Abstract
The Arabidopsis CBF transcriptional activators bind to the CRT/DRE regulatory element present in the promoters of many cold-regulated genes and stimulate their transcription. Expression of the CBF1 proteins in yeast activates reporter genes carrying a minimal promoter with the CRT/DRE as an upstream regulatory element. Here we report that this ability of CBF1 is dependent upon the activities of three key components of the yeast Ada and SAGA complexes, namely the histone acetyltransferase (HAT) Gcn5 and the transcriptional adaptor proteins Ada2 and Ada3. This result suggested that CBF1 might function through the action of similar complexes in Arabidopsis. In support of this hypothesis we found that Arabidopsis has a homolog of the GCN5 gene and two homologs of ADA2, the first report of multiple ADA2 genes in an organism. The Arabidopsis GCN5 protein has intrinsic HAT activity and can physically interact in vitro with both the Arabidopsis ADA2a and ADA2b proteins. In addition, the CBF1 transcriptional activator can interact with the Arabidopsis GCN5 and ADA2 proteins. We conclude that Arabidopsis encodes HAT-containing adaptor complexes that are related to the Ada and SAGA complexes of yeast and propose that the CBF1 transcriptional activator functions through the action of one or more of these complexes.
INTRODUCTION
The CBF1, CBF2 and CBF3 proteins of Arabidopsis thaliana (1,2), also known as DREB1B, DREB1C and DREB1A (3,4), respectively, are transcriptional regulatory proteins involved in cold-regulated (COR) gene expression. The CBF proteins, which contain AP2/EREBP DNA-binding domains (5), recognize the DNA regulatory element designated the CRT (C-repeat)/DRE (dehydration-responsive element) (6,7). The CRT/DRE is present in the promoters of multiple COR and dehydration-inducible genes, including COR15a, COR6.6/KIN2, COR47 and COR78/RD29A (8). In plants that are well irrigated and grown at normal temperatures (∼20°C) neither the CBF genes nor the target COR genes are expressed. However, within minutes of transferring plants to low non-freezing temperatures (∼4°C) the CBF genes are induced, followed at ∼2 h by expression of the COR genes (2). Constitutive overexpression of either CBF1 or CBF3 in transgenic Arabidopsis plants results in constitutive COR gene expression and an increase in freezing tolerance without a low temperature stimulus (3,9). These results indicate that the CBF proteins and the genes they regulate, the CBF regulon, have important roles in cold acclimation, the process whereby certain plants increase in freezing tolerance in response to low non-freezing temperatures.
The mechanism by which the CBF proteins activate expression of the COR genes is not known. Indeed, very little is known about how any transcriptional activator stimulates transcription in plants. In contrast, studies in yeast and animal systems have revealed two fundamental mechanisms by which transcriptional activators stimulate transcription. One is to help assemble the transcriptional apparatus at target promoters. Many transcriptional activation domains can interact directly with basal transcription factors, including the TATA-binding protein (TBP), TFIIA, TFIIB, TFIIH and components of the RNA polymerase II holoenzyme, and are thought to recruit these proteins to promoters (10–13). A second mechanism is to induce changes in chromatin structure that result in promoters becoming more accessible to RNA polymerase II and other components of the transcriptional apparatus. The changes in chromatin are accomplished by two major classes of protein complexes. One class comprises the SWI/SNF-related complexes, which alter the position of DNA in nucleosomes through a mechanism involving ATP hydrolysis (reviewed in 14,15). The second class comprises proteins that covalently modify the histone proteins in the nucleosome core (16). Most prominent among this second class are the histone acetyltransferase (HAT) proteins (reviewed in 15,17), which catalyze the addition of acetyl groups to specific lysine residues present in the N-terminal tails of core histones.
Expression of the CBF proteins in yeast activates reporter genes carrying a minimal promoter with the CRT/DRE as an upstream regulatory element (1,2). This finding indicates that the CBF proteins can interact with yeast proteins to stimulate transcription. Protein fusion experiments have demonstrated that the C-terminal half of CBF1 functions as an activation domain in both yeast and Arabidopsis (E.J.Stockinger, S.J.Triezenberg and M.F.Thomashow, unpublished results). This region of CBF1 (and homologous regions in CBF2 and CBF3), like many other activation domains from yeast and other eukaryotes, is rich in acidic amino acids. Some acidic activation domains, like those of the yeast activators Gcn4 and Pho4 and the herpes virus activator VP16, recruit HAT-containing complexes to promoters (18–20). The yeast Gcn5 protein is the HAT component of at least two distinct transcriptional adaptor complexes in yeast, designated Ada (mass ∼0.8 MDa) and SAGA (mass ∼1.8 MDa), that are capable of acetylating histone H3 in nucleosomes (21). These complexes include the transcriptional adaptor proteins Ada2 and Ada3, as well as other complex-specific proteins (22–25). Transcriptional activation by Gcn4 or Gal4–VP16 is significantly reduced in yeast mutants that do not produce either Ada2, Ada3 or Gcn5 (26–28).
The goal of this study was to explore the possibility that the CBF proteins might activate gene expression by recruiting HAT-containing adaptor complexes to promoters. Here we demonstrate that the ability of CBF1 to stimulate gene expression in yeast is dependent upon the activities of the Ada2, Ada3 and Gcn5 proteins. Moreover, we show that (i) Arabidopsis has homologs of both Ada2 and Gcn5; (ii) the Arabidopsis GCN5 protein has HAT activity; (iii) the Arabidopsis GCN5 protein can physically interact with Arabidopsis ADA2; (iv) the CBF1 transcriptional activator interacts with the Arabidopsis ADA2 and GCN5 proteins. Taken together, these results indicate that Arabidopsis encodes HAT-containing adaptor complexes that are related to the Ada and SAGA complexes of yeast and suggest that the CBF1 transcriptional activator functions through the action of one or more of these complexes.
MATERIALS AND METHODS
CBF1 activity in yeast
The yeast strains PSY316 (MATα ade2-101 his3-del.200 leu2-3,2-112 lys2 ura3-53) (26) and its descendant deletion mutant strains PSY316Δada2 (26), PSY316Δada3 (29) and PSY316Δtrp1Δgcn5 (30) were transformed with plasmid pEJS134 (1) and selected for uracil prototrophy. Plasmid pEJS134 bears two direct repeat copies of the CRT/DRE sequence (5′-GATCATTTCATGGCCGACCTGCTTTTT) upstream of the GAL1 TATA–lacZ reporter gene and integrates into the URA3 locus. The resulting strains are listed in Table 1. Into each of these reporter strain constructs we transformed plasmid pEJS251 (1), from which CBF1 was expressed under control of the ADC1 promoter. β-Galactosidase activity was assayed as described (31) for three independent colonies from each transformation event.
