Abstract
Neurofibromatosis type 1 (NF1) is a prevalent genetic disorder that affects growth properties of neural-crest-derived cell populations. In addition, approximately one-half of NF1 patients exhibit learning disabilities. To characterize NF1 function both in vitro and in vivo, we circumvent the embryonic lethality of NF1 null mouse embryos by generating a conditional mutation in the NF1 gene using Cre/loxP technology. Introduction of a Synapsin I promoter driven Cre transgenic mouse strain into the conditional NF1 background has ablated NF1 function in most differentiated neuronal populations. These mice have abnormal development of the cerebral cortex, which suggests that NF1 has an indispensable role in this aspect of CNS development. Furthermore, although they are tumor free, these mice display extensive astrogliosis in the absence of conspicuous neurodegeneration or microgliosis. These results indicate that NF1-deficient neurons are capable of inducing reactive astrogliosis via a non-cell autonomous mechanism.
Keywords: Neurofibromatosis, tumor suppressor, animal models, knockouts, astrogliosis
Von Recklinghausen's neurofibromatosis type 1 (NF1) is a common inherited neurological disease that affects 1 in 3500 individuals world wide (Riccardi 1999; Zhu and Parada 2001). Two major clinical features of NF1 are confined to the embryonic neural crest compartment and include formation of benign tumors along peripheral and optic nerves (neurofibromas and gliomas) as well as abnormal distribution of melanocytes (cafe-au-lait spots). NF1 patients also have increased risk of developing malignant tumors of neural origin (Gutmann et al. 1997; Riccardi 1999).
The human NF1 gene (Cawthon et al. 1990; Viskochil et al. 1990; Wallace et al. 1990) gene spans about 335 kb and contains 60 exons (Li et al. 1995; Viskochil 1999). The NF1 mRNA is ∼11–13 kb in length and encodes a 2818-amino-acid protein product that is designated neurofibromin (Buchberg et al. 1990; DeClue et al. 1991; Gutmann et al. 1991). Neurofibromin contains a functional GTPase-activating protein (GAP)-related domain (GRD; Ballester et al. 1990; Buchberg et al. 1990; Martin et al. 1990; Xu et al. 1990) that shares homology with the GAP family members, such as p120rasGAP in vertebrates (Trahey and McCormick 1987; Trahey et al. 1988; Vogel et al. 1988; Baba et al. 1995), ira1 and ira2 in yeast (Tanaka et al. 1990), and GAP1 in Drosophila (Gaul et al. 1992). Thus, neurofibromin GRD is both a structural and functional homolog of p120rasGAP.
Ras belongs to a superfamily of G proteins that cycle between active GTP-bound and inactive GDP-bound forms (Boguski and McCormick 1993). GAP proteins terminate ras-mediated signaling by stimulating conversion of the active GTP-bound form to the inactive GDP-bound form. Thus, the tumor-suppressor activity of neurofibromin is thought to be mediated via negative regulation of ras, which affects proliferation and differentiation in a variety of cell types (Bourne et al. 1990). Ras-activating mutations have been found in many human cancers (Bos 1989). Consistent with this, decrease or loss of neurofibromin activity is correlated with elevated ras-GTP levels in neurofibrosarcoma-derived cell lines (Basu et al. 1992; DeClue et al. 1992) and myeloid leukemia cells (Bollag et al. 1996). In addition, neurofibromin can negatively regulate p21 ras independent of its GAP activity in some contexts (Johnson et al. 1994).
Genotype analysis confirmed that NF1 acts as a classic tumor suppressor because the majority of mutations identified in NF1 patients are inactivating (Upadhyaya et al. 1994). Loss of heterozygosity (LOH) at the NF1 locus has been identified in pheochromocytomas (Xu et al. 1992), neurofibrosarcomas (Legius et al. 1993), and myeloid leukemias (Side et al. 1997). However, only a small percentage of benign neurofibromas, the hallmark feature of NF1, exhibit NF1 LOH (Colman et al. 1995; Sawada et al. 1996). This may in part be caused by the heterogeneous nature of neurofibromas, which are composed of Schwann cells, neurons, fibroblasts, and perineurial cells (Peltonen et al. 1988). Whether these heterogeneous tumors arise from NF1 LOH at one or all cell types remains to be determined.
NF1 homozygous (NF1−/−) mice die in utero because of abnormal cardiac development (Brannan et al. 1994; Jacks et al. 1994). NF1 heterozygous (NF1+/−) mice are viable and fail to develop features of human NF1, such as neurofibromas and cafe-au-lait spots, but do develop specific malignancies, including pheochromocytomas and myeloid leukemias in later life (Jacks et al. 1994). Thus, like many tumor suppressor knockout mice (Jacks 1996; Ghebranious and Donehower 1998; McClatchey and Jacks 1998), NF1+/− mice have failed to replicate the human condition with fidelity.
There are several nontumor syndromes that are associated with NF1 disease. For example, 35%–65% of NF1 children display specific learning disabilities (North 1999). Consistent with studies in human NF1, NF1+/− mice display spatial learning disability, which can be overcome by extended training (Silva et al. 1997). Guo et al. (2000) reported that Drosophila NF1 mutants have learning disabilities as well. The mechanisms underlying learning disability in both human and mouse NF1 brains remain unknown. However, these observations suggest that NF1 may play an important role in neuronal development and physiology.
To study the physiological role of NF1 in neural development and to generate a better model for human NF1, we have used the Cre/loxP system (Gu et al. 1994) to generate a conditional allele (NF1flox) at the NF1 locus. In this study, we describe the effectiveness of the NF1 conditional allele for studying the function of neurofibromin in differentiated neurons. The data support the model that loss of NF1 function in neurons can have non-cell autonomous consequences in vivo. This NF1 mouse model provides a valuable tool to study neuronal and glial phenotypes associated with human NF1 disease.
Results
Generation of NF1 conditional mutant mice by Cre/loxP
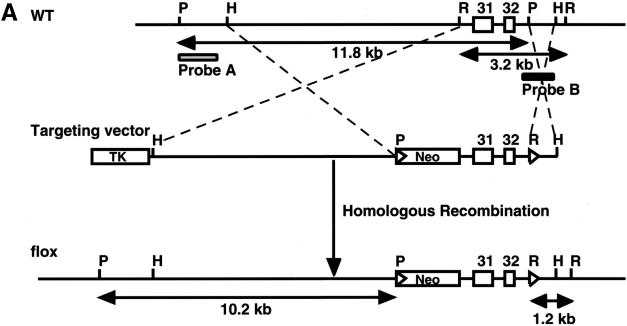
Exon 31 is essential for NF1 gene function in both humans and mice (Brannan et al. 1994). Therefore, we designed a gene-targeting strategy that flanked exons 31 and 32 with loxP sites, such that both exons can be deleted by Cre-mediated recombination (Fig. 1A). The targeting vectors were transfected into embryonic stem (ES) cells and 2 of 415 clones contained the targeted alleles as determined by Southern blot analysis (Fig. 1B). The homologous recombinant clones were injected into C57BL/6 blastocysts and subsequently transmitted into the germ line. Homozygous NF1flox/flox and compound heterozygous NF1flox/− mice were generated through appropriate crosses, and both strains are viable and fertile (Fig. 1C,D). To further characterize the NF1flox allele, we monitored a cohort of 35 NF1flox/flox and 29 NF1flox/− mice over a period of 14 mo. Unlike NF1flox/flox mice that exhibited a typical wild-type survival profile, NF1flox/− mice began to die at 10 mo of age, and only 80% survived beyond 14 mo (Fig. 1E). Thus, the NF1flox/− survival profile is indistinguishable from that of NF1+/− mice (Jacks et al. 1994). Based on this survival result, the NF1flox allele behaves like a wild-type allele despite the presence of two loxP sites and a neo cassette in the introns flanking exons 31 and 32.
Figure 1.
Generation of NF1 flox mice. (A) Schematic of the wild-type (WT) NF1 allele, targeting vector, and the flox allele. A loxP site with the pGKneo cassette was inserted into intron 30, and the second loxP site was introduced into intron 32. In this manner, exons 31 and 32 of the NF1 gene are flanked by two loxP sites. A 5′ external probe (probe A) and a 3′ internal probe (probe B) for Southern analysis are shown. P, PstI; H, HindIII; R, EcoRI. (B) (Left panel) Southern analysis with 5′ probe (probe A) shows a mutant (left lane) and a WT (right lane) ES clones. The mutant allele produces a 10.2-kb fragment; the WT allele produces an 11.8-kb fragment. (Right panel) Genomic DNA from the same ES clones hybridized with 3′ probe (probe B), confirming that the mutant ES clone has undergone homologous recombination in the NF1 locus. The mutant allele produces a 1.2-kb fragment; the WT allele produces a 3.2-kb fragment. (C) Southern analysis with the probes (as described above) on the littermates of NF1flox/+ intercross. (D) Southern analysis with 3′ probe shows different alleles of the NF1 gene: WT allele (3.2 kb), knockout (KO) allele (2.6 kb), and flox allele (1.2 kb). These mice were subjected to survival study. (E) Survival curve of NF1flox/flox and flox/KO mice. It shows the normal survival profile of NF1 flox allele. (F) A representative E13.5 NF1KO/Δ and WT littermate.
To determine whether Cre-mediated recombination in the NF1flox allele results in a null allele, we crossed NF1flox/flox mice with a transgenic strain that expresses Cre recombinase in the mouse germ line (Voiculescu et al. 2000). The resultant progeny harbor a recombined allele (NF1Δ) and a wild-type allele, NF1Δ/+. These mice were bred into the NF1 null background to generate compound heterozygous NF1Δ/− progeny. NF1Δ/− embryos are indistinguishable from NF1−/− embryos, which are characterized by an enlarged heart, peripheral edema, hemorrhage, small unpigmented eyes, and which die at E13.5 because of cardiac abnormalities (Fig. 1F; Brannan et al. 1994; Jacks et al. 1994). These results indicate that the NF1flox allele is susceptible to Cre-mediated recombination in vivo and that the recombined NF1flox allele (NF1Δ) is a phenocopy of the null allele. We conclude that the NF1flox allele represents a functional conditional allele.
