Abstract
Cholinergic neurotransmission in the central and autonomic nervous systems regulates immediate variations in and longer-term maintenance of cardiovascular function with acetylcholinesterase (AChE) activity that is critical to temporal responsiveness. Butyrylcholinesterase (BChE), largely confined to the liver and plasma, subserves metabolic functions. AChE and BChE are found in hematopoietic cells and plasma, enabling one to correlate enzyme levels in whole blood with hereditary traits in twins. Using both twin and unrelated subjects, we found certain single nucleotide polymorphisms (SNPs) in the ACHE gene correlated with catalytic properties and general cardiovascular functions. SNP discovery from ACHE resequencing identified 19 SNPs: 7 coding SNPs (cSNPs), of which 4 are nonsynonymous, and 12 SNPs in untranslated regions, of which 3 are in a conserved sequence of an upstream intron. Both AChE and BChE activity traits in blood were heritable: AChE at 48.8 ± 6.1% and BChE at 81.4 ± 2.8%. Allelic and haplotype variations in the ACHE and BCHE genes were associated with changes in blood AChE and BChE activities. AChE activity was associated with BP status and SBP, whereas BChE activity was associated with features of the metabolic syndrome (especially body weight and BMI). Gene products from cDNAs with nonsynonymous cSNPs were expressed and purified. Protein expression of ACHE nonsynonymous variant D134H (SNP6) is impaired: this variant shows compromised stability and altered rates of organophosphate inhibition and oxime-assisted reactivation. A substantial fraction of the D134H instability could be reversed in the D134H/R136Q mutant. Hence, common genetic variations at ACHE and BCHE loci were associated with changes in corresponding enzymatic activities in blood.
Introduction
Peripheral and central nervous system control of cardiovascular function mediated through the autonomic nervous system is critical in homeostatic maintenance of blood pressure and responsiveness to exercise, postural alterations, and stress (Suh et al., 1936; Vargas and Brezenoff, 1988; Buccafusco, 1996; Khan et al., 2002). Central cholinergic pathways in the spinal cord and higher brain centers modulate cardiovascular responses to influence basal blood pressure and baroreflex or pressor responses via the neurotransmitter acetylcholine (ACh). Increased arterial pressure activates stretch receptors in the aortic arch and carotid sinus. Baroreceptor activation initiates afferent impulses to the vasomotor center in the medulla of the brain stem. Stimulation of the vasomotor center increases vagal parasympathetic release of ACh largely at the sinoatrial node (pacemaker cell of the heart) and atrioventricular node, thereby decreasing heart rate and indirectly cardiac output (Irisawa et al., 1993). In turn, peripheral responses are controlled by nicotinic receptors in ganglia and the adrenal medulla as well as muscarinic receptor sites in postganglionic parasympathetic systems. ACh through nicotinic and muscarinic receptors elicits characteristic responses, whereas AChE plays a key role in modulating cholinergic transmission by efficiently catalyzing hydrolysis of released ACh (Nakahara et al., 1998).
The ChEs are serine hydrolase members of the α/β-hydrolase fold protein superfamily (Ollis et al., 1992). Mammalian ACHE gene is encoded within 7.5 kb, and in the human genome, it is located on chromosome 7q22 (Schumacher et al., 1986; Getman et al., 1992). Diverse molecular forms of the gene product AChE are differentially expressed in various tissues as a result of alternative splicing at the 3′ end of the open reading frame (Li et al., 1991, 1993). The butyrylcholinesterase (BCHE) gene spans over 70 kb of the human genome and is located on chromosome 3q26 (Allderdice et al., 1991; Gaughan et al., 1991). AChE and BChE share approximately 54% amino acid sequence identity. In many peripheral tissues, BChE is present in greater abundance (Lockridge et al., 1987). The catalytic sites for both AChE and BChE contain a catalytic triad of serine, histidine, and glutamic acid residues (Gibney et al., 1990; Sussman et al., 1991; Soreq et al., 1992). Whereas the specific function of AChE is directed to catalyzing the hydrolysis of ACh at cholinergic synapses, the evolved biological function of BChE in liver, plasma, and the intestine remains unclear, although it appears distinct from the immediate catalysis of neuronally released AChE.
Extensive polymorphisms in patients with succinylcholine apnea are well documented within the hBCHE gene (Darvesh et al., 2003; Mikami et al., 2008). hBChE mutations and activities have also been examined in farm workers exposed to pesticides (Howard et al., 2010). Although the ACHE gene is highly conserved across species, to date, 13 hACHE SNPs have been uncovered, of which 5 are nonsynonymous (Hasin et al., 2005). Human phenotypes associated with hACHE polymorphism include the YT blood group (Bartels et al., 1993), and those with hBCHE phenotypes include incidence of Alzheimer's disease (Scacchi et al., 2009).
Materials and Methods
Human Subjects.
Participants included 80 unrelated subjects from the San Diego area and twin subjects recruited by access to a population birth record-based twin registry (Cockburn et al., 2002). Twin subjects (n = 478 twin individuals) included n = 167 monozygotic pairs (33 male pairs, 134 female pairs) and n = 72 dizygotic pairs (14 male pairs, 39 female pairs, and 19 male/female dizygotic pairs). Twin zygosity assignment was based on self-identification, with further confirmation by the presence or absence of heterozygosity at the tyrosine hydrolase gene intron-A microsatellite (Zhang et al., 2004).
Ethnicities of all subjects were established by self-identification, as well as ethnicities reported for both parents and all four grandparents. Twin subjects consisted of 181 pairs of European ancestry, 12 pairs of African Americans, 15 pairs of Hispanics, 2 pairs of Filipino, 5 pairs of East Asians, 1 pair of Native American, and 23 pairs of mixed ancestry; unrelated subjects consist of 30 African Americans, 8 Hispanics (Mexican-American), 6 Filipinos, 2 East Asians, and 34 of European ancestry. Statistical analysis was restricted to twin subjects of European ancestry in genetic association tests. Twin ages were 14 to 84 years.
Within twin pairs, family histories for hypertension (in a first-degree relative before the age of 60 years) were as follows: 111 pairs were positive (one or both parents); 102 pairs were negative; and 26 pairs were indeterminate/unknown. There were 429 individuals that were normotensive, and 49 were hypertensive. No subjects had a history of renal failure. Each subject gave informed, written consent; our protocol was approved by the Institutional Review Board (Rana et al., 2009). Sequencing and genotyping, haplotype analysis, and physiological/autonomic phenotyping in vivo were performed as described earlier (Rana et al., 2009).
Measurements of In Vivo and Ex Vivo Cholinesterase Activity.
