Abstract
GATA-family transcription factors are critical to the development of diverse tissues. In particular, GATA-4 has been implicated in formation of the vertebrate heart. As the mouse Gata-4 knock-out is early embryonic lethal because of a defect in ventral morphogenesis, the in vivo function of this factor in heart development remains unresolved. To search for a requirement for Gata4 in heart development, we created mice harboring a single amino acid replacement in GATA-4 that impairs its physical interaction with its presumptive cardiac cofactor FOG-2. Gata4ki/ki mice die just after embryonic day (E) 12.5 exhibiting features in common with Fog2−/− embryos as well as additional semilunar cardiac valve defects and a double-outlet right ventricle. These findings establish an intrinsic requirement for GATA-4 in heart development. We also infer that GATA-4 function is dependent on interaction with FOG-2 and, very likely, an additional FOG protein for distinct aspects of heart formation.
Keywords: GATA-4, FOG cofactor, coronary vasculature, heart
Transcriptional activity of the GATA-factors is modulated through interaction with nuclear proteins, including zinc finger proteins of the Kruppel and FOG/U-shaped families, general coactivators (p300 and CBP), the myocardial-expressed protein Nkx2.5, and NF-AT3 (Durocher and Nemer 1998; Mackay and Crossley 1998; Blobel 2000; Molkentin 2000). Whereas the specificity and in vivo functional relevance of many of these interactions are incompletely defined, the association of GATA-1 with FOG-1 has been examined in detail. FOG-1 interacts with GATA-1 in hematopoietic cells and regulates the ability of GATA-1 to promote terminal differentiation of erythroid cells and megakaryocytes (Tsang et al. 1997). Mutation of specific residues within the conserved N-terminal zinc finger of GATA-1, such as V205G, disrupts binding to FOG-1, preserves DNA-binding properties of GATA-1, and renders GATA-1 unable to promote terminal differentiation of red blood cells (Crispino et al. 1999). Furthermore, mutation of Val 205 in humans leads to congenital dyserythropoietic anemia and thrombocytopenia (Nichols et al. 2000). Taken together, these findings demonstrate that direct physical association of GATA-1 and FOG-1 is essential for GATA-1's roles in transcription and, critical for the experiments reported herein, identifies a specific residue of the N finger that mediates cofactor interaction.
GATA-4, GATA-5, and GATA-6, nonhematopoietic expressed factors, are implicated in development of heart, endoderm, and intestinal epithelia, where they are expressed in an overlapping and dynamic fashion (Morrisey et al. 1997; Bossard and Zaret 1998; Gao et al. 1998; Charron and Nemer 1999; Koutsourakis et al. 1999; Parmacek and Leiden 1999; Molkentin 2000). GATA-4 has been extensively studied in the context of heart development, as it is present in precardiac splanchnic mesoderm and binds to and activates promoters and enhancers of numerous myocardial-expressed genes (Charron et al. 1999). In its absence, mouse embryos die by E7.0–9.5, with failure of ventral morphogenesis leading to cardiac bifida (Kuo et al. 1997; Molkentin et al. 1997). The death of the Gata4 null embryos before formation of a heart tube precludes analysis of the role of this factor in later cardiac organogenesis.
The second member of the FOG protein family, FOG-2, is expressed in cardiac and nervous system tissues and interacts with the N fingers of all GATA factors, including those expressed within the developing heart (GATA-4, GATA-5, and GATA-6; Lu et al. 1999; Svensson et al. 1999; Tevosian et al. 1999). As such, it is a candidate cofactor for these GATA factors in the heart. Mouse embryos lacking Fog2 die of heart failure between E12.5 and E15.5 (Svensson et al. 2000; Tevosian et al. 2000). Fog-2−/− hearts exhibit a constellation of defects, such as overriding aorta, subpulmonic stenosis, and subaortic ventricular septal defect (VSD), seen in the human congenital malformation Tetralogy of Fallot. In addition, despite formation of an intact epicardial layer and expression of epicardium-specific genes, initiation of coronary vasculature fails to take place. This is evidenced by the absence of induction of markers of coronary vessel development, including ICAM-2 and Flk-1 in Fog2-deficient hearts. These defects in coronary vasculature formation are secondary to lack of Fog2 expression specifically in myocardium, as demonstrated by transgenic rescue (Tevosian et al. 2000).