NF1 mutant dorsal root ganglion neurons gain neurotrophin independence because of direct loss of NF1 function
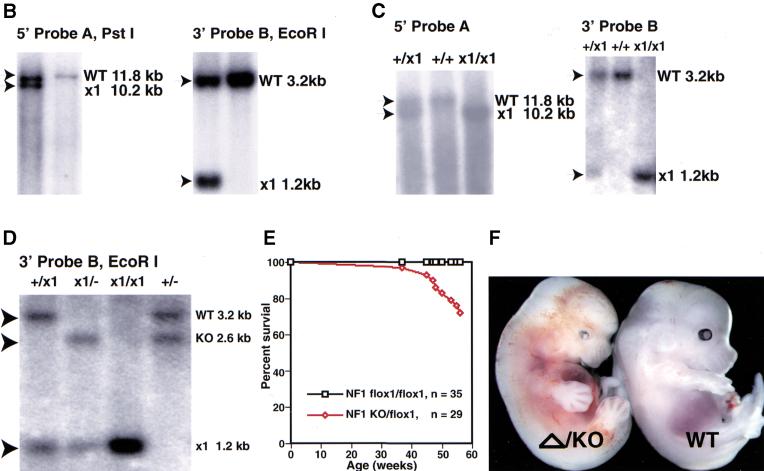
We showed previously that, unlike wild-type counterparts, cultured embryonic NF1−/− sensory and sympathetic neurons survive without added neurotrophins (Vogel et al. 1995). These data supported the model that loss of NF1 function results in constitutive activation of the ras pathway downstream from Trk receptors, even in the absence of neurotrophins. However, those studies could not exclude the possibility that loss of NF1 function developmentally altered neuronal precursors, enabling them to undergo differentiation without requiring neurotrophin signaling. To examine whether the neurotrophin independence that is acquired by NF1−/− neurons is direct, we repeated the neurotrophin survival experiments using dorsal root ganglion (DRG) neurons cultured from NF1flox/flox or NF1flox/− embryos. NF1flox/− cultured embryonic DRG neurons were infected with a cytomegalovirus (CMV) promoter-driven, Cre-expressing recombinant adenovirus at varying multiplicities of infection (MOI). Cre-mediated recombination was monitored by Southern blot analysis (Fig. 2A,B). MOI >20 was sufficient to mediate recombination of all the NF1flox alleles in cultured DRG neurons (Fig. 2B). Cre-adenovirus infected cultures (MOI = 50) of E13.5 NF1flox/flox or NF1flox/− DRG neurons were monitored for survival in the absence of neurotrophins. After 3 d of infection, NGF was removed by medium dilution and addition of neutralizing anti-NGF antibody. Only the cultures that were infected with Cre-adenovirus (resulting in NF1 deficiency) persisted in the absence of NGF (Fig. 2C). Uninfected cultures or cultures that were infected with a CMV LacZ-expressing adenovirus underwent classical, neurotrophin-deprivation cell death (Vogel et al. 1995). These data provide direct evidence that neurotrophin independence resulted from functional loss of NF1 in the differentiated neuronal cultures and further validate the functionality of the conditional NF1flox allele in primary cultures.
Figure 2.
NF1−/− neurons generated by Cre-mediated recombination survive in the absence of neurotrophins. (A) Schematic drawing of the NF1flox allele and the recombined allele resulted from Cre-mediated recombination. Southern analysis with 3′ probe (probe B) produces an 11.6-kb fragment from the flox allele and knockout (KO) allele, and a 7.2-kb fragment from the recombined (Δ) allele. Thus, the flox allele and recombined flox allele (Δ) can be distinguished by Southern analysis. Positions of primers (P1, P2, P3, and P4) are illustrated. The combination of P1, P3, and P4 primers is used to genotype NF1flox mice; the pair of P1 and P2 primers (R-Cre PCR) is used to detect Cre-mediated recombination, P1 and P4 primers are used to detect the presence of the flox allele (flox-PCR; see Materials and Methods). H, HindIII. (B) Southern analysis of NF1flox/KO dorsal root ganglion (DRG) after exposure to Cre-adenovirus. Genomic DNA from NF1flox/KO DRG cultures infected with Cre-adenovirus were hybridized with probe B. The multiplicity of infection (MOI) of each culture is illustrated. Of note, in this restriction analysis (HindIII), the KO allele produces a similar fragment (11.6 kb) as the flox allele. DRG cultures with MOI > 20 underwent complete deletion: The 11.6 kb band represents the KO allele; the 7.2-kb band represents the deleted (Δ) allele after Cre-mediated recombination. (C) Neurotrophin survival assay on E13.5 DRG cultures from NF1 flox/KO or flox/flox E13.5 embryos. Cultures were either mock infected or subjected to LacZ-, and Cre-adenovirus for 3 d in the presence of NGF (10 ng/mL). NGF was then removed by dilution of medium and addition of anti-NGF antibody. Cell counts were performed every 24 h for 5 d after plating. The results represent the mean of three independent experiments.
Synapsin I–Cre transgenic mice specifically express Cre recombinase in differentiated neurons
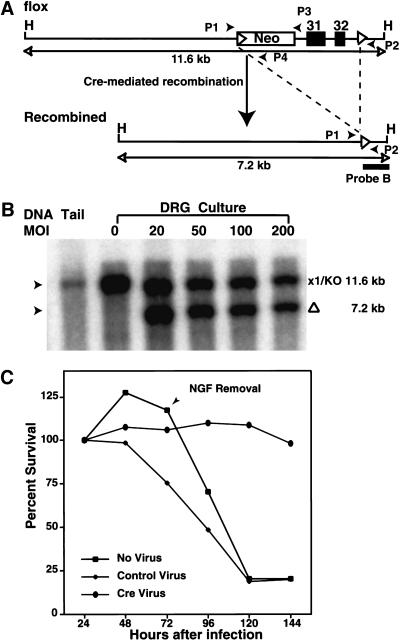
To assess the role of NF1 in neurons, we produced and characterized a neuronal-specific Cre recombinase transgenic (Tg) strain. In this strain, Cre expression is controlled by the rat Synapsin I promoter, which has been shown to drive transgene expression specifically in neuronal cells (Hoesche et al. 1993). To examine the temporal and spatial pattern of Cre activity, we crossed SynI–Cre Tg mice to a LacZ indicator Tg strain (β-actin/LacZ) that requires Cre recombinase activity to permit expression of the β-galactosidase (β-gal) gene (Tsien et al. 1996). We can detect Cre function as early as E12.5, when β-gal activity is restricted to brain, spinal cord, and DRG (Fig. 3A–D). Histological analysis showed that Cre activity is mainly distributed in differentiated neurons, outside the ventricular regions of the brain and spinal cord (data not shown). These data are consistent with endogenous Synapsin I gene expression (Melloni and DeGennaro 1994). To assess whether Cre activity remains restricted to neurons in adult mice, we first performed PCR assays on genomic DNA isolated from brain, spinal cord, heart, lung, kidney, and liver of NF1flox;SynI–Cre mice. Cre-mediated recombination was only detected in brain and spinal cord (data not shown). We next performed double immunohistochemistry using anti-LacZ (detecting Cre activity) and anti-NeuN (neuronal marker; Mullen et al. 1992) antibodies on brain and spinal cord sections from double transgenic SynI–Cre;LacZ mice. Figure 3E–P show representative examples of double labeling from the cortex, spinal cord, and hippocampus. NeuN immunoreactivity (red signal) essentially colocalized with LacZ reactivity (green signal), which indicates that Cre activity is mainly distributed in neurons. Unlike most areas of the brain, LacZ immunoreactivity is low in the cerebellum, where only few Purkinje cells remain positive (Fig. 3Q–S).
Figure 3.
Characterization of Synapsin I–Cre transgene (Tg) expression. E12.5 embryos (A) Cre Tg, (B) LacZ Tg, (C) and (D) Cre/LacZ double Tg were subjected to LacZ staining. Only the compound transgenic mice exhibit Cre-mediated activation of the β-galactosidase gene. Double-labeling immunoanalysis of Cre/LacZ double Tg with anti-NeuN and anti-LacZ antibodies for the cortex (E–J); spinal cord (K–M); hippocampus (N–P); and cerebellum (Q–S). Colocalization of LacZ and NeuN signals indicates that Cre-mediated recombination specifically occurs in differentiated neurons. Of note, the neuronal marker, NeuN is not expressed in the Purkinje cells. Ctx, cortex; SC, spinal cord; Hp, hippocampus; and Cb, cerebellum. Objective magnification: E–G and N–P, 20×; H–M and Q–S, 40×.
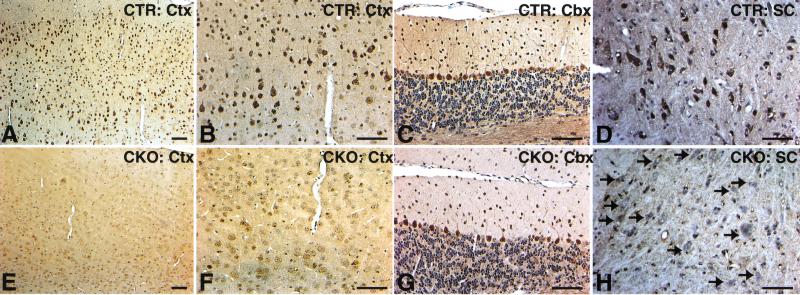
To directly examine the status of the floxed NF1 allele when exposed to Synapsin I-Cre expression, NF1flox/+;SynI–Cre mice were bred to NF1flox/flox mice, resulting in 25% of the offspring with NF1flox/flox;SynI–Cre genotype. We refer to the neuronal-specific NF1 mutant mice as, NF1SynIKO. To verify neuron-specific Cre activity, we performed immunohistochemistry using anti-NF1 antibody on brains and spinal cords from control and NF1SynIKO mutant mice. Although cortical and spinal cord neurons from control mice contain strong neurofibromin staining (Fig. 4A,B,D; Norland et al. 1993), neurofibromin immunoreactivity was completely lost in NF1SynIKO counterparts (Fig. 4E,F,H). However, consistent with the LacZ staining data, many mutant Purkinje cells remain positive for neurofibromin (Fig. 4G). Taken together, the data indicate that neuron-specific expression of Cre recombinase results in ablation of neurofibromin in most NF1SynIKO neurons.
Figure 4.
NF1SynIKO mice lose neurofibromin immunoreactivity in most neuronal populations. (A–D) Anti-NF1 immunoreactivity in the cortex (Ctx), cerebellum (Cbx), and spinal cord (SC) of control mice showing dark staining of neurons. (E–H) Loss of NF1 staining in the majority of neuronal populations in the mutant mice. An exception, however, is a subset of the Purkinje neurons in the Cbx (G), which retain NF1 immunoreactivity. A–C are from the same section, as are E–G. Scale bar, 50 μm.
