Abstract
Both the transcription of mRNAs from genes and their subsequent translation into proteins are inherently stochastic biochemical events, and this randomness can lead to substantial cell-to-cell variability in mRNA and protein numbers in otherwise identical cells. Recently, a number of studies have greatly enhanced our understanding of stochastic processes in gene expression by utilizing new methods capable of counting individual mRNAs and proteins in cells. In this review, we examine the insights that these studies have yielded in the field of stochastic gene expression. In particular, we discuss how these studies have played in understanding the properties of bursts in gene expression. We also compare the array of different methods that have arisen for single mRNA and protein detection, highlighting their relative strengths and weaknesses. In conclusion, we point out further areas where single-molecule techniques applied to gene expression may lead to new discoveries.
Keywords: molecular counting, single cell, single mRNA, single protein, random, mRNA, protein, cell-to-cell variability, gene expression noise, noisy gene expression
INTRODUCTION
Until relatively recently, scientists studying gene expression have measured the properties of gene expression on populations of cells rather than in individual cells, largely because of the technical challenges involved in making single-cell measurements. However, the advent of simple and accurate measurements of gene expression in individual cells has led researchers to find that the numbers of mRNAs and proteins can vary, sometimes dramatically, from cell to cell and that this variability is caused by the fundamentally stochastic nature of the biochemical events involved in gene expression.
Primary among these technical advances is the use of fluorescent proteins, such as GFP, whose importance in the field of stochastic gene expression is hard to overstate. Of course, even before fluorescent proteins were available, a few researchers still showed that gene expression was highly variable; such efforts include the pioneering work of Novick & Weiner (25), who used serial dilution and amplification of individual bacteria, and Ko et al. (19), who used single-cell enzymatic assays to show that levels of β-galactosidase expression varied significantly in individual mammalian cells. Yet, while these studies and others (35, 50) established the phenomenon, the ease with which GFP can be used to measure gene expression in individual cells led to an explosion in experimental work in stochastic gene expression that continues to this day, beginning with the seminal studies of Elowitz et al. (11) and Ozbudak et al. (26). These studies and the ones that have followed have shed light on many of the mechanisms that result in cell-to-cell variability in gene expression by using GFP and its variants in combination with time-lapse imaging, flow cytometry, and microscopy.
However, as researchers probe ever more deeply into the stochastic processes underlying gene expression variability, the limitations of GFP are becoming more and more apparent. One of the most serious limitations is sensitivity: When using conventional microscopy or flow cytometry, it is difficult to detect small numbers of fluorescent proteins. Given that stochastic effects are more prevalent at these low molecule numbers, sensitivity issues may make GFP an inappropriate choice of assay in some situations. Another problem is that GFP is typically measured in arbitrary fluorescence units rather than molecular units [with the notable exceptions of Rosenfeld et al. (34) and Gregor et al. (16)], thus limiting the ability to quantitatively evaluate increasingly sophisticated models of stochastic gene expression.
Ultimately, the ideal way to study stochastic gene expression would be to monitor the production, degradation, and functional states of individual biomolecules in real time in living cells. While such a goal may seem almost laughably unrealistic at first glance, the work highlighted in this review shows that researchers have made remarkable progress toward these seemingly unattainable ends. We begin by examining some recent work demonstrating the ability to count individual mRNAs within single cells, and then discuss developments in counting individual proteins. One of the key benefits of counting individual molecules is that it provides rigorous tests for stochastic models of gene expression, and we examine these connections, focusing in particular on the notion of bursts in transcription and translation, in which the production of mRNAs and proteins occurs in a pulsatile rather than continuous fashion. We conclude with some speculations about potential new areas in which single-molecule detection may drive the field of stochastic gene expression forward.
SINGLE-mRNA DETECTION
The detection of individual molecules of mRNA in single cells has the potential to dramatically enhance our understanding of transcription, not only in terms of its effects on cell-to-cell variability in gene expression but also in providing insights into the biochemical mechanisms involved. Using a variety of experimental methods, researchers have begun to understand some of these mechanisms, perhaps the most dramatic of which is transcriptional bursting.
Initially, stochastic models of gene expression assumed that mRNAs are produced and degraded according to the statistics of a Poisson process (42); that is, while the production and degradation happen at random, the probability of a transcript produced within any given time period is a constant that does not change in time (Figure 1). If one looks across a population of cells that are transcribing in this fashion, then one would expect to see a Poisson distribution of mRNA per cell:
where m is the number of mRNA molecules per cell and m̄ denotes the average mRNA number. The situation becomes more complex, however, when considering models in which mRNA production does not occur with a constant probability in time but rather occurs with much greater likelihood at some time periods than others (18, 28, 30). These transcriptionally active time periods are often referred to as transcriptional bursts. (In this review, we explicitly refer to bursts as being either transcriptional or translational to avoid confusion.)