Whole-blood samples obtained at mid-day from the 80 unrelated individual subjects and the 478 twin subjects were analyzed for ChE activity using a modification of the Ellman method (Ellman et al., 1961; Radić et al., 1993; Worek et al., 1999) with 0.5 mM acetylthiocholine as substrate at 37°C in 0.1 M phosphate buffer, pH 7.4. Each sample was assayed for total ChE activity and for specific AChE and BChE activities by including the selective inhibitors BW284c51 [1,5-bis(4-allyldimethylammoniumphenyl)-pentan-3-one dibromide] and ethopropazine (10-(2-diethylaminopropyl) phenothiazine; both from Sigma-Aldrich, St. Louis, MO), respectively, at a final concentration of 1 μM in the reaction mixture. Purified AChE activity was determined by the Ellman method at room temperature using 1 mM final concentration of acetylthiocholine.
Statistical Analyses.
Descriptive statistics (means ± S.D.), linear regression, and correlation statistics on the unrelated and twin panels, as well as analysis of variance and t-test analyses for all enzyme denaturation results, were conducted using GraphPad Prism (version 4; GraphPad Software, Inc., La Jolla, CA). Estimates of heritability (h2 ± VG/VP, where VG is additive genetic variance and VP is total phenotypic variance) were obtained using the variance-component methodology implemented in the SOLAR (“Sequential Oligogenic Linkage Analysis Routines”) package (Almasy and Blangero, 1998) (http://www.sfbr.org/solar/). This method maximizes the likelihood assuming a multivariate normal distribution of phenotypes in twin pairs (monozygotic versus dizygotic) with a mean dependent on a particular set of explanatory covariates. The null hypothesis (Ho) of no heritability was tested by comparing the full model, which assumes genetic variation (VG), and a reduced model, which assumes no genetic variation, using a likelihood ratio test. SOLAR was also used to evaluate whether an allelic variation at the locus contributed to a significant fraction of the trait heritability (i.e., locus-specific VG) by comparing models, including or excluding the genotype as a covariate.
Plasmids and Mutagenesis.
A cDNA encoding human AChE (hAChE) spliced from exons 4 to 6 (i.e., nerve/muscle form; Fig. 1; kindly provided by Dr. Oksana Lockridge, Eppley Institute, University of Nebraska Medical Center) was subcloned into a pcDNA3 vector for expression and purification. Soluble monomeric hAChE was generated by introducing an early stop codon at residue Glu548 as described previously for the soluble monomeric mouse AChE (Marchot et al., 1996) and is referred to as the wild-type form (wtT547). Single-base mutations were introduced into the wtT547 cDNA template using the Stratagene QuikChange mutagenesis kit (Stratagene, San Diego, CA) for the Arg3→Gln (R3Q) cSNP (SNP5), the Asp134→His (D134H) cSNP (SNP6), and the His322→Asn (H322N) cSNP (SNP9). A double mutant used the D134H cDNA template to generate the Arg136→Gln (D134H/R136Q) construct. Mutations were verified by DNA sequencing.
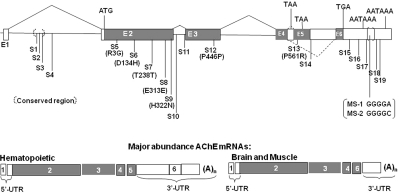
Fig. 1.
Human ACHE SNP discovery. Nineteen SNPs are mapped to the ACHE gene extending from 7.5 kb of the upstream proximal CAP site through the termination codon to the second polyadenylation signal (AATAAA). ACHE SNP discovery using a deep-resequencing strategy revealed 4 nonsynonymous cSNPs (5, 6, 9, 7, 13), including the well known YT blood group antigen (Bartels et al., 1993); 3 synonymous cSNPs; 12 SNPs in untranslated or intronic regions, including 3 SNPs (SNP1–3) in the enhanceosome region of intron 1, with its highly conserved sequence; and 2 microsatellite repeats in the bracketed 3′-untranslated region. Corresponding amino acid sequences are indicated in the parentheses under the identified SNP. The six exons encompass an untranslated exon 1, exons 2 through 4 of invariant splice order, and two alternatively spliced exons, 5 and 6. This yields three distinct carboxyl-terminal sequences from a direct read-though after exon 4, an exon 4 to 5 splice, and an exon 5 to 6 splice (Li et al., 1991, 1993). A truncated soluble species ends at amino acid 547, the 5′ end of exon 4. Supplemental Table 1 details the National Institutes of Health dbSNP rs number and allelic frequencies.
Cell Culture and Transfection.
Human embryonic kidney (HEK) 293 cells were maintained at 37°C and 10% CO2 in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Cells were transfected by Ca3(PO4)2 precipitation or with FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN) with 10 or 6 μg of mutant plasmid, respectively. Clonal selection was dependent on incorporation of the neomycin-resistance gene and selected by growth in G418. Clones with the highest relative AChE activity were selected for large scale protein production and purification.
Purification of Recombinant Enzymes and Determination of Catalytic Parameters.
Enzyme was purified from media of cells grown on multilayer plates as described by Radić et al. (1993). Catalytic constants, Km, kcat, and Kss were determined as described previously from substrate concentration dependences and titration of catalytic sites (Radić et al., 1993).
Thermal and Chemical Stability Assays.
Stability assays were performed on the wtT547 (wild-type, AChE truncated at amino acid 547) and mutant proteins using 1.0 μM enzyme concentration in 0.1 M phosphate buffer, pH 7, for the thermal denaturation assay and 1.0 μM enzyme in 3 M urea (UltraPure Urea; Invitrogen, Carlsbad, CA) for the chemical denaturation assay. Thermal stability experiments were conducted at 50°C for 60 min, and the urea stability experiments were conducted at room temperature. Aliquots were diluted 30-fold in phosphate buffer with 0.01% BSA at room temperature to end denaturation.
Cholinesterase Inhibition by Paraoxon.
AChE species (40 nM wtT547 or D134H) were incubated with 300 nM paraoxon (diethyl p-nitrophenyl phosphate; Sigma-Aldrich) in 100 mM phosphate buffer, pH 7.4, containing 0.01% BSA for up to 40 min at 25°C. Uninhibited enzyme samples were run in parallel as controls. Inhibition was stopped by the addition of 1.0 mM ATCh, and residual activity was measured. Inhibition constants were determined as described previously (Kovarik et al., 2003).
Oxime Reactivation of Phosphorylated Cholinesterase.
WtT547 and mutant D134H enzymes at 33 μM, inhibited with 45 μM paraoxon for 30 min at room temperature, were diluted 10 times and passed through a Sephadex G-50 spin column (Roche Diagnostics, Indianapolis, IN) to remove excess unconjugated organophosphate. The enzymes were reactivated using 1 mM 2-PAM (Sigma-Aldrich) in 100 mM phosphate buffer, pH 7.4, containing 0.01% BSA at 25°C. Equivalent control samples of uninhibited enzyme were passed through a parallel column, and activities measured in the presence of oxime at the same concentration used for the reactivation of the inhibited enzymes. Data analysis for reactivation has been described previously (Kovarik et al., 2004).
Results
Cholinesterase Phenotyping.