Table 1. Yeast strains.
| Strain |
Genotype |
Reference |
| PSY316 | MATα ade2-101 his3-del.200 leu2-3,2-112 lys2 ura3-53 | (26) |
| PSY316Δada2 | MATα ade2-101 his3-del.200 leu2-3,2-112 lys2 ura3-53 Δada2 | (26) |
| PSY316Δada3 | MATα ade2-101 his3-del.200 leu2-3,2-112 lys2 ura3-53 Δada3 | (29) |
| PSY316Δgcn5 | MATα ade2-101 his3-del.200 leu2-3,2-112 lys2 ura3-53Δtrp1 Δgcn5 | (30) |
| EJS274 | MATα ade2-101 his3-del.200 leu2-3,2-112 lys2 URA3::CRT-lacZ | This work |
| EJS276 | MATα ade2-101 his3-del.200 leu2-3,2-112 lys2 Δada2 URA3::CRT-lacZ | This work |
| EJS278 | MATα ade2-101 his3-del.200 leu2-3,2-112 lys2 Δada3 URA3::CRT-lacZ | This work |
| EJS280 | MATα ade2-101 his3-del.200 leu2-3,2-112 lys2 Δgcn5 Δtrp1 URA3::CRT-lacZ | This work |
Isolation of cDNAs encoding atADA2a, atADA2b and atGCN5
Arabidopsis cDNA libraries CD4-10 (donated by John Walker to the Arabidopsis Biological Resource Center funded by National Science Foundation/Department of Energy/US Department of Agriculture Collaborative Research in Plant Biology Program grant USDA 92-37105-7675), CD4-14 and CD4-15 (32) were obtained through the Arabidopsis Biological Resource Center. Oligonucleotides used for PCR or sequencing were synthesized either at the Michigan State University Macromolecular Structure Facility or in the USDA Plant Gene Expression Center in Albany, CA.
EST clone EST147D16 (GenBank accession no. T76166) was identified through a BLAST search of the Arabidopsis EST database (provided by the National Center for Biotechnology Information) using the yeast Ada2 protein sequence as a query. The EST clone obtained from Tom Newman (Michigan State University, MI) was used as a hybridization probe to isolate a full-length ADA2a cDNA clone from the size-selected Arabidopsis cDNA library CD4-14 (32).
The Arabidopsis ADA2b gene was initially identified by the EU Arabidopsis Genome Project sequencing of chromosome 4 (accession no. 2244998; 33). Forward (5′-GCGAGGATCCGCATGGGTCGCTCTCGAGGGAACTTCC; underlining indicates the start codon) and reverse (5′-GCGGCGGCCGCACAACACTACACAAGCCTTA; underlining indicates the stop codon) primers were used to amplify and clone the predicted open reading frame for the ADA2b gene from the Arabidopsis cDNA library CD4-10.
The cDNA sequence of the Arabidopsis homolog of yeast GCN5 was initially submitted to GenBank (accession no. AF037442; T.Tomihama, K.Shoji and T.Okano, unpublished results). Forward (5′-GATTTTCTCAATCACCTTCACTATACTGCT) and reverse (5′-CCTACAAAGTAAAAACTTAAACATTCTGAA) primers were used to amplify a cDNA fragment from the Arabidopsis cDNA library CD4-10. Sequencing of this PCR fragment indicated differences from the deposited sequence that implied changes in the frame of the predicted polypeptide. A cDNA fragment spanning the HAT and bromodomains of Arabidopsis GCN5 was amplified using forward (5′-CCAGTGCGGCCGCCATGGTGGAAAAGATGGTGG) and reverse (5′-GGCACGCGGCCGCCATGTATTCCCAGTTCCAATG) primers and was then cloned into the NotI site of pET28a (Novagen), resulting in plasmid pSTQ431. The insert fragment of pSTQ431 was subsequently used as a hybridization probe to screen the Arabidopsis cDNA library CD4-15 for an independent cDNA clone, pEJS427. The nucleotide sequences of the PCR-derived and library-derived cDNA clones agree completely in all regions of overlap.
Nucleotide sequences of Arabidopsis cDNAs described above have the following assigned GenBank accession nos: ADA2a, AF338769; ADA2b, AF338770; GCN5, AF338768. The nucleotide sequence of a genomic cosmid fragment encoding GCN5 has also been deposited as AF338771.
RNA blot analysis
RNA blot analysis was performed with 10 µg (per lane) total RNA prepared from roots or leaves of in vitro grown plants, or mature rosette leaves of cold acclimated, soil-grown plants. Total Arabidopsis RNA was isolated essentially as described (34) with an added phenol:chloroform extraction following precipitation with LiCl. Following electrophoresis and blotting to nylon membranes using standard procedures (35), RNAs were hybridized with radiolabeled cDNA probes encoding atADA2a, atADA2b or atGCN5 and subsequently rehybridized successively with probes for eIF-4A and COR15a. Cloned cDNA fragments encoding atADA2a, atADA2b or atGCN5 were radiolabeled with 32P nucleotides either by random priming or by directed priming with internal oligonucleotides (36). Membranes were hybridized with probes in Church buffer (37) and washed in 1× SSC, 0.1% SDS at 65°C.
Histone acetylation assays
DNA fragments encoding the full-length atGCN5(1–568) and an N-terminal truncation derivative atGCN5(155–568) were subcloned into the His6-tagged vector pET-28c (Novagen) and transformed into Escherichia coli strain BL21(DE3). Protein expression was induced for 16 h at 20°C using IPTG (40 µg/ml) in 500 ml of LB medium containing kanamycin (100 µg/ml). Cells expressing the recombinant proteins were collected, washed once with phosphate-buffered saline and resuspended in 15 ml of buffer A (50 mM sodium phosphate pH 7.9, 500 mM NaCl, 10% glycerol) containing two tablets of protease inhibitor cocktail (Boehringer Mannheim) and 1 mg/ml pepstatin. Resuspended cells were passed three times through a French press (∼8000 p.s.i.) at 4°C. To the French press lysate was added Triton X-100 (to 1%), DTT (to 10 mM) and imidazole (to 25 mM). Cell extracts were centrifuged for 15 min at 20 000 g. Recombinant GCN5 proteins were purified by incubating 10 ml of the supernatant with 0.8 ml of Ni–NTA resin (Qiagen) with gentle shaking for 1.5 h at 4°C. The Ni–NTA was washed eight times, 5 min each wash, at 4°C with washing solution (8 ml of buffer A containing 6 mM 2-mercaptoethanol, 0.2% Triton X-100, 1 mg/ml pepstatin, 50 mg/ml PMSF, 1 mg/ml leupeptin and 2 mg/ml aprotinin). Two additional brief washes with 8 ml of washing solution containing 25 mM imidazole were performed before eluting proteins binding to the Ni–NTA with 800 µl of elution buffer (washing solution containing 250 mM imidazole).