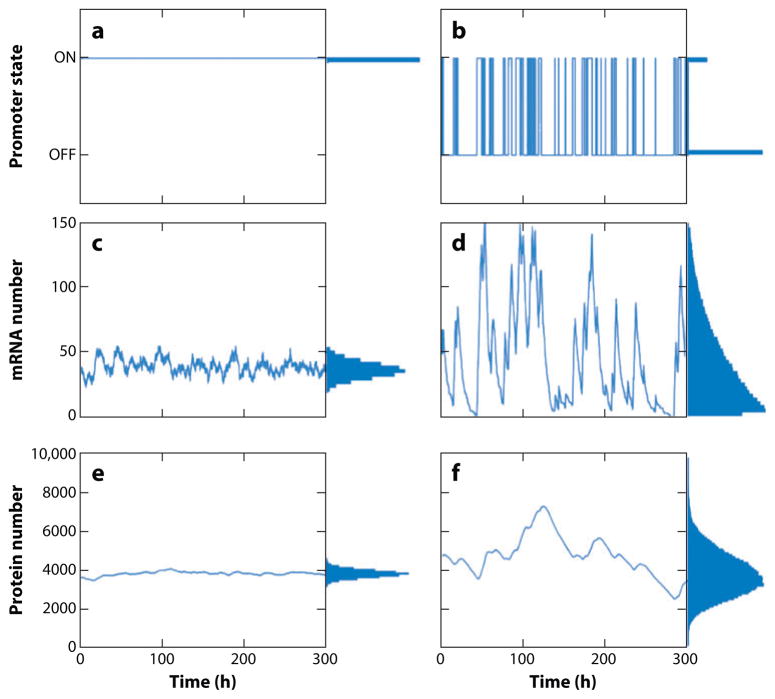
Figure 1.
(a) Promoter dynamics for a gene that is always in the active state (i.e., nonbursting) versus (b) promoter dynamics for a gene that switches between active and inactive states (i.e., bursty dynamics). (c) mRNA dynamics for nonbursting and (d) bursting genes. In the nonbursting case, one obtains a Poisson distribution of mRNAs per cell across the population, as shown in the marginal histogram, whereas the distribution of mRNAs per cell in the bursting case is much wider than a Poisson distribution despite having the same mean. Protein dynamics for (e) nonbursting and (f) bursting genes, again with the same mean. Although the underlying gene expression dynamics are bursty, the relatively long half-life of the protein results in a wide but Gaussian-looking population distribution, pointing out the need for single-molecule mRNA-counting approaches when studying bursty gene expression. The marginal histograms on the right of the time courses show the distribution of the promoter states, mRNAs, and proteins across a population.
One important consequence of transcriptional bursts is that they result in much higher variability in gene expression than the Poisson model predicts. However, experimentally distinguishing bursty transcription from nonbursty Poissonian transcription requires that measures of mRNA number per cell be made in molecular units. This requirement arises from the way in which variability scales as a function of mean mRNA number. For instance, in the Poisson model, as the mean increases, the relative variability about that mean should decrease, meaning that for large means, variability should be essentially negligible.
Transcriptional bursting, however, can lead to high variability even with high mean expression levels. In principle, it should thus be easy to tell the difference between these two situations, but the problem is that in the absence of molecular units, it is difficult to say whether an observation of high variability is the result of bursting or simply due to low levels of Poissonian transcription. Mathematically, one can encapsulate this argument through the use of the Fano factor, defined as the ratio of the variance to the mean. When measured in molecular units, the Fano factor for a Poisson distribution is exactly 1, whereas transcriptional bursts can result in Fano factors much larger than 1. (Some intuition can be gained from the fact that the Fano factor is approximately equal to the average number of transcripts produced during a burst, often referred to as the burst size.) However, when measured in arbitrary fluorescence units, the Fano factor contains an arbitrary scaling factor that makes such absolute numerical comparisons impossible (27), providing a strong rationale for counting the actual numbers of transcripts in individual cells.
It was against this theoretical backdrop that Golding et al. (15) began their beautiful study of the kinetics of transcription in Escherichia coli. Their main tool was the the MS2 mRNA detection technique developed simultaneously by Bloom and colleagues (4) and Singer and colleagues (6), which can be sensitive enough to visualize single mRNA molecules (14a). In the variant of the method used by Golding et al. (15), a gene is engineered to transcribe an mRNA containing 96 copies of a specific RNA hairpin in its untranslated region, each of which binds tightly to the coat protein of the bacteriophage MS2. This gene is then expressed in a cell that already expresses the MS2 coat protein fused to GFP. When 96 of the MS2-GFP proteins bind to an individual mRNA, enough fluorescent signal is generated that the individual mRNAs are detectable as diffraction-limited spots by conventional fluorescence microscopy. One can thus count the number of mRNAs in single cells by counting spots or, if the spots contain multiple mRNAs, by integrating the fluorescence in each spot. Upon performing this counting across an entire population of cells, they measured a Fano factor of roughly 4, which, being greater than 1, provided strong evidence for transcriptional bursting. Impressively, they went even further by measuring transcriptional activity in real time by using time-lapse microscopy. The authors found that transcription did indeed occur in bursts, with the gene itself switching randomly between transcriptionally active and inactive states. These switching events appeared to happen at exponentially distributed times, indicating that gene activation and inactivation were themselves Poisson processes, justifying the assumptions made in many models of transcriptional bursts (18, 28, 30). Moreover, using the temporal statistics of the switching events, they used a model of transcriptional bursts to predict the statistics of the population snapshots that they had measured experimentally, which showed a fairly good match between the two.