Cre recombinase is not expressed in astroglia
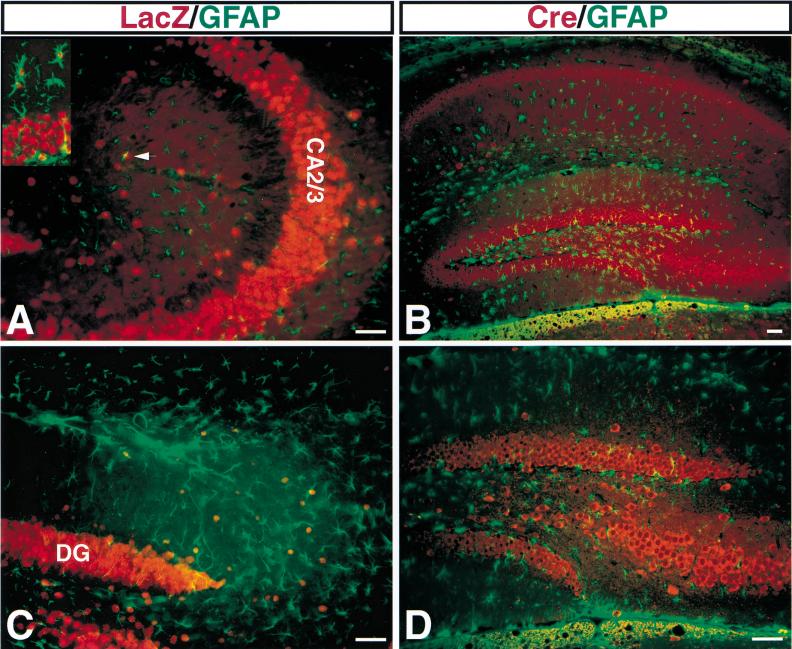
Most astrocytes do not normally express detectable NF1 (Daston et al. 1992). Therefore, to ensure that Cre-mediated recombination does not occur in NF1SynIKO astrocytes, we used two approaches. First, we crossed NF1SynIKO mice into two LacZ indicator strains (β-actin–LacZ and Rosa26–LacZ), which gave similar results. Rosa26–LacZ has been widely used and documented to express LacZ in all cells (Soriano 1999). Double labeling with antibodies directed against LacZ and astrocyte-specific glial fibrillary acidic protein (GFAP) was performed on SynI–Cre;Rosa26–LacZ brain sections. We focus on the hippocampus because it contains the highest levels of endogenous GFAP-positive astrocytes, although similar results were observed in all regions of the CNS. As shown in Figure 5A,C, LacZ immunoreactivity is mainly localized in the neuronal layer of CA2/3 (Fig. 5A) and dentate gyrus (Fig. 5C) and does not colocalize with GFAP. A positive control using a transgenic strain that expresses Cre recombinase both in neurons and glia, and therefore exhibits colocalization of LacZ and GFAP, is indicated in Figure 5A, inset. To rule out the possibility that Cre-mediated deletion may occur more efficiently in the floxed NF1 locus than in the floxed LacZ locus (which could result in astrocytes that are NF1 deficient but LacZ negative), we performed double labeling with anti-Cre and anti-GFAP (Fig. 5B,D). Like LacZ, Cre expression was confined to neuronal layers and did not coincide with GFAP-positive astrocytes. Thus, we conclude that most, if not all NF1SynIKO astrocytes do not undergo Cre-mediated recombination and that SynI–Cre Tg mice primarily and specifically express the Cre recombinase transgene in most populations of differentiated neurons. Consequently, the phenotypes described below are the result of NF1 loss in neurons.
Figure 5.
NF1SynIKO astrocytes do not express Cre recombinase. Double immunoanalysis with anti-LacZ/GFAP (A,C) and anti-Cre/GFAP (B,D). In the CA2/3 regions (A) and dentate gyrus (DG) (C) of the hippocampus, most of LacZ positive cells are localized in the neuronal layer with big nuclei. Of note, the inset shows the relative size of neuronal and small astrocytic nuclei and examples of colocalization of LacZ and glial fibrillary acidic protein (GFAP) signals. The arrow in A points out the apparent colocalization of LacZ and GFAP signals, which is actually the result of independent cells on two planes of the 50 μm section. (C) Some red nuclei are visible outside the DG. These are large nuclei that do not overlap with GFAP and likely represent interneuron nuclei. (D) High magnification of B, showing that Cre-positive cells are not GFAP positive. Scale bar, 50 μm.
Loss of NF1 in neurons affects cortical development
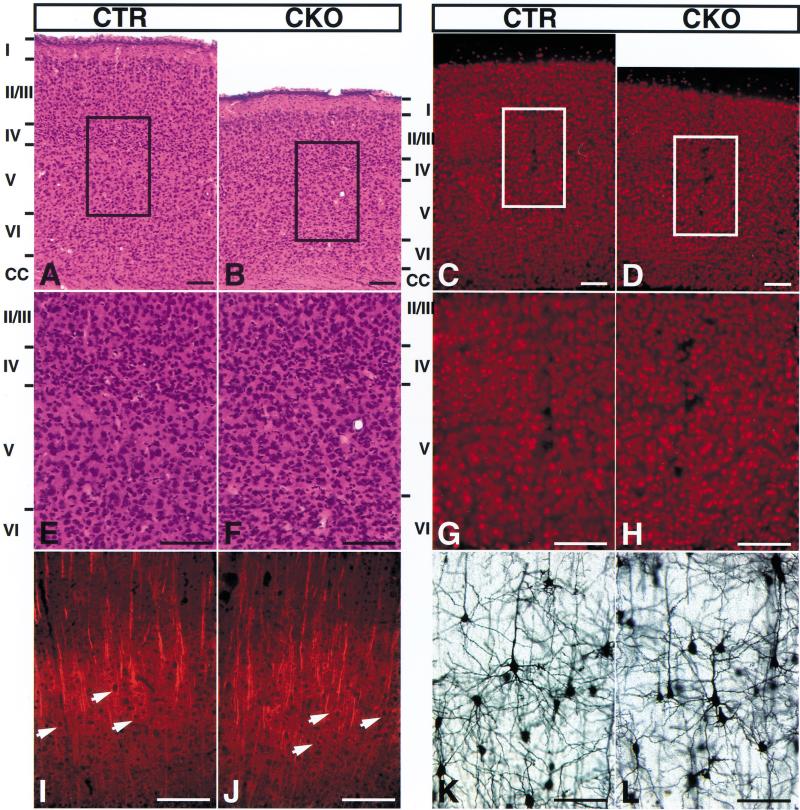
There was no apparent embryonic lethality occurring in NF1SynIKO mice, and newborn NF1SynIKO pups were indistinguishable from their control littermates (NF1flox/flox or flox/+;SynI–Cre; data not shown). These results indicate that neuronal NF1 function is not required for survival through birth. Three to four days after birth, NF1SynIKO mice began to exhibit growth retardation that was sustained into adulthood, with a reduction in both weight and size that stabilized at ∼50% of normal littermates (Table 1; data not shown). Otherwise, the NF1SynIKO mice appeared to be normal and were fertile. We monitored size and weight of internal organs in adult NF1SynIKO mice and found that the kidney, heart, liver, and spleen showed a proportional reduction in both weight and size (Table 1; data not shown). In contrast, we saw limited reduction in brain size (Table 1). Separate analysis of different structures of the brain revealed that reduced brain weight is limited to the forebrain (Table 1). Histological analysis of NF1SynIKO brains revealed a conspicuous defect in the cerebral cortex, which is characterized by a reduction in cortical thickness and an apparent increase in cell density (Fig. 6A–H). Morphometric analysis of serial coronal sections throughout the anterior-to-posterior length of NF1SynIKO and control brains indicated a significant reduction in mutant cortical thickness by ∼20% (Fig. 6A–D, 7A,B). This deficit appears to be specific to the cerebral cortex, given that the dorso–ventral and the medio–lateral axes (excluding the cortex), as well as the hippocampus and hindbrain, were not affected in NF1SynIKO brains (Fig. 7C).
Table 1.
Size and weight analysis of NF1SynIKO mice
| Genotype
|
Body
|
Forebrain
|
Hindbrain
|
Kidney
|
|---|---|---|---|---|
| Control | 23.13 ± 2.16 | 0.39 ± 0.02 | 0.14 ± 0.01 | 0.22 ± 0.01 |
| Mutant | 11.81 ± 1.98* | 0.33 ± 0.02* | 0.13 ± 0.01 | 0.13 ± 0.02* |
| Percent | 51.1% | 84.6% | 92.9% | 59.0% |
The reduction in NF1SynIKO body (48.9%) and kidney (41.0%) weight were statistically significant (P < 0.01). A separate evaluation of forebrain and hindbrain weight revealed a significant 15% reduction (P < 0.01) in forebrain, whereas the hindbrain was not significantly different from that of control littermates. Percent indicating weight percentage in mutants to controls.
P < 0.01.
Figure 6.
NF1SynIKO mice have reduced cerebral cortex thickness and increased cell density. Serial coronal sections of brains from control and mutant littermates were cut at 50 μm and analyzed by hematoxylin and eosin (H&E) staining, NeuN, MAP2, and Golgi staining. (A,E) Control cortex, H&E; (B,F) mutant cortex, H&E; (C,G) control cortex, NeuN; (D,H) mutant cortex, NeuN; the reduced cortical thickness is readily apparent in the mutant cortex. (I) Control cortex, MAP2; (J) mutant cortex, MAP2, arrows point out the presence of MAP2-positive pyramidal neurons in the mutant cortex; (K) control and (L) mutant cortex, Golgi stain. The morphology of the stained neurons appeared normal in the mutant cortex. Scale bar, 50 μm.
Figure 7.
