Abstract
A concrete example of the collaborative double-robust targeted likelihood estimator (C-TMLE) introduced in a companion article in this issue is presented, and applied to the estimation of causal effects and variable importance parameters in genomic data. The focus is on non-parametric estimation in a point treatment data structure. Simulations illustrate the performance of C-TMLE relative to current competitors such as the augmented inverse probability of treatment weighted estimator that relies on an external non-collaborative estimator of the treatment mechanism, and inefficient estimation procedures including propensity score matching and standard inverse probability of treatment weighting. C-TMLE is also applied to the estimation of the covariate-adjusted marginal effect of individual HIV mutations on resistance to the anti-retroviral drug lopinavir. The influence curve of the C-TMLE is used to establish asymptotically valid statistical inference. The list of mutations found to have a statistically significant association with resistance is in excellent agreement with mutation scores provided by the Stanford HIVdb mutation scores database.
Keywords: causal effect, cross-validation, collaborative double robust, double robust, efficient influence curve, penalized likelihood, penalization, estimator selection, locally efficient, maximum likelihood estimation, model selection, super efficiency, super learning, targeted maximum likelihood estimation, targeted nuisance parameter estimator selection, variable importance
1. Introduction
Targeted maximum likelihood estimation (TMLE) is a double robust method for estimating causal effects and their non-causal analogs, variable importance measures (van der Laan and Rubin, 2006). Observed data can be viewed as realizations of random variables arising from some true underlying data-generating distribution. An association or causal effect corresponds with some particular parameter of this unknown underlying distribution. TMLE is a two stage semi-parametric estimation methodology that minimizes bias in the estimate of this parameter of interest. Stage one estimates the density of the data-generating distribution. In stage two this initial density estimate is fluctuated in a manner specifically designed to provide the maximal change in the estimate of the target parameter. This targeted fluctuation is based on the efficient influence curve of the parameter, and involves estimating a nuisance parameter that can include a treatment mechanism, a missingness mechanism, and/or a censoring mechanism.
Collaborative targeted maximum likelihood estimation (C-TMLE) is an extension of TMLE that pursues an optimal strategy for nuisance parameter estimation. Theory advanced in van der Laan and Gruber (2010) provides the key insight that only the portion of the nuisance parameter that is not adequately accounted for in the first stage needs to be incorporated into the second stage fluctuation. Stage two of the C-TMLE approach exploits this collaborative double-robustness finding by creating a sequence of nuisance parameter estimates that grow increasingly larger, i.e., more and more non-parametric. Construction of the nuisance parameter estimates is guided data-adaptively based on the goodness-of-fit of the overall density estimate and its effect on the mean squared error of the estimate of the target parameter. In other words, each nuisance parameter estimate in the sequence is carefully constructed to provide the next in a series of fluctuations of the initial density estimate, and each fluctuation is carried out to create a series of candidate TMLE estimators. Likelihood-based cross-validation (possibly using the penalized likelihood) selects the best candidate for the given stage one estimator. This procedure can be carried out for multiple stage one estimators. The C-TMLE estimator is defined as the best among the set of candidate TMLE estimators indexed by stage one and stage two candidates, as chosen by likelihood-based cross-validation. In finite samples the C-TMLE estimator will often be more efficient than the standard TMLE estimator that utilizes an estimate of the entire nuisance parameter in the targeting step. C-TMLE enjoys all the properties of the standard TMLE estimator, namely, it is double robust and asymptotically efficient under appropriate regularity conditions..
C-TMLE is a general methodology that can be applied to a variety of estimators in many settings, including survival analysis, gene association studies, and longitudinal data structures. In this article we focus on one specific application of C-TMLE, non-parametric estimation of a point treatment effect. For simplicity, we limit the discussion of C-TMLE methodology to the simplest case in which there is only one stage one estimate. The extension to multiple stage one candidate estimators is straightforward. The performance of collaborative targeted maximum likelihood estimation methodology is compared with that of established estimation methods: G-computation, inverse probability of treatment weighting (IPTW), propensity score-based causal effect estimation, and unadjusted regression estimation of a point treatment effect. Results from three simulation studies designed to mimic commonly-encountered scenarios demonstrate the versatility of the CTMLE estimator. A fourth simulation contrasts C-TMLE with standard targeted maximum likelihood estimation (TMLE) introduced in van der Laan and Rubin (2006).
The paper is organized as follows. Section 2 describes TMLE and defines a new collaborative double robustness property that motivates the development of CTMLE. Other common estimators of causal effects and variable importance parameters found in the literature (G-computation, propensity score-based, unadjusted, and inverse probability of treatment weighted estimators) are reviewed in Section 3. A particular C-TMLE implementation and an influence curve-based method for obtaining inference is described in Section 4. Section 5 presents three simulations designed to offer a performance comparison across a variety of scenarios common to many analyses of real-world data. In Section 6 we compare the performance of the new C-TMLE estimator with standard TMLE. Variable importance estimates of the effect of HIV mutations on resistance to the anti-retroviral drug lopinavir are presented in Section 7. The paper concludes with a discussion in Section 8. An appendix provides a note on modification to the TMLE procedure that maintains its properties but also enforces Ψ(Qn) as an imputation estimator. In addition, R source code that implements and demonstrates applying C-TMLE to analyze simulated data is provided as supplemental materials.
2. Collaborative targeted maximum likelihood estimation
We review targeted maximum likelihood estimation of the additive treatment effect before defining the collaborative targeted maximum likelihood approach. Suppose we have a data set containing n independent and identically distributed observations, O1, . . . , On, of a random variable O = (W, A, Y), where W is a set of baseline covariates, A is a treatment variable, and Y is the outcome variable. For simplicity we focus on binary A, A = 1 denotes treatment, and A = 0 denotes control. The outcome variable can either be continuous or binary. Assume we are interested in estimating the marginal additive causal effect of treatment on the outcome. The parameter of interest of the probability distribution P0 of O is therefore defined non-parametrically as ψ0 = EW(E(Y | A = 1, W) − E(Y | A = 0, W)). Under the appropriate causal graph assumptions ψ0 corresponds with the G-computation formula for the marginal additive causal effect.
The probability distribution/density of O can be factored as P0(O) = Q0(O)g0(A | W), where Q0(O) = QY0(Y | A, W)QW0(W) and g0(1 | W) = P0(A = 1 | W). We used the notation QY for a conditional distribution of Y, given A, W, and QW for the marginal distribution of W. For notational convenience, let Q0(A, W) = E0(Y | A, W) be the true conditional mean of Y, given A, W, which is thus a parameter of QY 0. We note that ψ0 = Ψ (Q0) only depends on the data generating distribution P0 through its Q0-factor. The targeted maximum likelihood estimator of ψ0 is a particular substitution estimator
where Qn(A, W) is an estimated conditional mean of Y given A, W, and the marginal distribution QW0 is estimated with its empirical probability distribution.