Another method by which one can count single molecules of mRNAs in individual cells is fluorescence in situ hybridization (FISH) (12, 31). In this method, samples are fixed and then a hybridization is performed using a set of fluorescently labeled oligonucleotides, each complementary to a unique portion of the target mRNA. As with the MS2 method, the presence of sufficiently large numbers of fluorophores bound to an individual mRNA renders the molecule sufficiently fluorescent to be detected by fluorescence microscopy. One recent application of this method to the study of stochastic gene expression in bacteria was to the phenomenon of competence in Bacillus subtilis (22). B. subtilis has the remarkable property of being naturally competent (i.e., it takes up foreign DNA from the environment). This property only manifests itself, though, at the beginning of stationary phase, and only a small percentage (~15%) of the total population actually becomes competent. Maamar et al. (22) showed that noise in the expression of the transcription factor primarily responsible for competence, comK, underlies the stochastic decision to become competent or not: Occasionally, a stochastic accumulation of ComK protein will become large enough to allow the ComK protein to bind to its own promoter, dramatically upregulating its expression and resulting in cell competence. One difficulty in studying the expression of comK in noncompetent cells, however, is that ComK is lowly expressed, making it impossible to measure gene expression from the comK promoter with fluorescent proteins. Instead, the authors used single-molecule FISH to count the numbers of comK mRNAs in individual bacteria. They found that comK was indeed expressed at a low level in noncompetent cells (less than 1 transcript per cell), and that this expression level was modulated over time, resulting in a concomitant modulation in frequency of transition to competence. The authors also measured some of the statistical properties of fluctuations in mRNA numbers throughout the population, finding that the Fano factor was relatively close to 1 for the mRNAs they measured, indicating that for this gene bursting is not likely to be a significant source of variability.
Owing to the increased complexity of eukaryotic transcription, one might expect cell-to-cell variability in eukaryotic transcription to have stochastic properties different from those in prokaryotic transcription. Also utilizing a single-molecule FISH assay, Raj et al. (30) found that transcription in mammalian cells was extremely bursty, with short, infrequent bursts resulting in large variations in mRNA numbers from cell to cell, leading to Fano factors of 40 or higher. Their assay also showed a single, intensely bright spot in some cells (but not others) resulting from mRNAs that had not yet diffused away from an active site of transcription. Moreover, cells with these active transcription sites exhibited a larger percentage of nuclear mRNA than those without active transcription sites. Together, these facts present a picture of transcription in which short transcriptional bursts cause the production of large quantities of mRNAs, which are exported from the nucleus to the cytoplasm, where they slowly decay, highlighting the benefits of measuring the spatial locations of single mRNAs in individual cells. Further, the authors used multicolor FISH to visualize simultaneously two different mRNA transcripts, showing that transcription from genes located far apart from each other on the genome were expressed in uncorrelated transcriptional bursts, whereas those located near each other were expressed in strongly correlated transcriptional bursts.
Another completely different approach to counting the number of particular mRNAs within single cells is the use of single-cell quantitative reverse transcriptase polymerase chain reaction (RT-PCR). Bengtsson et al. (5) used such a method to show that gene expression in individual cells isolated from mouse pancreatic islets is subject to large fluctuations. Their assay, which involves isolating individual cells and performing RT-PCR on each cell, can yield absolute measures of transcript numbers with appropriate controls and standardization. The authors found that most population histograms of the numbers of mRNA per cell were close to lognormal distributions, which are distributions that appear Gaussian when a histogram is made of the log of the mRNA number. Although such distributions appear Gaussian in logarithmic coordinates, they can exhibit long tails in nonlogarithmic coordinates, similar to those observed by Raj et al. (30) and Warren et al. (49).
Another advantage of their assay is the ability to detect simultaneously the levels of five different target genes through the use of multiplex PCR. In their assay, Bengtsson et al. found that two related genes, Ins1 and Ins2, showed highly correlated expression between cells, whereas the other pairs of genes exhibited no significant correlations. Such correlations may arise from a number of sources, including fluctuations in common upstream gene expression factors. Thus, the analysis of correlations has the potential to uncover previously hidden regulatory connections between genes. Traditionally, the way to check if the expression of two genes is related would be to use an external trigger (such as a signaling molecule or some environmental change) and check if the mean levels of the two genes change concurrently. However, this presupposes the existence of such an external trigger, which might be available for the genes in question. By looking for correlations in cell-to-cell variations between the transcript levels of two different mRNAs, one might effectively perform a coexpression analysis without requiring any such trigger.
One interesting extension of the single-cell RT-PCR technique is so-called digital RT-PCR (49), which is a variation on digital PCR (47). In this assay, cDNA obtained from reverse-transcribing mRNA from a single cell is partitioned into many (potentially thousands) of individual PCR reactions. The result of this massive dilution of the cDNA is that each PCR reaction will contain either 0 or 1 cDNA molecules as a template, and the presence or absence of a single cDNA is then detected by the PCR itself in digital fashion. To facilitate the large amount of liquid handling required, the reactions are typically performed with a microfluidic device that fractionates the reactions into appropriately sized volumes. By providing a digital readout of gene expression, one can sidestep the need for the many careful controls necessary for quantifying mRNA counts by conventional single-cell RT-PCR. Warren et al. (49) used digital RT-PCR to examine variability in the expression of the transcriptional factor PU.1, which plays a central role in the process of hematopoiesis, the process by which blood stem cells differentiate into different blood cell types. Cell fate decisions in this process are thought to have a significant stochastic component, thus motivating measurements of variability in the expression of PU.1.
