Abstract
The CCCH zinc finger protein PIE-1 is an essential regulator of germ cell fate that segregates with the germ lineage during the first cleavages of the Caenorhabditis elegans embryo. We have shown previously that one function of PIE-1 is to inhibit mRNA transcription. Here we show that PIE-1 has a second function in germ cells; it is required for efficient expression of the maternally encoded Nanos homolog NOS-2. This second function is genetically separable from PIE-1's inhibitory effect on transcription. A mutation in PIE-1's second CCCH finger reduces NOS-2 expression without affecting transcriptional repression and causes primordial germ cells to stray away from the somatic gonad, occasionally exiting the embryo entirely. Our results indicate that PIE-1 promotes germ cell fate by two independent mechanisms as follows: (1) inhibition of transcription, which blocks zygotic programs that drive somatic development, and (2) activation of protein expression from nos-2 and possibly other maternal RNAs, which promotes primordial germ cell development.
Keywords: PIE-1, germ line, transcription, maternal mRNAs, CCCH fingers, P granules, Caenorhabditis elegans
In many animals, separation of soma and germ line occurs early during embryogenesis and is accompanied by marked differences in gene expression between the nascent somatic and germ lineages. For example, in C. elegans and Drosophila, embryonic germ cells initiate mRNA transcription later (Lamb and Laird 1976; Zalokar 1976; Kobayashi et al. 1988; Seydoux and Fire 1994) and maintain maternal RNAs longer than somatic cells (Wang and Lehmann 1991; Ding et al. 1993; Nakamura et al. 1996, Bashirullah et al. 1999). These characteristics are likely critical for the establishment of distinct germ-line and somatic fates. In particular, it has been proposed that transcriptional quiescence in early germ cells is essential to insulate the nascent germ line from transcriptional programs that promote somatic development (Seydoux et al. 1996). Whether and how transcriptional quiescence is coordinated with maternal RNA maintenance is not known.
In C. elegans, transcriptional quiescence in germ cells is dependent on the CCCH finger protein PIE-1 (Seydoux et al. 1996). PIE-1 is a maternal protein that segregates with the germ lineage during the first embryonic cleavages (Fig. 1; Mello et al. 1996). During the period that PIE-1 is present in the germ lineage, germ-line blastomeres do not transcribe mRNAs and lack a specific phosphoepitope (H5) on the carboxy-terminal domain (CTD) of RNA polymerase II (RNAPII). In the absence of PIE-1, germ-line blastomeres initiate mRNA transcription and become positive for RNAPII–H5 at the same time as somatic blastomeres (Seydoux and Dunn 1997). Germ-line blastomeres in pie-1 mutants produce only somatic descendents and no primordial germ cells (Mello et al. 1992). Expression of PIE-1 in HeLa cells has indicated that PIE-1 can inhibit mRNA transcription directly, possibly by targeting a complex that interacts with the CTD of RNAPII (Batchelder et al. 1999).
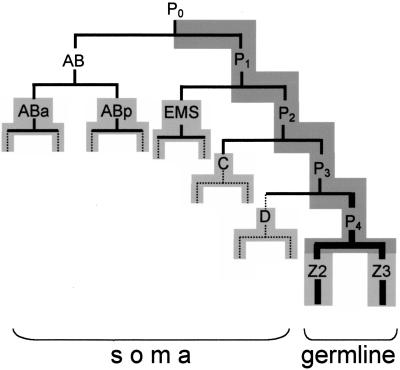
Figure 1.
Embryonic lineage. Abbreviated embryonic lineage showing the cleavages (horizontal bars) that give rise to the somatic and germ lineages (Sulston et al. 1983). Dark gray shading indicates cells with PIE-1 protein, and light gray shading indicates cells that have initiated mRNA transcription (Mello et al. 1996; Seydoux and Dunn 1997; Seydoux et al. 1996). Solid lines indicate cells with nos-2 RNA, broken lines indicates cells with no nos-2 RNA, and bold lines indicate cells with NOS-2 protein (P4, Z2 and Z3) (Subramaniam and Seydoux 1999).