Targeted maximum likelihood estimation involves obtaining an initial estimate of the true conditional mean of Y given A and W, and subsequently fluctuating this estimate in a manner designed to reduce bias in the estimate of the parameter of interest. Let be the initial estimate of the true conditional mean Q0(A, W). For example, if Y is binary, then one constructs a parametric (least favorable) model , fluctuating the initial estimate , where ɛ is the fluctuation parameter. The function h(A, W), known as the “clever covariate”, depends on the treatment assignment mechanism g0, and is given by
| (1) |
The theoretical basis for this choice of clever covariate is given in van der Laan and Rubin (2006). In particular, it has the bias-reduction property that if one estimates ɛ with the parametric maximum likelihood estimator, and one sets equal to the resulting update, then the resulting substitution estimator is asymptotically unbiased, even if the initial estimator is inconsistent. These results indicate that estimating g0 is crucial for reducing bias. However, the choice of an estimator gn should be evaluated by how it affects the mean squared error of the resulting targeted maximum likelihood estimator , making it a harder and different problem than estimating g0 itself.
TMLE has been shown to be double robust, i.e. the estimate is consistent if either the limits of or gn are correctly specified. When both are correct, the estimator is efficient (van der Laan and Rubin, 2006). Recent theoretical advances show that TMLE is also collaboratively double robust (van der Laan and Gruber, 2010). That is, if the initial estimator converges to a possibly misspecified Q, then gn needs to only converge to a conditional distribution of A that properly adjusts for a covariate that is a function of Q0 − Q. This result is intuitively a natural consequence of the fact that the clever covariate can only reduce bias if it is predictive of the outcome after taking into account the initial estimator. This collaborative double robustness property and a corresponding asymptotic linearity theorem are proven in a companion article in this issue.
A particular method for construction of a collaborative estimator gn involves building candidate treatment mechanism estimators that grow towards an unbiased estimator of the fully adjusted g0. In a departure from current practice, the construction of these candidates is guided by the log-likelihood loss function for Q0, thus not by the log-likelihood loss function for the conditional distribution of A given W, hence our use of the term “collaborative.”
Clever covariates based on these candidates give rise to a sequence of updated estimates, , each of which provides a candidate TMLE estimate of ψ0. The C-TMLE estimate is the best among these candidates, as determined by V-fold Q0-log-likelihood-based cross-validation.
3. Current methods for estimating marginal causal treatment effects
Current methods for estimating the marginal causal effect of a treatment A on outcome Y are compared with C-TMLE on simulated data below. The estimators under consideration are the G-computation estimator (Robins, 1986), the IPTW estimator (Hernan et al. (2000), Robins (2000b)), a double robust IPTW estimator (DR-IPTW), (Robins and Rotnitzky (2001); Robins et al. (2000); Robins (2000a)), a propensity score estimator (Rosenbaum and Rubin, 1983) that calculates the marginal treatment effect as the mean across strata defined by the conditional probability of receiving treatment, and an extension to propensity score estimators implemented in Matching, a publicly available R package (Sekhon (2008)).
Recall that our parameter of interest is given by: ψ0 = EW[E[Y | A = 1, W] − E[Y | A = 0, W]]. Each of the estimators we are considering rely on estimates of one or both of the following: Q0(A, W) ≡ E[Y | A, W] and g0(A, W) ≡ P(A | W). The first conditional distribution can be estimated by, for example, a regression of Y on A and W. The second, which we refer to as the treatment mechanism, is sometimes known, for example in a randomized trial. When the treatment mechanism is unknown it can be estimated by a logistic regression of A on W. Each estimator is defined below.
where refers to an initial estimate of Q0(A, W), refers to an updated targeted estimate of Q0(A, W), described in detail in the next section. For the propensity score method, si indicates a stratum of the propensity score of covariate vector Wi, and denotes an estimator of the true conditional mean E(Y | A = a, S = s) given treatment and propensity score. In the last equation mi indicates a set of matched observations to which subject i is assigned, where matches are based on minimizing a distance between the user supplied covariates W. Each set of matched observations indexed by m results in a corresponding mean regression representing an estimate of E(Y | A = a, M = m). The creation of the partitioning in sets of matched observations is only a function of the data (Wi, Ai), i = 1, . . . , n, thus ignoring the outcome data.
Regarding asymptotic properties of the estimators, the G-computation estimator relies on consistent estimation of Q0, the IPTW estimator relies on consistent estimation of g0, while the DR-IPTW estimator yields consistent estimates if one or both nuisance parameters are estimated consistently.
Notice that is a G-computation estimate. However, unlike G-computation, which is consistent only when Qn is a consistent estimator for Q0, C-TMLE estimates are consistent if either Q0 or g0 is estimated consistently. can equivalently be formulated as a double-robust IPTW estimator:
The propensity score method implemented uses the Deletion/Substitution/Addition (DSA) algorithm (Sinisi and van der Laan, 2004) to model conditional treatment probabilities given covariates W. This data-adaptive algorithm searches over a large space of polynomial models by adding, subtracting, or substituting terms, starting with a base user-specified regression model. The final model, selected by cross-validation with the L2 loss function, was used to estimate a propensity score for each observation. Observations were then divided into five strata based on the quantiles of these propensity scores. Regression of Y on A and strata indicator variables using the full model enabled the calculation of stratum-specific treatment effects, which were averaged to obtain the marginal effect. The Matching estimator generalizes the propensity score approach by carefully matching observations in the treatment and control groups in such a way that potential confounders are evenly distributed, across the matches.
The Matching procedure relies on the genetic algorithm (Holland and Reitman (1977)) to achieve this goal. This is a non-parametric approach for selecting weights on covariates that are in turn are used to determine which observations are matched. Candidate sets of matches are evaluated based on a loss function and a distance metric specified at run-time, and are used to generate successive sets of candidates that achieve good balance Sekhon (2008). The marginal treatment effect is the average effect across strata defined by the matches.
Propensity score methods are especially effective when overall match quality is a function of true confounders. Estimates can suffer even when overall match quality is high if a small subset of covariates responsible for introducing the most bias into the estimate is unevenly distributed between treatment and control groups. Because matches are made without regard to the outcome variable, these methods do not exploit all information available in the data and are known to be less than fully efficient (Abadie and Imbens, 2006). A violation of the experimental treatment assignment assumption, also called the positivity assumption, is known to reduce the quality of the match and introduce bias into the estimate, and can be detected once the matches have been specified. The lack of identifiability as measured by such an assumption results in potential bias for each method, but the augmented IPTW, targeted MLE, and G-computation method allow reliance on extrapolation.