The authors found that PU.1 does indeed display a large variability in all the different blood cell types examined, although the mean expression level was different in the various lineages. The authors also performed an experiment in which they presorted common myeloid progenitors [AU: Acronym not used at least 3 times in manuscript. OK] according to whether they displayed high or low levels of the cytokine receptor flk2 which has been correlated with differential functionality of common myeloid progenitors. They found that cells with high levels of flk2 displayed high expression of PU.1, whereas cells with low levels of flk2 showed low expression of PU.1. This discovery showed that variability in PU.1 expression is indeed correlated with functional distinctions between otherwise identical cells, a finding that has recently been extended by using microarrays (8).
From a methodological standpoint, each of the techniques used in these single-mRNA detection studies has various advantages and disadvantages (Table 1). For the MS2 technique, one major advantage is the ability to measure mRNA levels in real time—all the other methods except for molecular beacons (described below) require the use of fixed or lysed samples. Moreover, it yields spatial information on the locations of the individual mRNAs, which could prove invaluable in developmental studies in which positional information is critical (for an example of the use of MS2 in developmental systems, see Reference 13). The main problem, however, is that one must generate transgenes with large untranslated regions that may affect mRNA dynamics; for instance, Golding et al. (15) found that the incorporation of 96 protein-bound hairpins in the untranslated region of mRNAs rendered the mRNAs resistant to cellular nucleases. Also, the tendency of the MS2 coat protein to multimerize requires that one make a careful estimation of the total fluorescence within individual spots to determine the number of mRNAs contained therein (Table 2).
Table 1.
Comparison of different single mRNA detection methods
| Method | Endogenous mRNA detection? | Real-time measurements? | Detection of multiple mRNA species at once? | Other advantages | Other disadvantages | Reference (s) |
|---|---|---|---|---|---|---|
| MS2 | No | Yes | No | No need for external interventions (e.g., microinjection ), yields spatial information | mRNA tend to form clumps, requires long UTR sequence elements | (4 6, 15) |
| FISH | Yes | No | Up to 3 | Yields spatial information | Imaging can be difficult in small organisms | (12, 30, 31) |
| Single-cell RT-PCR | Yes | No | Up to 5 | Simple to perform with a large dynamic range | Requires careful standardizatio n, questions about efficiency and sensitivity at low numbers, no spatial information | (5) |
| Digital single-cell RT-PCR | Yes | No | Up to 2 | Easily interpretable signals, sensitive at low numbers of molecules | Requires microfluidics, questions about RT efficiency, no spatial information | (49) |
| Molecular beacons | No | Yes | Yes | No clumping of transcripts, yields spatial information | Requires microinjection or other invasive delivery methods, requires long UTR sequence elements |
Abbreviations: FISH, fluorescence in situ hybridization; RT, reverse transcriptase; RT-PCR, reverse transcriptase polymerase chain reaction; UTR, untranslated region
Table 2.
Comparison of different single protein detection methods
| Method | Endogenous protein detection? | Real-time measurements? | Detection of multiple proteins species at once? | Other advantages | Other disadvantages | Reference |
|---|---|---|---|---|---|---|
| β-galactosidase microfluidics | No | Yes | No | Works in a variety of cell types | Requires microfluidics, cells must be permeabilized, not as effective with large molecule numbers, no spatial information | (7) |
| Single fluorescent protein imaging | No | Yes | Potentially | Yields spatial information | Unlikely to work in present form in organisms larger than bacteria | (52) |
| Single-protein antibody labeling | Yes | No | Potentially | Simple to perform with a large dynamic range, works in many organisms | Requires microfluidics and complex optics, questions about antibody efficiency and sensitivity at low numbers, no spatial information | (17) |
Another method for the real-time detection of individual mRNAs is in vivo hybridization of target mRNAs with molecular beacons, which are single-stranded nucleic acid probes that only fluoresce upon hybridization to a target molecule (45, 46). The most comparable of the above methods is the MS2 technique. One advantage that molecular beacons possess is that they have no tendency to multimerize, thus simplifying the image analysis. One downside, however, is the delivery of the molecular beacons to the cell itself. The most commonly used methods are microinjection (46) and listeriolysin-O (33, 48), which may result in irregular doseages and decreased cell viability.
For FISH, the primary advantages in comparison to the MS2 method are the ability to detect endogenous transcripts, obviating the need for genetic manipulations that are often difficult to perform in many organisms, and the ability to detect simultaneously at least three separate transcripts (31). Meanwhile, FISH shares with MS2 the ability to provide spatial information. However, both FISH and MS2 also share the difficulty of counting transcripts when the mRNA density is high: If many mRNAs are in close spatial proximity (in bacteria, for instance), it is hard to distinguish individual fluorescent spots using conventional microscopy, although it is possible that the use of sophisticated subdiffraction-limit microscopy techniques can alleviate these problem (37, 38).
The RT-PCR-based methods are notable both for their potentially higher throughput and possibly simpler setup compared with FISH and MS2, and the data are less prone to subjective decisions in quantification than the fluorescence spot-finding algorithms required for FISH and MS2. Also, Bengtsson et al. (5) detected five different transcripts simultaneously within single cells, a feat difficult to perform with FISH. The two RT-PCR methods suffer, however, from uncertainties about the efficiency of the reverse transcriptase enzyme itself. Upon comparison, the single-cell RT-PCR experiments of Bengtsson et al. (5) are simpler to perform than the digital RT-PCR experiments of Warren et al. (49), which require the use of microfluidic devices to manage the large number of individual reactions. However, Bentgsson et al. (5) also note that their method is unable to detect transcripts at numbers below 10–20 copies per cell, whereas Warren et al. (49) counted mRNAs in individual cells at arbitrarily low copy numbers.