HAT assays were performed in 30 µl reactions containing 50 mM Tris–HCl pH 8.0, 10% glycerol, 1 mM DTT, 1 mM PMSF, 0.1 mM EDTA, 50 mM NaCl, 10 mM butyric acid, 20 µg calf thymus histones (Sigma), 200 µCi [3H]acetyl-CoA (NEN) and either 1.8 µg full-length atGCN5, 2.9 µg N-terminal truncated atGCN5 or 2.8 µg purified recombinant human TAF31 (as a negative control). The reaction mixture was incubated at 30°C for 30 min, after which the reactions were terminated by adding 20 µl of SDS–PAGE loading buffer. Twenty microliters of the terminated reaction mixtures were electrophoresed on 18% SDS–polyacrylamide gels. Gels were fixed (40% methanol, 10% acetic acid solution), soaked in autoradiographic enhancer (NEN Life Science Products), vacuum dried and exposed to X-ray film for 2 days.
In vitro protein interaction assays
Plasmids expressing GST fusion proteins bearing CBF1 or various fragments of atGCN5 (amino acids 1–568, 1–150 or 155–568) were constructed using pGEX vectors (Pharmacia). Recombinant proteins were expressed in E.coli strains XA90 or BL21(DE3) CodonPlus following induction with 40–60 µg/ml IPTG at 20°C for 20 h. Cells were lysed by French press in buffer B (10 mM Tris pH 7.9, 150 mM NaCl, 1 mM EDTA and 10% glycerol) with a protease inhibitor cocktail (Boehringer Mannheim) and 1 µg/ml pepstatin. Triton X-100 and DTT were added to 1% (v/v) and 10 mM final concentration, respectively, and the lysates were rocked at 4°C for 40 min. Following centrifugation at 20 000 g for 15 min, 1 ml of the supernatant was shaken together with 60 µl of glutathione–Sepharose beads (Pharmacia) for 90 min at 4°C. The beads were then washed eight times with 1 ml of buffer B containing 10 mM β-mercaptoethanol, 0.2% Triton X-100, 1 µg/ml pepstatin, 50 µg/ml PMSF, 1 µg/ml leupeptin and 2 µg/ml aprotinin. To check protein purity, the bead-bound protein was eluted with 2% SDS and visualized by silver staining after fractionation by SDS–PAGE.
To express the Arabidopsis ADA2a, ADA2b and GCN5 proteins in vitro the corresponding cDNAs were cloned into pET-28 vectors (Novagen). A plasmid expressing luciferase was provided by the manufacturer. Purified plasmid DNAs were used as templates in the TNT T7 transcription/translation system (Promega) containing [35S]methionine to radiolabel the expressed protein.
Depending on the specific experiment, either 1 or 2 µg GST or GST fusion protein bound to glutathione–Sepharose beads was incubated with 4 µl of radiolabeled translation mixture for 1 h at 25°C in 200 µl of binding buffer (40 mM HEPES pH 7.6, 100 mM NaAc, 5 mM MgCl2, 1 mM EDTA, 10% glycerol, 0.2% Triton X-100, 1 µg/ml pepstatin, 50 µg/ml PMSF, 1 µg/ml leupeptin and 2 µg/ml aprotinin) plus 8% bovine serum. After washing six times with 800 µl of binding buffer (with 300 or 400 mM NaCl) at 4°C or room temperature the bound proteins were eluted with 2% SDS and detected by autoradiography after SDS–PAGE.
RESULTS
CBF1 requires Ada2, Ada3 and Gcn5 to activate transcription in yeast
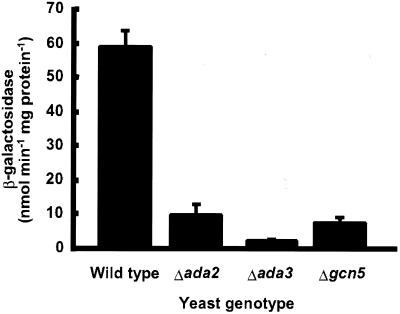
Our previous studies demonstrated that the Arabidopsis CBF1 transcription factor induces the expression of reporter genes in yeast that carry the CRT/DRE regulatory element in their promoters (1,2). If CBF1 functions in yeast by recruiting the Ada and/or SAGA complexes to promoters its activity should be impaired in strains with null mutations in either ADA2, ADA3 or GCN5. To test this hypothesis, CBF1 was expressed in yeast strains carrying the lacZ reporter gene under the control of a minimal promoter with the CRT/DRE as an upstream regulatory element. In addition, the strains carried deletions of either ADA2, ADA3 or GCN5. Assays of β-galactosidase activity indicated that the ability of CBF1 to induce expression of the lacZ reporter gene was greatly impaired in all three null mutants (Fig. 1). The effects of the ada2, ada3 and gcn5 mutations on CBF1 activity were similar in degree to their effects on the activities of the Gcn4 and VP16 transcriptional activators (26–28). The effects of the ada2, ada3 and gcn5 mutations on CBF1 activity do not reflect a general debilitation of the transcriptional machinery, for the activities of other transcriptional activators such as HAP4 are not affected by mutations in these genes (26,28; results not shown). Thus, we concluded that CBF1 functions through the action of the Ada2, Ada3 and Gcn5 proteins in yeast.
Figure 1.
Transcriptional activation by CBF1 in yeast requires the yeast ADA2, ADA3 and GCN5 genes. A reporter gene consisting of two CRT/DRE cis-acting sequences inserted 5′ to a GAL1 minimal promoter–lacZ gene fusion was integrated into the URA3 locus of the wild-type parental yeast strain PSY316 and the respective isogenic ada2, ada3 and gcn5 deletion mutant strains. These strains were transformed with a 2µ plasmid carrying the CBF1 coding sequence under control of the constitutive promoter ADC1 and were grown to mid-log phase (OD590 = 0.6 to 0.8) before cell extracts were assayed for β-galactosidase activity (31). Values for β-galactosidase activity were normalized to total yeast cellular protein. Error bars represent the standard deviation about the mean for three independent yeast cultures.
Arabidopsis has two genes encoding distinct Ada2 proteins
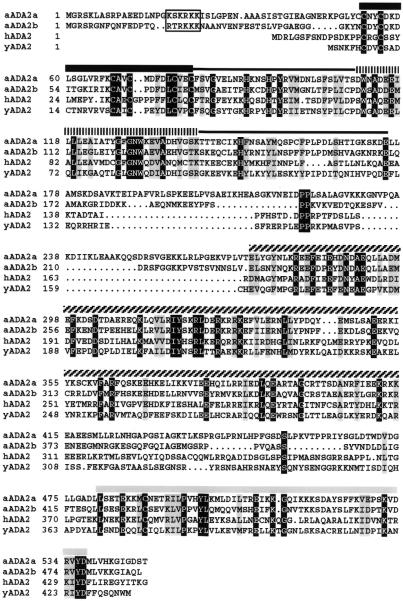
The observation that CBF1 activity in yeast requires the action of Ada2, Ada3 and Gcn5 raised the possibility that Arabidopsis might have transcriptional complexes related to Ada and/or SAGA and, more specifically, might encode homologs of Ada2, Ada3 and Gcn5. BLAST searches (38) of the Arabidopsis EST database (39) indicated the presence of a cDNA clone encoding an apparent Ada2 homolog (EST clone accession no. T76166). DNA sequencing and northern blot analysis indicated that the EST clone was missing a significant portion of the 5′-end of the Ada2 coding sequence. A cDNA encoding a full-length Arabidopsis Ada2-like polypeptide was isolated by screening a size-selected cDNA library (32) using the partial EST cDNA clone as probe (Fig. 2). We designate the Ada2-like polypeptide atADA2a and the gene ADA2a.