4. C-TMLE implementation
The general C-TMLE procedure is to create several stage 1 (non-targeted) density estimators and carry out stage 2 procedures for each of these. Penalized cross-validation is used to choose among the final candidate estimators that are indexed by stage 1 and stage 2 candidates. The implementation presented here is based on only one initial stage 1 estimate for simplicitly. We describe specific choices that were employed for the simulations and data analysis presented in the following sections, occasionally noting other implementation options.
Step 1: Obtain an estimate of Q0(A, W). A data-adaptive machine learning approach to obtaining this initial estimate is recommended. The super learner (SL) is a prediction algorithm that creates a weighted combination of predictions of many individual prediction algorithms, with weights selected using V-fold cross-validation (van der Laan et al., 2007). In practice, it is important to include algorithms in the SL library of predictors that cover different model spaces, e.g. support vector machines, splines, neural nets, etc., since the true best estimation method is unknown. If SL is not used, any particular data adaptive machine learning algorithm providing a consistent estimate is acceptable. For the simulations described in the next section, the DSA algorithm was used to provide the initial estimate of the the true regression of Y on treatment A and confounders W.
-
Step 2: Generate candidate second stage estimators . In the simulations forward selection was used to build a sequence of updates for g0 that are increasing in size.
Though not required, a sensible approach is to use the intercept model for g to construct a first clever covariate, h1, used to create the first targeted maximum likelihood candidate, .
This results in the first candidate second stage estimator , where ɛ1 is fitted by least-squares regression of Y on h1 with offset . Next we create an updated model for g by adding a main term to the intercept, and the resulting targeted MLE using the corresponding clever covariate is evaluated. The best main term is selected based on a penalized log-likelihood criterion for the targeted MLE fit. Additional terms are incorporated in the g-fit as long as they increase the overall penalized log-likelihood for the resulting Q0-targeted MLE fit. Thus the penalized likelihood is defined as the empirical sum of squared residuals at the resulting Q0-fit plus a penalty term proportional to the estimated variance of the target parameter, the empirical variance of D*, the main component of the efficient influence curve (see below), at the resulting Q0-fit and the candidate g-fit. In the event that no terms in the model for g increase the penalized likelihood of the resulting Q0-fit, the targeted MLE update is carried out with the clever covariate that provided the best penalized log-likelihood, and the above process is iterated with this new initial estimator and next clever covariate indexed by g fits that are still building on last g-fit. -
As an example suppose that in addition to the intercept term, m terms, ordered 1, . . . , m, are incorporated into the model for g, at which point no further increase of the penalized log likelihood is possible. We define candidate estimators through as:
where the corresponding models contains all the terms in the model for plus one additional term, i = 2, . . . , m. At this point is considered as a new “initial” estimate of the true regression, and the entire process starts over in order to build a second clever covariate augmenting the previous fit used in hm+1. To continue the example, . This process is iterated until all terms are incorporated into the final model for g. If the maximal number of terms that can be added is given by K, then this results in K candidate estimators , k = 1, . . . , K, corresponding with treatment mechanism estimators , k = 1, . . . , K. Note that the number of clever covariates in that are added to the initial estimator cannot be predicted, and depends on how many covariates can be added to the treatment mechanism estimator in each iteration before reaching the local maximum (not allowing a further increase of the penalized log-likelihood).Note that the model for g is not restricted to main terms only. For example, variables can be created that correspond to higher-order terms. In addition, a categorical or continuous covariate can be split into many binary covariates, thereby allowing for more nonparametric modeling of the effect of a single covariate. When there are many covariates it might be desirable in practice to terminate the procedure before all covariates have been incorporated into the model for g, though care must be taken to ensure that none of the candidates thereby excluded from the subsequent selection process potentially maximize the penalized log-likelihood criterion. SL can be integrated into the second stage as well. A series of increasingly non-parametric propensity score SL estimates can be obtained based on different adjustment sets. These SL fits are used as the main terms for the stage 2 forward selection to build candidate ĝ estimators.
The presented algorithm illustrates that the number of clever covariates used to update the initial estimator depends entirely on the likelihood and cannot be pre-determined. Terms are incorporated into the model for g for a single clever covariate until there is a decrease in the likelihood. At that point the estimate is updated from and the process iterates until all candidate TMLEs have been constructed.
We also note that we can represent these estimators and corresponding treatment mechanism estimators as mappings Q̂k and ĝk applied to the empirical distribution Pn: , , k = 1, . . . , K. These mappings Pn → Q̂k(Pn) represent our candidate estimators of the true regression Q0, and in the next step we use cross-validation to select among these candidate algorithms.
- Step 3: Select the estimator that maximizes the V-fold cross-validated penalized likelihood, where V was set to 5. Maximizing the penalized likelihood is equivalent to minimizing the residual sum of squares (RSS) plus a penalty term corresponding to the mean squared error (MSE), which can be decomposed into variance and bias terms:
These terms are defined as follows:
where v ranging from 1 to V indexes the validation set Val(v) for the vth fold, Ψ(Q) is a mapping from Q to the parameter of interest, and denotes the k-th C-TMLE applied to the corresponding training sample , containing n(1 − p) observations, with p = 1/V.
There are many variations for obtaining . For example, given an a priori set of candidate nuisance parameter estimators, ĝj, that includes highly nonparametric candidates we could construct clever covariates hj(g), and then use forward selection with this set of clever covariates, using the initial estimator as off-set, to build (second stage) model-fits for Q0 of increasing size, where each term in the model corresponds to one of the clever covariates. The number of clever covariates that are added in this forward-selection algorithm can be selected using likelihood-based cross-validation.
Note that in contrast with the algorithm described above, in which previous coefficients are used as fixed offsets in the regression, coefficients in front of each term are estimated by least squares, thereby solving the efficient influence equation corresponding to each ĝj, in particular the most non-parametric of these. Because these covariates are highly correlated, refitting all coefficients in front of clever covariates at each step in the forward selection algorithm is likely to result in highly variable coefficient estimates, and therefore less stability in the estimate of the parameter of interest.
Another alternative approach is to define , where is the targeted MLE updating the initial estimator with the final selected clever covariate defined by carrying out the k* moves in the above forward selection algorithm to obtain a g-fit, where k* is the optimal number of moves selected by likelihood-based cross-validation (exactly as above). This variation did not improve performance in simulation studies not presented in this article. We mention these alternatives only to underscore the fact that C-TMLE methodology can be implemented in a variety of ways, and is not limited to the specific implementation presented here.