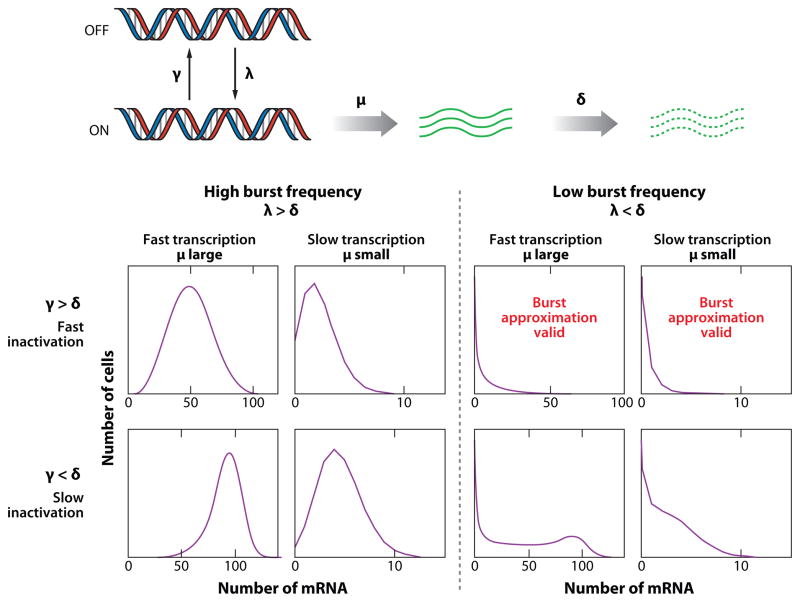
The studies described above have also contributed greatly to evaluating models of burst-like stochastic gene expression. The most common model was that first analyzed by Peccoud & Ycart (28) in which the gene itself transitions randomly between transcriptionally active and inactive states (Figure 2). Such a model contains four parameters: λ, the rate at which the gene transitions from the inactive to the active state; γ, the rate at which the gene transitions from the active to the inactive state; μ, the rate of transcription when the gene is in the active state; and δ, the rate of mRNA degradation. Peccoud & Ycart solved this model for the moments of steady-state distribution (28), which was extended to a complete analytic expression for the distribution by Raj et al. (30). This distribution can then be used to extract parameters from mRNA-counting experiments, potentially revealing new information about what parameters are subjected to regulation. For instance, Raj et al. (30) used this model to show that modulating the amount of transcription factor resulted in a modulation of the average burst size (μ/γ) while leaving the burst frequency fixed; more generally, it is possible for transcriptional regulation to occur through a change of any one (or combination) of the parameters μ, λ, and γ.
Figure 2.
Distributions resulting from different values of the parameters in the gene activation/inactivation model of Peccoud & Ycart (28). The top row corresponds to the parameter γ being larger than the mRNA decay rate δ. The left side of the figure corresponds to high burst frequency compared with δ, whereas the right side corresponds to low burst frequency. The transcription rate γ was also altered as indicated. As mentioned in the text, the burst approximation is only valid when the burst frequency is low and the inactivation rate is faster than the mRNA decay rate. In particular, the bimodal expression pattern that appears with high μ and small λ, and γ cannot appear when one uses the burst approximation.
One important parameter regime of this model is that of instantaneous bursts, which occur when the rate of gene inactivation γ is larger than both the rate of mRNA degradation δ and the rate of gene activation λ. Intuitively, the former condition allows one to effectively ignore mRNA degradation during the burst itself and the latter condition ensures that individual activation events are infrequent enough that their appearance is a Poisson process, thus allowing one to make the approximation that all the mRNAs are synthesized at the same time. The number of parameters is thus reduced by 1: The model now consists only of λ, which can be interpreted as the burst frequency, and μ/γ, which is the average burst size, with δ unchanged. The steady-state distribution of this reduced model can also be solved approximately (14, 30), and this model appears to apply well for certain situations (30). There are situations in which this model cannot apply, though, the most notable being cases of bimodal mRNA distributions, which are the result of long transcriptional bursts during which the mRNA level approaches a new steady state.
Implicit in this model is the assumption that the gene activation and inactivation events are random Poisson processes, in which case the time between events would be exponentially distributed. Recent theoretical work by Pedraza & Paulsson (29) showed that one would obtain similar (i.e., experimentally indistinguishable) distributions even if the times between the gene activation and inactivation events were not close to Poisson. This raises the possibility that parameters extracted from steady-state measurements do not correspond to anything physical, an option that cannot be excluded using snapshot data such as those obtained from fixed or lysed cells. However, real-time imaging of transcription in living cells has shown that, at least for certain genes in Escherichia coli (15) and Dictyostelium discoideum (9), the distributions of these events are indeed exponential. Nevertheless, owing to the complexities of transcriptional regulation in higher eukaryotes, researchers will have to obtain real-time observations of transcription in those organisms to specify exactly what sorts of models are applicable.
Yet while there is now a growing body of evidence supporting transcriptional bursts, their biological origins remain unclear. In prokaryotes, the results of Golding et al. (15) convincingly show that transcriptional bursts do indeed occur in E. coli, and the authors proposed a host of possible causes, including simple mechanisms such as transcription factor binding and unbinding as well as more complex processes such as DNA conformational changes and sigma factor retention resulting in pulsatile reinitiation of transcription. Indeed, the very presence of prokaryotic transcriptional bursts themselves may be gene specific, since Maamar et al. (22) found that the Fano factor for the mRNA distributions they measured were close to 1, thus arguing against transcriptional bursts in that particular case. Only further experimentation can provide answers to these questions.
