings suggest that RNA binding and the GSG domain are not required for the induction of cell death.
Figure 2.
The C-terminal 14 amino acids of QKI-7 induce apoptosis. (A) A schematic diagram of GFP fusion proteins is shown at left. The GSG, KH, NK, and CK regions are indicated. The striped box at the C terminus denotes the unique sequences of the QKI alternatively spliced isoforms. NLS represents the SV40 large T antigen nuclear localization signal. Expression plasmids encoding these GFP fusion proteins were transfected in NIH 3T3 cells. After 12 h (solid bars) and 36 h (open bars) the cells were fixed and stained with DAPI to visualize the apoptotic nuclei and expressed as a percentage of green cells. The localization of each protein at 12 h is indicated at right. (C) A predominant cytoplasmic localization; (N) a predominant nuclear localization. Each bar represents the mean + S.E. of 3 experiments (n > 450). (*) Values that differ significantly from GFP at 36 h (*, P < 0.01). (B) The amino acids of the unique sequences of the different QKI isoforms used. (C) C6 glioma cells were transfected with the indicated amount of the expression plasmids encoding GFP–QKI-7 or GFP:14. After 12 and 36 h the cells were fixed and stained with DAPI to visualize the apoptotic nuclei and expressed as a percentage of green cells. Each bar represents the mean + S.E. of three experiments (n > 300).
The QKI proteins are known to be alternatively spliced at their C termini resulting in at least four different isoforms (Fig. 2B). The most common isoforms as detected by Northern blot analysis are QKI-5, QKI-6, and QKI-7 (Ebersole et al. 1996). The deletion of the C-terminal 14 amino acids, unique to isoform QKI-7, was sufficient to prevent cell death (Fig. 2A; QKI:1–311). Larger deletions as in proteins QKI:1–205 and QKI:1–180 were also unable to induce apoptosis (Fig. 2A). These data suggest that the induction of apoptosis may be isoform specific and that the QKI-7 C-terminal 14 amino acids are required. To investigate whether the C-terminal 14 amino acids of QKI-7 were able to confer to heterologous proteins the ability to induce apoptosis, we fused the QKI-7 C-terminal 14 amino acids to nonapoptotic proteins such as GFP and C. elegans GLD-1 (Chen and Richard 1998). The expression of either GFP or GLD-1 with the C-terminal 14 amino acids of QKI-7 (GFP:14, GLD-1:14) was sufficient to induce apoptotic cell death to similar levels as QKI-7 (Fig. 2A). These findings demonstrate that the addition of amino acids EWIEMPVMPDISAH at the C terminus of cytoplasmic proteins is sufficient to induce apoptotic cell death. We next targeted QKI-7 to the nucleus with a strong nuclear localization signal (NLS) and asked whether nuclear QKI-7 could induce apoptosis in NIH 3T3 cells. The GFP fusion protein containing QKI-7:NLS was localized in the nucleus but was unable to induce cell death (Fig. 2A), suggesting that the C-terminal 14 amino acids of QKI-7 signal apoptotic cell death in the cytoplasm.
The ability of QKI-7 and GFP:14 to induce apoptosis in a cell line that expresses endogenous QKI proteins was investigated. We have shown previously that C6 glioma cells express three QKI isoforms (Chen and Richard 1998). C6 glioma cells were transfected with GFP–QKI-7 or GFP:14 and analyzed for apoptotic cell death. The transfection of GFP–QKI-7 induced apoptosis in a dose-dependent manner with minimal apoptosis observed with 1 μg of DNA (∼25%) and a maximal response with 3 μg of DNA (∼70%, Fig. 2C). In contrast, the expression of GFP:14 induced near maximal levels of apoptosis at 1 μg of DNA (∼60%) and increased minimally with 3 μg of DNA (∼70%). These data suggest that elevated concentrations of QKI-7 are required to induce apoptosis in cells expressing endogenous QKI isoforms. The fact that GFP:14 was more potent than QKI-7 at inducing apoptosis in C6 glioma cells suggests that C6 glioma cells, unlike NIH 3T3 cells where QKI-7 and GFP:14 were equipotent, are able to neutralize the ability of QKI-7 to induce apoptosis.
The QKI isoforms
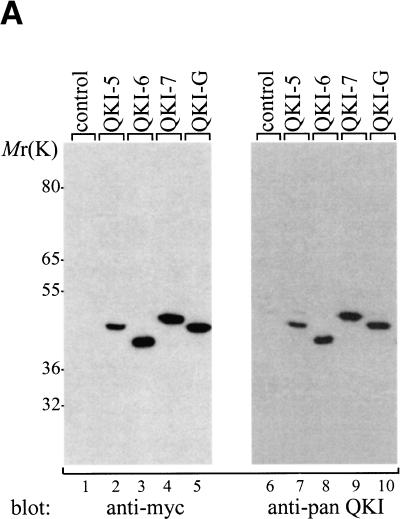
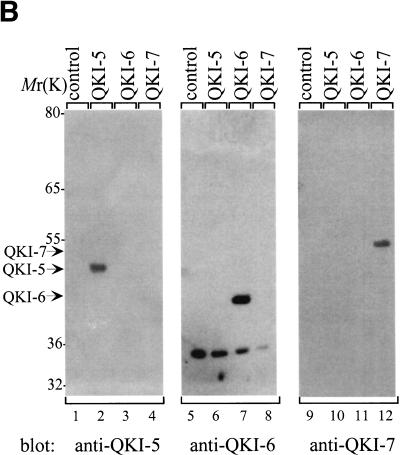
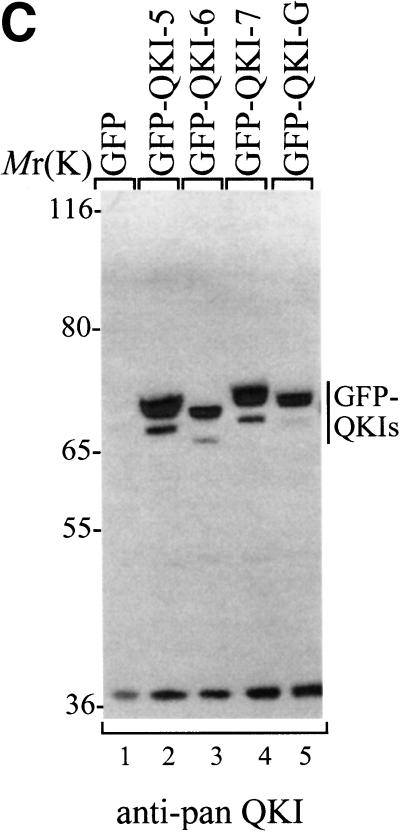
To evaluate the ability of the different QKI isoforms to induce apoptosis, we generated cDNAs for isoforms QKI-5, QKI-6, and QKI-G. The integrity of the QKI isoforms was verified by immunoblotting HeLa cell lysates transfected with myc epitope-tagged QKI proteins by using anti-myc, anti-‘pan’ QKI antibodies, anti-QKI-5-, anti-QKI-6-, or anti-QKI-7-specific antibodies. The anti-‘pan’ QKI antibody recognizes the KH domain of the QKI isoforms (Chen and Richard 1998) and the anti-QKI-5, -QKI-6, and -QKI-7 antibodies recognize the C-terminal specific sequence of each isoform (Hardy et al. 1996). The QKI isoforms migrated on SDS–polyacrylamide gels with molecular masses ranging from 40 to 47 kD and were all recognized with anti-myc and anti-‘pan’ QKI antibodies (Fig. 3A). The proteins encoded by the QKI-5, QKI-6, and QKI-7 cDNAs were also each recognized with anti-QKI-5, -QKI-6, and -QKI-7 antibodies, respectively (Fig. 3B). The QKI proteins were fused to GFP, transfected in NIH 3T3 cells, and analyzed by immunoblotting with the anti-‘pan’ QKI antibody. The GFP–QKI fusion proteins migrated with mobilities of ∼70 kD (Fig. 3C). Doublets were observed that most likely represent a second start site in the GFP coding region. Endogenous proteins with approximate molecular masses of 36 kD were observed by immunoblotting NIH 3T3 cells with anti-‘pan’ Qk1 or HeLa cells with anti-QKI-6 antibodies (Fig. 3B,C). The identity of these protein is unknown. In summary, these immunoblotting experiments demonstrate that the cDNAs encode the proper QKI isoforms.