4.1. Inference
The variance of the influence curve (IC) of the C-TMLE provides suitable inference, under certain regularity conditions, and assuming that the collaborative estimator gn converges to a g0 = g0(Q), where g0(Q) represents a true conditional distribution of A given W(Q) for a subset or reduction W(Q) of all covariates W, so that P0D* (Q, g0(Q), ψ0) = 0. For example, it suffices that the limit g0(Q) is a true conditional distribution of A, given W(Q), for a W(Q) such that (Q0 − Q)(1, W), (Q0 − Q)(0, W) only depend on W through W(Q). For details we refer to the companion paper van der Laan and Gruber (2010). The asymptotics theorem presented in that paper states that ψn is an asymptotically linear estimator of ψ0 with influence curve
where ICg0 denotes the influence curve of the linearization of P0D* (Q, gn, ψ0) viewed as an estimator of P0D* (Q, g0, ψ0). This additional term ICg0 represents the contribution to the influence curve from the estimator gn. The formula for the efficient influence curve/canonical gradient D* (Q, g0, ψ0) is given in the previous section for the particular causal effect parameter, ψ0 = EW[E[Y | A = 1, W] − E[Y | A = 0, W]].
In our application of C-TMLE, gn is a data adaptively selected logistic regression model fitted with maximum likelihood estimation. Thus, if we define {gα : α} as the logistic regression model selected, and αn is the MLE, then gn = gαn. We will approximate ICg0 with the influence curve of the asymptotic linearization of P0D* (Q, gαn, ψ0) − D* (Q, gα, ψ0). This ICg0 can now be determined with a straightforward application of the delta method. The formula for ICg0 we derived is given by:
where
The notation W⃗ is used to denote the vector of main terms that is included in the logistic regression model gαn. Note that a0 is a vector of the same dimension as W⃗.
This influence curve is estimated by its empirical analog, given by:
where
The standard error of the C-TMLE is now estimated as , where is the sample variance of the estimated influence curve. A 95% confidence interval (CI) is constructed as ψn ± 1.96SE(ψn). The bootstrap is an alternative valid method for asymptotically valid inference, but it is much more computationally intensive.
We remark that it is good practice to incorporate the additional term ICg0 in the influence curve, thereby targeting the true influence curve of the estimator. We can provide the following qualitative understanding of the contribution of ICg0 to the influence curve of the estimator. If converges to the true Q0, then the term ICg0 equals zero, and if is inconsistent, and gn converges to the fully adjusted g0, then ICg0 is known to reduce the variance of the influence curve (section 2.3.4 van der Laan and Robins (2003)). Based on these two facts, we suggest that ignoring the contribution ICg0 will typically result in asymptotically conservative confidence intervals. Empirical evidence presented in Section 6 using finite samples (n = 1000) supports this. However, from a theoretical point of view, there seems to be no guarantee that ICg0 always reduces the variance.
5. Simulation studies
For each simulation we have a data structure O = (W, A, Y), where W = (W1, . . . , W6) is a set of potential confounders of the relationship between binary treatment variable A and continuous outcome Y. Our parameter of interest is the marginal causal effect of treatment on the outcome: ψ = EW[E[Y | A = 1, W] − E[Y | A = 0, W]]. The simulations are designed to demonstrate estimator performance in the face of confounding of the relationship between treatment and outcome, complex underlying data-generating distributions, and practical violations of the Experimental Treatment Assumption (ETA), i.e., P(A = a | W) < α, for some small α, implying that there is very little possibility of observing both treated and untreated subjects for some combination of covariates present in the data.
These simulations are designed specifically to illustrate features of the C-TMLE estimator, and there are other simulations for which the relative performance of the estimators would differ. For example, when a correct model for the underlying data generating distribution is known, a parametric regression approach would be optimal. When the outcome is rare, as is often the case in safety analysis, we would not expect the initial fit, , to have much predictive power. In this case, the fully adjusted g0 is very likely needed for full bias reduction, so creating and evaluating intermediate candidates with C-TMLE may be needlessly computationally expensive. Standard TMLE might be a better approach. Adjusting for many confounders may lead to violations of the ETA assumption when n is small relative to the number of confounders or if the confounders are very strongly predictive of treatment. There are two ways to deal with this. First, one could extrapolate based on model assumptions to arrive at an estimate of the desired parameter. Secondly, one could acknowledge that the parameter of interest is not identifiable from the data, and choose an adjustment set that provides bias reduction without yielding an estimate with variance so large that it is essentially meaningless. This latter approach is taken by second stage of the C-TMLE procedure, by basing the selection of confounders based on the penalized log-likelihood, while the extrapolation approach is still present through the initial first stage estimator.
5.1. Data generation
Covariates W1, . . . W5 were generated as independent normal random variables. W6 is a binary variable.
Two treatment mechanisms were defined:
The observed outcome Y was generated as
with corresponding regression equations:
We consider three different data-generating distributions, (Q1,0, g1,0) in simulation 1, (Q2,0, g1,0) in simulation 2, and (Q2,0, g2,0) in simulation 3. Note that W6 is strongly correlated with treatment mechanism A in simulations 1 and 2 (corr=0.54), but is not an actual confounder of the relationship between A and Y. W1, W2, and W3 are confounders. The linear nature of the confounding due toW3 in simulation 1 differs from that in simulations 2 and 3, where the true functional form is quadratic. In this way simulations 2 and 3 mimic realistic data analysis scenarios in which the unknown underlying functional form is seldom entirely captured by the regression model used in the analysis. Finally, the treatment mechanism in simulations 1 and 2 leads to ETA violations (p(A = a | W) ranges between 9 × 10−7 and 0.9999978, approximately one-third of the probabilites are outside the range (0.05, 0.95)). In simulation 3 there are no ETA violations (0.11 < p(A = a | W) < 0.88). In each simulation the true value of the parameter of interest is 1.
5.2. Simulation
1000 samples of size n = 1000 were drawn from each data generating distribution. Marginal treatment effect estimates were calculated based on the unadjusted regression of Y on A, Gcomp, IPTW, DR-IPTW, propensity score and C-TMLE methods.
A main-effects model for Gcomp and DR-IPTW, Q̂, was obtained using the DSA algorithm with the maximum model size set to seven. A model for the treatment mechanism ĝ used in IPTW, DR-IPTW, propensity score, and Matching estimation was also selected by DSA, again restricted to main terms. The Matching function considered this treatment mechanism model as merely one additional covariate, indistinguishable from the other potential confounders, W. The procedure was run using default settings, except population size for each generation was increased to 200. In contrast, the C-TMLE algorithm includes an aggressive search through a larger space of models to obtain an initial estimate of the density. As a proxy for the super-learner algorithm we used the DSA algorithm to select a model for Q̂ containing at most six terms, allowing quadratic terms and two-way interactions.
We expect to see that the estimators that rely on consistent estimation of Q0 are unbiased in simulation 1, (Gcomp, DR-IPTW, C-TMLE), while estimators relying on consistent estimation of g0 are unbiased in simulation 3 (IPTW, DR-IPTW, propScore, Matching, C-TMLE).