Meanwhile, in eukaryotes in general and higher eukaryotes in particular, it seems as though transcriptional bursting is most certainly the norm, with most if not all noise studies in the field providing some evidence for pulsatile transcription. One early candidate for the cause of transcriptional bursts was chromatin remodeling. Eukaryotic genes are wrapped around histone proteins that form chromatin fibers, and chromatin can be remodeled from a tightly bound, transcriptionally inert structure to a more loosely bound, transcriptionally active conformation through the action of various chromatin-remodeling enzymes. Thus, random events of chromatin remodeling could result in random bursts of transcription. Yet, despite the clarity of this hypothesis, it has yet to be decisively proven or disproven; so far, the only studies providing any hints are those of Raser and O’Shea (32), in which the alteration of chromatin-remodeling enzymes resulted in changes in stochastic gene expression, and Raj et al. (30), in which genomic position (and thus chromatin context) appeared to have a strong effect on covariation in bursting between multiple genes. A conclusive test of the connection between chromatin remodeling and transcriptional bursting will also require single-molecule techniques, this time directed at the gene itself. Given that experiments monitoring chromatin remodeling in real time have already been carried out (44), a suitable combination of these different single-molecule techniques will likely settle the question.
Another consideration is the propagation of fluctuations in mRNA levels to those of proteins. Although no study has yet combined single-molecule mRNA detection with single-molecule protein detection, two of the single-molecule mRNA studies highlighted here have used conventional fluorescent proteins to examine these problems. Golding et al. (15) found that the mRNA and protein levels exhibited a linear correlation in single cells. They also found that the correlation was weakest in the time just following cell division, which they ascribed to the randomizing effects of the binomial partitioning of mRNAs and proteins upon cell division. Raj et al. (30) tried to examine the relationship between protein degradation rates and the correlation between mRNA and protein levels. They found that mRNA and protein levels correlated strongly when protein lifetime was short, but that this correlation decreased when protein lifetime was long, a finding also born out in models of stochastic protein and mRNA production. The authors found it generally difficult, however, to detect small numbers of protein molecules in individual eukaryotic cells owing to their large cellular volumes, making the development of single-molecule techniques to count the number of proteins in individual cells important. We outline some recent efforts toward this goal in the next section.
SINGLE-PROTEIN DETECTION
Ultimately, much of cellular function is carried out by the proteins encoded for by the mRNAs, and hence the enumeration of individual proteins is essential to a complete understanding of stochastic effects in gene expression. Unfortunately, achieving the required probe specificity is far more difficult with proteins than with nucleic acids. Nevertheless, new techniques are emerging that are giving researchers a glimpse into the protein content of individual cells (Table 2).
Recently, two exciting studies from the laboratory of X. Sunney Xie have detailed their efforts to detect individual protein molecules in living cells. Although both studies reaching strikingly similar biological conclusions, their approaches were rather different. In Cai et al. (7), the authors combined the high efficiency of the β-galacotosidase enzyme with microfluidics to count protein numbers by measuring enzymatic activity. β-galactosidase is efficient at cleaving substrates, and several reagents produce easily detectable substances upon enzymatic activity. However, such product molecules are usually quickly exported from the cell itself, thus diffusing the signal greatly, which is why such assays are typically performed on populations rather than single cells. To circumvent this problem, the authors confined each cell to a defined small volume using a microfluidic device, an approach based on previously described methods used to detect the activity of single enzymes (36). The concentration of the fluorescent product from a single enzyme is made high enough so that the fluorescent signal is easily detectable. Because the nonfluorescent substrate is present in saturating quantities, the increase in signal is linear, with the slope directly proportional to the number of β-galacotosidase enzymes present. Thus, by measuring changes in the signal slope, the authors detected the formation of single enzymes.
Yu et al. (52), used fluorescent proteins in a manner that allowed for the detection of single-protein molecules. The authors noted that individual fluorescent proteins generate enough fluorescence for detection given a long enough exposure time, but the problem is that they diffuse too rapidly to produce a localized signal within such time periods. To solve this problem, they fused a bright, fast-folding variant of yellow fluorescent protein (Venus YFP) to a peptide sequence that anchors itself to the membrane, thus dramatically reducing the mobility of the YFP molecules. Once anchored in this fashion, they directly imaged the molecules using a standard fluorescence microscope coupled with laser illumination.
Impressively, although these methods are different in character, the results obtained from both studies were almost identical. The main finding was that proteins were produced in short but infrequent bursts, presumably occurring during the lifetime of single, infrequently transcribed mRNAs. They parameterized their data using a model in with two parameters: a, referring to the burst frequency; and b, referring to the average burst size. (This model is in principle similar to models used for describing mRNA distributions arising from short transcriptional bursts.)The main assumption is that individual burst events are short compared with the protein lifetime, which, mathematically speaking, means that as the mRNA lifetime is much shorter than the protein lifetime, likely a valid approximation for a significant number of genes. During the lifetime of the mRNA, the number of proteins produced is taken from a geometric distribution, which has a nice biological interpretation: Once the mRNA is transcribed, it will be continuously translated into protein by ribosomes. The presence of the ribosomes also confers protection from various ribonucleases. However, every time a ribosome finishes translating and thus unbinds from the mRNA, there is a certain probability that a ribonuclease will bind rather than another ribosome. This process leads to the geometric distribution of burst sizes of mean size b (23), and when one combines this burst size distribution with the random appearances of bursts (parameterized by a), Cai et al. (7) found that the distribution of proteins across a population is given by the γ distribution. This model has been applied to small gene networks such as genetic autoregulation and transcriptional cascades by Friedman et al. (14), and a recent study has shown how to extend this work to find distributions in the presence of transcriptional bursts (40). In terms of the underlying rates of transcription, translation, and mRNA and protein degradation, parameter a is the rate of transcription and parameter b is the ratio of the translation rate to the mRNA degradation rate (42).