As expected for a protein that inhibits transcription, much of PIE-1 accumulates in the nuclei of germ-line blastomeres (Mello et al. 1996). Significant levels of PIE-1, however, are also detected in the cytoplasm, most notably in association with P granules (Mello et al. 1996). P granules are RNA-rich organelles specific to the germ line (Strome and Wood 1982; Seydoux and Fire 1994; Pitt et al. 2000). PIE-1 contains two CCCH motifs that belong to a class of zinc fingers implicated in binding to RNA (Bai and Tolias 1996; Barabino et al. 1997; Lai et al. 2000). Consistent with an RNA-binding function, PIE-1's second finger (ZF2) is sufficient to target PIE-1 to P granules (Reese et al. 2000). These observations led us to investigate whether PIE-1 might have a function outside of the nucleus, perhaps involved in regulating the stability or translation of RNAs on P granules.
One RNA associated with P granules is nos-2. nos-2 encodes a Nanos homolog essential for primordial germ cell development (Subramaniam and Seydoux 1999). Like many maternal RNAs (Class II RNAs; Seydoux and Fire 1994), nos-2 RNA is rapidly degraded in somatic lineages and is maintained only in the germ lineage (Fig. 1). NOS-2 protein is translated in the germ-line blastomere P4 and continues to be present in its daughters, the primordial germ cells Z2 and Z3 (Fig. 1). Depletion of NOS-2 by RNA interference leads to several defects in the development of Z2 and Z3, including a failure to associate with the somatic gonad (Subramaniam and Seydoux 1999).
In this report, we show that PIE-1 is required for nos-2 RNA maintenance and for NOS-2 protein expression. Our findings indicate that PIE-1 is a bifunctional protein that regulates both transcriptional repression and maternal RNA expression in germ-line blastomeres.
Results
PIE-1 is required to maintain maternal mRNAs in the germ-line blastomere P4
To determine whether pie-1 is needed to maintain nos-2 RNA in the germ lineage, we examined the distribution of nos-2 RNA in embryos derived from mothers homozygous for either of two null mutations in pie-1 [pie-1(zu127) and pie-1(zu154); hereafter referred to as pie-1(−) embryos]. The 1- to 4-cell pie-1(−) embryos showed the normal pattern of nos-2 RNA (Fig. 2C). At the 8-cell stage, nos-2 RNA was absent from somatic cells but was often detected in the germ-line blastomere P3, as is the case in wild type (Table 1). In contrast, most 28-cell and older pie-1(−) embryos completely lacked detectable nos-2 RNA (Fig. 2G; Table 1). We conclude that pie-1 is required to maintain nos-2 RNA in P4.
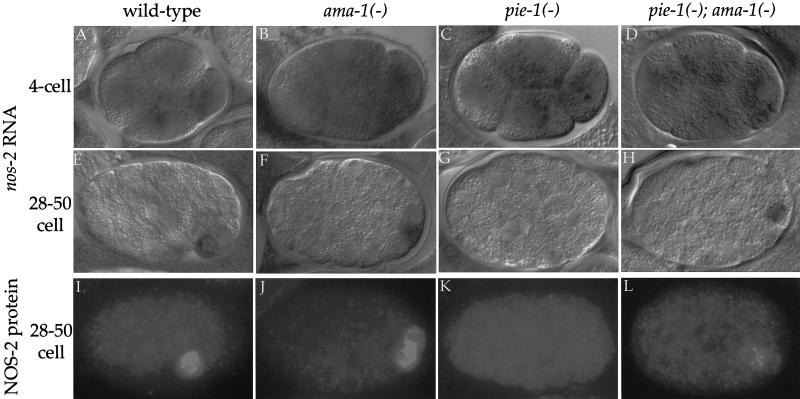
Figure 2.
PIE-1 is required for nos-2 mRNA stabilization in germ-line blastomeres and for NOS-2 protein expression. (A–D) 4-cell and (E–H) 28–50-cell embryos hybridized to nos-2 antisense probe. (A,E) Wild type. nos-2 RNA initially is uniformly distributed (A) and later is degraded in somatic lineages and maintained only in the germ lineage (E). (B,F) ama-1(RNAi). Same distribution as in wild type, confirming the maternal origin of the nos-2 RNA. (C,G) pie-1(zu127). nos-2 RNA is not maintained in P4. Identical results were obtained with pie-1(zu154) embryos. (D,H) pie-1(zu154); ama-1(RNAi). nos-2 RNA is restored in P4. (I–L) 28–50-cell embryos immunostained for NOS-2 protein. (I) Wild type, (J) ama-1(RNAi). NOS-2 protein is present in P4, confirming its maternal origin. (K) pie-1(zu127). NOS-2 protein is not expressed. Identical results were obtained with pie-1(zu154) embryos. (L) pie-1(zu154); ama-1(RNAi). NOS-2 protein is expressed, but at a lower level compared with wild-type and ama-1(RNAi) embryos.