5.3. Results
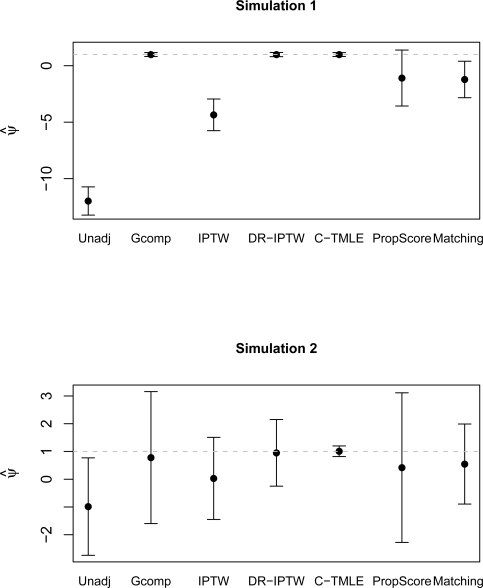
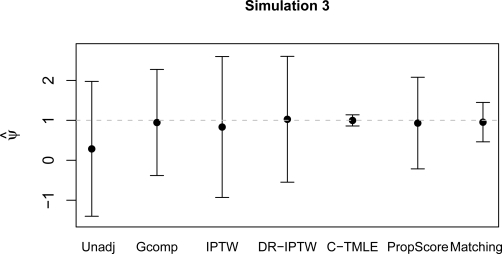
Mean estimates of the treatment effect and standard errors are shown in Table 1 for each simulation. Mean estimates and (0.025, 0.975) quantiles of the probability distribution of each estimator are plotted in Figures 1 and 2.
Table 1:
Mean estimate and standard error (SE) for each estimator based on 1000 iterations with sample size n = 1000. ψ0 = 1.
| Simulation 1
|
Simulation 2
|
Simulation 3
|
||||
|---|---|---|---|---|---|---|
| ψ̄n | SE | ψ̄n | SE | ψ̄n | SE | |
| Unadj | −11.97 | 0.64 | −0.98 | 0.91 | 0.29 | 0.86 |
| Gcomp | 0.99 | 0.09 | 0.76 | 1.22 | 0.95 | 0.68 |
| IPTW | −4.36 | 0.72 | 0.03 | 0.76 | 0.83 | 0.90 |
| DR-IPTW | 0.99 | 0.09 | 0.94 | 0.62 | 1.03 | 0.80 |
| C-TMLE | 0.99 | 0.09 | 1.00 | 0.10 | 1.00 | 0.07 |
| PropScore | −1.09 | 1.27 | 0.42 | 1.38 | 0.93 | 0.59 |
| Matching | −1.22 | 0.82 | 0.54 | 0.73 | 0.96 | 0.25 |
Figure 1:
Mean estimates and (0.025,0.975) quantiles for each estimation method, simulations 1 and 2. Dashed line is at true parameter value.
Figure 2:
Mean estimates and (0.025,0.975) quantiles for each estimation method, simulation 3. Dashed line is at true parameter value.
Figures 1 and 2 illustrate each estimator’s behavior. As expected, estimators relying on consistent estimation of Q0 are unbiased in simulation 1, estimators relying on consistent estimation of g0 are unbiased in simulation 3.
The unadjusted estimator yields biased results in all three simulations due to its failure to adjust for confounders.
The G-computation estimator performs well in simulation 1 when the model is correctly specified. We understand that misspecification (simulations 2 and 3) will often, though not always, lead to bias in the estimates. However the plots highlight another phenomenon that is easy to overlook. the inability of the misspecified model to adequately account for the variance in the outcome often leads to large residual variance of the estimator, and in practice would have low power to reject a null hypothesis.
Truncation bias due to ETA violations causes the IPTW estimator using truncated weights to fail in simulations 1 and 2. The estimate is not biased in simulation 3, but the variance is so large that even in this setting where we’d expect IPTW to be reliable it would fail to produce a significant result.
DR-IPTW estimates are unbiased and have low variance when the functional form is correctly modeled by the regression equation (simulation 1). Though we see little bias in the other two simulations, the variance is large due to misspecification of the treatment mechanism. Because W6 is a strong predictor of A and is indistinguishable from a true confounder of the relationship between Y and A it is always included in the treatment mechanism, behavior that does not help achieve an accurate estimate of the true treatment effect.
Propensity score estimators are known to perform poorly when there are ETA violations, e.g. simulations 1 and 2 (Sekhon (2008)). Researchers constructing the propensity score could observe this and choose an alternate propensity score model, but without using information about the outcome this choice would likely be made based on the predictive power of the model, not the potential bias reduction. Both propensity score-based methods do a reasonable job in simulation 3. Abadie and Imbens (2006) shows that matching estimators will not obtain the semi-parametric efficiency bound. This theory is borne out in simulation 3, where neither matching methodology confidence interval is as tight as that of the collaborative targeted maximum likelihood estimator.
6. Comparison of C-TMLE and TMLE
The double robust property of the targeted maximum likelihood estimator obviates the need for accurate estimation of both Q0 and g0 since correct specification of either one leads to consistent estimates of the parameter of interest. However, accurate estimates of both are needed to achieve the Cramer-Rao efficiency bound. Implementations of the standard targeted maximum likelihood estimator (TMLE) therefore strive for ideal estimates of both Q0 and g0.
In contrast, the collaborative nature of the second stage of the C-TMLE estimation algorithm leads to selection of an estimator, gn, that targets that portion of the treatment mechanism needed to reduce bias not already adequately addressed by the first stage estimator for Q0. For example, covariates included in the model for might not be selected into the model for gn because they do not increase the penalized log-likelihood. At the same time, confounders that are not adequately adjusted for in the initial density estimate are quickly added to model for gn unless the gain in bias reduction is offset by too great an increase in variance. When the initial estimate of the density is a very good fit for the true underlying density, TMLE and C-TMLE have similar performance w.r.t bias, but the C-TMLE will have smaller variance by selecting a gn that targets non fully adjusted g0, resulting in a possibly super efficient estimator. When the initial fit is less good, C-TMLE makes judicious choices regarding inclusion of covariates in the treatment mechanism. As predicted by theory, again, this might lead to lower variances when no covariates cause ETA violations. When inclusion of all confounding covariates does violate the ETA assumption, the C-TMLE estimator, in essence, targets a less ambitious data adaptively selected parameter that is identifiable. Data were simulated to illustrate these phenomena.
6.1. Data generation
Covariates W1, W2, and W3 were generated as independent random uniform variables over the interval [0, 1]. W4 and W5 are independent normally distributed random variables.