Results and Discussion
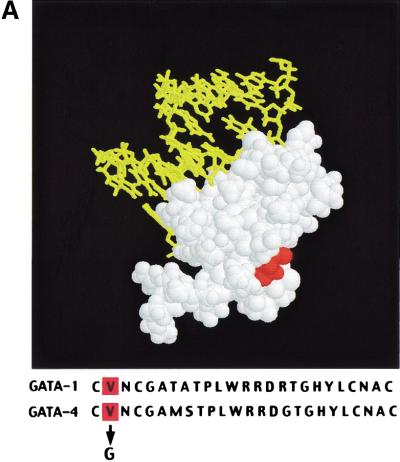
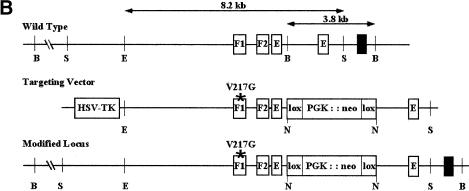
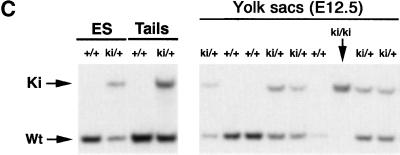
We sought to establish an intrinsic role for GATA-4 in heart development by generating mice harboring a knock-in mutation that cripples its interaction with FOG-2 or other FOG-factors. Residue 217 of GATA-4, which corresponds to Val 205 of GATA-1, was changed to glycine by gene targeting in embryonic stem (ES) cells. This residue faces away from DNA (Fig. 1A) and lies within the GATA–N finger: FOG interface. Substitution with glycine disrupts interaction with either FOG-1 or FOG-2 and leaves DNA-binding properties of GATA factors unperturbed. Targeted mutation of murine Gata4 was accomplished with the construct depicted in Figure 1B. A floxed neomycin resistance cassette was incorporated into an intron downstream of the exon containing V217. ES cells harboring both the V217G mutation and the floxed neomycin cassette were injected into host blastocysts to generate chimeras. Gata4+/ki mice appeared normal. No liveborn homozygotes resulted from interbreeding of heterozygous offspring, indicating the Gata4ki/ki embryos are embryonic lethal. Southern blotting of timed matings (Fig. 1C) demonstrated that Gata4ki/ki embryos die between E11.5 and E13.5. Their gross appearance was remarkably similar to that of Fog2−/− embryos (Svensson et al. 2000; Tevosian et al. 2000). Gata4ki/ki embryos are pale and edematous, compared with wild-type littermates, and peripheral hemorrhage is often observed (Fig. 2A,B). The neomycin cassette was removed in some strains by interbreeding with a Cre-recombinase-deleter strain (Mao et al. 1999). No differences in phenotype were observed between strains containing or lacking the neomycin cassette. Furthermore, heterozygotes were born at the expected frequency and displayed no detectable phenotype, arguing that the GATA-4 mutant protein does not act in a dominant negative manner.
Figure 1.