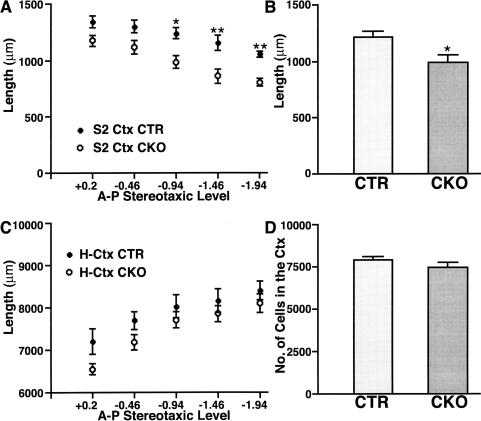
Increased cell density in the cerebral cortex of NF1SynIKO mice. Anterior-to-posterior morphometric analysis of the secondary somatosensory cortex (S2 Ctx) revealed a significant reduction in cortical thickness in NF1SynIKO mice compared to control (CTR) animals. The reduction was significant particularly at caudal levels; −0.94 (P < 0.05), −1.46, and −1.94 (P < 0.01) mm from Bregma (A). A comparison of the average cortical thickness in these mice showed an ∼20% decrease (P < 0.05) in the NF1SynIKO mice (B). Despite the reduced cortical thickness in mutant mice, the rest of the brain size is comparable to that in CTR mice. Measurement of the horizontal brain length, excluding the cortex (H-Ctx) in mutant mice showed no significant differences throughout the anterior-to-posterior axis when compared to CTR mice (C). Quantitative analysis of the total number of cells in the cortex failed to demonstrate any differences because of genotype (D). *, P < 0.05; **, P < 0.01.
We next examined the cortex of the mutant mice for total cell number (as determined by H&E counts) and neuron number using NeuN staining (Fig. 7D). These data indicated that negligible total cell or neuronal loss occurs in the mutant cortices. Thus, the decreased dimension of the neuronal NF1 mutant cerebral cortex is not likely to be the result of reduced cell number (Fig. 6A–H). The increase in cell density often resulted in less apparent lamination, although we detected intact layered structure in some regions of the NF1SynIKO mutant brains (Fig. 6B,F).
To evaluate neuronal differentiation in NF1SynIKO brains, we performed immunohistochemistry using anti-MAP2 (dendritic marker) antibody. As shown in Figure 6I,J, the pattern of MAP2 immunoreactivity was indistinguishable between control and mutant brains, except that the mutant cortex tended to show greater MAP2 immunoreactivity consistent with higher neuronal density. We also examined cortical, hippocampal, and Purkinje neurons in NF1SynIKO mutant brains using antibodies against several calcium-binding proteins, including calbindin, parvalbumin, and calretinin. We observed no apparent abnormalities (data not shown).
To test whether high neuronal density in the mutant cortex results in abnormal organization of dendrites and axons, we performed Golgi staining on control and mutant brains to permit visualization of individual neurons and their dendritic arbors within the cortex (Ramon y Cajal 1904). Golgi staining indicated that NF1SynIKO mutant neurons have normal arborization of dendrites and axons when compared to control counterparts (Fig. 6K,L). Collectively, these data show that NF1SynIKO mutant mice have normal neuronal development but display high cell density in a reduced cerebral cortex, which often results in less apparent lamination. The mechanistic basis underlying the cortical defect remains to be elucidated.
NF1SynIKO mutant brains display astrogliosis
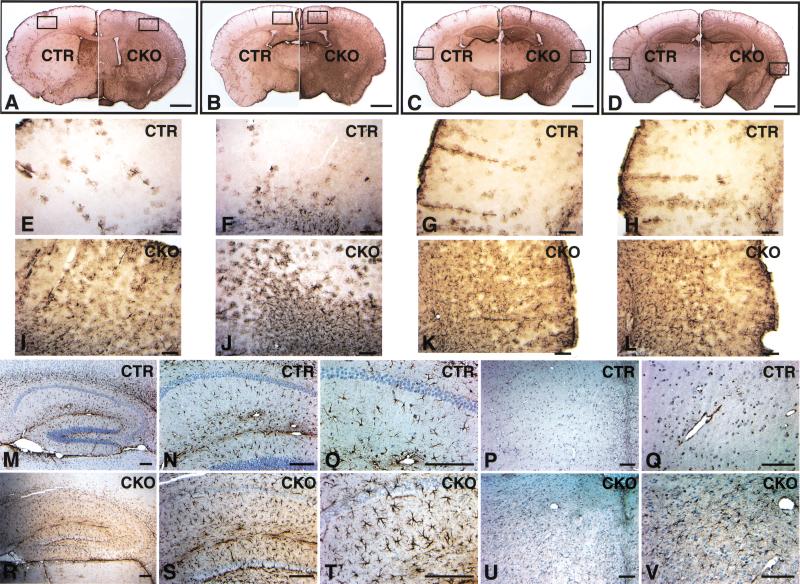
Because neuronal development in NF1SynIKO brains is relatively normal, we examined additional cell populations. Brains from adult control and mutant mice were serially sectioned and subjected to immunohistochemistry using the astrocyte-specific anti-GFAP antibody. Ten of ten NF1SynIKO mice displayed extensive GFAP immunoreactivity, which reflects astrogliosis throughout the cerebral cortex, hippocampus, and brainstem (Fig. 8). Figure 8A–D shows four levels of coronal brain sections from rostral to caudal. The mutant brains (right panels) showed significantly higher GFAP levels compared with control counterparts (left panels). Intense GFAP staining was especially prominent in the cerebral cortex, where endogenous GFAP immunoreactivity is normally low (Fig. 8A–D). High magnification showed the GFAP-positive cell morphology to be typical of reactive astrocytes (Fig. 8E–L; Ridet et al. 1997).
Figure 8.
NF1SynIKO mice display extensive astrogliosis. Serial coronal sections were cut at 50 μm thickness (cryostat) or 5 μm thickness (paraffin), and subjected to GFAP immunoanalysis. (A–D) Four sections from rostral to caudal, with the left hemispheres in each panel from control mice and the right hemispheres from mutant mice. Note the overall darker brown staining in the right hemispheres, indicating extensive astrogliosis in the mutant brains. (E–H) High magnification of the control cortex (the boxed area of the left hemisphere in A–D). (I–L) High magnification of the mutant cortex (the boxed area of the right hemisphere in A–D). (M–O) Control and (R–T) mutant hippocampus. Note the hypertrophic nature of the reactive astrocytes (compare T to O). Control (P,Q) and mutant (U,V) brainstem. Scale bar: A–D, 1 mm; E–V, 50 μm.
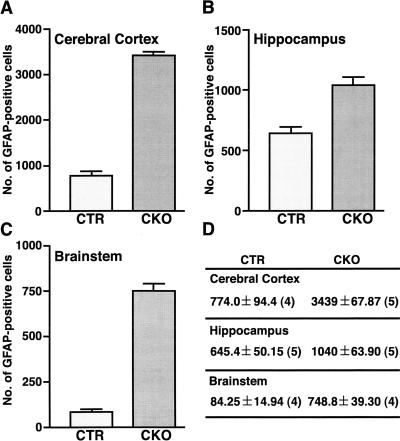
To confirm the reactive nature of NF1SynIKO astrocytes, we performed immunohistochemistry with a second independent anti-GFAP antibody on thin sections (see Materials and Methods). As shown in Figure 8M–O and Figure 8R–T, the mutant hippocampus showed increased numbers of GFAP-positive astrocytes that are hypertrophic with thicker and increased processes. Other brain regions also showed dramatic increase in the number of GFAP-positive astrocytes, including the brainstem (Fig. 8U,V), where there is little endogenous GFAP immunoreactivity (Fig. 8P,Q). To quantify the degree of GFAP-positive astrocyte increase, we counted GFAP-positive cells in the cerebral cortex, hippocampus, and brainstem. As indicated in Figure 9, there is 4.4-fold increase in the number of GFAP-positive astrocytes in the cerebral cortex, 1.6-fold increase in the hippocampus, and ninefold increase in the brainstem of NF1SynIKO mice as compared to the control counterparts.
Figure 9.
The number of glial fibrillary acidic protein (GFAP)-positive cells is significantly increased in the mutant brains. The quantification of GFAP-positive cells in the cortex (A), hippocampus (B), and brainstem (C). (D) Mean value ± SEM (number of brains examined).
NF1SynIKO brains do not develop neuronal degeneration or microgliosis
The data presented above support the notion that neuron specific loss of NF1 function leads to astrogliosis through an indirect mechanism. Because astrogliosis is often associated with neuronal degeneration (Ridet et al. 1997), it is possible that neuronal loss of NF1 causes neuronal degeneration, which leads to induction of astrogliosis. We examined NF1SynIKO brains for evidence of necrosis or apoptosis using Nissl staining and TUNEL assays and were unable to detect evidence of degenerating neurons (data not shown). A second normal response to CNS injury or cell death is the induction of microglia into the region. Microgliosis is characterized by cellular hypertrophy and increased proliferation (Streit et al. 1999). To examine whether NF1SynIKO brains develop microgliosis, we used an antibody against macrophage surface antigen (anti-mac1; Rabchevsky et al. 1998) to detect the morphology of microglia. Serial sections from all regions of NF1SynIKO brains were compared to normal counterparts, and no evidence of microgliosis was found (data not shown). Taken together, these results indicate that loss of NF1 in neurons does not elicit the classical morphological features of neuronal degeneration and microgliosis but, rather, may alter normal neuronal physiology, thus exerting an effect on surrounding astrocytes (Kraig et al. 1991; Steward et al. 1991).