In vivo cholinesterase activity was ascertained for the 80 unrelated samples and the 478 twin samples (167 monozygotic and 72 dizygotic twin pairs). The AChE activity mean for the unrelated samples was 42.92 (S.D. = 7.2) ΔA/min/μl with a 95% confidence interval (CI) of 41.32 to 44.52, and the BChE activity mean was 9.82 (S.D. = 2.4) ΔA/min/μl with a 95% CI of 9.28 to 10.36. AChE activity mean for the twin samples was 42.04 (S.D. = 7.2) ΔA/min/μl with a 95% CI of 41.39 to 42.69, and the BChE activity mean was 9.77 (S.D. = 2.3) ΔA/min/μl with a 95% CI of 9.56 to 9.98. Frequency distributions of AChE and BChE activity for both the unrelated panel and the twin panel displayed approximate Gaussian distributions (i.e., normal distributions).
Linear regression and correlation analyses of ChE activity were performed on the monozygotic and dizygotic twin pairs to obtain evidence of correlation within each type of pair. Blood AChE enzymatic activity differed by gender (P = 0.0001; male 45.1 ± 0.84 ΔA/min/μl versus female 41.5 ± 0.4 ΔA/min/μl) and by biogeographic ancestry (P < 0.05; white 42.9 ± 0.30 ΔA/min/μl, African American 37.1 ± 1.6 ΔA/min/μl, Hispanic 40.4 ± 1.1 ΔA/min/μl). By twin-pair variance congruent analyses in SOLAR (Table 1), the effects of several traits (as covariates) on the two cholinesterases were determined. Blood AChE enzymatic activity also varied (Table 1) with SBP (P = 0.0193). Blood BChE enzymatic activity varied (Table 1) by age (P = 0.003), weight (P < 0.0001), height (P = 0.0278), SBP (P = 0.033), and BMI (P < 0.0001).
TABLE 1.
ChE activity: heritability and association analysis
Trait heritability (H2r) and the associated P values and covariate P values are displayed for blood ChE activities, AChE and BChE, in the San Diego twin study. Genetic and environmental factors that contribute to the blood ChE heritability are listed (in boldface: P < 0.05). Heritability is the fraction of trait variance accounted for by additive genetic variance (h2 = VG/VP), estimated by variance components in the SOLAR package. Mean ± S.E.M. is the confidence interval for heritability. The P value column is the significance of the trait heritability estimate (initially considered in isolation; top row for each trait). The covariate P value is the significance of a particular covariate (in that row) for effects on additive genetic variance of that trait.
| SOLAR Heritability Analysis: San Diego Caucasian Twins ChE Activity | |||||
|---|---|---|---|---|---|
| Trait/Covariate | H2r | H2r S.E.M. | P Value | Covariate P Value | n |
| AChE | 0.4883 | 0.0610 | 9.35 × 10−11 | 364 | |
| BP status | 0.4873 | 0.0609 | 8.91 × 10−11 | 0.0791 | 364 |
| SBP (mm Hg) | 0.4784 | 0.0642 | 1.13 × 10−9 | 0.0193 | 343 |
| ACHE SNP-13 P561R | 0.5465 | 0.0737 | 2.42 × 10−8 | 0.0126 | 274 |
| ACHE SNP-16 3′-UTR | 0.6114 | 0.0638 | 1.57 × 10−10 | 0.0811 | 287 |
| ACHE haplotype 1 | 0.4775 | 0.0623 | 3.31 × 10−10 | 0.0394 | 362 |
| ACHE diplotype 2/4 | 0.4851 | 0.0618 | 1.68 × 10−10 | 0.0129 | 362 |
| ACHE diplotype 2/7 | 0.4763 | 0.0626 | 4.08 × 10−10 | 0.0514 | 361 |
| BChE | 0.814 | 0.0279 | 1.09 × 10−32 | 364 | |
| Age (years) | 0.8061 | 0.0292 | 4.37 × 10−31 | 0.0031 | 364 |
| Weight (kg) | 0.8015 | 0.0298 | 1.25 × 10−30 | 6.21 × 10−9 | 364 |
| Height (cm) | 0.8097 | 0.0286 | 7.91 × 10−32 | 0.0278 | 364 |
| SBP (mm Hg) | 0.8067 | 0.0311 | 1.63 × 10−26 | 0.0331 | 343 |
| BMI (kg/mg2) | 0.7921 | 0.0312 | 1.45 × 10−29 | 4.96 × 10−12 | 364 |
| BCHE SNP-1 intron-2 | 0.7822 | 0.0206 | 2.79 × 10−31 | 0.0021 | 423 |
| BCHE SNP-2 3′-UTR | 0.7894 | 0.0299 | 9.1 × 10−32 | 0.0322 | 420 |
BP status, hypertensive/normotensive; ACHE SNP13, rs1799806; ACHE SNP16, rs17228616; BCHE SNP1, rs1355534 (intron-2); BCHE SNP2, rs3495 (3′-UTR); ACHE diplotype 2/4, diploid haplotype composed of ACHE haplotypes 2 and 4; ACHE diplotype 2/7, diploid haplotype composed of ACHE haplotypes 2 and 7.
SNP Discovery and Genotyping.
Results of DNA resequencing of the AChE gene on the panel of 80 unrelated individuals identified 19 SNPs and 2 potential microsatellite variations in the 3′-untranslated region (UTR) (Fig. 1). Seven of the identified SNPs are in the coding region (cSNPs); four are nonsynonymous cSNPs encoding a different amino acid including the well known YT blood group antigen, and three are synonymous cSNPs encoding the same amino acid. Twelve SNPs are in untranslated regions of the gene with three in a highly conserved region of intron 1. Supplemental Table S1 identifies reference numbers for SNPs in NCBI dbSNP and their minor allelic frequencies in European Americans and African Americans computed in the unrelated panel for each SNP. Several SNPs reflected significant differences in the allelic frequency of African American individuals compared with the frequency for individuals of European ancestry.
ACHE Haplotype Structure.
Haplotype reconstruction using PHASE version 2 was performed on the twin subjects using SNPs 5 (R34Q), 8 (E259E), 9 (H343N), 12 (P477P), and 16 (3′-UTR). Haplotype pair (or “diplotype”) frequencies in the twins were determined with the most common pair being 1/1 followed by 2/4 and 2/7 (Table 2).
TABLE 2.