Targeting the GATA4–FOG-2 interaction in mice. (A) Structure of the N finger of chicken GATA-1 modeled with DNA. The essential valine is highlighted in red in both the illustration and in the sequence alignment of the murine factors, GATA-1 (V205) and GATA-4 (V217), shown below. Note the high conservation of the residues within the N finger of the two GATA proteins. (B) Partial restriction map of the murine Gata4 wild-type locus (top), the Gata4 knock-in targeting vector (middle), and targeted homologous recombination before excision of the selection cassette (bottom). The targeting construct contains the HSV-tk and neomycin resistance (neoR) genes under the control of the mouse phosphoglycerate kinase (PGK) promoter. Homologous recombination results in replacement of wild-type Gata4 with genomic DNA harboring a substitution of valine to glycine at position 217 in the N finger of GATA-4, as well as the incorporation of neomycin cassette. Gata4 coding exons are shown as empty boxes, whereas the exon used as a probe used for Southern blot analysis is highlighted by a black box. S, SacI; E, EcoRV; B, BglII; N, NotI. (C) Southern blot analysis of ES cell DNA and mouse tail DNA (left panel) showing the presence of heterozygous mutant animals (ki/+). Analysis of E12.5 embryos resulting from an intercross of Gata4 knock-in heterozygotes (ki/+), demonstrating the presence of all expected genotypes (right panel). The wild-type allele (WT) generated a 3.8-kb band after digestion of genomic DNA with BglII. In contrast, the knock-in mutated allele (Ki) generated a much larger fragment because of the replacement of the intronic BglII site with NotI.
Figure 2.
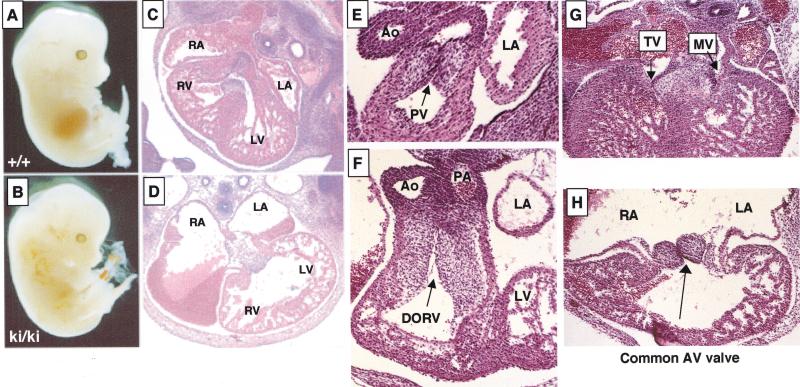
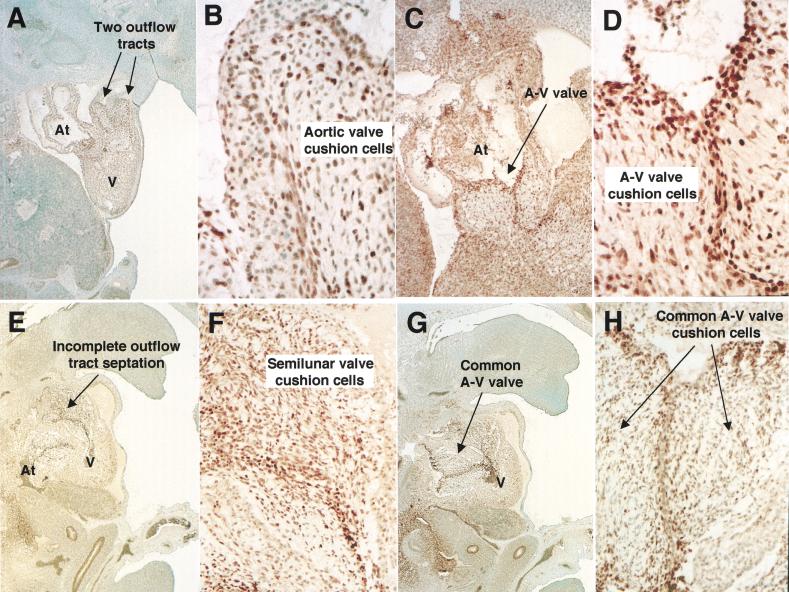
Heart defects in Gata4 mutant (ki/ki) embryos. (A,B) Wild-type (A) and mutant (B) embryos at E13.5 showing edema and peripheral hemorrhaging in a mutant. (C,D) Transverse sections through wild-type (C) and mutant (D) hearts at E13.5 at the level of the atrioventricular (AV) junction show enlarged atria, thin myocardium, and the absence of a ventricular septum. Original magnification, 40×. (E,F) Transverse sections of wild-type (E) and mutant (F) hearts at the level of the aortic and pulmonary outflow tracts. Gata4ki/ki hearts have a double outlet right ventricle, in which all blood exits the heart into both great arteries, the pulmonary artery and the aorta. The left ventricle, which normally delivers blood to the aorta, fails to communicate with an artery in the mutant. Also note the apparent increase in cellularity of both outflow tracts and semilunar valves in the mutant. Original magnification, 400×. (G,H) Transverse sections of wild-type (G) and mutant (H) hearts at the level of the AV junction. Gata4ki/ki hearts form a common AV valve that is situated between the left and right ventricles. For comparison, the mitral (MV) and tricuspid (TV) valves of the wild-type heart are indicated by arrowheads. Original magnification, 100×.