Table 1.
nos-2 RNA and NOS-2 protein expression
| Genotype
|
nos-2 RNA
|
NOS-2 protein P4
|
|
|---|---|---|---|
| P3
|
P4
|
||
| wild type | 88% (32) | 96% (26) | 100% (100) |
| ama-l(RNAi) | 100% (13) | 100% (41) | 91% (108) |
| pie-1(−) | 96% (28) | 14% (36) | 0% (21) |
| ama-l(RNAi); pie-l(−) | 100% (10) | 94% (35) | 57% (72)a |
Number of embryos scored (n) shown in parenthesis.
Often lower levels of NOS-2 staining (see Fig. 2) compared to wild type and ama-l(RNAi).
We also examined the pattern of two other Class II maternal RNAs, cey-2 and nos-1 (Seydoux and Fire 1994; Subramaniam and Seydoux 1999). As for nos-2 RNA, we found that cey-2 and nos-1 require pie-1 for maintenance in P4, but not in P3 or earlier blastomeres (data not shown). These observations indicate that pie-1 is required to maintain several Class II RNAs in the germ lineage after the 8-cell stage.
Maintenance of nos-2 mRNA in P4 depends in part on PIE-1's ability to inhibit mRNA transcription in the germ lineage
In the absence of pie-1, mRNA transcription is activated prematurely in the germ lineage (Seydoux et al. 1996). To determine whether activation of transcription is responsible for loss of maternal nos-2 RNA, we tested whether nos-2 RNA could be restored in P4 by blocking mRNA transcription in pie-1 mutants. We used RNA-mediated interference to deplete embryos of the large subunit of RNAPII (ama-1). ama-1(RNAi) embryos do not express embryonically transcribed GFP transgenes and show a significant reduction in RNAPII immunoreactivity (Materials and Methods; Powell-Coffman et al. 1996). We found that pie-1(−); ama-1(RNAi) embryos maintain nos-2 RNA in P4 as efficiently as wild-type embryos or ama-1(RNAi) controls (Fig. 2H; Table 1). We conclude that maintenance of nos-2 RNA in P4 depends in part (see below) on inhibition of mRNA transcription in the germ lineage.
In contrast to the situation in the germ lineage, inhibition of mRNA transcription was not sufficient to maintain nos-2 RNA in somatic blastomeres (Fig. 2F). This observation indicates that, although embryonic transcription can trigger maternal RNA degradation in the germ lineage, it is not the only mechanism that triggers maternal RNA degradation in somatic lineages.
PIE-1 has a second function that promotes NOS-2 protein expression in P4
In wild-type embryos, NOS-2 protein first appears in P4 and persists for a short time in the P4 daughters Z2 and Z3 (Subramaniam and Seydoux 1999). As expected from the RNA in situ analyses, no NOS-2 protein was detected in pie-1(−) mutants (Fig. 2K; Table 1). Unexpectedly, however, we found that pie-1(−); ama-1(RNAi) embryos often lacked or had reduced levels of NOS-2 protein (Fig. 2L; Table I) compared with wild-type or ama-1(RNAi) controls, even though these embryos had normal levels of nos-2 RNA (Fig. 2H). This discrepancy raised the possibility that PIE-1 has a second function in promoting NOS-2 protein expression that is independent of its role in repressing transcription.
To explore the possibility that PIE-1 has two independent functions, we sought mutations that would disrupt one function without affecting the other. The three available pie-1 alleles completely eliminate PIE-1 protein expression and thus are not suited for this analysis (see exception below). Therefore, we created new pie-1 mutants in vitro and tested their function in vivo using a transgenic rescue assay (Materials and Methods). In this assay, pie-1 transgenes were introduced into hermaphrodites homozygous for a pie-1(null) mutation. The ability of each transgene to restore pie-1 function was assayed by examining the progeny of transgenic mothers by use of several criteria. First, the ability of the transgene to inhibit transcription was determined by examining the expression pattern of an embryonically transcribed med-1∷gfp fusion. Embryos that lack pie-1 activity express MED-1∷GFP in the germ lineage and in the EMS lineage; in contrast, embryos with wild-type pie-1 activity express MED-1∷GFP only in the EMS lineage (Maduro et al. 2001). Second, the transgene's ability to promote NOS-2 protein expression was assayed by staining embryos with anti-NOS-2 antibody. Finally, the percentage of transgenic mothers able to generate viable and fertile progeny was also recorded. Mothers that lack pie-1 activity produce only dead embryos with excess intestine and pharynx and no germ cells (Mello et al. 1992).