Loss of NF1 in neurons results in activation of endogenous MAP kinase
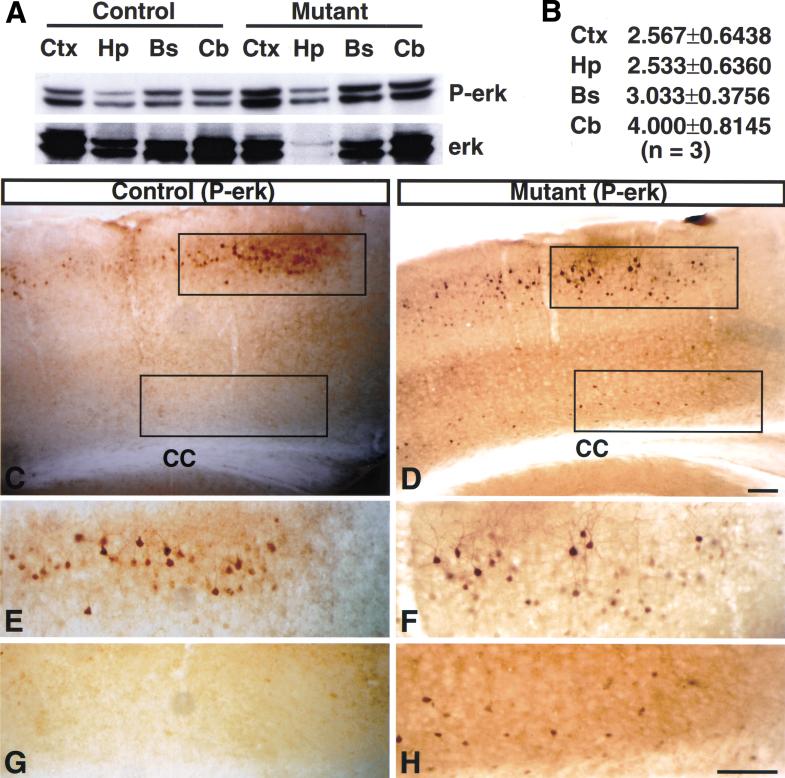
To investigate further the mechanism underlying astrogliosis in NF1SynIKO brains, we examined the activity of MAP kinase, a downstream component of ras/NF1 signaling that is activated in some human reactive astrocytes (Mandell and VandenBerg 1999). First, proteins were extracted from different regions of NF1SynIKO and control brains, including the cortex, hippocampus, brainstem, and cerebellum, and subjected to Western blot analysis with anti-phosphorylated erk (P-erk) antibody, which recognizes the active form of MAP kinase. All regions of NF1SynIKO brains showed significantly increased P-erk levels as compared with controls (Fig. 10A,B). We next localized activation of MAP-kinase using immunohistochemistry. Immunoanalysis revealed that NF1SynIKO mutant brains (Fig. 10D,F,H) contain significantly more P-erk positive cells than do control brains (Fig. 10C,E,G). In particular, Layers II/III and VI of the mutant cerebral cortex showed conspicuous astrogliosis (Fig. 8A–D) and always had a significant increase in number of P-erk positive cells (Fig. 10E–H). In all regions examined, the large majority of P-erk positive cells has unambiguous neuronal morphology (Fig. 10E,F,H) and coexpress a neuronal marker, NeuN (data not shown). Taken together, these results indicate that loss of NF1 in neurons resulted in activation of MAP kinase in these cells, with no evidence for similar activation in reactive astrocytes.
Figure 10.
Activation of MAP kinase in NF1SynIKO neurons. (A) (Top panel) Western blot analysis of tissues from different parts of control and mutant brains using anti-phosphorylated erk (P-erk) antibody. (Bottom panel) The same blot was stripped and reprobed with anti-P-erk antibody as a loading control. (B) P-erk and erk signals were quantified by densitrometry (see Materials and Methods). The level of P-erk in each sample is normalized to level of erk protein signal. Each number represents mean value ± standard error of the mean (SEM) of ratio of P-erk levels of three mutants to three controls. Sections of cerebral cortex from control (C) and mutant mice (D) were analyzed with anti-P-erk antibody. The mutant cortex exhibits greater number of phospho-erk positive cells corresponding to neurons and not to reactive astrocytes. E and G are high magnification of the top and bottom boxed area of C; F and H are high magnification of the top and bottom boxed area of D. Ctx, cerebral cortex; Hp, hippocampus; Bs, brainstem; Cb, cerebellum; CC, corpus callosum. Scale bar, 100 μm.
Finally, to examine whether the observed increase in the number of GFAP-positive astrocytes results from excess proliferation of glial precursors or mature astrocytes, we employed in vivo BrdU incorporation assays. We examined three neonatal and three adult (5-month-old) NF1SynIKO and control brains and failed to detect an increase in proliferation (data not shown). The results suggest that increased proliferation may not play a major role in this astrogliosis model (see Discussion).
NF1SynIKO mice are tumor free
Although extensive astrogliosis was found in NF1SynIKO brains, we failed to detect the formation of optic gliomas and astrocytomas. In total, 29 NF1SynIKO mice were monitored over a period of 14 mo, followed by complete necropsy and histological analysis of the CNS. No evidence of tumor appearance was uncovered (data not shown). In the periphery, we dissected entire spinal cords together with intact DRG from five 14-month-old NF1SynIKO mice that were subjected to complete histological analysis and found no sign of neurofibroma formation (data not shown). Thus, loss of NF1 in differentiated neurons, although able to induce astrogliosis, is not sufficient to induce the formation of gliomas or neurofibromas. However, it remains undetermined whether neuronal loss of NF1 can contribute to tumorigenesis in combination with other genetic and/or epigenetic factors.
Discussion
The generation of knockout mice has provided insight into the role of NF1 in developmental processes, such as cardiac development and neuronal survival (Brannan et al. 1994; Vogel et al. 1995). However, the early lethality of NF1−/− mice limited these studies from extending into later developmental stages or adulthood. The generation of NF1 conditional mutants now permits more detailed examination of the physiological function of NF1 both in vitro and in vivo.
In vitro
Previously, NF1 function has been studied in neurons, Schwann cells, and fibroblasts through isolation of primary cells from NF1−/− embryos (Kim et al. 1995; Rosenbaum et al. 1995; Vogel et al. 1995). The interpretation of these studies was hampered by the inability to distinguish a direct functional defect in mature cells from an indirect effect resulting from developmental abnormalities in precursor cells. With the conditional mutant, neurons or Schwann cells can be permitted to develop and differentiate in the wild-type state, and ablation of NF1 can be performed in mature cells, thus separating steady-state defects from developmental effects. NF1−/− neurons generated by Cre-mediated recombination in culture display neurotrophin-independent survival indistinguishable from germ-line NF1 mutant DRG. This confirms that neurotrophin independence of NF1−/− neurons directly results from loss of NF1 function rather than abnormal differentiation of NF1−/− neural crest. Another useful feature of the NF1 conditional system is that it permits pooling of tissues or cells from entire litters, thus providing adequate amounts of material for performing biochemical assays. For example, we were able to show that loss of NF1 in primary Schwann cells results in increased basal levels of MAP kinase activity (Y. Zhu and L.F. Parada, unpubl.) as previously reported in fibroblasts and neurons (van der Geer et al. 1997; Klesse and Parada 1998). Recent studies from Drosophila suggest that NF1 may also regulate cAMP-mediated signaling pathways (Guo et al. 1997; The et al. 1997). For the reasons cited above, the NF1 conditional system should provide effective means of addressing the role of NF1 in cAMP-mediated signaling pathways in primary cells, such as neurons and Schwann cells.
NF1 in the CNS
To date, we have observed two CNS phenotypes in NF1SynIKO mice (cortical compression and astrogliosis). In contrast to other brain regions, NF1SynIKO cerebral cortices are statistically smaller by 20% than the controls. One study suggested that brains of 10 NF1 patients have abnormal cortical lamination (Rosman and Pearce 1967). Whether less apparent lamination in both mouse and human results from increased neuronal density remains to be determined. Nonetheless, the compression that is found uniquely in the cerebral cortex indicates a unique role for NF1 in cortical development. This feature may be of relevance to a diverse set of disorders termed “micrencephally”, that result in reduced size of forebrain and cerebellum (Hatten 1999). In this context, recent reports of an FGF-2 mouse knockout indicated reduction in cortical size that is apparently not associated with neurodegeneration (Dono et al. 1998). Although the similarity of the phenotypes may be more apparent than real, it remains formally possible that FGF-2 signaling may in someway relate to NF1 function in cortical development.
The role of NF1 in astrogliosis
In response to infection, injury, or neurodegeneration, CNS astrocytes undergo hypertrophy, possible proliferation, and altered gene expression, including up-regulation of astrocytic GFAP (Ridet et al. 1997). One postmortem case study reported three NF1 patients with extensive astrogliosis. However, the fact that two of the patients had undergone chemotherapy has confounded the interpretation of the underlying cause (Nordlund et al. 1995). Two recent reports have suggested that some NF1 heterozygous mutant mice may have increased GFAP-positive astrocytes (Gutmann et al. 1999; Rizvi et al. 1999). In one instance, mild increases (40%) were reported throughout the brain (Gutmann et al. 1999), whereas a second study reported significant increases only in the periaqueductal gray and nucleus accumbens with little evidence in other structures (Rizvi et al. 1999). We were unable to detect evidence of significantly increased GFAP-positive cells in heterozygous NF1+/− mice (see Materials and Methods). Our results provide the first direct evidence that loss of both NF1 alleles in neurons is sufficient and required to induce astrogliosis in the CNS. These data do not support the idea that haploinsufficency could lead to this phenotype.
Critical to our conclusions is the specificity of the synapsin promoter–Cre transgenic mouse that is employed in this study. We therefore employed detailed and exhaustive immunohistochemical assays to assess the expression of lacZ reporters, of endogenous NF1, and of Cre recombinase in the context of two independently derived LacZ indicator mouse strains (β-actin/LacZ and Rosa26–LacZ), as well as in NF1SynIKO mice.
Finally, activation of MAP kinase was only observed in neurons, not astrocytes, thus further supporting that ras activation only occurred in neurons of NF1SynIKO mice. In an ongoing study, we have ablated NF1 in neural precursor stem cells, which results in NF1 loss in both mature neurons and glia (Y. Zhu, P. Ghosh, A. Messing, and L.F. Parada, in prep.). The morphological phenotypes in the CNS of these mice are distinct from that of this study, and importantly, the astrocytes display conspicuous MAPK activation, which is never observed in NF1SynIKO astrocytes. We therefore conclude that loss of NF1 is neuron specific and results in non-cell autonomous consequences.
A unique model for astrogliosis and learning disability
To date, NF1SynIKO mice give no evidence of neurofibromas or optic gliomas that are common features of human NF1 (Riccardi 1999). This suggests that loss of NF1 in differentiated neurons is not sufficient to induce tumor development despite the ability to induce astrogliosis in the CNS. Ras-mediated signaling has been implicated in human gliomagenesis, given that activated mutations or amplification of the epidermal growth factor receptor (EGFR) gene have been found in a subset of human gliomas (Schlegel et al. 1994; Hayashi et al. 1997). However, astrogliosis in NF1SynIKO mice is likely to be mediated through pathways other than ras, given that MAPK activation was only present in neurons and not in reactive astrocytes. The simplest model for the observed astrogliosis is that a paracrine effect, mediated by NF1-deficient neurons, induces astroglial hypertrophy and GFAP induction (Rabchevsky et al. 1998). Alternatively, loss of NF1 may induce synaptic hyperactivity, which leads to astrocyte reactivity (Kraig et al. 1991; Steward et al. 1991). One interesting feature in this astrogliosis model is the absence of neuronal degeneration and microgliosis. The fact that most known cytokines, such as CNTF and interleukin-6 (IL-6), can induce both astrogliosis and microgliosis suggests the existence of a unique signal from NF1-deficient neurons that only activates astrocytes, but not microglia (Ridet et al. 1997). Among the known transgenic models, TGFα is the only identified cytokine that, when overexpressed in brain, leads to astrogliosis but not to microgliosis (Rabchevsky et al. 1998). The fact that TGFα binding to EGFR presumably activates the downstream MAPK pathway in reactive astrocytes suggests that TGFα is not likely to be the inducer for astrogliosis in this context. Furthermore, the observation that there is no increase in proliferation during postnatal development and in adults suggests that astrogliosis in this model is likely to be a consequence of hypertrophy and up-regulation of GFAP in preexisting differentiated astrocytes. This notion is consistent with the following two observations: There is no increase in the total number of cells and neurons in the mutant cortex; and the areas that contain lower numbers of endogenous GFAP-positive astrocytes, such as the cortex and brainstem, show more dramatic astrogliosis than those areas that have higher numbers of GFAP-positive astrocytes, such as the hippocampus. However, it remains possible that increased proliferation occurs in mutant brains some time between P0 and 5 mo of age. These observations suggest that neuronal loss of NF1 leads to the activation of MAP kinase, exerting a signal to activate surrounding astrocytes, including up-regulation of GFAP. Future identification of this unique inducer will shed new insight into the underlying mechanism of astrogliosis and neuronal/glial interactions.