Wild-type and the GATA-4ki/ki embryonic hearts at E12.5 were examined in serial sections, cut in the transverse plane, and inspected from the cephalic to the caudal aspect of the specimens (Fig. 2C–H). Gata4ki/ki hearts revealed a double-outlet right ventricle where both great arteries arise from the right ventricle (Fig. 2F). The ventricular septal defect is the only outlet for the left ventricle (Fig. 2F). In addition, the endocardial cushion cells of both the pulmonary and aortic outflow tracts appear more numerous than in wild-type embryos. Furthermore, robust mitotic activity is evident in these areas, and the pulmonary and aortic outflow tracts comingle in some planes of section. In wild-type embryos, however, the pulmonary and aortic outflow tracts display distinct endocardial cushions (Fig. 2E). Right and left atria of the mutant are massively dilated and freely communicate (Fig. 2D,H), and a common atrioventricular valve opens into the center of a large ventricular cavity, which lacks a ventricular septum (Fig. 2H). In contrast to a wild-type heart, in which two distinct atrioventricular valves (the tricuspid and mitral valves; Fig. 2G) develop in continuity with the septum and with the aortic valve, respectively (Fig. 2E), the common AV valve in the mutant heart has no fibrous continuity with the aortic valve. In addition, Gata4ki/ki myocardium appears thin (cf. Fig. 2G,H). These abnormalities were consistently seen in the GATA-4ki/ki embryos. Similarly, complete penetrance was also observed in FOG-2−/− embryos (Tevosian et al. 2000).
To validate that the observed phenotype results from expression of a qualitatively different GATA-4 protein rather than from an altered pattern, or level, of expression, we examined GATA-4 protein expression. Wild-type and Gata4ki/ki E12.5 hearts were sectioned in the sagittal plane and immunostained for expression of GATA-4 (Fig. 3A,B). At this stage, GATA-4 protein is present in both endocardium and myocardium of developing atria and ventricles. Immunostaining is strongest in surface endothelial cells and mesenchymal cells of the endocardial cushion tissues, which develop into the semilunar (Figs. 3B,F) and atrioventricular valves (Fig. 3D,H). GATA-4 protein is also present in the embryonic liver and gut epithelium in both wild-type and mutant embryos, as expected (Fig. 3A,E; Arceci et al. 1993). Taken together, these studies demonstrate that GATA-4ki protein is expressed comparably to wild-type protein.
Figure 3.
Expression of Gata4 in the heart. Sagittal sections of wild-type (A–D) and Gata4 ki/ki (E–H) embryos at E12.5 were stained with an α-GATA-4 antibody. Both wild-type and mutant hearts display similar staining within the semilunar and AV valve cells. Note the staining of outflow tracts in both the wild-type and the mutant heart. At, atrium; AV, atrioventricular; V, ventricle. Original magnification, A,E,G, 40×; B,D,F,H, 400×; C, 100×.