We first tested mutations predicted to disrupt zinc coordination by the CCCH fingers (Fig. 3A). Mutations in the first finger (ZF1) cause low levels of PIE-1 to accumulate in somatic blastomeres (Reese et al. 2000). In our rescue assay, however, the ZF1 mutant behaved similarly to a wild-type transgene (Fig. 3), indicating that ZF1 is not essential for pie-1 activity. We obtained a different result when mutating the second finger (ZF2). The ZF2 mutant transgene was impaired in its ability to promote NOS-2 protein expression (Fig. 3B), although it showed essentially wild-type localization (Materials and Methods) and was able to inhibit MED-1:GFP expression almost as efficiently as the wild-type and ZF1 transgenes (Fig. 3B). The frequency and intensity of NOS-2 staining in embryos expressing the ZF2 mutant transgene was reminiscent of pie-1(−); ama-1(RNAi) embryos. This similarity suggests that, like the pie-1(−); ama-1(RNAi) embryos, embryos that express the ZF2 mutant have a pie-1-dependent defect in NOS-2 expression that cannot be attributed to a defect in transcriptional repression. We conclude that ZF2 is not essential for transcriptional repression but is required for full NOS-2 expression.
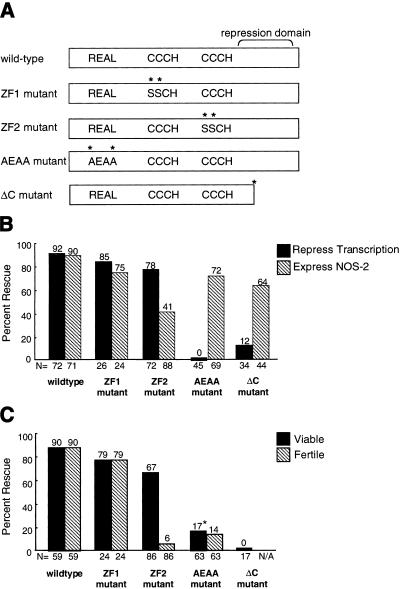
Figure 3.
Functional analysis of pie-1 transgenes and ΔC mutant. (A) Diagrams showing the location of sequence motifs in PIE-1. The transcriptional repression domain spans amino acids 223–304 (Batchelder et al. 1999), and the ΔC truncation is at codon 243. Mutations are indicated by asterisks. The ZF1, ZF2, and AEAA mutants are transgenes, whereas ΔC is a genetic mutant. (B,C) Bars represent the percent of animals rescued for the specific phenotype tested. The percent rescue is indicated at the top of each bar and the number of embryos examined (N) is shown at the bottom. Embryos derived from transgenic mothers were scored for transcriptional repression (solid bars) and NOS-2 protein expression (hatched bars). Note that transcriptional repression was assayed by scoring MED-1:GFP in the transgenic animals, and by scoring RNAPII–H5 in ΔC animals. (B) Transgenic mothers were scored for their ability to produce live progeny that reached adulthood (solid bars) and fertile progeny (hatched bars). (*) These mothers generated significantly fewer progeny than mothers carrying the wild-type, ZF1 mutant, or ZF2 mutant transgenes. Presumably, these few escapers correspond to rare embryos in which transcription was sufficiently repressed to rescue viability. See Materials and Methods for details.
We also examined the activity of a transgene with mutations in both fingers. Most embryos expressing the double mutant transgene failed to inhibit MED-1:GFP and failed to express NOS-2 (data not shown). These observations suggest that, in addition to their independent functions, ZF1 and ZF2 may be required redundantly for transcriptional repression.