Treatment mechanism g0 was designed so that W3 is highly predictive of treatment:
The observed outcome Y was generated as
with corresponding regression equation:
6.2. Simulation
C-TMLE and TMLE estimates of the parameter of interest, again defined as ψ = EW[E[Y | A = 1, W] − E[Y | A = 0, W]], were obtained for 1000 samples of size n = 1000 drawn from data generating distribution (Q0, g0). For this study we deliberately select a misspecified main-terms only model for Q0 by running the DSA algorithm on 100,000 observations drawn from that same distribution. P(A = a | W) for these observations ranges from 0.004 to 0.996. Approximately 17% of the observations have covariates indicating that the probability of receiving treatment is less than 0.05, indicating that practical ETA violations in finite samples will cause unstable TMLE estimates.
For each iteration an initial regression, , was obtained by fitting the DSA-selected model, Y = A + W1 + W2, on n observations in the sample. We expect that any estimate of ψ based solely on this model is likely to be incorrect because the model fails to take into account the effect on the outcome of the missing interaction term, and also fails to adjust for the confounding effect of W5. The targeting step for both targeted maximum likelihood estimators reduces this bias.
In order to construct the covariate used to target the parameter of interest in the updating step of the TMLE algorithm we obtain an estimate gn of g0 by running the DSA algorithm, allowing quadratic terms and two-way interaction terms to enter the model. This model was not fixed over the 1000 iterations; the model selection process was carried out each time a sample was drawn from the population. Similarly, covariates that were candidates for inclusion in the model for gn in the second stage of the C-TMLE estimation algorithm include (W1, . . . , W5, ), and all two-way interaction terms (WiWj), where i ≠ j.
6.3. Results
Results of the simulation are shown in Table 2. A small number of aberrant TMLE estimates were major contributers to the variance of that estimator. The three highest TMLE estimates of the treatment effect were (771.91, 37.22, 9.52). It is likely that these high values arise from atypical samples containing observations that presented unusually strong ETA issues. In contrast, all C-TMLE estimates calculated from those same samples range between 0.307 and 1.698. Both estimators’ average treatment effect estimates are not far from the true value, ψ0 = 1. As expected, the variance of the TMLE estimator is many times larger than that of the C-TMLE estimator.
Table 2:
Comparison of C-TMLE and TMLE estimators at different levels of truncation. Mean estimate and variance based on 1000 iterations.
| truncation level | # obs truncated | ψ̄n | variance | |
|---|---|---|---|---|
| C-TMLE | ∞ | 0 | 0.98 | 0.04 |
| TMLE | ∞ | 0 | 1.73 | 597.52 |
| 40 | 1 | 1.36 | 162.38 | |
| 10 | 2 | 0.94 | 1.99 | |
| 5 | 9 | 0.92 | 1.68 | |
Not surprisingly, W3, the strong predictor of treatment that is not a true confounder of the relationship between treatment and outcome, is included in every one of the 1000 models for gn selected by the DSA algorithm, but it is included in only 35 of the models constructed in the second stage of the C-TMLE algorithm. At the same time, the interaction term W4W5 is included in only two out of 1000 models for g0 selected by DSA, but is present in 576, more than half, of the collaborative models.
This clearly demonstrates the differences between TMLE’s reliance on an external estimate of g0 and the collaborative approach to estimating the treatment mechanism used by C-TMLE. However, we note that the degradation of TMLE performance under sparsity is due to the unboundedness of the fluctuation function, and can be mitigated by employing an alternative fluctuation function that respects known bounds on the data model. Though a full discussion is beyond the scope of this paper, details may be found in Gruber and van der Laan (2010).
6.4. Confidence intervals
The variance of the influence curve provides the basis for calculation of a 95% confidence interval for the C-TMLE estimate.
Two sets of confidence intervals were constructed for each of the 1000 iterations in simulation 4, with misspecified by a main-terms only regression model. As described above, one set of CIs is based on D* (Q, g), the first term of the IC. The second set is based on the variance of D* (Q, g) + ICg, which includes the contribution from the estimation of gn. Table 3 shows that CIs based on D* alone are conservative when the model for is misspecified, as expected. In contrast, observed coverage closely approximates the nominal 95% coverage rate when the contribution from the ICg term is taken into account.
Table 3:
Empirical coverage of 1000 confidence intervals constructed at a nominal 95% level. SE calculated as , where the IC was estimated with and without ICg.
| Coverage | ||
|---|---|---|
| D* (Q, g0) | D* (Q, g0) + ICg | |
| misspecified | .979 | .943 |
| correct | .932 | .933 |
Confidence intervals were also created for an additional 1000 samples from the same data generating distribution that were analyzed using a correct model for . Coverage rates for these confidence intervals are given in Table 3. When is correctly specified we observe little difference in the coverage rate whether or not we take the contribution from ICg into account, indicating zero contribution to the variance from the estimate of gn. Attaining the nominal rate indicates that inference is reliable even when the estimator is super efficient.
7. Data analysis
We apply the C-TMLE estimator to an observational dataset previously analyzed with the goal of identifying HIV mutations that affect response to the antiretroviral drug lopinavir. (Bembom et al., 2009, 2008) The data includes observations on O = (W, A, Y), where the outcome, Y, is the change in log10 viral load measured at baseline and at follow-up after treatment has been initiated. If follow-up viral load was beneath the limit of detection Y was set to the maximal change seen in the population. A ∈ {0, 1} is an indicator of the presence or absence of a mutation of interest, taking on the appropriate value for each of the 26 candidate mutations in 26 separate analyses. W consists of 51 covariates including treatment history, baseline characteristics, and indicators of the presence of additional HIV mutations. Practical ETA violations stemming from high correlations among some of the covariates and/or low probability of observing a given mutation of interest make it difficult to obtain stable low variance estimates of the association between A and Y. Bembom used a targeted maximum likelihood estimation approach incorporating data-adaptive selection of an adjustment set that relies on setting a limit on the maximum allowable truncation bias introduced by truncating treatment probabilities less than α to some specified lower limit. Covariates whose inclusion in the adjustment set introduces an unacceptable amount of bias are not selected. That study’s findings showed good greement with Stanford HIVdb mutation scores, values on a scale of 0 to 20 (http://hivdb.stanford.edu, as of September, 2007, subsequently modified), where 20 indicates evidence exists that the mutation strongly inhibits response to drug treatment and 0 signifies that the mutation confers no resistance. Because the C-TMLE method includes covariates in the treatment mechanism only if they improve the targeting of the parameter of interest without having too adverse an effect on the MSE, we expect similar performance without having to specify truncation levels or an acceptable maximum amount of bias.