Yet, although these two techniques are undeniably elegant, it is unclear how well they will translate to other types of organisms in which the protein count is much higher and the cellular volume is much larger. Moreover, the use of various reporter gene constructs presupposes the ability to perform genetic manipulations, which are often difficult or impossible to perform in many organisms. To circumvent these problems, Huang et al. (17) used a combination of microfluidics, immunofluorescent labeling, and new optics to count the number of endogenous protein molecules in organisms both large and small. Their approach was to lyse cells in small microfluidic chambers and then use fluorescently conjugated antibodies to label the target protein. They then flowed the now fluorescent proteins over a confocal microscope to image the individual proteins. The imaging step is one of the principal difficulties in this type of method, since the field of illumination is usually much smaller than the channels through which the proteins flow, thus making it hard to detect all the proteins as they pass by the objective. The authors solved this problem by utilizing cylindrical optics, thereby illuminating the entire cross section of the protein channel.
They then used their system to measure the number of β-adrenergic receptors in individual insect cells and found that the numbers of proteins fluctuated wildly from cell to cell, with numbers as low as 2000 and as high as 60,000. They also measured the numbers of the constituents of the phycobilisome (i.e., the complex responsible for harvesting light energy from the sun) in individual cyanobacteria. Huang et al. found that expression of this complex was much more variable in nitrogen-starved conditions than in nitrogen-rich conditions. Some caveats to this method include the limits of its sensitivity (which the authors estimated to be around seven molecules in their cyanobacteria experiments) and its throughput, but it is nevertheless a promising and general methodology for measuring cell-to-cell variability in endogenous protein levels.
FUTURE DIRECTIONS
The application of single-molecule techniques to the measurement of gene expression in single cells has provided many new insights into the field of stochastic gene expression, and the utilization of these and other methods not yet invented for progressively more complex biological problems will undoubtedly lead to further discoveries. Such research may move toward studying all the individual stochastic biochemical reactions involved in gene expression, rather than just counting and monitoring mRNA and protein numbers. Some work has already been done along these lines, with the study of Elf et al. (10) examining the kinetics of individual transcription factors searching for their DNA binding site in living E. coli; further work may reveal the contribution of these random binding and unbinding events to stochastic expression of the target genes. In higher eukaryotes, the notion that chromatin remodeling is responsible for transcriptional bursting has still not been proven directly and remains a ripe target for combining single-molecule DNA measurements with transcriptional measurements. More generally, virtually all the enzymatic activities involved in gene expression can be subjected to single molecule scrutiny to determine exactly which individual processes are the most important in making gene expression stochastic. Candidates include the stepping behavior of individual RNA polymerases; splicing and other posttranscriptional mRNA processing; the nuclear export of mRNAs, including gene translocation to the nuclear periphery (39); the activity(ies) of ribosomes, ribonucleases, and proteases. These are but a smattering of the many important elements involved in gene expression, and studying how these individual molecules function in vivo will almost certainly change our conception of stochastic gene expression.
Another avenue of inquiry in which single-molecule techniques may provide fresh insights is the biological consequences of noise. Thus far, the field has focused primarily on cases in which noise in gene expression can be beneficial, providing useful phenotypic variability in genetically identical populations (2, 20, 21, 43, 51). Often this variability is enhanced by thresholding and amplification of noise by genetic feedback loops (1, 22, 41). Maamar et al. (22) used single-mRNA detection to try to infer thresholding behavior at the protein level, but it is likely that direct observation of individual protein molecules in real time will be required to truly observe the actions of such feedbacks. Less well studied (but perhaps more important in general) are instances in which noise is detrimental to robust function, an example of which is development in multicellular organisms. In such cases single-molecule detection may be of primary importance in detecting low numbers of important biomolecules during developmental processes (31), thus allowing researchers to gauge the extent to which noise is tolerated in such systems.
Biological insights may also arise from parallelization of single-molecule gene expression measurements to a genomic scale, i.e., measuring the detailed stochastic properties of gene expression (such as mRNA and protein burst frequency and size) for most genes in an organism. These sorts of measurements can lead to insights into the nature and consequences of noise, as demonstrated by using GFP in yeast (3, 24). Single-molecule measurements allow for the detection of many potentially interesting genes whose expression levels are below the GFP detection limit, and would allow for more careful measurements of important parameters that can only be inferred by GFP measurements. It remains to be seen how easily such methods can be parallelized to facilitate such studies, but the capacity for new insights is great.
In conclusion, we feel that these trailblazing single-molecule stochastic gene expression experiments are pointing in the direction to which the rest of the field will head. The ability of these methods to yield quantitative data raises several exciting possibilities that seemed impossible only a few years ago. We look forward to expecting the unexpected as the combination of single-molecule detection and molecular biology breathe new life into the still-young field of stochastic gene expression.