Figure 2.
Arabidopsis encodes two apparent homologs of the ADA2 proteins of human and yeast. Amino acid sequences for ADA2 homologs from Arabidopsis (a), human (h) and yeast (y) were deduced from cDNA sequences and aligned using the PILEUP program of the GCC sequence analysis package. Amino acids identical in all four sequences are shown as white letters on a black background, whereas amino acids with similar side chains in all four sequences are shown as black letters with a gray background. Three regions of sequence similarity are indicated by black, hatched or gray lines above the sequences, including a Cys-rich region (thick black line) and a myb motif (vertical stripes). Potential nuclear localization signals in both Arabidopsis ADA2 sequences are boxed.
Subsequent to our identification of the cDNA encoding atADA2a, a gene encoding a second putative Arabidopsis Ada2-like protein was identified by the EU Arabidopsis Genome Project (GenBank accession no. 2244998; 33). The gene, which is located on chromosome 4, was predicted to have 11 exons and 10 introns and span ∼4.6 kb of DNA. Attempts to amplify the corresponding cDNA based on the predicted exons were unsuccessful. However, inspection of the genomic sequence revealed an alternative first exon that included putative coding sequences quite similar to atADA2a. Based on this insight, we successfully amplified fragments from a cDNA library resulting in isolation of a cDNA encoding a second Ada2-like protein. The polypeptide was designated atADA2b and the gene ADA2b (Fig. 2).
The deduced atADA2a and atADA2b polypeptides have predicted molecular weights of 62 and 58 kDa, respectively, and share 49% amino acid sequence identity. In Figure 2 the primary structures of ADA2a and ADA2b are aligned with known Ada2 homologs from yeast and humans. Strong similarities between the four polypeptides are found primarily in three separate regions. The most N-terminal region of sequence similarity includes a Cys-rich module and a motif resembling a portion of the myb oncoprotein, both recognized initially in the yeast Ada2 sequence (26). The corresponding region of yeast Ada2 is capable of binding in vitro to both yeast Gcn5 and the transcriptional activation domain of VP16 (30,40,41). The second region of sequence similarity includes a domain which in yeast Ada2 was sufficient for in vitro interaction with yeast Ada3 (30,41). A third region of sequence similarity is present near the C-terminus; deletions of this region in the yeast homolog had no discernable effect in vivo or in vitro (41). An additional feature of both Arabidopsis ADA2 polypeptides is the presence of an N-terminal extension absent in the yeast and human Ada2 proteins. This region harbors a cluster of arginine and lysine residues that resembles a typical nuclear localization sequence.
Arabidopsis encodes a Gcn5 homolog with HAT activity
A BLAST search of the Arabidopsis EST database identified a cDNA sequence encoding a potential homolog of Gcn5 (GenBank accession no. AF037442). The predicted ORF was somewhat shorter than that of the yeast Gcn5 protein. We amplified, cloned and sequenced a putative full-length Gcn5 cDNA fragment from an Arabidopsis cDNA library and used the cDNA as a hybridization probe to screen a second Arabidopsis cDNA library for an independent cDNA clone. The DNA sequences of both of these cDNAs lacked a cytidylate residue reported at position 519 of the original EST clone and contained an additional adenylate residue between positions 1676 and 1677 of the original sequence. These sequences have been confirmed in a genomic clone of the Arabidopsis GCN5 locus (Y.Mao, M.F.Thomashow and S.J.Triezenberg, unpublished results). These differences in sequence result in a much longer deduced ORF (568 amino acids) than originally reported (418 amino acids). Of the 12 additional differences between our nucleotide sequence and the originally deposited sequence, six result in changes in predicted amino acids.
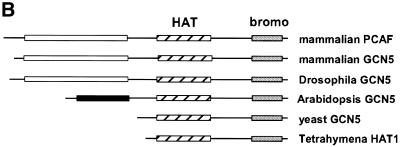
Alignments of the putative Arabidopsis Gcn5 protein, designated atGCN5, with homologs from yeast, humans, Drosophila and Tetrahymena are presented in Figure 3. Two regions of atGCN5 share a high degree of sequence identity with the other Gcn5 proteins. In the HAT domain itself the Arabidopsis homolog is 60% identical and 72% similar to yeast Gcn5. Nearly all of the key amino acids or clusters essential for yeast Gcn5 activity in vitro and in vivo (19,42) are conserved in the Arabidopsis atGCN5 protein. In addition, the C-terminal region of the atGCN5 polypeptide includes a conserved bromodomain, a domain that is thought to promote protein binding to chromatin (43,44). A third feature characteristic of all described metazoan Gcn5 homologs is the presence of a large N-terminal extension absent in the Gcn5 proteins from the lower eukaryotes yeast and Tetrahymena. The Arabidopsis homolog also has an N-terminal extension, but this region is significantly smaller than, and has no obvious sequence similarity with, the metazoan N-terminal extensions.
Figure 3.
The predicted Arabidopsis GCN5 protein sequence reveals HAT and bromodomains that are similar to regions of GCN5 proteins from other organisms. (A, above) Amino acid sequences predicted from cDNA or gene sequences for GCN5 homologs were aligned using the PILEUP program. Prefixes to protein names indicate species: h, human; m, mouse; d, Drosophila; a, Arabidopsis; y, Saccharomyces cerevisiae; Tt, Tetrahymena. The Arabidopsis GCN5 sequence is shown in bold. Amino acids identical in all eight sequences are indicated by white letters with a black background; amino acids with similar side chains in all eight sequences are shown on a gray background. The HAT domain is represented by a black bar above the sequences. The Glu residue critical for catalysis by yeast Gcn5 is marked by a black triangle. The bromodomain is represented by a gray bar above the sequences. (B, opposite) Schematic representation of regions of sequence similarity among GCN5 homologs. The HAT domain is represented by hatched bars, the bromodomain is denoted by shaded bars and the N-terminal extensions found in animal and Arabidopsis GCN5 proteins are shown as open and filled bars, respectively.
Histone acetylation assays were conducted to determine whether Arabidopsis atGCN5 has the HAT activity characteristic of Gcn5 proteins. N-terminal polyhistidine tags were fused to both full-length (amino acids 1–568) and N-terminal truncated (amino acids 155–568) versions of atGCN5 and the proteins were expressed in E.coli. The His-tagged proteins were purified using a Ni–NTA affinity matrix and histone acetylation was assayed by incubating the proteins with calf thymus core histones and [3H]acetyl CoA. The reaction products were resolved on SDS–polyacrylamide gels and the acetylated histones were detected by fluorography. The results indicated that both forms of atGCN5 effectively acetylated histone H3 (Fig. 4). Another His-tagged recombinant protein (in this case human TAF32) purified in a similar manner showed no HAT activity, as expected. We conclude that the atGCN5 protein not only resembles a HAT in primary structure but has HAT activity with similar specificity to other Gcn5 proteins. The gene encoding the atGCN5 protein was designated GCN5.