Mutations in ZF2 disrupt primordial germ cell positioning in embryos
Most embryos expressing the ZF2 mutant transgene developed into viable but sterile adults (Fig. 3C). This is in contrast to pie-1 mutant embryos, which do not survive embryogenesis (Mello et al. 1992), and to embryos expressing the wild-type and ZF1 mutant transgenes, which were both viable and fertile (Fig. 3C). To investigate the cause of sterility, we immunostained embryos with a P granule antibody to visualize germ cells. In wild-type embryos that have completed gastrulation, the primordial germ cells Z2 and Z3 are nestled inside a membrane-bound structure (the somatic gonad) situated approximately two-thirds of the way along the anterior–posterior axis of the embryo (Fig. 4A–F). In contrast, in embryos expressing the ZF2 mutant transgene, Z2 and/or Z3 were often found at an ectopic location outside of the somatic gonad. In most cases, Z2 and Z3 were found near the posterior end of the elongating embryo (Fig. 4G,H). In other embryos, one of the primordial germ cells appeared to be in its normal position, whereas the other was found in a more posterior location (Fig. 4I,J). Strikingly, in some instances, Z2 and Z3 appeared to be outside of the embryo, floating freely within the confines of the egg shell (Fig. 4K,L). We conclude that ZF2 function in PIE-1 is required for proper localization of Z2 and Z3 in embryos.
Figure 4.
Primordial germ cells are mislocalized in embryos that express the PIE-1-ZF2 mutant. (A–F) Wild-type and (G–L) mutant embryos stained with anti-P granule antibody to show position of germ cells.
Mutations that affect PIE-1 distribution in nuclei interfere with transcriptional repression but not with NOS-2 expression
Beginning in the 2-cell stage, PIE-1 accumulates in the nuclei of germ-line blastomeres (Mello et al. 1996; Tenenhaus et al. 1998). We reasoned that mutations that affect this localization without affecting PIE-1 levels in the cytoplasm might be good candidates for mutations that disrupt transcriptional repression without affecting NOS-2 expression. Although PIE-1 does not contain a classical nuclear localization signal (NLS), a short sequence near the amino terminus was shown recently to be essential for maximal nuclear localization of a PIE-1: GFP fusion (Reese et al. 2000; K. Reese and G. Seydoux, unpubl.). Mutations in this sequence (REAL to AEAA) reduce PIE-1 levels in nuclei without significantly changing cytoplasmic levels (Fig. 5; Materials and Methods).
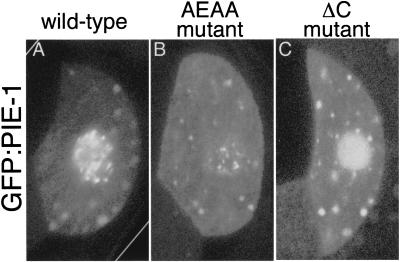
Figure 5.
Subcellular localization of wild-type and mutant PIE-1:GFP fusions. (A–C) Confocal images showing GFP fluorescence in the P2 germ-line blastomere of embryos expressing wild-type GFP:PIE-1 (A), GFP:PIE-1(AEAA) (B), and GFP:PIE-1(ΔC) (C).
Embryos expressing the AEAA mutant failed to repress transcription in germ-line blastomeres (Fig 3B). This finding is consistent with previous studies that showed that transcriptional repression requires high levels of PIE-1 in the germ lineage (Tenenhaus et al. 1998). Surprisingly, the AEAA mutant transgene was able to promote NOS-2 expression almost as well as the ZF1 transgene and significantly better than the ZF2 mutant transgene. These results indicate that, when PIE-1 is present at normal levels in the cytoplasm, activation of transcription does not necessarily interfere with NOS-2 expression.