Figure 3.
Expression of the QKI isoforms. (A) Untransfected (control) or myc–QKI-5, myc–QKI-6, myc–QKI-7, and myc–QKI-G were expressed in HeLa cells. The cell lysates were separated by SDS-PAGE and analyzed by immunoblotting with anti-myc (lanes 1–5) or anti-pan QKI antibodies (lanes 6–10). The molecular mass markers are shown at left in kD. (B) HeLa cells were transfected with the indicated myc–QKI as in A and analyzed by immunoblotting with anti-QKI-5 (lanes 1–4), anti-QKI-6 (lanes 5–8), and anti-QKI-7 (lanes 9–12) antibodies. The migration of QKI-5, QKI-6, and QKI-7 is indicated. (C) NIH 3T3 cells were transfected with expression plasmids expressing the indicated GFP fusion protein. The cells were lysed and analyzed by immunoblotting with anti-pan QKI antibodies.
Localization of the QKI isoforms
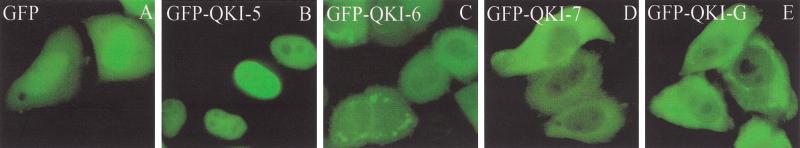
The cellular localization of GFP–QKI-5, GFP–QKI-6, GFP–QKI-7, and GFP–QKI-G was determined. HeLa cells were transfected with expression vectors encoding the QKI isoforms for 12 h and visualized by fluorescence microscopy. GFP–QKI-5 was predominantly nuclear (Fig. 4B), consistent with the presence of a nuclear localization signal at its C terminus (Wu et al. 1999). GFP–QKI-6- and GFP–QKI-G-expressing cells contained the GFP fusion protein in the nucleus and the cytoplasm (Fig. 4C,E). QKI-7 was predominantly cytoplasmic (Fig. 4D). Some cytoplasmic punctate staining was observed with QKI-6, QKI-7, and QKI-G that may represent focal adhesion structures. In summary, QKI-7 is predominantly cytoplasmic, QKI-5 is predominantly nuclear, and the QKI isoforms QKI-6 and QKI-G are localized in both the cytoplasm and the nucleus.
Figure 4.
The localization of the transfected QKI isoforms in HeLa cells. Expression plasmids expressing GFP (A), GFP–QKI-5 (B), GFP–QKI-6 (C), GFP–QKI-7 (D), or GFP–QKI-G (E) were transfected in HeLa cells. After 12 h the cells were fixed and visualized using fluorescence microscopy.
The different QKI isoforms and apoptosis
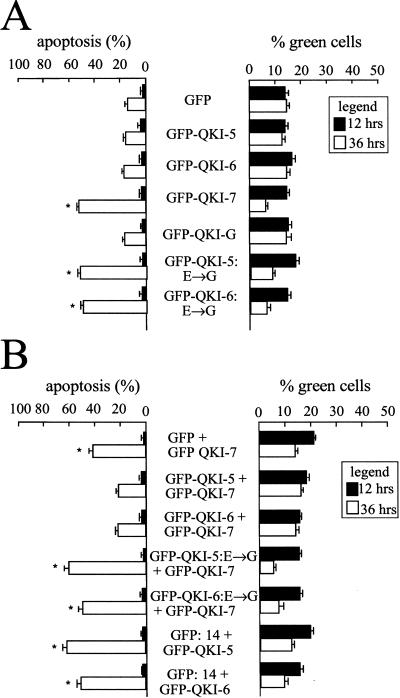
The ability of the different QKI isoforms to induce apoptosis was examined by transfecting NIH 3T3 cells with expression vectors encoding the GFP–QKI isoforms and analyzing the green cells for the presence of apoptosis by fluorescence microscopy at 12 and 36 h after transfection. GFP–QKI-7 was the only wild-type isoform able to induce apoptosis at 36 h (Fig. 5A). Approximately 60% of the GFP–QKI-7 transfected cells were apoptotic 36 h after DNA transfection. In contrast, background levels of apoptosis (∼15%) were observed at 36 h with cells transfected with GFP–QKI-5, GFP–QKI-6, GFP–QKI-G, or GFP alone (Fig. 5A). These findings demonstrate that the QKI isoforms do not have a general toxic effect but that QKI-7 has the unique ability to signal to the cell death machinery.
Figure 5.
Characterization of the apoptosis induced by the QKI isoforms. (A) QKI-7 is the only QKI apoptotic inducer. NIH 3T3 cells were transfected with expression vectors encoding the indicated GFP fusion protein. After 12 h (solid bars) and 36 h (open bars) the cells were fixed and stained with DAPI. The % green cells represents the number of transfected cells as a percentage of the total number of cells as visualized by DAPI. Each bar represents the mean + S.E. of 3 experiments (n > 450). (*) Values that differ significantly from GFP at 36 h (P < 0.01). (B) Dimerization is required for QKI-5 or QKI-6 to suppress the apoptosis induced by QKI-7. NIH 3T3 cells were cotransfected with expression plasmids encoding the GFP proteins indicated and the data were expressed as in A.
The ethylnitrosourea-induced mutation qkkt4 altering glutamic acid 48 to glycine (E48G) (Justice and Bode 1986; Ebersole et al. 1996) was introduced in the different isoforms and the resulting proteins were examined for their ability to induce apoptosis in NIH 3T3 cells. Surprisingly, all QKI isoforms including QKI-5, QKI-6, QKI-7, and QKI-G containing the E48G substitution were now able to induce apoptosis when expressed in NIH 3T3 cells (Fig. 5A; data not shown). The number of green cells remaining 36 h after DNA transfection was also reduced, consistent with cell death. These data suggest that the apoptosis observed with the substitution of QKI glutamic acid 48 to glycine may represent a gain-of- function that is independent of the C-terminal unique sequences.