Figure 4.
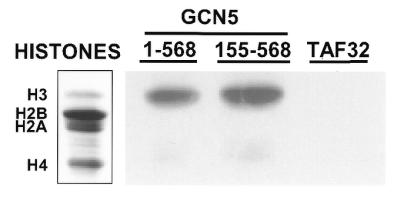
Arabidopsis GCN5 selectively acetylates histone H3 in vitro. Full-length atGCN5(1–568), a truncated form of atGCN5 (comprising residues 155–568, which retains the conserved HAT domain) and human TAF32 were expressed as polyhistidine-tagged fusion proteins in E.coli and subsequently purified using a Ni–NTA affinity matrix. Purified recombinant proteins (1.8 µg full-length atGCN5; 2.9 µg truncated atGCN5; 2.8 µg hTAF32) were incubated for 30 min at 30°C with 20 µg calf thymus core histones in the presence of [3H]acetyl-CoA. Following fractionation by 18% SDS–PAGE, gels were fixed and either stained with Coomassie blue (left) or exposed to X-ray film for fluorography (right).
Arabidopsis ADA2a, ADA2b and GCN5 are transcribed in leaves and roots
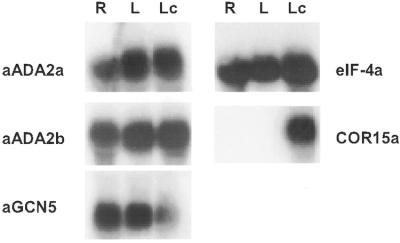
Transcripts for the ADA2a, ADA2b and GCN5 genes of Arabidopsis were detected in leaf and root tissues at normal growth temperatures (Fig. 5). Low temperature treatment did not affect the transcript levels of the genes in leaf tissue (the slight decrease in GCN5 transcript levels observed in Figure 5 was not consistently observed).
Figure 5.
Arabidopsis ADA2a, ADA2b and GCN5 are transcribed in roots and leaves. Northern filters were prepared with total RNA (20 µg) isolated from roots (R) or leaves (L) of plants grown in vitro at 22°C or from leaves of cold acclimated plants grown in soil (Lc). Filters were hybridized with cDNA probes for either ADA2a, ADA2b or GCN5 and then stripped and hybridized in succession with cDNA probes for eIF-4A and COR15a. Autoradiography for the ADA2a, ADA2b, GCN5 and eIF-4A probes was for 22 h using a single intensifying screen at –80°C; exposure for the COR15a probe was for 4 h at room temperature.
The Arabidopsis atADA2 and atGCN5 polypeptides interact directly
Protein–protein interaction experiments were conducted to determine whether the Arabidopsis atADA2 polypeptides can interact directly with the atGCN5 protein. To test this, plasmids were constructed to express GST fusion proteins containing either full-length atGCN5(1–568), an N-terminal fragment atGCN5(1–150) or N-terminally truncated atGCN5(155–568). Equivalent amounts of each fusion protein or GST alone were bound to glutathione–Sepharose beads and incubated with Arabidopsis ADA2a or ADA2b polypeptides that had been expressed and radiolabeled in vitro. After washing off non-binding material, polypeptides that bound to the glutathione–Sepharose beads were eluted, fractionated by SDS–PAGE and detected by autoradiography. The results presented in Figure 6 indicate that neither atADA2a nor atADA2b interacted with GST alone or GST fused to the N-terminal portion of atGCN5, but that they both interacted efficiently with GST fused to full-length atGCN5 or N-terminally truncated atGCN5 polypeptide. We conclude that both Arabidopsis ADA2 proteins interact directly with atGCN5 protein and that the interaction does not require the N-terminal region of the atGCN5 polypeptide.
Figure 6.
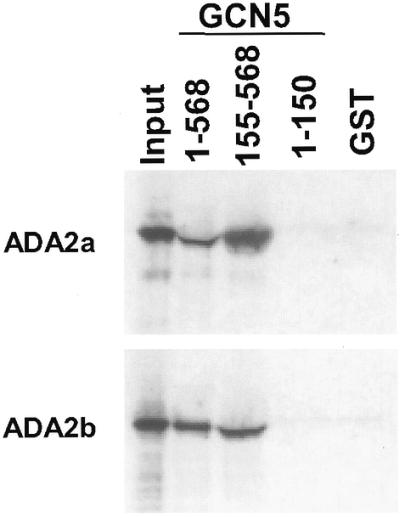
In vitro interaction of Arabidopsis GCN5 with atADA2a and atADA2b. Portions of the atGCN5 protein were expressed in E.coli as GST fusions and bound to glutathione–Sepharose beads. His-tagged atADA2a and atADA2b were expressed and radiolabeled using in vitro transcription–translation reactions. The translation reactions were incubated with the GST–GCN5 fusion proteins, washed and then eluted in a SDS buffer. The bound proteins were detected by autoradiography after fractionation by SDS–PAGE. The lanes from left to right contain: 50% ADA2 protein input; eluate from GST–GCN5(1–568); eluate from GST–GCN5(155–568); eluate from GST–GCN5(1–150); eluate from GST alone.
CBF1 interacts with atADA2 and atGCN5
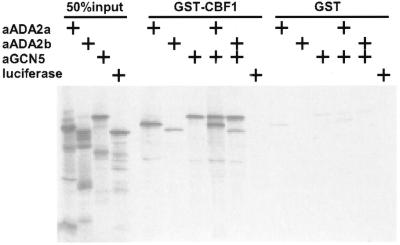
The results presented above raised the possibility that CBF1 might function in Arabidopsis by recruiting to promoters an adaptor complex(s) that includes the atADA2 and atGCN5 proteins. If true, then CBF1 might directly interact with these proteins. To test this possibility a GST fusion protein bearing the entire CBF1 polypeptide was expressed in E.coli and bound to glutathione–Sepharose beads. The Arabidopsis atADA2a, atADA2b and atGCN5 proteins, as well as luciferase as a negative control, were then transcribed and translated in vitro in the presence of [35S]methionine. The radiolabeled proteins were incubated with the GST–CBF1 fusion protein or GST alone as a negative control. After washing, proteins that were bound to the glutathione–Sepharose beads were eluted, fractionated by SDS–PAGE and detected by autoradiography. The results presented in Figure 7 show that all three Arabidopsis adaptor proteins were effectively retained on beads carrying the GST–CBF1 fusion protein, but not on the matrix containing GST alone. The slight retention observed on the GST matrix could be eliminated using a more stringent wash condition that did not affect protein retention on the GST–CBF1 matrix (data not shown). Combinations of atADA2a or atADA2b with atGCN5 did not increase retention of the proteins on the GST–CBF1 matrix. The luciferase protein was not retained on the GST–CBF1 fusion protein, indicating that CBF1 does not avidly bind proteins in general. Likewise, some of the minor products present in preparations of the radiolabeled proteins (presumably reflecting incomplete translation or partial proteolytic products) were not retained on the CBF1 matrix.