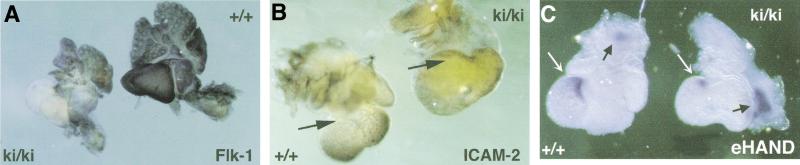
In several respects, the morphology of Gata4ki/ki hearts resembles that seen of Fog2−/− embryos. A distinctive feature of Fog2−/− hearts is the absence of coronary vasculature (Tevosian et al. 2000). To examine the status of their coronary vasculature, Gata4ki/ki hearts were immunostained for two endothelial cell markers. Flk-1, the receptor for vascular endothelial growth factor (VEGF), is not detectable in Gata4ki/ki hearts, though lung tissue stains intensely (Fig. 4A). Similarly, abundance of the intracellular adhesion molecule-2 (ICAM-2) is reduced in Gata4ki/ki hearts; note the absence of the developing capillary plexus (Fig. 4B, arrows). These findings are indistinguishable from those reported previously for Fog2−/− hearts. As such, they provide strong evidence that FOG-2 serves as a transcriptional cofactor for GATA-4 in myocardium as an essential step in the initiation of coronary vascular development.
Figure 4.
Aberrant expression of coronary vessel and myocardial transcripts. (A) Staining of E12.5 wild-type (+/+) and mutant (ki/ki) hearts with an α-Flk-1 antibody (dorsal view). Staining of the mutant is vastly reduced within the heart, but lung tissue stained with equal intensity. (B) Immunostaining of wild-type and mutant hearts at E12.5 using the α-ICAM-2 antibody (dorsal view). Note the absence of a well-developed vascular tree in the mutant heart. (C) Whole-mount RNA in situ staining of eHand in E11.5 hearts. eHand expression is down-regulated in the outer myocardial layer (white arrows), whereas there is more intense staining in the outflow tract of the mutant (dark arrows). Note that the direction of the outflow tract relative to the heart is altered in the mutant, consistent with the pathological findings.
Of numerous myocardial-expressed genes assayed in Fog2−/− hearts, only the basic–helix–loop helix eHand and dHand genes were altered in their expression on loss of Fog2 (Tevosian et al. 2000). To assess this phenotype in Gata4ki/ki hearts, the expression of Hand genes was examined by RNA in situ hybridization. Expression of eHand is reduced in the myocardium of Gata4ki/ki hearts relative to that seen in wild-type hearts (Fig. 4C). Interestingly, whereas the expression of eHand in the outer curvature of the ventricles is reduced (Fig. 4C, white arrows), its expression in the outflow tract is increased (Fig. 4C, dark arrows). This increased staining likely results from the increased cellularity within this region of the Gata-4 mutant hearts and was not observed in Fog2-deficient hearts (Fig. 2F). In contrast, the expression of dHand is only marginally reduced in the mutant heart (data not shown).