7.1. Analysis description
The dataset consists of 401 observations on 372 subjects. Correlations due to the few subjects contributing more than one observation were ignored. Separate analyses was carried out for each mutation. In each, an initial density estimate, , was obtained using DSA restricted to addition moves only to select a main-terms model containing at most 20 terms, where candidate terms in W include pre-computed interactions detailed in Bembom et al. A was forced into the model. An estimate of the effect on change in viral load was recorded for each mutation. Influence curve-based variance estimates incorporating the contribution from estimating g given by the ICg term, was used to construct 95% confidence intervals.
7.2. Results
Table 4 lists the Stanford mutation score associated with each of the HIV mutations under consideration, as well as the C-TMLE estimate of the adjusted effect of mutation on lopinavir resistance. 95% confidence intervals were constructed based on the variance of the IC. Confidence intervals entirely above zero indicate a mutation increases resistance to lopinavir. Eight of the twelve mutations having a mutation score of 10 or greater fall into this category. Point estimates for the remaining four mutations were positive, but the variance was too large to produce a significant result. Only one of the six mutations thought to confer slight resistance to lopinavir was flagged by the procedure, though with the exception of p10FIRVY point estimates were positive. Stanford mutation scores of 0 for four of the five mutations found to have a significantly negative effect on drug resistance support the conclusion that these mutations do not increase resistance, but are not designed to offer confirmation that a mutation can decrease drug resistance. However, Bembom et al. report that there is some clinical evidence that two of these mutations, 30N and 88S, do indeed decrease lopinavir resistance.
Table 4:
Stanford score (2007), C-TMLE estimate and 95% confidence interval for each mutation. Starred confidence intervals do not include 0.
| mutation | score | estimate | 95% CI |
|---|---|---|---|
| p50V | 20 | 1.703 | (0.760, 2.645)* |
| p82AFST | 20 | 0.389 | (0.091, 0.688)* |
| p54VA | 11 | 0.505 | (0.241, 0.770)* |
| p54LMST | 11 | 0.369 | (0.002, 0.735)* |
| p84AV | 11 | 0.099 | (−0.139, 0.337) |
| p46ILV | 11 | 0.046 | (−0.222, 0.315) |
| p82MLC | 10 | 1.610 | (1.377, 1.843)* |
| p47V | 10 | 0.805 | (0.282, 1.328)* |
| p84C | 10 | 0.602 | (0.471, 0.734)* |
| p32I | 10 | 0.544 | (0.325, 0.763)* |
| p48VM | 10 | 0.306 | (−0.162, 0.774) |
| p90M | 10 | 0.209 | (−0.063, 0.481) |
| p33F | 5 | 0.300 | (−0.070, 0.669) |
| p53LY | 3 | 0.214 | (−0.266, 0.695) |
| p73CSTA | 2 | 0.635 | (0.278, 0.992)* |
| p24IF | 2 | 0.229 | (−0.215, 0.674) |
| p10FIRVY | 2 | −0.266 | (−0.545, 0.012) |
| p71TVI | 2 | 0.019 | (−0.243, 0.281) |
| p23I | 0 | 0.822 | (−0.014, 1.658) |
| p36ILVTA | 0 | 0.272 | (−0.001, 0.544) |
| p16E | 0 | 0.239 | (−0.156, 0.633) |
| p20IMRTVL | 0 | 0.178 | (−0.111, 0.467) |
| p63P | 0 | −0.131 | (−0.417, 0.156) |
| p88DTG | 0 | −0.426 | (−0.842,–0.010)* |
| p30N | 0 | −0.440 | (−0.853,–0.028)* |
| p88S | 0 | −0.474 | (−0.781,–0.167)* |
Our findings are quite consistent with the Stanford mutation scores and with the results from the previous analysis using the data-adaptively selected adjustment set targeted maximum likelihood estimation approach. The C-TMLE method was able to achieve these results without relying on ad hoc or user-specified tuning parameters.
8. Discussion
Simulation studies demonstrate the collaborative double robustness and efficiency of C-TMLE methodology, which allows for consistent efficient estimation in situations when other estimators can fail to perform adequately. In practice these failures may lead to biased estimates and to confidence intervals that fail to attain the correct coverage, as suggested by the IPTW results in simulations 1 and 2, where weights depend on a variable highly predictive of treatment that is not a true confounder of the relationship between Y and A. It is worth noting that the unadjusted estimator applied to data from a randomized controlled trial in which randomization fails to evenly distribute confounders across treatment arms will also yield (finite sample) biased results, as we saw in simulations 1,2, and 3.
As simulations 2 and 3 demonstrate, a misspecied parametric model not only results in biased estimates, but can also easily fail to adequately explain the variance in the outcome. Therefore estimates of the parameter of interest will have a larger variance than the semiparametric information bound achieved by an efficient estimator, such as C-TMLE. Such misspecified parametric models can easily result in the construction of a confidence interval that contains 0, and therefore a failure to reject a false null hypothesis, even when the point estimate is close to the true value of the parameter of interest. Since misspecied parametric models are the rule rather than the exception, in the analysis of data from an unknown data-generating distribution, using C-TMLE combined with super learning for the initial estimator, is a prudent course of action.
Estimators that rely on nuisance parameter estimation (IPTW, DR-IPTW, TMLE, propensity score-based estimation) break down when there are ETA violations, failing to reduce bias, or even increasing bias, while incurring high variance that renders estimates meaningless (no statistical signicance). An effort to reduce variance through truncation introduces bias into the estimate, and requires a careful trade-off. C-TMLE addresses these issues, in the sense that it is able to utilize the covariates for effective bias reduction, avoiding harmful bias reduction efforts. As a targeted-MLE, the bias-variance tradeoff is targeted towards the estimation of the parameter of interest, not the estimate of the entire density. The collaborative nature of the estimation of the treatment mechanism in the C-TMLE confers three advantages:
The treatment mechanism model will exclude covariates that are highly predictive of treatment but do not truly confound the relationship between treatment and the outcome.
The treatment mechanism model will include only covariates that help adjust for residual bias remaining after stage 1 adjustment.
Cross-validation based on a penalized log-likelihood will not select a treatment mechanism model that includes a term that leads to violations of the ETA assumption and thereby large variance of the corresponding targeted MLE without the benefit of a meaningful bias reduction.
Influence-curve based inference is theoretically sound, and achieves the desired coverage rate across a wide range of simulations, in addition to the ones presented. An implementation of C-TMLE methodology written for the R statistical environment that includes a demonstration based on the data-generating scheme for simulation 2 is available as supplemental materials.
Supplementary Material
C-TMLE implementation and demo (R source code)
Appendix.