ACHE haplotype reconstruction using five ACHE SNPs (5, 8, 9, 12, and 16)
ACHE haplotypes and diplotypes (diploid haplotypes) in 491 San Diego twins. Haplotype inferences were done in the program Phase <http://depts.washington.edu/ventures/UW_Technology/Express_Licenses/PHASEv2.php>. Nonsynonymous cSNPS: SNP5–R34Q (C/T) SNP9–H353N (G/T); synonymous cSNPs: SNP8–E344E (C/T) SNP12–P477P (C/T); noncoding SNP: SNP16–3′-UTR (C/A).
| Haplotype No. | SNPs 5, 8, 9, 12, and 16 | No. of Chromosomes | Diploid Haploytpe (Diplotype) | No. of Individuals |
|---|---|---|---|---|
| 1 | CCGCC | 891 | 1/1 | 439 |
| 2 | CCGCA | 36 | 1/2 | 1 |
| 3 | CCGTC | 6 | 1/3 | 5 |
| 4 | CCGTA | 15 | 1/6 | 3 |
| 5 | CCTCA | 1 | 1/7 | 1 |
| 6 | CCTTC | 3 | 1/8 | 3 |
| 7 | CCTTA | 26 | 2/2 | 1 |
| 8 | CTGCC | 3 | 2/4 | 10 |
| 9 | TCGCC | 1 | 2/5 | 1 |
| 2/7 | 22 | |||
| 3/9 | 1 | |||
| 4/4 | 2 | |||
| 4/7 | 1 | |||
| 7/7 | 1 |
Cholinesterase Heritability and Allelic Association Studies.
Implementing variance components from twin pair data in SOLAR software, both heritability and association analyses were conducted on the complete twin sample set (n = 478) and again on a subset of 362 twins of European ancestry (Table 1). Analysis was repeated on the twins of European ancestry to address any analytic bias arising from allelic and physiological differences due to different ethnic backgrounds. Because of the limited number of twins of other ethnicities, analysis based on one ethnicity could be performed only on the twins of European ancestry. Studies on both twin panels revealed a significant degree of heritability (h2) of the biochemical phenotypes (ChE activity) along with significant associations with several cardiovascular and metabolic phenotypic traits. Heritability studies on the complete twin panel set revealed an h2 of approximately 52% for AChE activity and an h2 of approximately 80% for BChE activity (all ethnicities included; data not shown). Restricting the same analysis to the twins of European ancestry gave similar values for ChE heritability, with an h2 of 48.8 ± 6.1% for AChE activity and an h2 of 81.4 ± 2.8% for BChE activity (Table 1).
By SOLAR, significant ACHE trait genetic marker-on-trait associations (Table 1) were found in the Caucasian twins for ACHE SNP13 (P561R, P = 0.0126 by generalized estimating equation; Fig. 2), haplotype 1 (P = 0.0394), and haplotype pair 2/4 (P = 0.0129). BChE activity was also associated with systolic BP (P = 0.0193). Significant BCHE trait genetic marker-on-trait associations (Table 1) were found in the Caucasian twins for BCHE SNP1 (intron 2, P = 0.002), and SNP2 (3′-UTR, P = 0.03). BChE activity was significantly associated with multiple metabolic syndrome traits (Table 1): weight, height, systolic BP, and BMI. At the ACHE locus, we discovered only one nonsynonymous variant with a high minor allele frequency: R561P (rs1799806; Supplemental Table 1), at approximately 50%; thus, the common genetic variation governing interindividual differences in blood AChE expression may well be regulatory or noncoding.
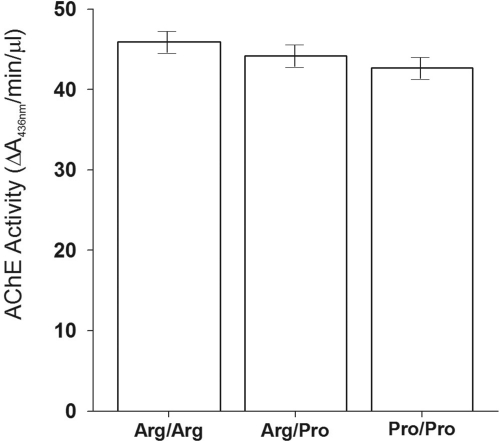
Fig. 2.
Association with blood AChE activity stratified by genotypes of residue R561P in the twin subjects (SOLAR: P = 0.0126; generalized estimating equation for descriptive statistics; Hardy-Weinberg equilibrium: P = 0.075). Three different diploid genotypes arose from the SNP13 P561R variant: Arg/Arg, 46 ± 1 units/μl (n = 63); Pro/Arg, 44 ± 1 (n = 154); and Pro/Pro, 43 ± 1 (n = 60). AChE activities of Arg/Arg and Pro/Arg, as well as Pro/Pro and Pro/Arg genotypes, were not significantly different (P > 0.05), whereas activities of homozygote Arg/Arg and Pro/Pro genotypes were significantly different (P = 0.05). The results, adjusted for age and sex, were obtained from analyses in twins of European ancestry. Change in blood AChE activity by genotype (approximately 3 units/μl in Arg/Arg versus Pro/Pro homozygotes) was comparable to that found across sex (approximately 4 units/μl, male versus female) and biogeographic ancestry groups (approximately 5 units/μl, black versus white).
Temperature Sensitivity of the hAChE D134H Mutant Enzyme.
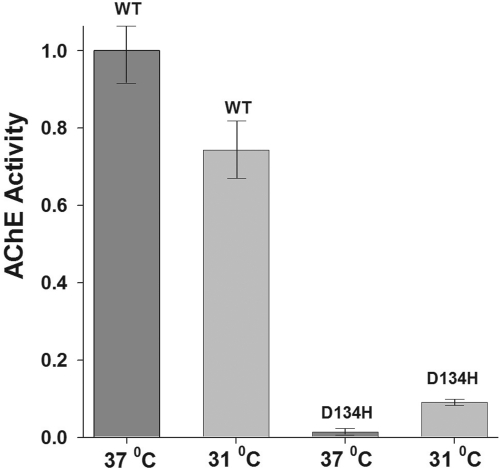
The low D134H protein expression rendered it difficult initially to collect sufficient quantities of this mutant for purification. To determine whether low expression of D134H is due to temperature sensitivity of protein folding, cells expressing wild-type enzyme or D134H mutant after washing and transfer to serum-free medium were incubated at 37°C or 31°C for a total of 60 h. Temperature-sensitivity experiments were conducted on both transiently and stably transfected cells. Serum-free medium was collected and tested by Ellman assay for enzymatic activity at 24-, 48-, and 60-h intervals. Results were normalized to the wtT547 protein at 37°C for each time interval, and a representative graph of the stable plates at 48 h is shown in Fig. 3. An expected fall in activity measured from media collected from cells expressing the wtT547 protein at 31°C compared with 37°C. For the D134H mutant, media collected from cells at 37°C showed virtually no activity, whereas media collected from cells incubated at 31°C expressed activity levels that were at least 5-fold higher but still significantly lower than wild type activity. Cells expressing the D134H mutant were then incubated at 31°C and yielded sufficient quantities of enzyme for subsequent protein production.
Fig. 3.