Our data point to essential roles for the GATA-4:FOG-2 interaction in heart morphogenesis and coronary vascular development. The power of our analysis rests on the exquisite specificity of the knock-in mutation within the N finger of GATA-4. The residue we have chosen to modify is required for physical interaction with FOG-like proteins and does not influence the DNA-binding specificity of the GATA-protein (Crispino et al. 1999). Although displaying many similar features, Gata4ki/ki hearts are distinguished from Fog2−/− hearts, however, by the presence of a double-outlet right ventricle and defects in the semilunar valves and outflow tracts. As immunostaining confirmed that GATA-4 is expressed at wild-type levels in the semilunar valves of the Gata4 mutant heart, it is likely that another FOG, or FOG-like protein, that functions as a cofactor for GATA-4 in transcription is expressed in these valve cells. Though high-level expression of the only other known vertebrate FOG-like factor, FOG-1, has not previously been observed by in situ RNA hybridization, FOG-1 transcripts are present at low levels in Northern blots of total heart RNA (A.P. Tsang and S.H. Orkin, unpubl.). Thus, it is quite possible that disruption of the physical interaction between GATA-4 and FOG-1, or a novel, undefined FOG protein, is responsible for impaired development of the semilunar valves and the appearance of a double-outlet right ventricle. Given the profound effects of mutation of either GATA-4 or FOG-2 proteins on heart morphogenesis in mice, it is worth considering their potential relevance to human congenital heart defects, such as the Tetralogy of Fallot or the double-outlet right ventricle (Walters et al. 2000). Whereas mutation of several genes, such as Jmj (Jumanji), Sox4, and Egfr/Shp2, give rise to the double-outlet right ventricle defect in mice (Chen et al. 1996; Ya et al. 1998; Lee et al. 2000), and defects in other genes for transcription factors, such as FOG-2, NF1, neurotrophin 3, and RXRα, result in all or a subset of the Tetralogy of Fallot (Jacks et al. 1994; Donovan et al. 1996; Gruber et al. 1996; Lee et al. 1997; Tevosian et al. 2000), the consistent and combined phenotype seen in the Gata4ki/ki mice is unique. Although it is sometimes difficult to distinguish between double-outlet right ventricle with associated pulmonary stenosis and the Tetralogy of Fallot, it is clear that the defects in the GATA-4ki/ki hearts are different in the outflow tracts than those observed in Fog2-deficient embryos.
Through the use of an altered specificity mutant, we demonstrate that GATA-4 very likely requires both FOG-2 and an additional FOG, or FOG-like protein, as cofactors for distinct aspects of heart development. Interaction with FOG-2 is essential for the initiation of coronary vasculature and for some morphogenetic events, whereas interaction with a distinct FOG protein appears to be required for formation of cardiac valves. Our results are surprising in that two other GATA-factors, GATA-5 and GATA-6, are also expressed in myocardium, and indirect data have suggested that they might compensate for the absence of GATA-4. For example, previous studies show that GATA-4 is dispensable for terminal differentiation of cardiomyocytes and that Gata4−/− ES cells contribute to all layers of the heart. In these experiments, it has been suggested that GATA-5 or GATA-6 functionally replace GATA-4 (Narita et al. 1996). It is possible that proper expression of the GATA-4ki/ki protein, as distinguished from the absence of GATA-4 in the knock-out situation, precludes compensation by other GATA factors. Indeed, immunostaining with an α-GATA-6 antibody demonstrated that GATA-6 expression, though similar to that of GATA-4, is not up-regulated in the Gata4ki/ki hearts (data not shown). In addition, staining with α-GATA-5 antibody revealed a normal pattern in Gata4ki/ki hearts (data not shown). As GATA-5 is no longer expressed within the ventricles of the heart at E12.5, it is unlikely that it would compensate for the absence of functional GATA-4 (Morrisey et al. 1997).
Our findings implicate GATA-4 as the principal GATA factor relevant to heart morphogenesis and coronary vasculature development and as the primary partner for FOG proteins in the heart. This represents the second example of transcriptional regulation involving GATA–FOG protein complexes and argues for their broad involvement as key regulators of multiple developmental pathways.
Materials and methods
Targeted mutagenesis of the murine Gata4 gene
An 8.2-kb EcoRV−SacI fragment of murine Gata4 genomic DNA containing the N finger of GATA-4 was subcloned into pBluescript II KS (+/−) phagemid (Stratagene). By site-directed mutagenesis, Val 217 was changed to glycine (the codon GGC was changed to GTC; GeneEditor, Promega). An intronic BglII site was changed to a NotI site to facilitate introduction of a floxed neomycin expression cassette. HSV-tk was cloned into a SalI site 5′ of the homology region. The targeting construct was linearized with PvuI and electroporated into TL1 ES cells. The Gata4 gene from two independently generated, targeted ES clones was amplified by PCR, sequenced, and found to be correctly mutated in one clone. This clone was injected into C57BL/6 blastocysts to generate chimeras. Genotyping was done thereafter by Southern blot analysis, as described in the legend of Figure 1C.