Suppression of apoptosis by coexpressing QKI-5 and QKI-6
The observation that elevated levels of QKI-7 were required to induce apoptosis in C6 glioma cells and the fact that the E48G substitution induces apoptosis implied that dimerization plays a role in the regulation of apoptosis. To determine the role of dimerization in QKI-mediated apoptosis, GFP–QKI-7 was cotransfected with either GFP alone, GFP–QKI-5, or GFP–QKI-6 and assessed for apoptotic cell death by using the nuclear stain DAPI. The expression of GFP–QKI-5 or GFP–QKI-6 with GFP–QKI-7 suppressed the ability of GFP–QKI-7 to induce apoptosis (Fig. 5B). This suppressive effect was not observed when GFP alone was transfected with GFP–QKI-7. We next examined the ability of GFP–QKI-5:E→G or GFP–QKI-6:E→G to suppress the apoptosis mediated by GFP–QKI-7 (Fig. 5B). GFP–QKI-5:E→G and GFP–QKI-6:E→G failed to suppress the apoptosis induced by GFP–QKI-7 demonstrating that dimerization is essential for preventing cell death. To further establish that dimerization is essential for the suppression of apoptosis, we investigated the ability of QKI-5 and QKI-6 to suppress the apoptosis induced by GFP:14, a protein that cannot dimerize. Thirty-six hours after transfection, ∼60% of the cells expressing GFP:14 were apoptotic regardless of whether GFP–QKI-5 or GFP–QKI-6 was expressed (Fig. 5B). These data suggest that QKI-5 and QKI-6 suppress apoptosis by forming heterodimers with QKI-7.
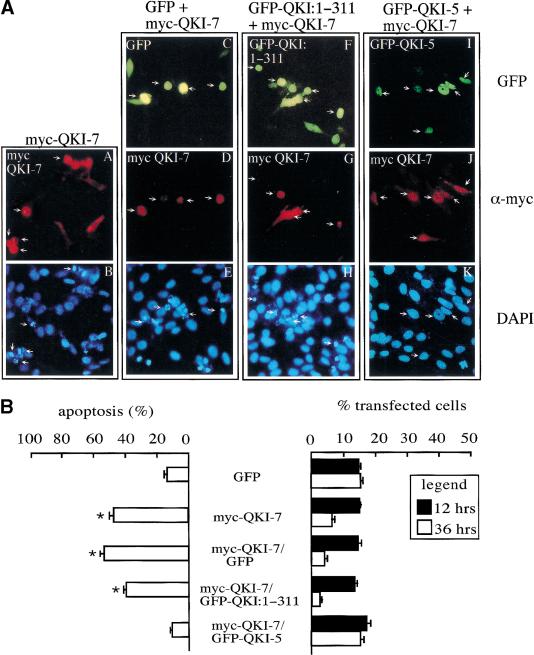
Our major concern with the cotransfection studies in Figure 5B was the difficulty in assessing whether the green cells expressed both isoforms. To identify the cells that were transfected with two isoforms GFP alone, GFP–QKI:1–311, or GFP–QKI-5 were cotransfected with myc–QKI-7 in NIH 3T3. The expression of myc–QKI-7 was visualized by indirect immunofluorescence by using anti-myc antibodies followed by a rhodamine-conjugated secondary antibody. The cells that were both green and red were visualized for apoptotic cell death by using the nuclear stain DAPI. Representative fields are shown (Fig. 6A) and the data were quantitated and expressed as percentage apoptosis (Fig. 6B). The cells expressing myc–QKI-7 alone, myc–QKI-7/GFP, or myc–QKI-7/GFP–QKI:1–311 exhibited characteristics of apoptosis including cell shrinkage, cytoplasm condensation (Fig. 6A, panels A, D, and G, respectively), and irregular, condensed, and fragmented nuclei (Fig. 6A, panels B, E, and H, respectively). The myc–QKI-7-expressing cells displayed ∼50% apoptosis and ∼5% of the transfected cells remained attached to the dish 36 h after transfection (Fig. 6B). The cells that were cotransfected with GFP–QKI-5 and myc–QKI-7 displayed a healthy and normal appearance 36 h post-transfection (Fig. 6A, panels I–K). These cells had background apoptosis at 36 h (∼10%, Fig. 6B, left) and most of the cells transfected survived or remained on the dish (Fig. 6B, right). We also noted that the majority of cells that expressed one isoform also expressed the other (Fig. 6A). These findings suggested that our cotransfection procedure resulted in a high percentage of cells that expressed both isoforms, implying that the majority of the green cells in Figure 5B contained both GFP fusion proteins. The experiments demonstrate the ability of isoforms QKI-5 and QKI-6 (data not shown), but not QKI:1–311, to suppress the apoptosis induced by QKI-7. QKI:1–311 is expressed exclusively in the cytoplasm (Fig. 2A) and was unable to rescue the apoptosis induced by QKI-7 (Fig. 6). These findings suggest that the expression of a QKI isoform or protein fragment able to heterodimerize with QKI-7 is not sufficient to suppress the apoptosis, but that the heterodimerizing QKI isoform or protein fragment must have access to the nucleus.
Figure 6.
QKI-7-induced apoptosis is suppressed by QKI-5. (A) NIH 3T3 cells were transfected with an expression plasmid that encodes myc–QKI-7 alone (A,B) or with expression vectors for GFP (C–E), GFP–QKI:1–311 (F–H), or GFP–QKI-5 (I–K). After 36 h the cells were fixed and visualized by indirect immunofluorescence by using the anti-myc antibody followed by rhodamine-conjugated secondary antibody. DAPI was used to visualize the nuclei. Each column represents the same field of cells as visualized under the green (top), red (middle), and blue (bottom) filters. The arrows are used to align the panels. (B) Quantitation of the apoptosis suppressed by QKI-5. NIH 3T3 cells were transfected as in A. For the cells expressing both a GFP fusion protein and a myc epitope-tagged protein, the apoptotic cells were expressed as a percentage of the cells that were both green and red. The panel at right indicates the percentage of cells that were transfected from the total number of cells. Each bar represents the mean + S.E. of 3 experiments (n > 450). (*) Values that differ significantly from GFP at 36 h (P < 0.01).
Heterodimerization of the QKI isoforms in mammalian cells
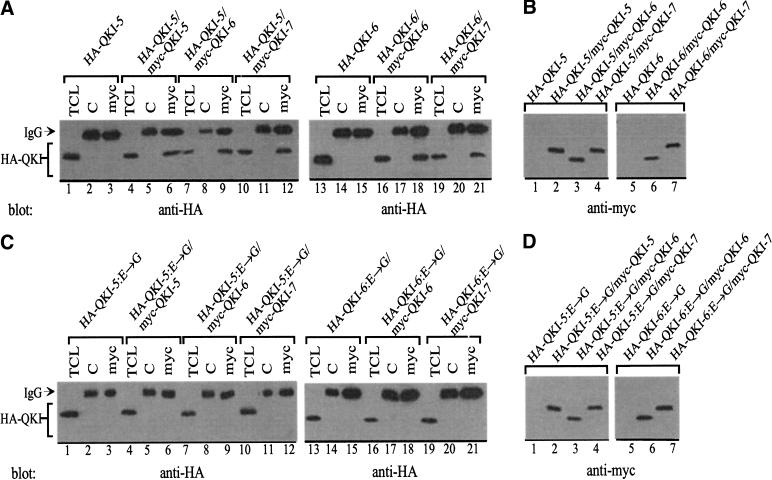
We noticed that QKI-7 entered the nucleus in the presence of QKI-5 (Fig. 6A, panel J). These findings suggested that heterodimerization was regulating the localization of QKI-7. To confirm that the QKI isoforms were forming heterodimers in mammalian cells, we cotransfected the different isoforms in HeLa cells and performed coimmunoprecipitation studies. Hemagglutin (HA)-tagged QKI-5 was transfected alone or cotransfected with myc–QKI-5, myc–QKI-6, or myc–QKI-7. The cells were lysed, the lysates were divided equally, and the proteins immunoprecipitated with control immunoglobulin G (IgG) and anti-myc antibodies. The proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-HA antibodies. HA–QKI-5 coimmunoprecipitated with myc-QKI-5, myc–QKI-6, and myc–QKI-7 (Fig. 7A, lanes 1–12), demonstrating that heterodimerization occurs in vivo. Similar findings were obtained with HA–QKI-6 (Fig. 7A, lanes 13–21). These observations show that heterodimerization occurs between QKI isoforms that display predominant cytoplasmic and nuclear localizations such as QKI-7 and QKI-5. We next examined whether the E48G substitution could prevent heterodimerization. HA-tagged QKI-5:E→G or QKI-6:E→G were cotransfected with myc–QKI-5, myc–QKI-6, or myc–QKI-7 and analyzed as described above. HA–QKI-5:E→G and HA–QKI-6:E→G did not coimmunoprecipitate with any of the wild-type isoforms (Fig. 7B, lanes 1–21). These results suggest that the heterodimerization domain is similar to the homodimerization domain located in the NK region of the GSG domain.