Temperature sensitivity of the D134H mutant expression in cultured cells. Stably transfected HEK cells in serum-free medium expressing either the wild-type enzyme or the D134H mutant were incubated at 37°C and 31°C for 48 h. Results of six experiments have been normalized to the wild-type protein at 37°C and presented with the associated standard deviation. Difference between AChE activities at 31°C and 37°C was significant for both wild type (P = 0.017) and D134H mutant (P = 3.4 × 10−7). Both the wild-type and mutant protein sequences are truncated at amino acid 547 to yield soluble protein.
Kinetic Characterization of the hAChE Mutants.
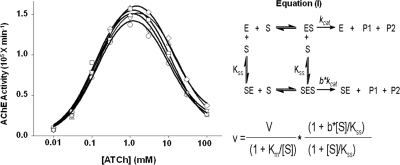
To determine whether the nonsynonymous cSNPs affect enzymatic activity of the mutant proteins, the catalytic parameters, Km, Kss, kcat, and b, of all the mutant enzymes were determined (Fig. 4). No significant differences in catalytic properties, reflected in the substrate dependence for catalysis and inhibition, were noted the mutant enzymes compared with the wtT547 enzyme (Supplemental Table S2).
Fig. 4.
Catalytic parameters of the mutant (○, R3Q; ▵, D134H; ▿, H322N; ♢, D134H/R136Q) and wild-type (□, T547) AChE. Activities were measured with 0.01 to 100 mM acetylthiocholine (ATCh) as substrate. Curves were generated by nonlinear fit of measured AChE activity to the Equation (I) (resulting constants are given in Supplemental Table S2).
Thermal and Chemical Stability Analysis of the hAChE Mutants.
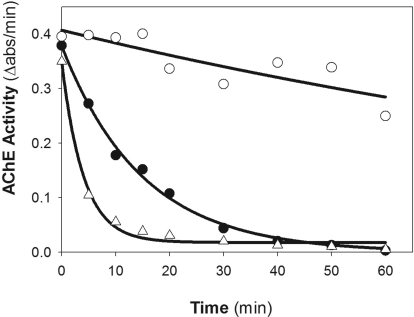
To delineate the stability parameters of AChE (R3Q, D134H, H322N), thermal and chemical denaturation assays of AChE mutants were conducted alongside the wild-type enzyme in triplicate. The findings primarily showed significant differences in decay times between the wtT547 enzyme and for the D134H mutant enzyme.
One-way analysis of variance performed for both R3Q and H322N mutant proteins revealed a small but statistically insignificant difference (P > 0.05) in stability compared with the wild-type species. However, the D134H mutant showed significant differences from the wtT547 enzyme in stability both in the thermal assay (P < 0.01) and in the chemical assay (P < 0.05) (Fig. 5; Table 3).
Fig. 5.
Time course of 52°C thermal denaturation of wild-type (○), D134H (▵), and D134H/R136Q (●) T547 AChE.
TABLE 3.
AChE activity decay rates influenced by temperature and urea perturbations for wild-type and mutant AChEs
Rate constants ± S.E. are calculated from triplicate experiments. Values in the columns are reported as first-order rate constants and half-times, t50. Significance of the differences between mutant decay rates and wtT547 and between D134H and D134H/R136Q mutants is indicated.
| Enzyme | Decay Rate Constant at 52°C | 52°C t50 Ratio+ | Decay Rate Constant in Urea | Urea t50 Ratio+ |
|---|---|---|---|---|
| min−1 | min−1 | |||
| wtT547 | 0.011 ± 0.002 | 1.0 | 0.040 ± 0.003 | 1.0 |
| R3Q | 0.024 ± 0.008 | 0.36* | 0.049 ± 0.005 | 0.82* |
| H322N | 0.022 ± 0.004 | 0.48** | 0.060 ± 0.01 | 0.63* |
| D134H | 0.30 ± 0.07 | 0.036*** | 0.25 ± 0.04 | 0.17*** |
| D134H/R136Q | 0.070 ± 0.02 | 0.18**†† | 0.096 ± 0.006 | 0.42***††† |
t50 mutant/t50 wild type.
P > 0.05 (vs. wtT547).
P = 0.05 (vs. wtT547).
P < 0.05 (vs. wtT547).
P = 0.05 (vs. D134H).
P < 0.05 (vs. D134H).
Rescue of the D134H Single Mutant by the Double Mutant D134H/R136Q.
We hypothesized that the instability of catalytic activity found for D134H mutant may be related to local alterations in protein conformation leading to a metastable protein state. Using the human AChE crystal structure (Protein Data Bank code 1B41) (Kryger et al., 2000), we modeled the substituted histidine mutant at residue 134 and found it to be in close proximity to Arg136. The change in charge from negative to positive and the close proximity of the His to another positively charged amino acid could adversely affect proper folding. This could be attributed to both charge repulsion and steric hindrance because the molecular mass for the histidine side chain is greater than that for aspartic acid (81 Da versus 59 Da).
To test this hypothesis using the single D134H cDNA as a template, a double mutant (D134H/R136Q) was constructed in an attempt to rescue the D134H mutant. To alleviate electrostatic repulsion afforded by the close proximity of two cationic amino acid side chains, the uncharged glutamine (Gln) was substituted for the Arg at position 136 (Arg136→Gln). In theory, this would eliminate both the charge repulsion and minimize steric hindrance because Gln is uncharged with a side chain smaller than the Arg.
Upon successful incorporation and verification of the substitution of Arg136→Gln by site-directed mutagenesis and sequencing, HEK cells were stably transfected and cultured at 37°C. It is noteworthy that cells were able to produce the double mutant enzyme with expressed protein yields from harvested media equivalent to wtT547 production. This demonstrated at least a fractional rescue by the R136Q substitution in that temperature sensitivity of the single D134H mutant in cultured cells was partially corrected.
Characterization of the double mutant was conducted as outlined above for the single mutation gene products. Kinetic parameters (Km, Kss, b factor, and kcat) determined for the D134H/R136Q mutant enzyme did not differ significantly from the wtT547 or other single mutant forms of AChE (see Fig. 4 and Supplemental Table S2). We were also able to confirm that the double mutant did reverse a significant fraction of instability in both the temperature and chemical denaturation assays of the single Asp134→His mutation (Fig. 5; Table 3).
Organophosphate Inhibition of Cholinesterases.
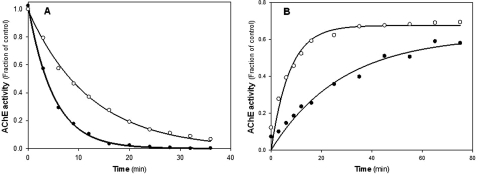
The first-order rate inhibition constants for 1 μM paraoxon determined in triplicate were as follows: wtT547 kobs = 0.20 min−1; D134H kobs = 0.09 min−1 (Fig. 6). Inhibition of the D134H mutant enzyme was approximately two times slower than the wild-type enzyme.
Fig. 6.
Time course of alkyl phosphate inhibition of wild-type (●) and D134H (○) hAChE activity by exposure to 1.0 μM paraoxon (A) and reactivation by 1.0 mM 2-PAM (B). Curves were generated by nonlinear regression as described previously (Kovarik et al., 2003, 2004).