TMLE as an imputation estimator
Consider the goal of estimating EY1. It is desirable for the estimator Qn to satisfy 0 = ∑i{EQn(Y1|Oi) − PQn(Y1 = 1)}, which makes Ψ(Qn) an imputation estimator. In other words, averaging the imputed values EQn(Y1|Oi) for Y1 under Qn for i = 1, .., n, gives the same estimator as the mean of Y1 under Qn. Below, we show that this holds if Qn solves the score equation ∑i Ai(Yi − EQn(Y |Ai, Wi)). With this insight, we can now compute a targeted maximum likelihood estimator that is also an imputation estimator, by using a bivariate ɛ fluctuation function involving both the clever covariate and the term A. Similarly, we can construct a targeted maximum likelihood estimator that is also an imputation estimator for the additive causal effect parameter EY (1) − Y (0). This problem was presented in Rubin and van der Laan (2008).
Details of this derivation follow. Let O = (W, A, Y), with Y binary. Consider the score D(Q)(O) = EQ(Y1 | O) − EQY1. We note that
We wish to find the component of D(Q) that is in the tangent space of the conditional distribution of Y, given A, W. We have
We decompose
The first component can be written as
which reduces to A(Y − EQ(Y | A, W)).
Consider now the score D(Q)(O) = EQ(Y0 | O) − EQY0. We note that
We wish to find the component of D(Q) that is in the tangent space of the conditional distribution of Y, given A, W. We have
We decompose
We can write this first component as
which reduces to (1 − A)(Y − EQ(Y | A, W)).
Suppose now that one wishes to solve the score equation of score D(Q) = EQ(Y1 − Y0 | Oi) − EQ(Y1 − Y0). We follow the same proof as above. Firstly,
We wish to find the component of D(Q) that is in the tangent space of the conditional distribution of Y, given A, W. We have E(D(Q) | A, W) = EQ(Y | A = 1, W) − EQ(Y | A = 0, W)). We decompose
We can write this first component as
which reduces to (2A − 1)(Y − EQ(Y | A, W).
Therefore, if we want to be an imputation estimator for both EY0, EY1, then we wish to have an estimator that solves the bivariate score equation
This can be arranged by applying the targeted MLE update with the fluctuation function corresponding with covariate-extension ɛ(1, A, hg(A, W)), where hg denotes the clever covariate for the target parameter (say) EY1 − EY0. If one wishes to solve the score equation of score D(Q) = EQ(Y1 − Y0 | Oi) − EQ(Y1 − Y0), then, one uses the bivariate extension ɛ1(2A − 1) + ɛ2hg.
Contributor Information
Susan Gruber, University of California, Berkeley.
Mark J. van der Laan, University of California, Berkeley
References
- Abadie A, Imbens GW. Large sample properties of matching estimators for average treatment effects. Econometrica. 2006;74:235–67. doi: 10.1111/j.1468-0262.2006.00655.x. [DOI] [Google Scholar]
- Bembom O, Fessel JW, Shafer RW, van der Laan MJ. Data-adaptive selection of the adjustment set in variable importance estimation. Technical report, DigitalCommons@Florida Atlantic University [ http://digitalcommons.fau.edu/cgi/oai2.cgi] (United States), 2008. URL http://www.bepress.com/ucbbiostat/paper231.
- Bembom O, Petersen ML, Rhee S-Y, Fessel WJ, Sinisi SE, Shafer RW, van der Laan MJ. Biomarker discovery using targeted maximum likelihood estimation: Application to the treatment of antiretroviral resistant HIV infection. Statistics in Medicine. 2009;28:152–72. doi: 10.1002/sim.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S, van der Laan MJ. A targeted maximum likelihood estimator of a causal effect on a bounded continuous outcome. Technical report 265, Division of Biostatistics, University of California, Berkeley, May 2010. [DOI] [PMC free article] [PubMed]
- Hernan MA, Brumback B, Robins JM. Marginal structural models to estimate the causal effect of zidovudine on the survival of HIV-positive men. Epidemiology. 2000;11(5):561–570. doi: 10.1097/00001648-200009000-00012. [DOI] [PubMed] [Google Scholar]
- Holland JH, Reitman JS. Cognitive systems based on adaptive algorithms. SIGART Bull. 1977;63:49–49. ISSN 0163-5719. http://doi.acm.org/10.1145/1045343.1045373. [Google Scholar]
- Robins JM, Rotnitzky A. Comment on the Bickel and Kwon article, “Inference for semiparametric models: Some questions and an answer”. Statistica Sinica. 2001;11(4):920–936. [Google Scholar]
- Robins JM, Rotnitzky A, van der Laan MJ. Comment on “On Profile Likelihood” by S.A. Murphy and A.W. van der Vaart. Journal of the American Statistical Association – Theory and Methods. 2000;450:431–435. doi: 10.2307/2669381. [DOI] [Google Scholar]
- Robins JM. Robust estimation in sequentially ignorable missing data and causal inference models. Proceedings of the American Statistical Association. 2000a.
- Robins JM. A new approach to causal inference in mortality studies with sustained exposure periods - application to control of the healthy worker survivor effect. Mathematical Modelling. 1986;7:1393–1512. doi: 10.1016/0270-0255(86)90088-6. [DOI] [Google Scholar]
- Robins JM. Statistical models in epidemiology, the environment, and clinical trials (Minneapolis, MN, 1997) Springer, New York: 2000b. Marginal structural models versus structural nested models as tools for causal inference; pp. 95–133. [Google Scholar]
- Rosenbaum PR, Rubin DB. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70:41–55. doi: 10.1093/biomet/70.1.41. [DOI] [Google Scholar]
- Rubin DB, van der Laan MJ. Doubly robust ecological inference. Technical report 236, Division of Biostatistics, University of California, Berkeley, May 2008.
- Sekhon JS. Multivariate and propensity score matching software with automated balance optimization: The matching package for R. Journal of Statistical Software. 2008. Forthcoming.
- Sinisi S, van der Laan MJ. The Deletion/Substitution/Addition algorithm in loss function based estimation: Applications in genomics. Journal of Statistical Methods in Molecular Biology. 2004;3(1) doi: 10.2202/1544-6115.1069. [DOI] [PubMed] [Google Scholar]
- van der Laan MJ, Gruber S. Collaborative double robust penalized targeted maximum likelihood estimation. The International Journal of Biostatistics. 2010. [DOI] [PMC free article] [PubMed]
- van der Laan MJ, Robins JM. Unified methods for censored longitudinal data and causality. Springer; New York: 2003. [Google Scholar]
- van der Laan MJ, Rubin D. Targeted maximum likelihood learning. The International Journal of Biostatistics. 2006;2(1) doi: 10.2202/1557-4679.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan MJ, Polley E, Hubbard A. Super learner. Statistical Applications in Genetics and Molecular Biology. 2007;6(25) doi: 10.2202/1544-6115.1309. ISSN 1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
C-TMLE implementation and demo (R source code)