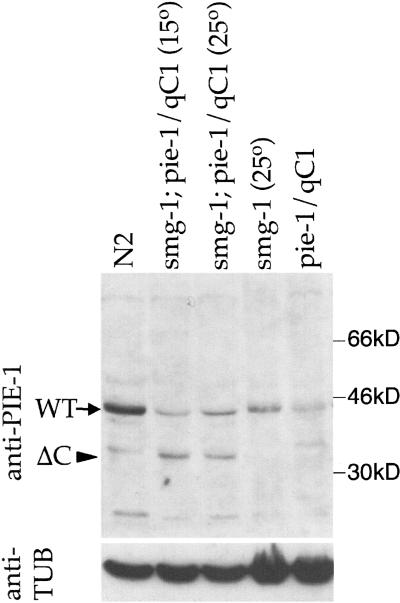
We obtained similar results with a second mutation that alters PIE-1 localization in nuclei. The pie-1(zu154) mutation introduces a premature stop codon after ZF2 (Fig. 3A; Materials and Methods). Due to nonsense-mediated RNA decay, this allele produces no protein (Tenenhaus et al. 1998) and behaves like a null by genetic criteria (Mello et al. 1992). However, when combined with the smg-1(cc545) mutation, which inactivates nonsense-mediated RNA decay (Pulak and Anderson 1993; A. Fire, pers. commun.), zu154 produces a truncated protein that can be detected with anti-PIE-1 antibody (Fig. 6). This truncated protein localizes to the germ lineage, but accumulates in nuclei in a pattern distinct from that of wild-type PIE-1 (Fig. 5C). Specifically, the protein does not accumulate in the nuclear foci in which wild-type PIE-1 is normally found (Fig. 5, cf. A and C). We found that embryos that express this truncated product express NOS-2 but fail to inhibit transcription in the germ lineage (Fig. 3B). We conclude that the carboxyl terminus of PIE-1 is essential for transcriptional repression but not for NOS-2 expression.
Figure 6.
Western analysis of pie-1(ΔC). Protein extracts were prepared from a mixed stage population (homozygotes and heterozygotes) of the following strains: (lane 1) N2 (wild type), (lane 2) smg-1(cc545ts) unc-54(r293); pie-1(zu154) unc-25 (e156)/qC1 grown at 15°C, (lane 3) smg-1(cc545ts) unc-54(r293); pie-1(zu154) unc-25(e156)/qC1 grown at 25°C, (lane 4) smg-1(cc545ts) unc-54(r293) I grown at 25°C, (lane 5) pie-1(zu154) unc-25(e156)/qC1. Western analysis was performed by use of anti-PIE-1 antibody. (Arrow) Wild-type PIE-1; (arrowhead) truncated PIE-1 (ΔC). To control for loading, the same blot was stripped and reprobed with an anti-tubulin antibody.
We have proposed that embryos that lack pie-1 activity are inviable because premature activation of transcription in the germ lineage causes cell fate transformations that eliminate essential lineages (Mello et al 1992; Seydoux et al. 1996). Consistent with this postulate, we found that embryos expressing the AEAA mutant transgene or truncated PIE-1(ΔC) are mostly inviable (Fig. 3C). Furthermore, embryos expressing truncated PIE-1(ΔC) exhibited cell fate transformations in the germ lineage identical to those observed in pie-1(null) mutants (Materials and Methods). We conclude that inhibition of transcription is essential to preserve germ cell fate.
Notably, the rare survivors obtained among embryos expressing the AEAA transgene developed into fertile adults (Fig. 3C). This is in contrast to embryos expressing the ZF2 mutant transgene, which grew up to be mostly sterile. This observation further supports the view that the sterility associated with the ZF2 mutant transgene is due to a defect in a PIE-1 function that is independent of transcriptional repression.
Discussion
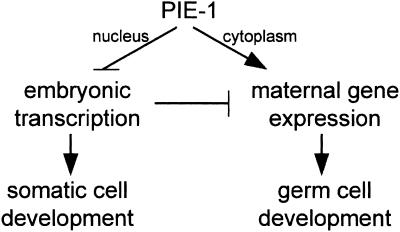
In previous studies, we have shown that PIE-1 inhibits mRNA transcription in germ-line blastomeres. In this study, we show that PIE-1 also affects the stability and expression of maternal RNAs in these cells. On the basis of our findings, we propose the following model for PIE-1 function (Fig. 7). In the nuclei of germ-line blastomeres, the main function of PIE-1 is to inhibit mRNA transcription. This inhibition has two consequences. First, it blocks maternally encoded transcription factors (e.g., SKN-1) from activating zygotic genes (e.g., med-1) that promote somatic fates. This block is essential to preserve germ cell fate. Second, inhibition of mRNA transcription also prevents degradation of maternal RNAs in germ-line blastomeres. Parallel to its role in nuclei, PIE-1 also performs a separate function in the cytoplasm of germ-line blastomeres. PIE-1's cytoplasmic function promotes the expression of maternally encoded factors essential for primordial germ cell development. These factors include NOS-2 and likely other maternal proteins that regulate the localization of primordial germ cells during embryonic development.
Figure 7.