The fact that 30%–65% of NF1 children have learning disabilities (North 1999) and that NF1 is highly expressed in both embryonic and adult neurons (Daston and Ratner 1992; Daston et al. 1992; Huynh et al. 1994) suggests that NF1 may have an important role in CNS development. Preliminary results indicate that NF1SynIKO mice also display severe learning disability (Y. Zhu, R. Costa, L.F. Parada, and A.J. Silva, unpubl.). It will be of interest to determine whether learning disability is the consequence of activated ras/MAPK pathway in neurons or of astrogliosis interfering with synaptic function. Thus, NF1SynIKO mice may provide a unique model for one aspect of human NF1—learning disability.
Finally, through the use of two novel mouse strains (NF1flox and Syn–Cre Tg), this study underscores the importance of revisiting retrospective studies of astrogliosis in individuals with NF1. Correlation of this phenotype to those patients with learning disabilities may provide a key to dissecting the physiological basis for this important problem.
Materials and methods
Generation of NF1 flox mice
Two oligos were designed to generate a SacI–XbaI linker containing a 34-bp loxP site and an EcoRI site (5′ primer, 5′ to 3′: CGGAATTCCGATAACTTCGTATAGCATACATTATACGA AGTTAT; 3′primer, 5′ to 3′:
CTAGATAACTTCGTATAATGTATGCTATACGAAGTTAT CGGAATTCCGACT). The SacI–XbaI linker was subcloned into the pJax34 plasmid containing a pGKneo cassette (Jackson lab). The EcoRI fragment containing the pGKneo and a loxP site retrieved from the above construct was subcloned into the EcoRI site of an 8.6-kb HindIII–PstI NF1 genomic DNA (Fig. 1A). The resultant construct is designated pYZ05. The same SacI–XbaI linker was subcloned into 5′ of 1.0-kb PstI–HindIII NF1 genomic DNA and designated pYZ04. The insert of the pYZ04 construct was modified at both ends with NotI linkers and subcloned into the NotI site of pYZ05. The resultant construct is designated pYZ07. The 11.6-kb HindIII fragment from the pYZ07 construct was subcloned into a modified pBS-KSII vector containing a thymidine kinase cassette. The final construct is designated pYZ08D (Fig. 1A). ClaI linearized targeting vector was transfected into R1 ES cells (Zhu et al. 1998).
Molecular analysis
NF1flox mice were genotyped by either Southern analysis (Fig. 1C,D) or a three-oligonucleotide primer PCR assay. Two primers (P1 and P3) from intron 30 of the NF1 gene were designed to detect a 480-bp band from the WT allele; a third primer (P4) from the neo gene and P1 were used to amplify a 350-bp band from the flox allele (Fig. 2A). PCR was performed under the following conditions: 0.25 mM of dNTPs, 3.0 mM of MgCl2, 0.2 μM of primers, and 2 units of Taq polymerase (GIBCO); and the program: 1 cycle of 94°C for 3 min; 35 cycles of 94°C for 1 min; 55°C for 2 min; and 72°C for 3 min; and followed by 72°C for 4 min. A sensitive PCR assay for detecting Cre-mediated recombination and completion of deletion was established as the following: Two primers (P1 and P2; Fig. 2A) were used to detect a 280-bp band that resulted from Cre-mediated recombination in the NF1flox allele. The P1 and P4 primers were used to detect the completion of Cre-mediated recombination. The PCR conditions are identical to that for NF1flox genotyping. Genotyping the Cre transgene was carried out by a PCR assay under the following conditions: 1.25 mM of dNTPs, 1.5 mM of MgCl2, 0.4 μM of primers, and 2 units of Taq polymerase (GIBCO); and the program: 1 cycle of 94°C for 1 min; 30 cycles of 94°C for 30 sec; 65°C for 30 sec; and 72°C for 1 min; and followed by 72°C for 2 min. The sequences for primers (5′ to 3′) are: P1, CTTCAGAC TGATTGTTGTACCTGA; P2, CATCTGCTGCTCTTAGAG GAACA; P3, ACCTCTCTAGCCTCAGGAATGA; P4, TGATT CCCACTTTGTGGTTCTAAG; Cre1, CATGTTTAGCTGGC CCAAATGTTGCTG; and CreNukATG, CGACCATGCCCAA GAAGAAGAGGAAGGTG.
Primary culture
E13.5 NF1flox/flox or flox/− embryos were dissected, and DRG neurons were isolated and cultured in the presence of NGF (10 ng/mL, GIBCO) as described (Vogel et al. 1995). Four hours after plating, the DRG cultures were either infected with LacZ- and Cre-adenovirus at MOI = 50 or mock infected. Three days later, NGF was removed from the culture by serial dilution and addition of anti-NGF neutralizing antibody (150 μg/mL, Boehringer Mannheim). Neuronal counts were performed every 24 hr for 5 d after plating.
Production and characterization of SynI–Cre Tg mice
The SynI–Cre transgene was constructed using the SalI–XhoI 4.4-kb fragment of pBL4.3Syn–CAT, kindly provided by Dr. Manfred Kiliman (Hoesche et al. 1993), containing the rat Synapsin I promoter and 100 bp of 5′ untranslated CAT sequence. This was placed immediately 5′ of a modified bacteriophage P1 Cre recombinase gene that included translational consensus and nuclear localization signals. The human growth hormone gene was placed 3′ of Cre to provide transcription termination and polyadenylation signals. The transgene was isolated from plasmid sequence by NotI digestion and injected into pronuclei of fertilized C57BL/6 × CBA (F2) zygotes. Four independent lines were produced (667, 671, 675, 679) and maintained in the C57BL/6 background for more than three generations. E11.5, E12.5, and E13.5 embryos from the cross of LacZ reporter and SynI–Cre Tg mice were dissected and subjected to β-gal histochemical analysis as described (Zhu et al. 1998). SynI–Cre/LacZ double heterozygotes were anesthetized and perfused with PBS followed by cold 4% paraformaldehyde. Brains were dissected, fixed in 4% paraformaldehyde at 4°C overnight, followed by dehydration in 30% sucrose at 4°C for 48 h. Cryostat sections were prepared and subjected to double-labeling immunohistochemistry.
The control and conditional mutant mice
The control mice that were used in this report are the pool of phenotypically indistinguishable mice with three genotypes: NF1flox/flox, NF1+/+;SynI–Cre and NF1flox/+;SynI–Cre. The conditional mutant mice include both NF1flox/flox;SynI–Cre and NF1flox/KO;SynI–Cre mice, which also have similar phenotypes.
Size and weight analysis of NF1SynIKO mice
NF1SynIKO and control littermates were used to determine overall body size (horizontal and vertical dimensions in cm) and weight (g), as well as size of several internal organs including kidney, liver, and spleen. The brain was separated into forebrain and hindbrain for analysis.
Golgi-Cox staining
Animals were perfused with 0.9% buffered saline and their brains immersed in 40 mL of Golgi-Cox solution (Glaser and Van der Loos 1981) for 14 d. The brains were then vibratome-sectioned at 100 μm in PBS, mounted, and stained following a modified protocol (Kolb and McClimans 1986).
Immunohistochemistry
Free-floating sections were either cut at 50 μm on a freezing microtome or cut at 75 μm on a vibrotome. Paraffin sections were cut at 5 μm, deparaffinized and rehydrated. The color visualization of the primary antibodies was performed with a horseradish peroxidase system (Vectastain ABC kit, Vector). Slides were counterstained with hematoxylin, washed in H2O, dehydrated with 100% EtOH three times for 1 min each, with xylene three times for 1 min each, and then mounted with permount. For immunofluorescence, slides were incubated with either Cy3-conjugated anti-rabbit/mouse or Cy2-conjugated anti-rabbit secondary antibodies at 1:200 dilution for 45 min at room temperature. Slides were then washed with PBS four times each and mounted with immu-mount (Shandon). The dilutions of primary antibodies used in this study were: LacZ (1:100, 5′ and 3′), NeuN (1:500, Chemicon), MAP2 (1:100, Sigma), calbindin (1:1000, SWant), parvalbumin (1:2000, SWant), GFAP (anti-mouse, 1:100, Pharmingen), GFAP (anti-rabbit, 1:5000, DAKO), mac1 (anti-mouse, 1:250, Chemicon), Cre (1:200, Novaden), NF1 (a gift from Nancy Ratner, 1:2000; David Gutmann, 1:100), P-erk (Biolabs, 1:200), and S100 (Signet, 1:1000).
Morphometric analysis of cerebral cortex
Cryostat sections were serially cut at 50-μm thickness and subjected to H&E staining and NeuN immunoanalysis. Five coronal brain sections per animal (n = 5 per genotype) were evaluated at 500-μm intervals from 0.2 mm to −1.94 mm with respect to Bregma according to a stereotaxic atlas of the mouse brain (Franklin and Paxinos 1997). The thickness of the S2 somatosensory cerebral cortex, as well as the horizontal distance from left–right corpus callosum at mid dorsal-to-ventral level, were measured using an ocular micrometer. A subset of these tissue sections was digitized at 20× magnification (three frames per section) throughout the entire cerebral cortex, and the total cell number was quantified manually from printouts.