Oxime-Assisted Reactivation of Phosphorylated Cholinesterases.
To compare reactivation rates for both paraoxon-inhibited wtT547 and the D134H enzymes, the oxime 2-PAM was used in the reactivation to remove the conjugated OP. Four independent reactivation experiments were run using 1.0 mM 2-PAM; two were run at 37°C and two at 25°C to account for the temperature sensitivity of the D134H mutant protein. The D134H mutant protein had a rate of reactivation that was at least four times faster than the rate of reactivation for the wtT547 protein. Taking an average of the calculated results from experiments performed at 25°C, the first-order reactivation rate constants were determined as follows: wtT547 kobs = 0.027 min−1; D134H kobs = 0.11 min−1 (Fig. 6B). Additional reactivation experiments that were run using a range of 2-PAM concentrations (0.1–1.5 mM) showed that the reactivation rate of the D134H mutant was always faster than the rate of wtT547 reactivation (see Supplemental Fig. S1).
Discussion
Heritability and Association of ACHE and BCHE in Twin Pairs.
We find that AChE measured by enzymatic activity in blood is substantially heritable, with additive genetic variance accounting for 48.8 ± 6.1% of trait variability (P = 9.35 × 10−11) (Table 1). In addition, common allelic variation at the ACHE locus influenced enzymatic activity, with a significant influence at SNP13 (P561R, P = 0.0126; Fig. 2). Thus, genetic variation at the ACHE locus may be a determinant of AChE expression and secretion (i.e., ACHE can serve as a cis-QTL for the AChE activity trait). Because P561R lies in exon 5 (Fig. 1), we cannot test the effects of this variant in the recombinant nerve/muscle system; we have not extended the recombinant system to the hematopoietic version of AChE, which does include exon 5. AChE activity also was associated with systolic BP (P = 0.0193) and appeared to be influenced by biogeographic ancestry group (European American > Hispanic > African American) and gender (male > female).
BChE enzymatic activity was substantially more heritable (at 81.4 ± 2.8%, P = 1.09 × 10−32) and influenced by allelic variation in intron 2 (SNP1, P = 0.0021) and the 3′-UTR (SNP2, P = 0.0322). Thus, the BCHE locus is a cis-QTL for expression of the enzyme, providing confirmation and extension of our genome-wide linkage results on chromosome 3q26 (LOD = 3.0) for the BCHE trait (Valle et al., 2006). In addition, as previously reported for BCHE in the metabolic syndrome (Valle et al., 2006), BChE activity varied (Table 1) with age, weight, height, SBP, and BMI. In a previous report on genome-wide linkage for the BCHE trait (Valle et al., 2006), we also identified a novel trans-QTL on chromosome 5q (LOD = 3.34).
AChE Structure, Activity, and Stability: Effects of Genetic Polymorphism.
We demonstrate how a single nucleotide substitution, giving rise to the D134H mutation, affects protein structure and conformation. In turn, this may alter catalytic properties under stress-related conditions, possibly because of local misfolding and formation of a metastable conformation. Because OP-based pesticides and nerve agents react covalently with the active site serine of AChE, individuals carrying this mutation in the AChE protein display different rates of OP inhibition or reactivation of the conjugate. Although functional consequences of the identified nonsynonymous mutations as studied in vitro were unequivocal, their low minor allele frequencies in the general population (Supplemental Table 1) precluded appropriate statistical power in the evaluation of their effects on twin traits, given the sample size of our twin set. Although direct evidence in the population thus is lacking because of the low mutation frequency and hence inadequate statistical power, individuals carrying the D134H mutation might be expected to show altered sensitivity to organophosphorus pesticides or chemical weapons targeting the AChE enzyme.
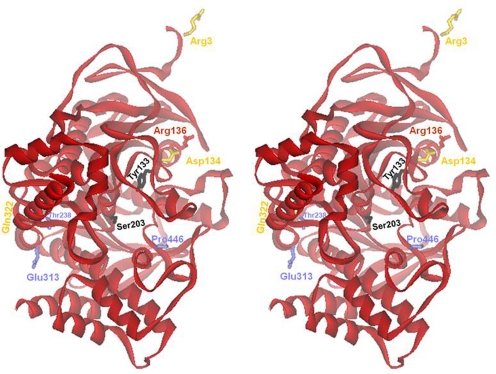
The position of an amino acid encoded by a nonsynonymous cSNP on the AChE surface may also affect interaction of monomers in association with other molecules and could possibly produce antigenic responses as with the YT antigen (Bartels et al., 1993) (Fig. 7). Again, functional consequences of nonsynonymous cSNPs in ACHE need further investigation.
Fig. 7.
Locations of SNPs mapped onto human AChE three-dimensional structure of the truncated soluble form containing the N-terminal 547 AChE amino acid residues (see Fig. 1). Synonymous SNPs are indicated by blue amino acid side chains and blue labels, whereas nonsynonymous SNPs are indicated as yellow side chains and yellow labels. The active center serine (Ser203) and Tyr133 are shown in black.
Another critical consideration is the tissue location of AChE. AChE is readily accessible in whole-blood samples for activity measurements, whereas the tissues where activity most probably affects physiologic function, such as brain and skeletal muscle, are typically not accessible. However, our study should reveal intrinsic differences in AChE catalytic parameters or AChE stability, encoded by nonsynonymous cSNPs so that AChE parameters intrinsic to the sequence should result in comparable phenotypes in the CNS, skeletal muscle, or autonomic nervous system. On the other hand, gene expression controlled by transcription, translation, and certain post-translational events may be observed in only certain tissues. For example, an upstream enhancer that controls expression rather uniquely in skeletal muscle and platelets is driven by a conserved sequence found in the first intron. SNPs 1–3 in Fig. 1 would be expected to only exert an influence in skeletal muscle and the platelet precursor, the megakaryocyte (Camp et al., 2008, 2010).
The instability of the D134H, a nonsynonymous cSNP, was found in cell culture indicating a trafficking and secretion deficiency, probably arising from protein folding. On the other hand, the enzyme, when isolated under low temperature conditions, did not exhibit altered kinetic parameters under physiologic conditions. This instability could be attributed to the position of the amino acid substitution on the surface of the mature protein as this mutation is far from the catalytic triad (Ser203, His447, and Glu334) (Gibney et al., 1990; Sussman et al., 1991; Shafferman et al., 1992) that is located deep within the aromatic gorge of the enzyme structure. On the other hand, amino acid residue 134 is in the proximity of the oxyanion hole (Gly121, Gly122, and Ala204) connected by a regional protein backbone lacking a well defined secondary structure. Accordingly, this region could be more fluid and, therefore, more sensitive to polymorphic residue differences. The substitution from a negative residue (Asp134) to an ionizable cation (His134) places the cationic side chain in close proximity to Arg136 that contains a permanent positive charge that may influence side chain position or its pKa. In addition, Arg136 is close to the surface loop near the N terminus. Placing two positive residues in close proximity could create an electrostatic field of charge repulsion affecting protein trafficking and interactions with chaperones. Although catalytic properties of the mutant enzyme are unaltered, we do see additional heat and chemical denaturation susceptibility along with altered rates of OP inactivation and reactivation.