PIE-1 has two functions in germ-line blastomeres. PIE-1 in nuclei inhibits embryonic transcription, thereby preventing transcription factors such as SKN-1 from inducing somatic development in germ-line blastomeres. Indirectly, this inhibition also prevents degradation of maternal RNAs. PIE-1 in the cytoplasm promotes maternal gene expression independently, possibly by interacting directly with maternal RNAs on P granules. This latter function is required to activate protein expression from nos-2, and possibly other maternal RNAs, which regulate the localization of primordial germ cells during embryogenesis.
Inhibition of mRNA transcription by nuclear PIE-1 is essential for germ cell fate
Several observations support the view that the main function of PIE-1 in nuclei is to inhibit transcription. First, analysis of PIE-1 function in HeLa cells has shown that the carboxy-terminal domain of PIE-1 can inhibit transcription when targeted to promoters by a heterologous DNA-binding domain (Batchelder et al. 1999). Second, a mutant lacking most of this domain fails to repress transcription in embryos and fails to localize to the nuclear foci in which wild-type PIE-1 is found (Figs. 3 and 5). Third, a mutant (AEAA mutant) that reduces PIE-1 levels in nuclei severely impairs PIE-1's ability to repress transcription (Figs. 3 and 5). We conclude that nuclear PIE-1 is essential to inhibit transcription.
We have proposed previously that inhibition of transcription by PIE-1 maintains germ cell fate by protecting germ-line blastomeres from transcription factors that can activate somatic development. One of these factors is SKN-1, a maternally encoded transcription factor present in EMS and P2, but that acts only in EMS to specify its fate (Bowerman et al. 1992). Presumably, SKN-1 is prevented from also acting in P2 by the general transcriptional block imposed on that cell by PIE-1 (Mello et al. 1996; Seydoux et al. 1996). In support of this hypothesis, we have found that mutants that specifically disrupt PIE-1's ability to repress transcription cause (1) activation of the SKN-1 target med-1 in P2 descendents (Fig. 3B), (2) activation of a marker of gut (E) fate in P2 descendents (Materials and Methods), and (3) embryonic lethality, as expected for a P2 to EMS transformation (Fig. 3C). These findings confirm that inhibition of transcription in germ-line blastomeres is essential to preserve germ cell fate.
PIE-1 has a second function important for maternal NOS-2 protein expression and fertility
The most significant finding of this study is that PIE-1 has an additional activity along with inhibiting transcription. Three independent lines of evidence support this claim. First, depletion of RNAPII by RNA interference is sufficient to rescue some but not all aspects of the pie-1(−) phenotype; in particular, it is not sufficient to restore normal NOS-2 protein levels in primordial germ cells. Second, two mutants (AEAA and ΔC) that block PIE-1's ability to inhibit transcription do not impair significantly PIE-1's ability to promote NOS-2 expression. Third, a mutant (ZF2 mutant) that impairs PIE-1's ability to promote NOS-2 expression does not affect PIE-1's ability to inhibit transcription. These data show that PIE-1's effects on transcription and NOS-2 expression can be separated genetically. We conclude that the two effects are mediated by distinct PIE-1 activities.
Embryos that express the ZF2 mutant are viable but exhibit a striking defect in primordial germ cell positioning that results in nearly 100% sterility. In these mutants, Z2 and Z3 do not enter the somatic gonad but instead occupy more posterior and superficial locations in the embryo, occasionally exiting the embryo entirely. In C. elegans, as in many other animals, primordial germ cells associate with intestinal cells in the inside of the embryo before entering the somatic gonad (Sulston et al. 1983). Our finding that Z2 and Z3 are often found on the outside of embryos expressing the ZF2 mutant transgene suggests that pie-1 activity may be required for Z2 and Z3 to adhere to the developing gut. Loss of nos-2 activity by RNA interference also leads to ectopic primordial germ cells (Subramaniam and Seydoux 1999), but that phenotype is different from the one reported here. In nos-2(RNAi) embryos, the ectopic germ cells are always located inside the embryo near a gut cell anterior to the somatic gonad, suggesting that, in those embryos, only the interaction with the somatic gonad has been disrupted. These phenotypic differences suggest that PIE-1 regulates the expression of additional factors beside NOS-2 to allow Z2 and Z3 to make specific cellular contacts during embryogenesis. It will be interesting to determine whether these factors include members of the integrin and cadherin families of adhesion proteins, which have been shown to be required for primordial germ cell migration and adhesion in mice (Anderson et al. 1999; Bendel-Stenzel et al. 2000).