Cell counts
For astrocyte counts, cryostat sections from brains (4–6 mo old) were serially cut at 50-μm thickness and subjected to GFAP immunohistochemistry as described above. The number of GFAP-positive cells was counted in one out of each seven sections. For the cerebral cortex, the regions containing motor and somatosensory cortex were chosen; for the hippocampus, a rectangle covering both CA1 and dentate gyrus was introduced, and the number of GFAP-positive cells was determined in the rectangle of each section. For the brainstem, the regions containing the facial nucleus were analyzed.
Western blot analysis
Brains were dissected into regions including cortex, hippocampus, brainstem, and cerebellum and frozen in liquid nitrogen. Proteins were extracted and subjected to Western blot analysis as previously described (Klesse and Parada 1998). The dilutions of primary antibodies used were: P-erk (1:1000, Biolabs) and erk (1:5000, Santa Cruz). Densitometry analysis was performed with the Molecular Dynamics Computing Densitometry (300A).
Statistical analysis
The values were compared using a two-way ANOVA (Super ANOVA program, Abacus Concepts) to evaluate for differences caused by phenotype and anterior-to-posterior level. One-way ANOVA with these two variables combined, followed by Student-Newman-Keuls post-hoc test, was used to determine differences in individual groups. P-values < 0.05 were considered to be significant.
BrdU assay and tumor analysis
Neonatal mutant and control littermates (P0) were pulsed with BrdU for 2 h and adult (5-month-old) mice were pulsed with BrdU five times at 2-h intervals. The dose of BrdU was 50 μg/g (body weight). BrdU-treated mice were fixed and processed as described above. BrdU immunohistochemistry was performed as described previously (Zhu et al. 1998). The dilution of primary BrdU antibody was 1:50 (Becton Dickinson).
In total, 29 NF1SynIKO mice were monitored over a period of 14 mo. Complete necropsy was performed under a dissecting microscope, and the brains were subjected to histological analysis (see above). Five of these 29, 14-month-old mice were perfused as described above, and spinal column was postfixed in 4% paraformaldehyde at 4°C overnight. The next day, the entire spinal cords with DRGs were dissected and fixed in 4% paraformaldehyde at 4°C overnight. Fixed tissues were then processed and paraffin embedded. Paraffin sections were serially cut at 5-μm thickness, stained with H&E, and examined under light microscope.
Acknowledgments
We thank David Anderson for gift of the β-actin/LacZ reporter mice and Nancy Ratner and David Gutmann for providing NF1 antibody. We thank Steve Kernie for critical reading of the manuscript and all members of the Parada Lab for helpful discussions and support. We thank Tammy Oliver for assistance with histology. L.F.P acknowledges support from: NINDS-R01-NS34296-05; DAMD 17-97-1-7343; the National Neurofibromatosis Foundation, and the Texas Neurofibromatosis Foundation. We thank Thierry Hennet and Michael Durnin for assistance in generating the Syn1-Cre transgenic mice. J.D.M. acknowledges support from NIH grant DK 48247. J.D.M. is an Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL luis.parada@utsouthwestern.edu; FAX (214) 648-1960.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.862101.
References
- Baba H, Fuss B, Urano J, Poullet P, Watson JB, Tamanoi F, Macklin WB. GapIII, a new brain-enriched member of the GTPase-activating protein family. J Neurosci Res. 1995;41:846–858. doi: 10.1002/jnr.490410615. [DOI] [PubMed] [Google Scholar]
- Ballester R, Marchuk D, Boguski M, Saulino A, Letcher R, Wigler M, Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990;63:851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Bollag G, Clapp DW, Shih S, Adler F, Zhang YY, Thompson P, Lange BJ, Freedman MH, McCormick F, Jacks T, et al. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet. 1996;12:144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- Bos JL. ras oncogenes in human cancer: A review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: A conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- Brannan CI, Perkins AS, Vogel KS, Ratner N, Nordlund ML, Reid SW, Buchberg AM, Jenkins NA, Parada LF, Copeland NG. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes & Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- Buchberg AM, Cleveland LS, Jenkins NA, Copeland NG. Sequence homology shared by neurofibromatosis type-1 gene and IRA-1 and IRA-2 negative regulators of the RAS cyclic AMP pathway. Nature. 1990;347:291–294. doi: 10.1038/347291a0. [DOI] [PubMed] [Google Scholar]
- Cawthon RM, Weiss R, Xu GF, Viskochil D, Culver M, Stevens J, Robertson M, Dunn D, Gesteland R, O'Connell P, et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62:193–201. doi: 10.1016/0092-8674(90)90253-b. [DOI] [PubMed] [Google Scholar]
- Colman SD, Williams CA, Wallace MR. Benign neurofibromas in type 1 neurofibromatosis (NF1) show somatic deletions of the NF1 gene. Nat Genet. 1995;11:90–92. doi: 10.1038/ng0995-90. [DOI] [PubMed] [Google Scholar]
- Daston MM, Ratner N. Neurofibromin, a predominantly neuronal GTPase activating protein in the adult, is ubiquitously expressed during development. Dev Dyn. 1992;195:216–226. doi: 10.1002/aja.1001950307. [DOI] [PubMed] [Google Scholar]
- Daston MM, Scrable H, Nordlund M, Sturbaum AK, Nissen LM, Ratner N. The protein product of the neurofibromatosis type 1 gene is expressed at highest abundance in neurons, Schwann cells, and oligodendrocytes. Neuron. 1992;8:415–428. doi: 10.1016/0896-6273(92)90270-n. [DOI] [PubMed] [Google Scholar]
- DeClue JE, Cohen BD, Lowy DR. Identification and characterization of the neurofibromatosis type 1 protein product. Proc Nat Acad Sci. 1991;88:9914–9918. doi: 10.1073/pnas.88.22.9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeClue JE, Papageorge AG, Fletcher JA, Diehl SR, Ratner N, Vass WC, Lowy DR. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell. 1992;69:265–273. doi: 10.1016/0092-8674(92)90407-4. [DOI] [PubMed] [Google Scholar]
- Dono R, Texido G, Dussel R, Ehmke H, Zeller R. Impaired cerebral cortex development and blood pressure regulation in FGF-2-deficient mice. EMBO J. 1998;17:4213–4225. doi: 10.1093/emboj/17.15.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KBJ, Paxinos G. The mouse brain in stereotaxic coordinates. San Diego, CA: Academic Press; 1997. [Google Scholar]
- Gaul U, Mardon G, Rubin GM. A putative Ras GTPase activating protein acts as a negative regulator of signaling by the Sevenless receptor tyrosine kinase. Cell. 1992;68:1007–1019. doi: 10.1016/0092-8674(92)90073-l. [DOI] [PubMed] [Google Scholar]
- Ghebranious N, Donehower LA. Mouse models in tumor suppression. Oncogene. 1998;17:3385–3400. doi: 10.1038/sj.onc.1202573. [DOI] [PubMed] [Google Scholar]
- Glaser EM, Van der Loos H. Analysis of thick brain sections by obverse-reverse computer microscopy: Application of a new, high clarity Golgi-Nissl stain. J Neurosci Methods. 1981;4:117–125. doi: 10.1016/0165-0270(81)90045-5. [DOI] [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase β gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- Guo HF, The I, Hannan F, Bernards A, Zhong Y. Requirement of Drosophila NF1 for activation of adenylyl cyclase by PACAP38-like neuropeptides. Science. 1997;276:795–798. doi: 10.1126/science.276.5313.795. [DOI] [PubMed] [Google Scholar]
- Guo HF, Tong J, Hannan F, Luo L, Zhong Y. A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature. 2000;403:895–898. doi: 10.1038/35002593. [DOI] [PubMed] [Google Scholar]
- Gutmann DH, Aylsworth A, Carey JC, Korf B, Marks J, Pyeritz RE, Rubenstein A, Viskochil D. The diagnostic evaluation and multidisciplinary management of neurofibromatosis 1 and neurofibromatosis 2. J Am Med Assoc. 1997;278:51–57. [PubMed] [Google Scholar]
- Gutmann DH, Loehr A, Zhang Y, Kim J, Henkemeyer M, Cashen A. Haploinsufficiency for the neurofibromatosis 1 (NF1) tumor suppressor results in increased astrocyte proliferation. Oncogene. 1999;18:4450–4459. doi: 10.1038/sj.onc.1202829. [DOI] [PubMed] [Google Scholar]
- Gutmann DH, Wood DL, Collins FS. Identification of the neurofibromatosis type 1 gene product. Proc Natl Acad Sci. 1991;88:9658–9662. doi: 10.1073/pnas.88.21.9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME. Central nervous system neuronal migration. Annu Rev Neurosci. 1999;22:511–539. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Ueki K, Waha A, Wiestler OD, Louis DN, von Deimling A. Association of EGFR gene amplification and CDKN2 (p16/MTS1) gene deletion in glioblastoma multiforme. Brain Pathol. 1997;7:871–875. doi: 10.1111/j.1750-3639.1997.tb00890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoesche C, Sauerwald A, Veh RW, Krippl B, Kilimann MW. The 5′-flanking region of the rat synapsin I gene directs neuron-specific and developmentally regulated reporter gene expression in transgenic mice. J Biol Chem. 1993;268:26494–26502. [PubMed] [Google Scholar]
- Huynh DP, Nechiporuk T, Pulst SM. Differential expression and tissue distribution of type I and type II neurofibromins during mouse fetal development. Dev Biol. 1994;161:538–551. doi: 10.1006/dbio.1994.1052. [DOI] [PubMed] [Google Scholar]
- Jacks T. Tumor suppressor gene mutations in mice. Annu Rev Genet. 1996;30:603–636. doi: 10.1146/annurev.genet.30.1.603. [DOI] [PubMed] [Google Scholar]
- Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- Johnson MR, DeClue JE, Felzmann S, Vass WC, Xu G, White R, Lowy DR. Neurofibromin can inhibit Ras-dependent growth by a mechanism independent of its GTPase-accelerating function. Mol Cell Biol. 1994;14:641–645. doi: 10.1128/mcb.14.1.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HA, Rosenbaum T, Marchionni MA, Ratner N, DeClue JE. Schwann cells from neurofibromin deficient mice exhibit activation of p21ras, inhibition of cell proliferation and morphological changes. Oncogene. 1995;11:325–335. [PubMed] [Google Scholar]