Acknowledgments
We would like to thank Ido Golding for many helpful comments on the manuscript. We also apologize to any authors whose work we were unable to mention due to space constraints. A.v.O was supported by NSF grant PHY-0548484 and NIH grants R01-GM068957 and R01-GM077183. A.R. was supported by NSF Fellowship DMS-0603392 and a Burroughs Wellcome Fund Career Award at the Scientific Interface.
ACRYONYMS
- Fano factor
mathematically defined as the variance of a distribution divided by the mean
- FISH
fluorescence in situ hybridization
- Molecular beacon
a single-stranded hairpin-shaped nucleic acid probe with a fluorophore and a quencher that fluoresces upon hybridization to a single-stranded target nucleic acid
- MS2
a bacteriophage whose coat protein binds strongly with a particular RNA hairpin
- RT-PCR
reverse transcriptase polymerase chain reaction
- Steady-state distribution
the distribution of mRNA per cell across a population that is equilibrated in the sense that the distribution will not change over time
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–32. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- 2.Acar M, Mettetal JT, van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nat Genet. 2008;40:471–75. doi: 10.1038/ng.110. [DOI] [PubMed] [Google Scholar]
- 3.Bar-Even A, Paulsson J, Maheshri N, Carmi M, O’Shea E, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–43. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 4.Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol. 1999;9:569–78. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- 5.Bengtsson M, Stahlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15:1388–92. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–45. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 7.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–62. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 8.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–47. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16:1018–25. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–94. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–86. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 12.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–90. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 13.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13:1159–68. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 14.Friedman N, Cai L, Xie XS. Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys Rev Lett. 2006;97:168302. doi: 10.1103/PhysRevLett.97.168302. [DOI] [PubMed] [Google Scholar]
- 14a.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living cells. Curr Biol. 2003;13:161–67. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–36. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 16.Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007;130:153–64. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang B, Wu H, Bhaya D, Grossman A, Granier S, et al. Counting low-copy number proteins in a single cell. Science. 2007;315:81–84. doi: 10.1126/science.1133992. [DOI] [PubMed] [Google Scholar]
- 18.Kepler TB, Elston TC. Stochasticity in transcriptional regulation: origins, consequences, and mathematical representations. Biophys J. 2001;81:3116–36. doi: 10.1016/S0006-3495(01)75949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko MS, Nakauchi H, Takahashi N. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J. 1990;9:2835–42. doi: 10.1002/j.1460-2075.1990.tb07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–78. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 21.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–29. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci USA. 1997;94:814–19. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–46. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 25.Novick A, Weiner M. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci USA. 1957;43:553–66. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 27.Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–18. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- 28.Peccoud J, Ycart B. Markovian modelling of gene product synthesis. Theor Popul Biol. 1995;48:222–34. [Google Scholar]
- 29.Pedraza JM, Paulsson J. Effects of molecular memory and bursting on fluctuations in gene expression. Science. 2008;319:339–43. doi: 10.1126/science.1144331. [DOI] [PubMed] [Google Scholar]
- 30.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raj A, Van Den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–79. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–14. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhee WJ, Santangelo PJ, Jo H, Bao G. Target accessibility and signal specificity in live-cell detection of BMP-4 mRNA using molecular beacons. Nucleic Acids Res. 2008;36:e30. doi: 10.1093/nar/gkn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–65. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 35.Ross IL, Browne CM, Hume DA. Transcription of individual genes in eukaryotic cells occurs randomly and infrequently. Immunol Cell Biol. 1994;72:177–85. doi: 10.1038/icb.1994.26. [DOI] [PubMed] [Google Scholar]
- 36.Rotman B. Measurement of activity of single molecules of beta-D-galactosidase. Proc Natl Acad Sci USA. 1961;47:1981–91. doi: 10.1073/pnas.47.12.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–95. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–6. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat Struct Mol Biol. 2007;14:1049–55. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 40.Shahrezaei V, Swain PS. Analytical distributions for stochastic gene expression. Proc Natl Acad Sci USA. 2008;105:17256–61. doi: 10.1073/pnas.0803850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–19. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 42.Thattai M, van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA. 2001;98:8614–19. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thattai M, van Oudenaarden A. Stochastic gene expression in fluctuating environments. Genetics. 2004;167:523–30. doi: 10.1534/genetics.167.1.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol. 1999;145:1341–54. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–08. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 46.Vargas DY, Raj A, Marras SA, Kramer FR, Tyagi S. Mechanism of mRNA transport in the nucleus. Proc Natl Acad Sci USA. 2005;102:17008–13. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA. 1999;96:9236–41. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W, Cui ZQ, Han H, Zhang ZP, Wei HP, et al. Imaging and characterizing influenza A virus mRNA transport in living cells. Nucleic Acids Res. 2008;36:4913–28. doi: 10.1093/nar/gkn475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci USA. 2006;103:17807–12. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White MR, Masuko M, Amet L, Elliott G, Braddock M, et al. Real-time analysis of the transcriptional regulation of HIV and hCMV promoters in single mammalian cells. J Cell Sci. 1995;108(Pt. 2):441–55. doi: 10.1242/jcs.108.2.441. [DOI] [PubMed] [Google Scholar]
- 51.Wolf DM, Vazirani VV, Arkin AP. Diversity in times of adversity: probabilistic strategies in microbial survival games. J Theor Biol. 2005;234:227–53. doi: 10.1016/j.jtbi.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 52.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–03. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]