Histological analysis
Mouse embryos were isolated from Gata4 heterozygous knock-in matings between E10.5 and 13.5. Embryos were fixed in Bouin's, dehydrated into increasing concentrations of ethanol, transferred into xylene, and sectioned in paraffin at 6 μm and stained with hematoxylin and eosin.
Immunohistochemistry
Murine tissue used for immunohistochemistry was fixed in 10% formalin overnight, processed, and paraffin embedded using standard histologic techniques. Tissue sections 4 μ thick were dewaxed in xylene then rehydrated by passage through graded alcohol solutions. Sections were immersed in 10 mmole/L citrate buffer (pH 6.0) in a thermoresistant container and heated in a microwave oven (800 W, General Electric) at 199°F for 30 min. A rabbit polyclonal antibody to GATA-4 (sc-9053, Santa Cruz Biotechnology) was used at a 1:50 dilution and detected by the Rabbit DAKO Envision Plus System, Peroxidase DAB (Dako). Negative controls were performed by substituting the primary antibody with species- and isotype-matched, nonimmune immunoglobulins. Additional controls included omission of the primary antibody as well as substitution of the primary antibody of interest with one of differing specificity. Whole-mount staining of hearts with the α-Flk-1 and α-ICAM-2 antibodies (obtained from Pharmingen) was performed as described (Tevosian et al. 2000).
In situ hybridization analysis
Whole-mount hybridization was performed using riboprobes labeled with digoxigenin-UTP as described (Tevosian et al. 2000).
Acknowledgments
We thank S. Tevosian, Y. Fujiwara, and C.Yu for helpful discussion and C. Browne, A. Chapdelaine, S. Galusha, M. Hamblen, and C. Quigley for their expertise and technical assistance. This work was supported in part by a Special Fellowship of the Leukemia and Lymphoma Society to J.D.C. and an NIH-NHLBI, Medical Student Research Training Fellowship to M.B.L. S.H.O. is an Investigator of the Howard Hughes Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL orkin@rascal.med.harvard.edu; FAX (617) 355-7262.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.875201.
References
- Arceci RJ, King AA, Simon MC, Orkin SH, Wilson DB. Mouse GATA-4: A retinoic acid–inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA. CREB-binding protein and p300: Molecular integrators of hematopoietic transcription. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- Charron F, Nemer M. GATA transcription factors and cardiac development. Semin Cell Dev Biol. 1999;10:85–91. doi: 10.1006/scdb.1998.0281. [DOI] [PubMed] [Google Scholar]
- Charron F, Paradis P, Bronchain O, Nemer G, Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol Cell Biol. 1999;19:4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KS, Paladugu A, Aldaz CM, Gould MN. Cloning and chromosomal localization of the rat Stat5 and Yy1 genes. Cytogenet Cell Genet. 1996;74:277–280. doi: 10.1159/000134434. [DOI] [PubMed] [Google Scholar]
- Crispino JD, Lodish MB, MacKay JP, Orkin SH. Use of altered specificity mutants to probe a specific protein–protein interaction in differentiation: The GATA-1:FOG complex. Mol Cell. 1999;3:219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- Donovan MJ, Hahn R, Tessarollo L, Hempstead BL. Identification of an essential nonneuronal function of neurotrophin 3 in mammalian cardiac development. Nat Genet. 1996;14:210–213. doi: 10.1038/ng1096-210. [DOI] [PubMed] [Google Scholar]