Figure 7.
Heterodimerization of the QKI isoforms in fibroblasts is abolished by the E→G amino acid substitution. (A,C) HeLa cells were transfected with myc–QKI and HA–QKI isoforms as indicated. The cells were lysed and immunoprecipitated with IgG as a control (C) or anti-myc antibodies (myc). The bound proteins as well as an aliquot of total cellular lysate (TCL) were separated by SDS-PAGE and analyzed by immunoblotting with anti-HA antibodies. The migration of the heavy chain of IgG and the HA–QKI isoforms is shown. (B,D) The total cellular lysate of panels A and C were confirmed for the equivalent expression of the myc epitope-tagged QKI proteins.
Heterodimerization controls QKI-7 localization
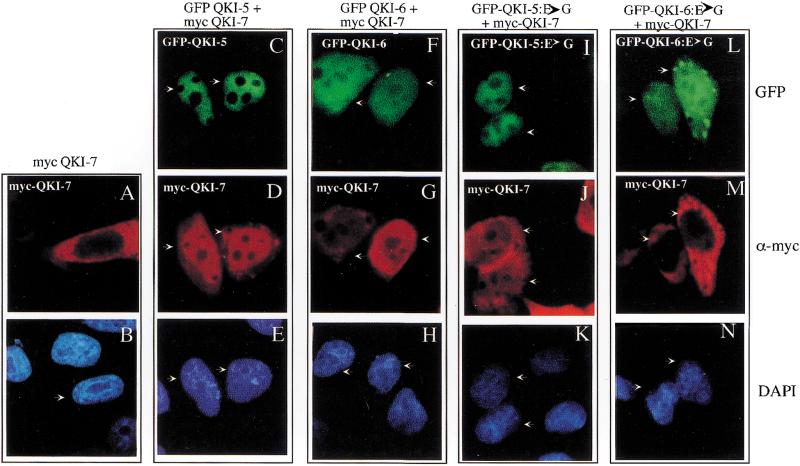
The relocalization of the QKI isoforms was further investigated in HeLa cells, a cell type where it could be easily observed. Myc–QKI-7 was transfected or cotransfected with GFP–QKI-5 or GFP–QKI-6 in HeLa cells and visualized by using fluorescence microscopy 12 h post-transfection. The 12-h time point was chosen because no apoptosis was observed at this time. Myc–QKI-7 localized exclusively to the cytoplasm of HeLa cells (Fig. 8A,B). However, myc–QKI-7 staining could be observed throughout the cell, with prominent nuclear localization, when coexpressed with GFP–QKI-5 (Fig. 8C–E). Interestingly, the localization of QKI-5 did not relocalize to the cytoplasm with the coexpression of QKI-7 (Fig. 8, C–E). The coexpression of GFP–QKI-6 also relocalized myc–QKI-7 to the nucleus (Fig. 8F–H), but less nuclear staining was observed as compared with GFP–QKI-5 (Fig. 8, cf. D and G). Although significant cytoplasmic staining of QKI-7 was observed with QKI-6 (Fig. 8G), QKI-6 was able to suppress the apoptosis by QKI-7 (Fig. 5B). These findings suggest that in addition to relocalizing QKI-7 to the nucleus, QKI-6 may also inactivate cytoplasmic QKI-7 by forming heterodimers.
Figure 8.
Nuclear translocation of QKI-7 with QKI-5 or QKI-6 but not with QKI-5:E→G or QKI-6:E→G. Myc–QKI-7 was either transfected alone (A,B) or cotransfected with GFP–QKI-5 (C–E), GFP–QKI-6 (F–H), GFP–QKI-5:E→G (I–K), or GFP–QKI-6:E→G (L–N) into HeLa cells. After 12 h the cells were fixed, immunostained with an anti-myc antibody followed by a rhodamine-conjugated secondary antibody, and mounted onto a glass slide in the presence of the nuclear stain DAPI. The cells were visualized by fluorescence microscopy. Each column represent the same field of cells as visualized under the green (top), red (middle), and blue (bottom) filters. The arrows are used to align the panels.
If dimerization is required for relocalization, then the E48G substitution should impair the relocalization. HeLa cells were cotransfected with myc–QKI-7 and GFP–QKI-5:E→G or GFP–QKI-6:E→G and visualized by fluorescence microscopy. Myc–QKI-7 failed to relocalize to the nucleus with the coexpression of GFP–QKI-5:E→G or GFP–QKI-6:E→G (Fig. 8I–N). These results suggest that the nuclear localization of QKI-7 is controlled by heterodimerization.
QKI-7 adenovirus induces apoptosis in primary rat OLs
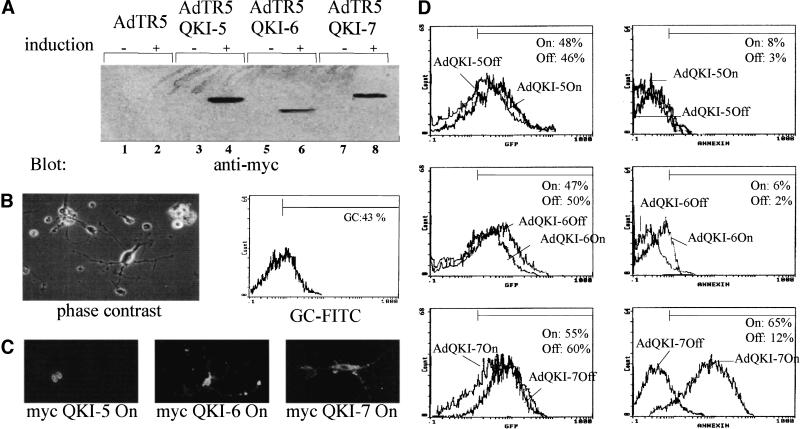
The dysmyelination phenotype observed in the qkv mice suggests that the QKI isoforms are involved in the normal physiology of the OLs (Hogan and Greenfield 1984). We wanted to examine whether QKI-7 was a potent apoptotic inducer in a cell type where it is known to have a physiological role. We constructed adenoviruses that express tetracycline-inducible QKI-5, QKI-6, and QKI-7. The QKI adenoviruses also express GFP constitutively and it is a marker of infection. Initially, we performed experiments with HeLa cells to verify that the adenoviruses were expressing the QKI isoforms in an inducible manner. HeLa cells were coinfected with two adenoviruses: one supplying constitutive levels of the tetracycline-regulated transactivator (tTA) and the other expressing the desired QKI isoform under a tetracycline-inducible promoter. Induced and noninduced infected HeLa cells were lysed, and the proteins were separated by SDS-PAGE and immunoblotted with anti-myc antibodies. QKI-5, QKI-6, and QKI-7 were expressed in a tetracycline-inducible manner (Fig. 9A). The expression of the QKI isoforms was tightly regulated, as little or no expression was observed in the noninduced cultures.