Figure 7.
In vitro interaction of Arabidopsis CBF1 with atADA2a, atADA2b and atGCN5. A GST–CBF1 fusion protein and GST alone were expressed in E.coli and bound to glutathione–Sepharose beads. The Arabidopsis ADA2a, ADA2b and GCN5 proteins as well as luciferase (as a negative control) were expressed and radiolabeled using in vitro transcription–translation reactions. The translation reactions were incubated with GST or the GST–CBF1 fusion protein, washed and then eluted in SDS. The bound proteins were detected by autoradiography after fractionation by SDS–PAGE. Lanes on the left (50% input) contain 50% of the radiolabeled protein that was incubated with each GST protein. The middle lanes (GST–CBF1) show proteins, singly or in pairs, that were bound and eluted from GST–CBF1. Lanes on the right (GST) show proteins bound and eluted from GST alone.
DISCUSSION
A recent fundamental advance in understanding transcriptional control mechanisms is the discovery of HAT-containing adaptor complexes (15–17). These complexes, which are recruited to promoters by transcriptional activators, are thought to remodel chromatin by acetylating specific histones and by making the promoters more accessible to the transcriptional apparatus. Here we present evidence that plants have HAT-containing adaptor complexes that are related to the Ada and SAGA chromatin-modifying complexes of yeast. Specifically, we establish that (i) Arabidopsis encodes homologs of the yeast Gcn5 and Ada2 proteins; (ii) the Arabidopsis GCN5 protein has HAT activity; (iii) the Arabidopsis GCN5 and ADA2 proteins physically interact with each other. Furthermore, we show that the activity of the Arabidopsis transcriptional activator CBF1 is dependent on the activities of the Gcn5, Ada2 and Ada3 proteins in yeast and that CBF1 physically interacts with the Arabidopsis GCN5 and ADA2 proteins in vitro. These results suggest that CBF1 stimulates transcription, in part, through the recruitment of Ada/SAGA-like chromatin-modifying complexes to the promoters of its target genes.
The two Arabidopsis ADA2 genes encode proteins that strongly resemble the previously described counterparts from yeast and humans (26,30). Regions of yeast Ada2 implicated in interaction with Gcn5 and with Ada3 are notably conserved in both plant homologs (Fig. 2). The most N-terminal region of sequence similarity includes two potential zinc fingers comprising six conserved cysteines in the Cys-x2-Cys ‘ZZ’ zinc finger configuration (45) with an additional two flanking conserved histidine residues. This motif is also found in the transcriptional adaptor/co-activator protein p300 and the CREB (cAMP response element-binding protein)-binding protein, CBP. The N-terminal region of yeast Ada2 protein was sufficient for in vitro interactions with yeast Gcn5 and with the transcriptional activation domain of VP16 (30,40). Deletion of the Cys-rich region from yeast Ada2 reduced co-immunoprecipitation of Gcn5, but had little discernable phenotype in vivo (41). The N-terminal region of Ada2 also contains one copy of a motif present three times in the DNA-binding domain of the Myb protein family (46). Deletion of the myb motif did not affect co-precipitation of Gcn5, but nevertheless crippled the ability of Ada2 to support transcriptional activation in vivo (41). A central region of these proteins also shows considerable sequence similarity. Deletions in this region of the yeast Ada2 protein disrupted the in vitro interaction with yeast Ada3 and caused debilitating phenotypes in vivo. Despite the considerable sequence similarity in a third region near the C-terminus, deletions in this region of the yeast Ada2 protein had no significant effects in vivo or in vitro (41). Interestingly, the N-termini of the Arabidopsis ADA2 proteins, which are very similar to each other, show little similarity with the N-termini of the fungal or human Ada2 proteins. This region of the atADA2 proteins includes a stretch of basic amino acids that resembles nuclear localization sequences. The significance of this difference between the Arabidopsis and other Ada2 proteins is not known.
The significance of Arabidopsis having two ADA2 homologs is also unknown. It is possible that the two genes are functionally redundant. Indeed, both atADA2 proteins can interact with recombinant atGCN5 in vitro with similar apparent affinities and no gross differences in expression of the two genes were observed (Fig. 6). However, we cannot yet exclude the possibility that the two atADA2 proteins have different roles. The two genes may be expressed in different cell types within leaves, roots or other tissues or differentially in response to developmental or environmental cues. It is also possible that the two atADA2 proteins may be components of distinct adaptor protein complexes that mediate distinct regulatory signals. Additional experimentation will be required to resolve these issues.
The Arabidopsis GCN5 polypeptide shows significant similarities in amino acid sequence to the HAT catalytic domains and bromodomains of previously described Gcn5 homologs from yeast, Tetrahymena, Drosophila, mice and humans. Moreover, the Arabidopsis GCN5 protein has HAT activity specific for histone H3, the hallmark of the Gcn5 proteins. Thus, the role of atGCN5 in gene regulation is likely to be similar to that proposed for the other Gcn5 proteins; remodeling chromatin to facilitate assembly of the transcriptional apparatus. Interestingly, however, the N-terminal region of Arabidopsis GCN5 is notably different from those found in metazoan Gcn5 proteins and is completely absent in the lower eukaryote yeast and Tetrahymena Gcn5 proteins. It will be of interest to determine whether this region of atGCN5 interacts with other proteins and affects the nature of the Ada/SAGA-like complex. In animal systems this domain may contribute to recognition of chromatin substrates (47) and may also carry other functions relevant to transcriptional activation (19), but no definitive explanation of the function of this domain has yet been elucidated.
To our knowledge this is the first description of genes encoding plant transcriptional adaptor proteins with putative roles in chromatin remodeling. In maize, an enzyme with histone acetyltransferase activity has been described at the biochemical level and its cDNA sequence reported (48,49). However, this enzyme is a cytoplasmic type B histone acetyltransferase, which presumably acetylates newly synthesized histone H4 for transport to the nucleus and incorporation into nucleosomes; it is unlikely to mediate chromatin modification for transcriptional regulation. An Arabidopsis N-acetyltransferase designated HOOKLESS has also been described (50). Mutations in the HOOKLESS gene result in developmental defects, notably a lack of the differential cell growth that leads to apical hook formation in hypocotyls. The deduced sequence of the HOOKLESS protein indicates that it is a member of the Gcn5-related superfamily of acetyltransferases. However, the protein is more closely related to enzymes that acetylate small metabolites such as spermine than to HAT proteins and thus, again, is unlikely to have a role in modifying chromatin structure and transcriptional activation.