Conclusions and Perspectives.
The structure of cholinesterase enzymes and polymorphism of their genes may provide additional insight into susceptibility to cardiovascular diseases, hypersensitivity to pesticides, and individual risks associated with chemical terrorism agents directed to the cholinergic nervous system.
A complete analysis of cholinesterase phenotypes in humans is limited by accessibility of the gene product where it is expressed in the central, autonomic, and somatic motor systems. For example, an upstream intron selectively controls expression in skeletal muscle (Camp et al., 2008, 2010), and an alternatively spliced sequence influences the processing of AChE in CNS areas such as the striatum. The accessible gene product found in erythrocytes is not necessarily influenced by this intron or the splice option expressed primarily in brain (Dobbertin et al., 2009; Bernard et al., 2011). On the other hand, the nonsynonymous cSNP found in the open-reading frame of the gene and reflected in a change in stability of the gene product can be expected to influence expression, activity, and turnover of the enzyme in all regions of the central, autonomic and somatic motor nervous systems.
Supplementary Material
Acknowledgments
We thank Drs. Nicholas Schork and Philip Bourne for assistance in pursuing this study and for helpful discussions. The expert assistance of Shelley Camp in recombinant DNA techniques and helpful discussions is gratefully acknowledged.
This work was supported in part by the National Institutes of Health National Heart, Lung, and Blood Institute [Grant P01-HL058120-10]; the National Institutes of Health National Institute of General Medical Sciences [Grants R01-GM018360-39, T32-GM07752]; the National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant U01-NS58046]; and the National Institutes of Health National Institute of Environmental Health Sciences [Grant P42-ES010337-10].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.111.180091.
 The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
- AChE
- acetylcholinesterase
- BChE
- butyrylcholinesterase
- SNP
- single-nucleotide polymorphism
- cSNP
- coding SNP
- ACh
- acetylcholine
- hAChE
- human AChE
- h2
- heritability
- 2-PAM
- pyridine-2-aldoxime methyl methanesulfonate
- SOLAR
- sequential oligogenic linkage analysis
- BP
- blood pressure
- kb
- kilobase
- wtT547
- wild-type monomeric hAChE truncated at amino acid 547
- SBP
- systolic BP
- BMI
- body mass index
- UTR
- untranslated region
- HEK
- human embryonic kidney
- BSA
- bovine serum albumin
- QTL
- quantitative trait locus
- LOD
- logarithm of the odds
- CI
- confidence interval.
Authorship Contributions
Participated in research design: Taylor, O'Connor, Valle, Rana, and Radić.
Conducted experiments: Valle, Rana, Mahboubi, Shih, and Rao.
Performed data analysis: Valle, Rana, Wessel, Mahboubi, Shih, Rao, and Radić.
Wrote or contributed to the writing of the manuscript: Valle, Taylor, O'Connor, Rana, and Radić.
References
- Allderdice PW, Gardner HA, Galutira D, Lockridge O, LaDu BN, McAlpine PJ. (1991) The cloned butyrylcholinesterase (BCHE) gene maps to a single chromosome site, 3q26. Genomics 11:452–454 [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels CF, Zelinski T, Lockridge O. (1993) Mutation at codon 322 in the human acetylcholinesterase (ACHE) gene accounts for YT blood group polymorphism. Am J Hum Genet 52:928–936 [PMC free article] [PubMed] [Google Scholar]
- Bernard V, Girard E, Hrabovska A, Camp S, Taylor P, Plaud B, Krejci E. (2011) Distinct localization of collagen Q and PRiMA forms of acetylcholinesterase at the neuromuscular junction. Mol Cell Neurosci 46:272–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccafusco JJ. (1996) The role of central cholinergic neurons in the regulation of blood pressure and in experimental hypertension. Pharmacol Rev 48:179–211 [PubMed] [Google Scholar]
- Camp S, De Jaco A, Zhang L, Marquez M, De la Torre B, Taylor P. (2008) Acetylcholinesterase expression in muscle is specifically controlled by a promoter-selective enhancesome in the first intron. J Neurosci 28:2459–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp S, Zhang L, Krejci E, Dobbertin A, Bernard V, Girard E, Duysen EG, Lockridge O, De Jaco A, Taylor P. (2010) Contributions of selective knockout studies to understanding cholinesterase disposition and function. Chem Biol Interact 187:72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn M, Hamilton A, Zadnick J, Cozen W, Mack TM. (2002) The occurrence of chronic disease and other conditions in a large population-based cohort of native Californian twins. Twin Res 5:460–467 [DOI] [PubMed] [Google Scholar]
- Darvesh S, Hopkins DA, Geula C. (2003) Neurobiology of butyrylcholinesterase. Nat Rev Neurosci 4:131–138 [DOI] [PubMed] [Google Scholar]
- Dobbertin A, Hrabovska A, Dembele K, Camp S, Taylor P, Krejci E, Bernard V. (2009) Targeting of acetylcholinesterase in neurons in vivo: a dual processing function for the proline-rich membrane anchor subunit and the attachment domain on the catalytic subunit. J Neurosci 29:4519–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellman GL, Courtney KD, Andres V, Jr, Featherstone RM. (1961) A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmacol 7:88–95 [DOI] [PubMed] [Google Scholar]
- Gaughan G, Park H, Priddle J, Craig I, Craig S. (1991) Refinement of the localization of human butyrylcholinesterase to chromosome 3q26.1-q26.2 using a PCR-derived probe. Genomics 11:455–458 [DOI] [PubMed] [Google Scholar]
- Getman DK, Eubanks JH, Camp S, Evans GA, Taylor P. (1992) The human gene encoding acetylcholinesterase is located on the long arm of chromosome 7. Am J Hum Genet 51:170–177 [PMC free article] [PubMed] [Google Scholar]
- Gibney G, Camp S, Dionne M, MacPhee-Quigley K, Taylor P. (1990) Mutagenesis of essential functional residues in acetylcholinesterase. Proc Natl Acad Sci USA 87:7546–7550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin Y, Avidan N, Bercovich D, Korczyn AD, Silman I, Beckmann JS, Sussman JL. (2005) Analysis of genetic polymorphisms in acetylcholinesterase as reflected in different populations. Curr Alzheimer Res 2:207–218 [DOI] [PubMed] [Google Scholar]