Figure 9.
Adenoviruses expressing QKI-7 induce apoptosis in primary rat OLs. (A) The generation of QKI-inducible adenoviruses. HeLa cells were coinfected at an M.O.I. of 10 with an adenovirus that constitutively expresses the tTA and a tetracycline-inducible adenovirus expressing myc–QKI-5, myc–QKI-6, or myc–-QKI-7. (+) Absence of doxycyclin; (−) addition of 1 μg/mL of doxycyclin. Essentially 100% of the cells were green 36 h after infection (not shown). The cells were harvested, lysed in sample buffer, separated by SDS-PAGE, and immunoblotted with anti-myc antibodies. pAdTR5 denotes an adenovirus generated with vector alone. pAdTR5–QKI-5, pAdTR5–QKI-6, and pAdTR5–QKI-7 represent the adenoviruses expressing QKI-5, QKI-6, and QKI-7, respectively. (B) Phase contrast photograph of a typical primary rat OLs preparation 2 d after maturation in growth factors (left panel). Uninfected OLs were stained live with anti-galactocerebroside FITC-conjugated antibodies and analyzed by flow cytometry. (C) Primary rat OLs were coinfected as in A, fixed, permeabilized, and stained with anti-myc antibodies followed by a rhodamine-conjugated secondary antibody. The cells were visualized by fluorescence microscopy. (D) Primary rat OLs were coinfected as in A, stained live with annexin V–phycoerythrin, and analyzed by flow cytometry. The percentage of GFP and annexin V positive cells is indicated for each condition. The experiment shown is representative of three experiments.
Primary rat OLs were isolated from newborn rats as described (Lubetzki et al. 1991). After 2 d of maturation in the presence of growth factors, the cells displayed an OL appearance with little or no contaminating astrocytes (Fig. 9B, left). Approximately 43% of the cells stained with anti-galactocerebroside antibody conjugated to FITC, a well-known marker for mature OLs (Fig. 9B, right). Thus, our cultures contained ∼40% mature OLs and ∼60% immature OLs. These cells were coinfected as described above with a tTA-expressing adenovirus and the QKI-inducible adenoviruses and could be visualized by indirect immunofluorescence by using the anti-myc antibody. The myc epitope-tagged QKI isoforms were expressed in their respective compartments in the OLs. QKI-5 was nuclear, QKI-6 localized throughout the cell, and QKI-7 was cytoplasmic (Fig. 9C). We next examined whether the overexpression of the QKI isoforms resulted in cell death. The induced and noninduced infected cells were stained live with annexin V–phycoerythrin to detect early signs of apoptosis. The stained cells were analyzed by flow cytometry for the expression of GFP and annexin V–phycoerythrin. Approximately ∼65% of the cells infected with the QKI-7 adenovirus in the ON state stained positive for annexin V–phycoerythrin (Fig. 9D, bottom right). The infection with QKI-5 or QKI-6 induced and noninduced had no significant increase in staining with annexin V (Fig. 9D, top right). All OLs cultures were infected at equivalent MOIs as observed with the expression of the GFP marker (Fig. 9D, left). These data suggest that QKI-7 is an apoptotic inducer in mature and immature primary rat OLs.
Discussion
In this study, we show that a balance of the QKI isoforms regulates the activity of the QKI-7 apoptotic inducer. The expression of the different QKI isoforms including QKI-5, QKI-6, and QKI-G in fibroblasts or primary rat OLs did not induce apoptosis. The coexpression of either QKI-5 or QKI-6 with QKI-7 caused the nuclear translocation of QKI-7 and suppressed the apoptosis normally observed with the expression of QKI-7. These data suggest that heterodimerization causes the nuclear translocation of QKI-7 and may be the major mechanism responsible for its inactivation.
The region necessary for the induction of apoptosis was mapped to the unique C-terminal 14 amino acids of QKI-7. This represents a new functional domain in the QKI proteins. Other functional domains characterized previously in QKI include the GSG domain required for dimerization and RNA binding (Chen and Richard 1998) and an NLS in the unique sequences of QKI-5 (Wu et al. 1999). The finding that the C-terminal sequences of QKI-7 are sufficient to confer the ability to induce apoptosis to GFP and GLD-1 suggests that the 14 amino acids of QKI-7 signal to the apoptotic machinery independent of other functional domains. Thus, the RNA-binding activity of QKI-7 is not required for induction of cell death. Mutations or deletions within the QKI-7 GSG domain did not suppress the apoptosis (data not shown). Because the 14 amino acids of QKI-7 confer the ability to induce apoptosis to heterologous proteins, we propose the name ‘killer sequence’ for this sequence. Database searches using the 14-amino-acid killer sequence EWIEMPVMPDISAH did not reveal a novel protein module. However, a core motif (underlined) was found for Drosophila, C. elegans, and mammalian proteins in GenBank. The proteins that contained this motif included C. elegans CED-9 (the histidine is replaced with a lysine) and hypothetical protein C09H10.9 (accession no. T19165), human and murine Hect2/rjs protein (accession no. AAC31433), Drosophila CG11958 and CG9906 gene products (accession nos. AAF57631 and AAF48618). The mechanism by which the killer sequence QKI-7 communicates to the apoptotic machinery is unknown.
The suppression of apoptosis by the coexpression of QKI-5 or QKI-6, but not QKI:1–311 suggests that heterodimerization with QKI-7 is not sufficient for the suppression. It has been shown that QKI-5 is capable of nucleoplasmic shuttling (Wu et al. 1999). Thus, QKI-5 and other QKI isoforms such as QKI-6 and QKI-G may shuttle into the nucleus as a heterodimer with QKI-7. Because QKI:1–311 has an exclusive cytoplasmic localization, QKI-7/QKI:1–311 heterodimers would remain cytoplasmic resulting in apoptosis. Our data suggest that cytoplasmic but not nuclear QKI-7 signals to the apoptotic machinery.
The qkv mice have been shown to contain an enhancer/promoter deletion in the qk gene (Ebersole et al. 1996). It has been shown that the qkv OLs express QKI-5, but not QKI-6 and QKI-7 (Hardy et al. 1996). How an enhancer/promoter deletion prevents the production of the QKI-6 and QKI-7 isoforms in OLs is unknown. It is thought that the absence of QKI-6 and QKI-7 prevents the proper maturation of the OLs and/or the process of myelination (Hardy et al. 1996). Our findings suggest that the balance in the QKI isoforms is critical for the normal function of the QKI proteins and cell viability. Based on our findings, the loss of QKI-6 and QKI-7 should not affect the viability of the OLs in the qkv mice and indeed normal quantities and hyperplasia of OLs have been reported (Friedrich 1975). We believe that the loss of QKI-6 and QKI-7 would provide an imbalance in the QKI isoforms that would impair the differentiation and maturation of the OLs resulting in the dysmyelination phenotype observed in the qkv mice. A balance between QKI isoforms (HOW; held-out-wings) has been reported for the export of the stripe mRNA in Drosophila melanogaster (Nabel-Rosen et al. 1999). This supports the hypothesis that the balance in the QKI isoforms regulates their function.