Sequence homologs of the yeast Ada2 and Gcn5 proteins have been identified in humans (30), mouse (47), Drosophila (51), Tetrahymena (52) and now Arabidopsis (this report; 33). The wide distribution amongst these diverse phyla suggests that these proteins play important roles in transcriptional activation, and perhaps other processes, in all eukaryotes. The precise functions of these proteins in the growth and development of these organisms, however, remain to be clarified. In yeast, mutations in ADA2 and GCN5 are not lethal, even though the genes are unique (i.e. are single copy genes). Inactivation of either ADA2 or GCN5, however, produces pleiotropic effects. Yeast carrying null mutations in either of these genes grow slowly, especially in minimal media, and are both cold- and high-temperature-sensitive (26,28). Microarray analysis indicated that under standard growth conditions expression of ∼5% of all yeast genes is affected by inactivation of GCN5 (53). The affected genes showed no obvious specialization, being involved in a wide range of cell processes. No similar comprehensive analysis has yet been performed in a higher eukaryote. However, it has recently been demonstrated that in mice, homozygous GCN5 null mutations result in an embryonic lethal phenotype (54,55). Interestingly, knockout mutations of the closely related PCAF gene had no discernable phenotype (55). To date, no ADA2 mutations have been described in higher eukaryotes. The ability to use reverse genetics in Arabidopsis to test gene function should help to determine whether the ADA2 and GCN5 genes of Arabidopsis participate broadly in controlling gene expression or are involved in specific functions, including the regulation of genes involved in environmental stress tolerance.
NOTE ADDED IN PROOF
A report published after submission of this manuscript indicates that Arabidopsis thaliana encodes proteins related to the p300 and CBP transcriptional coactivator proteins from animals (56).
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge gifts of yeast strains, plasmids and experimental advice from Dr Shelley Berger and colleagues (Wistar Institute, Philadelphia) and from Dr Fred Winston, Harvard University. We thank Dr Tom Newman for assistance with searching the Arabidopsis EST database and for plasmids. Dr Yuri Nedialkov provided the recombinant TAF32 protein used in Figure 4. Other members of the Thomashow and Triezenberg laboratories are thanked for discussions, experimental assistance and comments on the manuscript. S.J.T. thanks Peter Quail, Jim Tepperman and colleagues at the UDSA Plant Gene Expression Center (Albany, CA) for their gracious hospitality and scientific advice during a sabbatical. This work was supported by NSF grant MCB-9728462 and the Michigan Agricultural Experiment Station.
DDBJ/EMBL/GenBank accession nos: AF338768–AF338771
References
- 1.Stockinger E.J., Gilmour,S.J. and Thomashow,M.F. (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl Acad. Sci. USA, 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilmour S.J., Zarka,D.G., Stockinger,E.J., Salazar,M.P., Houghton,J.M. and Thomashow,M.F. (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J., 16, 433–442. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q., Kasuga,M., Sakuma,Y., Abe,H., Miura,S., Yamaguchi-Shinozaki,K. and Shinozaki,K. (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell, 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinwari Z.K., Nakashima,K., Miura,S., Kasuga,M., Seki,M., Yamaguchi-Shinozaki,K. and Shinozaki,K. (1998) An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun., 250, 161–170. [DOI] [PubMed] [Google Scholar]
- 5.Riechmann J.L. and Meyerowitz,E.M. (1998) The AP2/EREBP family of plant transcription factors. Biol. Chem., 379, 633–646. [DOI] [PubMed] [Google Scholar]
- 6.Baker S.S., Wilhelm,K.S. and Thomashow,M.F. (1994) The 5′ region of Arabidopsis thalianacor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol., 24, 701–713. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi-Shinozaki K. and Shinozaki,K. (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low temperature, or high salt stress. Plant Cell, 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomashow M.F. (1998) Role of cold-responsive genes in plant freezing tolerance. Plant Physiol., 118, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaglo-Ottosen K.R., Gilmour,S.J., Zarka,D.G., Schabenberger,O. and Thomashow,M.F. (1998) ArabidopsisCBF1 overexpression induces COR genes and enhances freezing tolerance. Science, 280, 104–106. [DOI] [PubMed] [Google Scholar]
- 10.He S. and Weintraub,S.J. (1998) Stepwise recruitment of components of the preinitiation complex by upstream activators in vivo. Mol. Cell. Biol., 18, 2876–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi N., Horn,P.J., Sullivan,S.M., Triezenberg,S.J., Boyer,T.G. and Berk,A.J. (1998) DNA-complex assembly activity required for VP16C transcriptional activation. Mol. Cell. Biol., 18, 4023–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaudreau L., Keaveney,M., Nevado,J., Zaman,Z., Bryant,G.O., Struhl,K. and Ptashne,M. (1999) Transcriptional activation by artificial recruitment in yeast is influenced by promoter architecture and downstream sequences. Proc. Natl Acad. Sci. USA, 96, 2668–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nevado J., Gaudreau,L., Adam,M. and Ptashne,M. (1999) Transcriptional activation by artificial recruitment in mammalian cells. Proc. Natl Acad. Sci. USA, 96, 2674–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cairns B.R. (1998) Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem. Sci., 23, 20–25. [DOI] [PubMed] [Google Scholar]
- 15.Workman J.L. and Kingston,R.E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- 16.Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 17.Kuo M.H. and Allis,C.D. (1998) Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays, 20, 615–626. [DOI] [PubMed] [Google Scholar]
- 18.Gregory P.D., Schmid,A., Zavari,M., Lui,L., Berger,S.L. and Horz,W. (1998) Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol. Cell, 1, 495–505. [DOI] [PubMed] [Google Scholar]
- 19.Kuo M.H., Zhou,J., Jambeck,P., Churchill,M.E. and Allis,C.D. (1998) Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev., 12, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda K., Steger,D.J., Eberharter,A. and Workman,J.L. (1999) Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol., 19, 855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant P.A., Duggan,L., Cote,J., Roberts,S.M., Brownell,J.E., Candau,R., Ohba,R., Owen-Hughes,T., Allis,C.D., Winston,F., Berger,S.L. and Workman,J.L. (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev., 11, 1640–1650. [DOI] [PubMed] [Google Scholar]
- 22.Grant P.A., Schieltz,D., Pray-Grant,M.G., Steger,D.J., Reese,J.C., Yates,J.R. and Workman,J.L. (1998) A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell, 94, 45–53. [DOI] [PubMed] [Google Scholar]
- 23.Grant P.A., Schieltz,D., Pray-Grant,M.G., Yates,J.R. and Workman,J.L. (1998) The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell, 2, 863–867. [DOI] [PubMed] [Google Scholar]