- Durocher D, Nemer M. Combinatorial interactions regulating cardiac transcription. Dev Genet. 1998;22:250–262. doi: 10.1002/(SICI)1520-6408(1998)22:3<250::AID-DVG7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Gao X, Sedgwick T, Shi YB, Evans T. Distinct functions are implicated for the GATA-4, -5, and -6 transcription factors in the regulation of intestine epithelial cell differentiation. Mol Cell Biol. 1998;18:2901–2911. doi: 10.1128/mcb.18.5.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber PJ, Kubalak SW, Pexieder T, Sucov HM, Evans RM, Chien KR. RXR α deficiency confers genetic susceptibility for aortic sac, conotruncal, atrioventricular cushion, and ventricular muscle defects in mice. J Clin Investig. 1996;98:1332–1343. doi: 10.1172/JCI118920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes & Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Lee RY, Luo J, Evans RM, Giguere V, Sucov HM. Compartment-selective sensitivity of cardiovascular morphogenesis to combinations of retinoic acid receptor gene mutations. Circ Res. 1997;80:757–764. doi: 10.1161/01.res.80.6.757. [DOI] [PubMed] [Google Scholar]
- Lee Y, Song AJ, Baker R, Micales B, Conway SJ, Lyons GE. Jumonji, a nuclear protein that is necessary for normal heart development. Circ Res. 2000;86:932–938. doi: 10.1161/01.res.86.9.932. [DOI] [PubMed] [Google Scholar]
- Lu JR, McKinsey TA, Xu H, Wang DZ, Richardson JA, Olson EN. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol Cell Biol. 1999;19:4495–4502. doi: 10.1128/mcb.19.6.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23:1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc Natl Acad Sci. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD. The zinc finger–containing transcription factors GATA-4, -5, and -6: Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes & Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Tang Z, Lu MM, Parmacek MS. GATA-5: A transcriptional activator expressed in a novel temporally and spatially-restricted pattern during embryonic development. Dev Biol. 1997;183:21–36. doi: 10.1006/dbio.1996.8485. [DOI] [PubMed] [Google Scholar]
- Narita N, Bielinska M, Wilson DB. Cardiomyocyte differentiation by GATA-4-deficient embryonic stem cells. Development. 1996;122:3755–3764. doi: 10.1242/dev.124.19.3755. [DOI] [PubMed] [Google Scholar]
- Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, Weiss MJ. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000;24:266–270. doi: 10.1038/73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmacek MS, Leiden JM. GATA transcription factors and cardiac development. In: Harvey RP, Rosenthal N, editors. Heart Development. San Diego: Academic Press; 1999. pp. 291–307. [Google Scholar]
- Svensson EC, Tufts RL, Polk CE, Leiden JM. Molecular cloning of FOG-2: A modulator of transcription factor GATA-4 in cardiomyocytes. Proc Natl Acad Sci. 1999;96:956–961. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson EC, Huggins GS, Lin H, Clendenin C, Jiang F, Tufts R, Dardik FB, Leiden JM. A syndrome of tricuspid atresia in mice with a targeted mutation of the gene encoding Fog-2. Nat Genet. 2000;25:353–356. doi: 10.1038/77146. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, Deconinck AE, Cantor AB, Rieff HI, Fujiwara Y, Corfas G, Orkin SH. FOG-2: A novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc Natl Acad Sci. 1999;96:950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, Izumo S, Fujiwara Y, Orkin SH. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Walters HL, III, Mavroudis C, Tchervenkov CI, Jacobs JP, Lacour-Gayet F, Jacobs ML. Congenital heart surgery nomenclature and database project: Double outlet right ventricle. Ann Thorac Surg. 2000;69:S249–S263. doi: 10.1016/s0003-4975(99)01247-3. [DOI] [PubMed] [Google Scholar]
- Ya J, Schilham MW, de Boer PA, Moorman AF, Clevers H, Lamers WH. Sox4-deficiency syndrome in mice is an animal model for common trunk. Circ Res. 1998;83:986–994. doi: 10.1161/01.res.83.10.986. [DOI] [PubMed] [Google Scholar]