The ethylnitrosourea-induced point mutation altering glutamic acid 48 to glycine prevents homo- and heterodimerization of all QKI isoforms (Fig 7; Chen and Richard 1998; Wu et al. 1999). Moreover, we showed that all QKI isoforms containing the E48G amino acid substitution are able to induce apoptosis, regardless of whether they contained the killer sequence. These data suggest that the QKI:E48G represents a gain-of-function amino acid substitution that is separate from the QKI-7 killer sequence. Thus, the QKI proteins have multiple regions that can induce apoptosis: QKI-7 has the killer sequence and all isoforms have an NK region that can be altered to induce cell death. These findings further support our hypothesis that the QKI isoforms are critical proteins for maintenance of cell viability and cell death. It is possible that QKI isoforms other than QKI-7 are apoptosis inhibitors or growth inducers themselves. The fact that QKI-5:E48G is a potent inducer of apoptosis suggests that the embryonic lethality observed in embryos containing the qkkt4 allele (Justice and Bode 1986; Ebersole et al. 1996) is caused by apoptotic cell death.
There are several known genes that give rise to alternatively spliced transcripts encoding proteins with opposing cell death functions. The genes for bcl-x (Boise et al. 1993) and C. elegans ced-4 (Shaham and Horvitz 1996) each encode a long and a short transcript. The longer transcripts encode proteins that protect against cell death and the shorter transcripts encode proteins that promote cell death. Two genes encoding caspases including Ich-1 (Wang et al. 1994) and interleukin-1β-converting enzyme (Alnemri et al. 1995) have been shown to produce alternatively spliced transcripts that have positive and negative influences on programmed cell death. In the examples mentioned above, it has been proposed by the authors that it is the balance between the activator and repressor of programmed cell death, controlled by factors that influence splicing, which determines whether a cell will live or die. It has been shown that the two splicing factors SC35 and hnRNP A1 have opposing roles in the alternative splicing of the Ich-1 or caspase 2 gene (Jiang et al. 1998).
In summary, we characterized four different QKI isoforms and their ability to induce apoptosis. The expression of the cytoplasmic QKI-7 isoform is sufficient to induce apoptosis in the absence of other signals. The expression of the other alternatively spliced QKI isoforms induced cell survival by protecting the cell against the QKI-7 apoptotic inducer. The balance between QKI-7 and the QKI isoforms was required for cell survival. If QKI-7 is the major isoform or if it is overexpressed, QKI-7 localizes to the cytoplasm and induces cell death. If the QKI isoforms are expressed with QKI-7 such that an appropriate balance is achieved, the proteins heterodimerize and cause the nuclear translocation of QKI-7 resulting in cell survival. In addition, our studies identify a short peptide sequence that could be used to tag proteins and create chimeric proteins able to induce cell death. Our findings support the hypothesis that factors that influence splicing can regulate cell death and survival.
Materials and methods
DNA constructions
The plasmid constructs myc–Bluescript QKI-7, QKI-7:E→G, QKI-7:81–325, QKI:1–205, QKI:1–180, GFP–QKI-7, GFP–QKI-7:E→G, and myc–pcDNA QKI-7 were described previously (Chen and Richard 1998). The corresponding GFP proteins were obtained by digesting the myc–Bluescript plasmids with EcoRI and subcloning in the EcoRI site of pEGFP-C1 (Clontech). The plasmids GFP–QKI:1–311, GFP–QKI-7:NLS, GFP–QKI-6, GFP–-QKI-G were constructed by amplifying plasmid myc–QKI-7 with the T7 promoter primer and the following reverse primers: 5′-CAAGAATTCATAACACACCACTGGGTTC-3′ (QKI:1–311), 5′-CGTGAATTCACACTTTCTTTTTCTTCTTTGGAT GGGCTGAAATATCAGGCATG-3′ (QKI-7:NLS), 5′-TTTGAA TTCACCCTTTTGTCGGAAAAGCCATCCCTAACACACC ACTGGGTTCAATAGG-3′ (QKI-6), 5′-GACGAATTCACAT TGTACAACTATCATACTTCCCTAACACACCACTGGGTT CAATAGG-3′ (QKI-G). Plasmid GFP–GLD1:14 was constructed by a two-step subcloning strategy: The GLD-1 DNA sequence was first subcloned into pEGFP-C1 using PCR amplification of myc–GLD-1 as the DNA template with the T7 promoter and the oligonucleotide 5′-TACAAGCTTGAAAGAGGTGTTGTTGAC-3′ as primer. The amplified DNA fragment was digested with BglII and HindIII and subcloned into the corresponding sites of pEGFP-C1, generating GFP–GLD-1. The QKI-7 C-terminal tail was then amplified using GFP–QKI-7 as a DNA template using a GFP reverse primer and the forward primer 5′-CATAAGCTTGAGTGGATTGAAATGCCA-3′. The amplified DNA fragment was digested with HindIII and BamHI and subcloned into the corresponding sites of GFP–GLD-1, generating GFP–GLD-1:14. The plasmid GFP:14 was constructed by inverse PCR using GFP–QKI-7 as a DNA template with the following oligonucleotides as forward and reverse primers containing a BglII site: 5′-GGTAGATCTGAGTGGATTGAAATG GCCAGTC-3′ and 5′-ATGGAATTCTATCTGTAGGTGC CATTCAG-3′. The amplified plasmid was digested with BglII and then ligated with T4 DNA ligase. Plasmid myc–QKI-5 was constructed by a two-step strategy: The C terminus of QKI-5 was amplified from mouse EST T83554 using the following primers 5′-CAATTCTAGAGTATCCTATCCTATTGAACCT AGT-3′ and 5′-CCAGAGCTCGAATTCAGCTCGCTGCACT GACGA-3′. The amplified DNA fragment was subcloned directionally in XbaI and SacI sites of Bluescript SK+ generating pXS. Subsequently, the N-terminal common sequences of QKI were amplified by using myc–QKI-7 as template and the T7 promoter primer and the reverse primer 5′-GTACTCTAGAATTGATG TAGCTGGTGCCA-3′. The DNA fragment flanked with XbaI sites (one from the polylinker and the other introduced by the reverse primer) was digested with XbaI and subcloned in pXS. The introduction of the XbaI site was such that no amino acid was altered. Moreover, at the 3′ end of QKI-5 an EcoRI site was introduced before the SacI site. Thus, the entire QKI-5 coding sequences were subcloned in pEGFP-C1 using EcoRI. Plasmid GFP–QKI-7:205–325 was constructed by amplifying sequences from GFP–QKI-7 using 5′-CATGAATTCACCAGCCCTT GCCTTTTC-3′ and a GFP reverse primer. The amplified sequences were digested with EcoRI and subcloned in pEGFP-C1. Myc- and HA-epitope-tagged QKI isoforms were constructed by digesting the corresponding GFP–QKI isoform plasmid with EcoRI and subcloning the DNA fragments in myc–Bluescript or HA–Bluescript. The substitution of E→G was introduced in the isoforms as follows: myc–QKI-7 contains an internal KpnI site downstream of the E→G mutation and a KpnI site is found in the polylinker at the 3′ end of the cDNA. Thus, QKI-7:E→G was digested with KpnI and the different C termini from the other isoforms were introduced in the KpnI. The myc–QKI:E→G constructs were then subcloned into the pEGFP-C1 or HA–BS using EcoRI. The pCEP4–Bcl-2 expression vector was a gift of Walter Nishioka (Vical Inc., San Diego, CA). The transfer vectors expressing the tetracycline-inducible QKI-5, QKI-6, and QKI-7 proteins were constructed by subcloning the cDNAs encoding myc-tagged QKI isoforms from myc–BS QKI-5, QKI-6, and QKI-7 into the BglII site of pADTR5–K7–GFPq (Massie et al. 1998, 1999). These plasmids each express a different QKI isoform under the regulation of a tetracycline-inducible promoter and a second cassette that constitutively expresses GFP. The latter cassette serves as a marker for infection. Recombinant adenoviruses were generated, purified, and titered as described by the manufacturer (Quantum Biotechnology). The adenovirus that expresses constitutively the trans-acting factor (tTA) was a gift of Bernard Massie (Biotechnology Research Institute, Montréal).