- Klesse LJ, Parada LF. p21 ras and phosphatidylinositol-3 kinase are required for survival of wild-type and NF1 mutant sensory neurons. J Neurosci. 1998;18:10420–10428. doi: 10.1523/JNEUROSCI.18-24-10420.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb B, McClimans J. Cryostat sectioning of Golgi-Cox tissue. Stain Technol. 1986;61:379–380. doi: 10.3109/10520298609113588. [DOI] [PubMed] [Google Scholar]
- Kraig RP, Dong LM, Thisted R, Jaeger CB. Spreading depression increases immunohistochemical staining of glial fibrillary acidic protein. J Neurosci. 1991;11:2187–2198. doi: 10.1523/JNEUROSCI.11-07-02187.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legius E, Marchuk DA, Collins FS, Glover TW. Somatic deletion of the neurofibromatosis type 1 gene in a neurofibrosarcoma supports a tumour suppressor gene hypothesis. Nat Genet. 1993;3:122–126. doi: 10.1038/ng0293-122. [DOI] [PubMed] [Google Scholar]
- Li Y, O'Connell P, Breidenbach HH, Cawthon R, Stevens J, Xu G, Neil S, Robertson M, White R, Viskochil D. Genomic organization of the neurofibromatosis 1 gene (NF1) Genomics. 1995;25:9–18. doi: 10.1016/0888-7543(95)80104-t. [DOI] [PubMed] [Google Scholar]
- Mandell JW, VandenBerg SR. ERK/MAP kinase is chronically activated in human reactive astrocytes. Neuroreport. 1999;10:3567–3572. doi: 10.1097/00001756-199911260-00019. [DOI] [PubMed] [Google Scholar]
- Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, Conroy L, Clark R, O'Connell P, Cawthon RM, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- McClatchey AI, Jacks T. Tumor suppressor mutations in mice: The next generation. Curr Opin Genet Dev. 1998;8:304–310. doi: 10.1016/s0959-437x(98)80086-5. [DOI] [PubMed] [Google Scholar]
- Melloni RH, Jr, DeGennaro LJ. Temporal onset of synapsin I gene expression coincides with neuronal differentiation during the development of the nervous system. J Comp Neurol. 1994;342:449–462. doi: 10.1002/cne.903420311. [DOI] [PubMed] [Google Scholar]
- Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- Nordlund M, Gu X, Shipley MT, Ratner N. Neurofibromin is enriched in the endoplasmic reticulum of CNS neurons. J Neurosci. 1993;13:1588–1600. doi: 10.1523/JNEUROSCI.13-04-01588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund ML, Rizvi TA, Brannan CI, Ratner N. Neurofibromin expression and astrogliosis in neurofibromatosis (type 1) brains. J Neuropathol Exp Neurol. 1995;54:588–600. doi: 10.1097/00005072-199507000-00013. [DOI] [PubMed] [Google Scholar]
- North K. Cognitive function and academic performance. In: Friedman JM, Gutmann DH, MacCollin M, Riccardi VM, editors. Neurofibromatosis: Phenotype, natural history, and pathogenesis. Baltimore, MD: Johns Hopkins Press; 1999. pp. 162–189. [Google Scholar]
- Peltonen J, Jaakkola S, Lebwohl M, Renvall S, Risteli L, Virtanen I, Uitto J. Cellular differentiation and expression of matrix genes in type 1 neurofibromatosis. Lab Invest. 1988;59:760–771. [PubMed] [Google Scholar]
- Rabchevsky AG, Weinitz JM, Coulpier M, Fages C, Tinel M, Junier MP. A role for transforming growth factor alpha as an inducer of astrogliosis. J Neurosci. 1998;18:10541–10552. doi: 10.1523/JNEUROSCI.18-24-10541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon y Cajal S. Histology of the nervous system. New York and UK: Oxford University Press; 1904. [Google Scholar]
- Riccardi VM. Historical background and introduction. In: Friedman JM, Gutmann DH, MacCollin M, Riccardi VM, editors. Neurofibromatosis: phenotype, natural history, and pathogenesis. Baltimore, MD: Johns Hopkins Press; 1999. pp. 1–25. [Google Scholar]
- Ridet JL, Malhotra SK, Privat A, Gage FH. Reactive astrocytes: Cellular and molecular cues to biological function. Trends Neurosci. 1997;20:570–577. doi: 10.1016/s0166-2236(97)01139-9. [DOI] [PubMed] [Google Scholar]
- Rizvi TA, Akunuru S, de Courten-Myers G, Switzer RC, III, Nordlund ML, Ratner N. Region-specific astrogliosis in brains of mice heterozygous for mutations in the neurofibromatosis type 1 (Nf1) tumor suppressor. Brain Res. 1999;816:111–123. doi: 10.1016/s0006-8993(98)01133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum T, Boissy YL, Kombrinck K, Brannan CI, Jenkins NA, Copeland NG, Ratner N. Neurofibromin-deficient fibroblasts fail to form perineurium in vitro. Development. 1995;121:3583–3592. doi: 10.1242/dev.121.11.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosman NP, Pearce J. The brain in multiple neurofibromatosis (von Recklinghausen's disease): A suggested neuropathological basis for the associated mental defect. Brain. 1967;90:829–838. doi: 10.1093/brain/90.4.829. [DOI] [PubMed] [Google Scholar]
- Sawada S, Florell S, Purandare SM, Ota M, Stephens K, Viskochil D. Identification of NF1 mutations in both alleles of a dermal neurofibroma. Nat Genet. 1996;14:110–112. doi: 10.1038/ng0996-110. [DOI] [PubMed] [Google Scholar]
- Schlegel J, Merdes A, Stumm G, Albert FK, Forsting M, Hynes N, Kiessling M. Amplification of the epidermal-growth-factor-receptor gene correlates with different growth behaviour in human glioblastoma. Int J Cancer. 1994;56:72–77. doi: 10.1002/ijc.2910560114. [DOI] [PubMed] [Google Scholar]
- Side L, Taylor B, Cayouette M, Conner E, Thompson P, Luce M, Shannon K. Homozygous inactivation of the NF1 gene in bone marrow cells from children with neurofibromatosis type 1 and malignant myeloid disorders. N Engl J Med. 1997;336:1713–1720. doi: 10.1056/NEJM199706123362404. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Frankland PW, Marowitz Z, Friedman E, Lazlo G, Cioffi D, Jacks T, Bourtchuladze R. A mouse model for the learning and memory deficits associated with neurofibromatosis type I. Nat Genet. 1997;15:281–284. doi: 10.1038/ng0397-281. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Steward O, Torre ER, Tomasulo R, Lothman E. Neuronal activity up-regulates astroglial gene expression. Proc Natl Acad Sci. 1991;88:6819–6823. doi: 10.1073/pnas.88.15.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit WJ, Walter SA, Pennell NA. Reactive microgliosis. Prog Neurobiol. 1999;57:563–581. doi: 10.1016/s0301-0082(98)00069-0. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nakafuku M, Satoh T, Marshall MS, Gibbs JB, Matsumoto K, Kaziro Y, Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990;60:803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- The I, Hannigan GE, Cowley GS, Reginald S, Zhong Y, Gusella JF, Hariharan IK, Bernards A. Rescue of a Drosophila NF1 mutant phenotype by protein kinase A. Science. 1997;276:791–794. doi: 10.1126/science.276.5313.791. [DOI] [PubMed] [Google Scholar]
- Trahey M, McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987;238:542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Trahey M, Wong G, Halenbeck R, Rubinfeld B, Martin GA, Ladner M, Long CM, Crosier WJ, Watt K, Koths K, et al. Molecular cloning of two types of GAP complementary DNA from human placenta. Science. 1988;242:1697–1700. doi: 10.1126/science.3201259. [DOI] [PubMed] [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, Tonegawa S. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- Upadhyaya M, Shaw DJ, Harper PS. Molecular basis of neurofibromatosis type 1 (NF1): Mutation analysis and polymorphisms in the NF1 gene. Hum Mutat. 1994;4:83–101. doi: 10.1002/humu.1380040202. [DOI] [PubMed] [Google Scholar]
- van der Geer P, Henkemeyer M, Jacks T, Pawson T. Aberrant Ras regulation and reduced p190 tyrosine phosphorylation in cells lacking p120-Gap. Mol Cell Biol. 1997;17:1840–1847. doi: 10.1128/mcb.17.4.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viskochil DH. The structure and function of the NF1 gene: Molecular pathology. In: Friedman JM, Gutmann DH, MacCollin M, Riccardi VM, editors. Neurofibromatosis: Phenotype, natural history, and pathogenesis. Baltimore, MD: Johns Hopkins Press; 1999. pp. 119–141. [Google Scholar]
- Viskochil D, Buchberg AM, Xu G, Cawthon RM, Stevens J, Wolff RK, Culver M, Carey JC, Copeland NG, Jenkins NA, et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–192. doi: 10.1016/0092-8674(90)90252-a. [DOI] [PubMed] [Google Scholar]
- Vogel KS, Brannan CI, Jenkins NA, Copeland NG, Parada LF. Loss of neurofibromin results in neurotrophin-independent survival of embryonic sensory and sympathetic neurons. Cell. 1995;82:733–742. doi: 10.1016/0092-8674(95)90470-0. [DOI] [PubMed] [Google Scholar]
- Vogel US, Dixon RA, Schaber MD, Diehl RE, Marshall MS, Scolnick EM, Sigal IS, Gibbs JB. Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature. 1988;335:90–93. doi: 10.1038/335090a0. [DOI] [PubMed] [Google Scholar]
- Voiculescu O, Charnay P, Schneider-Maunoury S. Expression pattern of a Krox-20/Cre knock-in allele in the developing hindbrain, bones, and peripheral nervous system. Genesis. 2000;26:123–126. doi: 10.1002/(sici)1526-968x(200002)26:2<123::aid-gene7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, Saulino AM, Fountain JW, Brereton A, Nicholson J, Mitchell AL, et al. Type 1 neurofibromatosis gene: Identification of a large transcript disrupted in three NF1 patients. Science. 1990;249:181–186. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- Xu GF, O'Connell P, Viskochil D, Cawthon R, Robertson M, Culver M, Dunn D, Stevens J, Gesteland R, White R, et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- Xu W, Mulligan LM, Ponder MA, Liu L, Smith BA, Mathew CG, Ponder BA. Loss of NF1 alleles in phaeochromocytomas from patients with type I neurofibromatosis. Genes Chromosomes Cancer. 1992;4:337–342. doi: 10.1002/gcc.2870040411. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Parada LF. Neurofibromin, a tumor suppressor in the nervous system. Exp Cell Res. 2001;264:19–28. doi: 10.1006/excr.2000.5138. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Richardson JA, Parada LF, Graff JM. Smad3 mutant mice develop metastatic colorectal cancer. Cell. 1998;94:703–714. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]