- Howard TD, Hsu FC, Grzywacz JG, Chen H, Quandt SA, Vallejos QM, Whalley LE, Cui W, Padilla S, Arcury TA. (2010) Evaluation of candidate genes for cholinesterase activity in farmworkers exposed to organophosphorus pesticides: association of single nucleotide polymorphisms in BCHE. Environ Health Perspect 118:1395–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irisawa H, Brown HF, Giles W. (1993) Cardiac pacemaking in the sinoatrial node. Physiol Rev 73:197–227 [DOI] [PubMed] [Google Scholar]
- Khan IM, Singletary E, Alemayehu A, Stanislaus S, Printz MP, Yaksh TL, Taylor P. (2002) Nicotinic receptor gene cluster on rat chromosome 8 in nociceptive and blood pressure hyperresponsiveness. Physiol Genomics 11:65–72 [DOI] [PubMed] [Google Scholar]
- Kovarik Z, Radić Z, Berman HA, Simeon-Rudolf V, Reiner E, Taylor P. (2003) Acetylcholinesterase active centre and gorge conformations analysed by combinatorial mutations and enantiomeric phosphonates. Biochem J 373:33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik Z, Radić Z, Berman HA, Simeon-Rudolf V, Reiner E, Taylor P. (2004) Mutant cholinesterases possessing enhanced capacity for reactivation of their phosphonylated conjugates. Biochemistry 43:3222–3229 [DOI] [PubMed] [Google Scholar]
- Kryger G, Harel M, Giles K, Toker L, Velan B, Lazar A, Kronman C, Barak D, Ariel N, Shafferman A, et al. (2000) Structures of recombinant native and E202Q mutant human acetylcholinesterase complexed with the snake-venom toxin fasciculin-II. Acta Crystallogr D Biol Crystallogr 56:1385–1394 [DOI] [PubMed] [Google Scholar]
- Li Y, Camp S, Rachinsky TL, Getman D, Taylor P. (1991) Gene structure of mammalian acetylcholinesterase. Alternative exons dictate tissue-specific expression. J Biol Chem 266:23083–23090 [PubMed] [Google Scholar]
- Li Y, Camp S, Taylor P. (1993) Tissue-specific expression and alternative mRNA processing of the mammalian acetylcholinesterase gene. J Biol Chem 268:5790–5797 [PubMed] [Google Scholar]
- Lockridge O, Bartels CF, Vaughan TA, Wong CK, Norton SE, Johnson LL. (1987) Complete amino acid sequence of human serum cholinesterase. J Biol Chem 262:549–557 [PubMed] [Google Scholar]
- Marchot P, Ravelli RB, Raves ML, Bourne Y, Vellom DC, Kanter J, Camp S, Sussman JL, Taylor P. (1996) Soluble monomeric acetylcholinesterase from mouse: expression, purification, and crystallization in complex with fasciculin. Protein Sci 5:672–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami LR, Wieseler S, Souza RL, Schopfer LM, Nachon F, Lockridge O, Chautard-Freire-Maia EA. (2008) Five new naturally occurring mutations of the BCHE gene and frequencies of 12 butyrylcholinesterase alleles in a Brazilian population. Pharmacogenet Genomics 18:213–218 [DOI] [PubMed] [Google Scholar]
- Nakahara T, Kawada T, Sugimachi M, Miyano H, Sato T, Shishido T, Yoshimura R, Miyashita H, Sunagawa K. (1998) Cholinesterase affects dynamic transduction properties from vagal stimulation to heart rate. Am J Physiol 275:R541–R547 [DOI] [PubMed] [Google Scholar]
- Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J. (1992) The alpha/beta hydrolase fold. Protein Eng 5:197–211 [DOI] [PubMed] [Google Scholar]
- Radić Z, Pickering NA, Vellom DC, Camp S, Taylor PP. (1993) Three distinct domains in the cholinesterase molecule confer selectivity for acetyl- and butyrylcholinesterase inhibitors. Biochemistry 32:12074–12084 [DOI] [PubMed] [Google Scholar]
- Rana BK, Wessel J, Mahboubi V, Rao F, Haeller J, Gayen JR, Eskin E, Valle AM, Das M, Mahata SK, et al. (2009) Natural variation within the neuronal nicotinic acetylcholine receptor cluster on human chromosome 15q24: influence on heritable autonomic traits in twin pairs. J Pharmacol Exp Ther 331:419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacchi R, Gambina G, Moretto G, Corbo RM. (2009) Variability of AChE, BChE, and ChAT genes in the late-onset form of Alzheimer's disease and relationships with response to treatment with donepezil and rivastigmine. Am J Med Genet B Neuropsychiatr Genet 150B:502–507 [DOI] [PubMed] [Google Scholar]
- Schumacher M, Camp S, Maulet Y, Newton M, MacPhee-Quigley K, Taylor SS, Friedmann T, Taylor P. (1986) Primary structure of Torpedo californica acetylcholinesterase deduced from its cDNA sequence. Nature 319:407–409 [DOI] [PubMed] [Google Scholar]
- Shafferman A, Kronman C, Flashner Y, Leitner M, Grosfeld H, Ordentlich A, Gozes Y, Cohen S, Ariel N, Barak D. (1992) Mutagenesis of human acetylcholinesterase. Identification of residues involved in catalytic activity and in polypeptide folding. J Biol Chem 267:17640–17648 [PubMed] [Google Scholar]
- Soreq H, Gnatt A, Loewenstein Y, Neville LF. (1992) Excavations into the active-site gorge of cholinesterases. Trends Biochem Sci 17:353–358 [DOI] [PubMed] [Google Scholar]
- Suh TH, Wang CH, Lim RKS. (1936) The effect of intracisternal application of acetylcholine and the localization of the pressor centre and tract. Chinese J Physiol 10:61–78 [Google Scholar]
- Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. (1991) Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science 253:872–879 [DOI] [PubMed] [Google Scholar]
- Valle A, O'Connor DT, Taylor P, Zhu G, Montgomery GW, Slagboom PE, Martin NG, Whitfield JB. (2006) Butyrylcholinesterase: association with the metabolic syndrome and identification of 2 gene loci affecting activity. Clin Chem 52:1014–1020 [DOI] [PubMed] [Google Scholar]
- Vargas HM, Brezenoff HE. (1988) Suppression of hypertension during chronic reduction of brain acetylcholine in spontaneously hypertensive rats. J Hypertens 6:739–745 [DOI] [PubMed] [Google Scholar]
- Worek F, Mast U, Kiderlen D, Diepold C, Eyer P. (1999) Improved determination of acetylcholinesterase activity in human whole blood. Clin Chim Acta 288:73–90 [DOI] [PubMed] [Google Scholar]
- Zhang L, Rao F, Wessel J, Kennedy BP, Rana BK, Taupenot L, Lillie EO, Cockburn M, Schork NJ, Ziegler MG, et al. (2004) Functional allelic heterogeneity and pleiotropy of a repeat polymorphism in tyrosine hydroxylase: prediction of catecholamines and response to stress in twins. Physiol Genomics 19:277–291 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.