Cell culture and transient transfection
NIH 3T3 cells were transfected with DNA plasmids encoding GFP alone, GFP fusion proteins, or myc epitope QKI-7 using LipofectAMINE PLUS reagent (Life Technologies-BRL). The cells were plated 12 h before transfection typically at a density of 105 cells/22 mm2 cover slip. The transfection of HA- and myc–Bluescript QKI isoforms into HeLa cells was carried out with the T7 vaccinia virus system as described previously (Chen et al. 1997). For the experiments with the caspase-3 inhibitor, 50 μM Z-DEVD.fmk (Calbiochem) or DMSO was added to the media after the LipofectAMINE transfection.
Apoptosis assays, transfection efficiency, and immunostaining
After transfection (12 or 36 h), the cells were fixed with 4% paraformaldehyde in 1× PBS for 10 min and permeabilized with 1% Triton X-100 in 1× PBS for 5 min, and the nuclei were stained with 3 μg/mL 4,6-diamidino-2-phenylindole (DAPI). The morphology of transfected cells and the localization of QKI isoforms were examined by fluorescence microscopy. Cells with characteristic morphological features such as nuclear condensation and fragmentation were considered apoptotic. The transfection efficiency was calculated as a percentage of transfected cells (green cells/total cells). For immunostaining, the fixed cells were incubated with the anti-myc 9E10 antibody (1:1000) at room temperature for 1 h and then followed by incubation of a rhodamine-conjugated goat anti-mouse secondary antibody (Jackson Laboratories; 1:200) for 30 min. The nuclei were stained with DAPI.
Protein expression
For protein expression, the cells were lysed in Laemmli buffer and the proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted. Immunoblotting was performed using anti-myc 9E10, or anti-hemagglutinin (HA), anti-pan QKI (Chen and Richard 1998), anti-QKI-5, anti-QKI-6, and anti-QKI-7 (Hardy et al. 1996) rabbit polyclonal antibodies, followed by a horseradish peroxidase-conjugated secondary antibody and developed using chemiluminescence. Coimmunoprecipitations were performed as described previously (Chen and Richard 1998).
Primary rat OLs cultures and cell viabilities
OLs progenitor cells were purified from newborn Sprague-Dawley rats essentially as described using a percoll gradient (Lubetzki et al. 1991). These cells were incubated for 2 d in media supplemented with 10% fetal calf serum, 2.5 ng/mL platelet-derived growth factor-AA, 2.5 ng/mL basic fibroblast growth factor, and 10 nM tri-iodothyronine. These OLs were infected with the indicated QKI expressing adenovirus and an adenovirus AdCMV–tTA that expresses the tTA. Each virus was added at an M.O.I. of 10.
OL cells survival assays were measured by staining with annexin V–phycoerythrin as described by the manufacturer (Pharmingen). Doxycyclin (1 μg/mL) was added at the same time as the adenoviruses to represent the uninduced cultures. The doxycyclin binds to the tTA and represses the tTA resulting in a noninduced culture. The induced and noninduced OLs were gated for GFP green expression and for annexin V–phycoerythrin red. The annexin V positive cells represented the cells undergoing early signs of programmed cell death and infection was calculated as the percentage of GFP positive cells.
Acknowledgments
We thank Karen Artzt for providing the isoform-specific anti-QKI antibodies, Bernard Massie for providing pAdTR5-K7-GFPq and the adenovirus expressing the tTA, and Walter Nishioka for the Bcl-2 expression vector. We are grateful to Taiping Chen for performing some of the initial C-terminal QKI deletions. This work was supported by a grant from the Multiple Sclerosis Society of Canada. J.P. is a recipient of the Research Studentship from the Multiple Sclerosis Society of Canada and S.R is a Scholar of the Canadian Institutes of Health Research of Canada.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sricha@po-box.mcgill.ca; FAX (514) 340-8295.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.860301.