- 24.Ogryzko V.V., Kotani,T., Zhang,X., Schlitz,R.L., Howard,T., Yang,X.J., Howard,B.H., Qin,J. and Nakatani,Y. (1998) Histone-like TAFs within the PCAF histone acetylase complex. Cell, 94, 35–44. [DOI] [PubMed] [Google Scholar]
- 25.Eberharter A., Sterner,D.E., Schieltz,D., Hassan,A., Yates,J.R., Berger,S.L. and Workman,J.L. (1999) The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 6621–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger S.L., Pina,B., Silverman,N., Marcus,G.A., Agapite,J., Regier,J.L., Triezenberg,S.J. and Guarente,L. (1992) Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell, 70, 251–265. [DOI] [PubMed] [Google Scholar]
- 27.Georgakopoulos T. and Thireos,G. (1992) Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J., 11, 4145–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcus G.A., Silverman,N., Berger,S.L., Horiuchi,J. and Guarente,L. (1994) Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J., 13, 4807–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piña B., Berger,S., Marcus,G.A., Silverman,N., Agapite,J. and Guarente,L. (1993) ADA3: a gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol. Cell. Biol., 13, 5981–5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Candau R., Moore,P.A., Wang,L., Barlev,N., Ying,C.Y., Rosen,C.A. and Berger,S.L. (1996) Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol. Cell. Biol., 16, 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose M. and Botstein,D. (1983) Construction and use of gene fusions to lacZ (β-galactosidase) that are expressed in yeast. Methods Enzymol., 101, 167–180. [DOI] [PubMed] [Google Scholar]
- 32.Kieber J.J., Rothenberg,M., Roman,G., Feldmann,K.A. and Ecker,J.R. (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell, 72, 427–441. [DOI] [PubMed] [Google Scholar]
- 33.Bevan M., Bancroft,I., Bent,E., Love,K., Goodman,H., Dean,C., Bergkamp,R., Dirkse,W., Van Staveren,M., Stiekma,W., Drost,L., Ridley,O., Hudson,S.-A., Patel,K., Murphy,G., Piffanelli,P., Wedler,H., Wedler,E., Wambutt,R., Weitzenegger,T., Pohl,T.M., Terryn,N., Gielen,J., Villarroel,R., De Clerck,R., Van Montague,M., Lecharny,A., Auborg,S., Gy,I., Kreis,M., Lao,N., Kavanagh,T., Hempel,S., Kotter,P., Entian,K.-D., Rieger,M., Schaeffer,M., Funk,B., Mueller-Auer,S., Silvey,M., James,R., Montfort,A., Pons,A., Puigdomenech,P., Douka,A., Voukelatou,E., Miliono,D., Hatzopoulos,P., Piravandi,E., Obermaier,B., Hilbert,H., Düsterhöft,A., Moores,T., Jones,J.D.G., Eneva,T., Palme,K., Benes,V., Rechman,S., Ansorge,W., Cooke,R., Berger,C., Delseny,M., Voet,M., Volckaert,G., Mewes,H.-W., Klosterman,S., Schueller,C. and Chalwatzis,N. (1998) Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature, 391, 485–491. [DOI] [PubMed] [Google Scholar]
- 34.Puissant C. and Houdebine,L.-M. (1990) An improvement of the single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Biotechniques, 8, 148–149. [PubMed] [Google Scholar]
- 35.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Feinberg A.P. and Vogelstein,B. (1983) A technique for radiolabelling restriction endonuclease fragments to high specific activity. Anal. Biochem., 132, 6–13. [DOI] [PubMed] [Google Scholar]
- 37.Church G.M. and Gilbert,W. (1984) Genomic sequencing. Proc. Natl Acad. Sci. USA, 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul S., Gish,W., Miller,W., Meyers,E. and Lipman,D. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 39.Newman T., de Bruijn,F.J., Green,P., Keegstra,K., Kende,H., McIntosh,L., Ohlrogge,J., Raikhel,N., Somerville,S., Thomashow,M., Retzel,E. and Somerville,C. (1994) Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol., 106, 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barlev N.A., Candau,R., Wang,L., Darpino,P., Silverman,N. and Berger,S.L. (1995) Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J. Biol. Chem., 270, 19337–19344. [DOI] [PubMed] [Google Scholar]
- 41.Candau R. and Berger,S.L. (1996) Structural and functional analysis of yeast putative adaptors: evidence for an adaptor complex in vivo. J. Biol. Chem., 271, 5237–5245. [DOI] [PubMed] [Google Scholar]
- 42.Wang L., Liu,L. and Berger,S.L. (1998) Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev., 12, 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeanmougin F., Wurtz,J.M., Le Douarin,B., Chambon,P. and Losson,R. (1997) The bromodomain revisited. Trends Biochem. Sci., 22, 151–153. [DOI] [PubMed] [Google Scholar]
- 44.Dhalluin C., Carlson,J.E., Zeng,L., He,C., Aggarwal,A.K. and Zhou,M.M. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- 45.Ponting C.P., Blake,D.J., Davies,K.E., Kendrick-Jones,J. and Winder,S.J. (1996) ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci., 241, 11–13. [PubMed] [Google Scholar]
- 46.Lane T., Ibanez,C., Garcia,A., Graf,T. and Lipsick,J. (1990) Transformation by v-myb correlates with trans-activation of gene expression. Mol. Cell. Biol., 10, 2591–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu W., Edmondson,D.G. and Roth,S.Y. (1998) Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol. Cell. Biol., 18, 5659–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eberharter A., Lechner,T., Goralik-Schramel,M. and Loidl,P. (1996) Purification and characterization of the cytoplasmic histone acetyltransferase B of maize embryos. FEBS Lett., 386, 75–81. [DOI] [PubMed] [Google Scholar]
- 49.Lusser A., Eberharter,A., Loidl,A., Goralik-Schramel,M., Horngacher,M., Haas,H. and Loidl,P. (1999) Analysis of the histone acetyltransferase B complex of maize embryos. Nucleic Acids Res., 27, 4427–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lehman A., Black,R. and Ecker,J.R. (1996) HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell, 85, 183–194. [DOI] [PubMed] [Google Scholar]
- 51.Smith E.R., Belote,J.R., Schiltz,R.L., Yang,X.-J., Moore,P.A., Berger,S.L., Yoshihiro,N. and Allis,C.D. (1998) Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res., 26, 2948–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brownell J.E., Zhou,J., Ranalli,T., Kobayashi,R., Edmondson,D.G., Roth,S.Y. and Allis,C.D. (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell, 84, 843–851. [DOI] [PubMed] [Google Scholar]
- 53.Holstege F.C.P., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,E.S. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- 54.Xu W., Edmondson,D.G., Evrard,Y.A., Wakamiya,M., Behringer,R.R. and Roth,S.Y. (2000) Loss of gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nature Genet., 26, 229–232. [DOI] [PubMed] [Google Scholar]
- 55.Yamauchi T., Yamauchi,J., Kuwata,T., Tamura,T., Yamashita,T., Bae,N., Westphal,H., Ozato,K. and Nakatani,Y. (2000) Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl Acad. Sci. USA, 97, 11303–11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bordoli L., Netsch,N., Lüthi,U., Lutz,W. and Eckner,R. (2001) Plant orthologs of p300/CBP: conservation of a core domain in metazoan p300/CBP acetyltransferase-related proteins. Nucleic Acids Res., 29, 587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]