References
- Adams JM, Cory S. The Bcl-2 protein family: Arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- Alnemri ES, Fernandes-Alnemri T, Litwack G. Cloning and expression of four novel isoforms of human interleukin-1 β converting enzyme with different apoptotic activities. J Biol Chem. 1995;270:4312–4317. doi: 10.1074/jbc.270.9.4312. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Dixit VM. Death receptors: Signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- Baehrecke EH. who encodes a KH RNA binding protein that functions in muscle development. Development. 1997;124:1323–1332. doi: 10.1242/dev.124.7.1323. [DOI] [PubMed] [Google Scholar]
- Boise LH, Gonzales-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- Chen T, Richard S. Structure-function analysis of QKI: A lethal point mutation in mouse quakingprevents homodimerization. Mol Cell Biol. 1998;18:4863–4871. doi: 10.1128/mcb.18.8.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Damaj B, Herrerra C, Lasko P, Richard S. Self-association of the single-KH domain family members Sam68, GRP33, GLD-1 and Qk1: Role of the KH domain. Mol Cell Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RD, Hugill A, Shedlovsky A, Noveroske JK, Best S, Justice MJ, Lehrach H, Dove WF. Contrasting effects of ENU induced embryonic lethal mutations of the quakinggene. Genomics. 1999;57:333–341. doi: 10.1006/geno.1999.5804. [DOI] [PubMed] [Google Scholar]
- Di Fruscio M, Chen T, Bonyadi S, Lasko P, Richard S. The identification of two DrosophilaKH domain proteins: KEP1 and SAM are members of the Sam68 family of GSG domain proteins. J Biol Chem. 1998;273:30122–30130. doi: 10.1074/jbc.273.46.30122. [DOI] [PubMed] [Google Scholar]
- Ebersole TA, Chen Q, Justice MA, Artzt K. The quakinggene unites signal transduction and RNA binding in the developing nervous system. Nat Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- Evan G, Littlewood T. A matter of life and cell death. Science. 1998;281:1317–1321. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- Francis R, Maine E, Schedl T. Gld-1: A cell-type specific tumor suppressor gene in C. elegans. Genetics. 1995;139:607–630. doi: 10.1093/genetics/139.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich VLJ. Hyperplasia of oligodendrocytes in quakingmice. Anat Embryol. 1975;147:259–271. doi: 10.1007/BF00315075. [DOI] [PubMed] [Google Scholar]
- Gibson TJ, Thompson JD, Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Letts. 1993;324:361–366. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- Gross A, McDonnell JM, Korsmeyer SJ. BCL-2 family members and the mitochondria in apoptosis. Genes & Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- Hardy RJ, Loushin CL, Friedrich VL, Jr, Chen Q, Ebersole TA, Lazzarini RA, Artzt K. Neural cell type-specific expression of QKI proteins is altered in the quakingviable mutant mice. J Neurosci. 1996;16:7941–7949. doi: 10.1523/JNEUROSCI.16-24-07941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan EL, Greenfield S. Animal models of genetic disorders of myelin. In: Morell P, editor. Myelin. New York: Plenum; 1984. pp. 489–534. [Google Scholar]
- Jan E, Motzny CK, Graves LE, Goodwin EB. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 1999;18:258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z-H, Zhang W-J, Rao Y, Wu JY. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc Natl Acad Sci. 1998;95:9155–9160. doi: 10.1073/pnas.95.16.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Schedl T. Mutations in GLD-1, a female germ cell-specific tumor suppressor gene in C. elegans, affect a conserved domain also found in Sam68. Genes & Dev. 1995;9:1491–1504. doi: 10.1101/gad.9.12.1491. [DOI] [PubMed] [Google Scholar]
- Justice M, Bode V. Induction of new mutations in a mouse t-haplotype using ethylnitrosourea mutagenesis. Genet Res. 1986;47:187–192. doi: 10.1017/s0016672300023119. [DOI] [PubMed] [Google Scholar]
- Justice MJ, Bode VC. Three ENU-induced alleles of the murine quakinglocus are recessive embryonic lethal mutations. Genet Res. 1988;51:95–102. doi: 10.1017/s0016672300024101. [DOI] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: Puzzle and paradigm. Genes & Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Kondo T, Furuta T, Mitsunaga K, Ebersole TA, Shichiri M, Wu J, Artzt K, Yamamura K, Abe K. Genomic organization and expression analysis of the mouse qkIlocus. Mamm Genome. 1999;10:662–669. doi: 10.1007/s003359901068. [DOI] [PubMed] [Google Scholar]
- Livingstone LR, White A, Sprouse J, Livanos E, Jacks T, Tisty TD. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- Lubetzki C, Goujet-Zalc C, Gansmuller A, Monge M, Brillat A, Zalc B. Morphological, biochemical, and function characterization of bulk isolated glial progenitor cells. J Neurochem. 1991;56:671–680. doi: 10.1111/j.1471-4159.1991.tb08202.x. [DOI] [PubMed] [Google Scholar]
- Massie B, Couture F, Lamoureux L, Mosser DD, Guilbault C, Jolicoeur P, Belanger F, Langelier Y. Inducible overexpression of a toxic protein by an adenovirus vector with a tetracycline-regulatable expression cassette. J Virol. 1998;72:2289–2296. doi: 10.1128/jvi.72.3.2289-2296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massie B, Mosser DD, Koutroumanis M, Vitté-Mony I, Lamoureux L, Couture F, Paquet L, Guilbault C, Dionne J, Chahla D, et al. New adenovirus vectors for protein production and gene transfer. Cytotechnology. 1999;28:1–12. doi: 10.1023/A:1008013211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezquita J, Pau M, Mezquita C. Four isoforms of the signal-transduction and RNA-binding protein Qk1 expressed during chicken spermatogenesis. Mol Reprod Devel. 1998;50:70–78. doi: 10.1002/(SICI)1098-2795(199805)50:1<70::AID-MRD9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Nabel-Rosen H, Dorevitch N, Reuveny A, Volk T. The balance between two isoforms of the DrosophilaRNA-binding protein How controls tendon cell differentiation. Mol Cell. 1999;4:573–584. doi: 10.1016/s1097-2765(00)80208-7. [DOI] [PubMed] [Google Scholar]
- Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- Porter AG. Protein translocation in apoptosis. Trends Cell Biol. 1999;9:394–401. doi: 10.1016/s0962-8924(99)01624-4. [DOI] [PubMed] [Google Scholar]
- Reed JC. Bcl-2 family proteins. Oncogene. 1998;17:3225–3236. doi: 10.1038/sj.onc.1202591. [DOI] [PubMed] [Google Scholar]
- Saccomanno L, Loushin C, Jan E, Punkay E, Artzt K, Goodwin EB. The STAR protein Qk1–6 is a translational repressor. Proc Natl Acad Sci. 1999;96:12605–12610. doi: 10.1073/pnas.96.22.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S, Horvitz HR. An alternatively spliced C. elegans ced-4RNA encodes a novel cell death inhibitor. Cell. 1996;86:201–208. doi: 10.1016/s0092-8674(00)80092-6. [DOI] [PubMed] [Google Scholar]
- Shedlovsky A, King TR, Dove WF. Saturation germ line mutagenesis of the murine t region including a lethal allele of the quakinglocus. Proc Natl Acad Sci. 1988;85:180–184. doi: 10.1073/pnas.85.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1 has characteristics of an RNA binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Abe K, Kim CH. Cloning and expression of the quakinggene in the Zebrafish embryo. Mech Develop. 1997;69:209–213. doi: 10.1016/s0925-4773(97)00164-0. [DOI] [PubMed] [Google Scholar]
- Tartaglia LA, Goeddel DV. Two TNF receptors. Immunol Today. 1992;13:151–153. doi: 10.1016/0167-5699(92)90116-O. [DOI] [PubMed] [Google Scholar]
- Vernet C, Artzt K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997;13:479–484. doi: 10.1016/s0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]
- Wan L, Dockendorff TC, Jongens TA, Dreyfuss G. Characterization of dFMR1, a Drosophilamelanogaster homolog of the fragile X mental retardation protein. Mol Cell Biol. 2000;20:8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Ich-1, an Ice/ced-3-related gene encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- Wu J, Zhou L, Tonissen K, Tee R, Artzt K. The quakingI-5 (QKI-5) has a novel nuclear localization signal and shuttles between the nucleus and the cytoplasm. J Biol Chem. 1999;274:29202–29210. doi: 10.1074/jbc.274.41.29202. [DOI] [PubMed] [Google Scholar]
- Zaffran S, Astier M, Gratecos D, Semeriva M. The held out wings (how) Drosophilagene encodes a putative RNA binding protein involved in the control of muscular and cardiac activity. Development. 1997;124:2087–2098. doi: 10.1242/dev.124.10.2087. [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen X. MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G2–M. Mol Cell Biol. 2000;20:5602–5618. doi: 10.1128/mcb.20.15.5602-5618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, Krieg PA. The KH domain protein encoded by quaking functions as a dimer and is essential for notochord development in Xenopusembryos. Genes & Dev. 1997;11:2176–2190. doi: 10.1101/gad.11.17.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, Grow M, Patterson KD, Ebersole TA, Chen Q, Artzt K, Krieg PA. Remarkable sequence conservation of transcripts encoding amphibian and mammalian homologues of quakinga KH domain RNA-binding protein. Gene. 1997;188:199–206. doi: 10.1016/s0378-1119(96)00795-0. [DOI] [PubMed] [Google Scholar]