Role of CCCH finger proteins in the embryonic germ lineage
PIE-1 belongs to a large family of proteins characterized by two CCCH fingers (Mello et al. 1996). In C. elegans, this family includes POS-1 and MEX-1, two maternally encoded proteins that, like PIE-1, segregate with the germ lineage where they associate with P granules (Guedes and Priess 1997; Tabara et al. 1999). Unlike PIE-1, however, POS-1 and MEX-1 are exclusively cytoplasmic and are not detected in nuclei. Accordingly, POS-1 and MEX-1 do not share PIE-1's inhibitory effect on zygotic transcription but, instead, appear to function only in the regulation of maternal gene expression (Guedes and Priess 1997; Tabara et al. 1999). In particular, POS-1 regulates the expression of APX-1, a Delta-like signal translated from maternal RNA in the P1 and P2 germ-line blastomeres (Mickey et al. 1996). In pos-1 mutants, apx-1 RNA is present but APX-1 protein is not, suggesting that translational activation of apx-1 requires POS-1 (Tabara et al. 1999). In contrast, APX-1 protein is expressed, albeit at reduced levels, in pie-1 mutants (Mickey et al. 1996). NOS-2 is not expressed in pos-1 and mex-1 mutants (C. Tenenhaus, unpubl.), although this defect may be secondary to a defect in PIE-1 localization also present in these mutants (Tenenhaus et al. 1998). Together, these observations support the idea that MEX-1, POS-1, and PIE-1 all contribute to maternal gene expression in germ-line blastomeres, although they likely act on different targets. Consistent with this hypothesis, pos-1, mex-1, and pie-1 mutants share little phenotypic similarity except for a lack of primordial germ cells (Mello et al. 1992; Guedes and Priess 1997; Tabara et al. 1999).
CCCH proteins in other organisms have been implicated in binding to RNA (Bai and Tolias 1996; Barabino et al. 1997; Carballo et al. 1998). For example, the CCCH fingers in mammalian TTP are required for sequence-specific binding to the TNF-α 3′ UTR (Lai et al. 2000). One possibility, therefore, is that POS-1, MEX-1, and PIE-1 affect maternal RNA expression by directly binding to target RNAs on P granules. Consistent with this possibility, the second finger (ZF2) in POS-1, MEX-1, and PIE-1 is sufficient for P granule localization (Reese et al. 2000). Alternatively or additionally, POS-1, MEX-1, and PIE-1 could affect maternal RNA expression indirectly by modifying P granule components or other factors that affect maternal protein synthesis and/or stability. We propose that PIE-1 is unique in having acquired the additional function of repressing transcription in the nucleus.
Coordination of transcriptional quiescence and maternal RNA maintenance in the embryonic germ lineage
In the absence of mRNA transcription, germ cells must rely on maternal RNAs to regulate their early development. What are the mechanisms that maintain maternal RNAs in germ cells? Our findings show that, in C. elegans, maternal RNA maintenance in germ cells is, in part, a consequence of transcriptional repression. Studies in Drosophila have implicated both maternally and zygotically encoded programs in the degradation of maternal RNAs (Bashirullah et al. 1999). Our findings support the view that both types of programs also operate in C. elegans and that inhibition of a zygotic degradation program by PIE-1 contributes to maternal RNA maintenance in germ cells. In addition, PIE-1 itself promotes translation of at least one maternal RNA (nos-2), possibly through a direct interaction on P granules. We conclude that PIE-1's influence on germ cell fate is twofold and involves both (1) blocking zygotic programs that drive somatic development and (2) activating maternal programs that drive germ cell development.
Material and methods
Strains
C. elegans strains were derived from the wild-type Bristol strain N2 and cultured by standard techniques as described in Brenner (1974). The following genotypes were used: dpy-18(e364) pie-1(zu127)/qC1, dpy-18(e364) pie-1(zu127)/qC1; axIs1101 [med-1:GFP], pie-1(zu154) unc-25(e156)/qC1, pie-1(zu154) unc-25 (e156)/qC1; axIs1101 [med-1:GFP], dpy-18(e364) pie-1(zu154) unc-25(e156); eDp6, unc-25(e156), dpy-18(e364), smg-1(cc545ts) unc-54(r293) I; pie-1(zu154) unc-25(e156)/qC1 III, unc-119(ed4) III, smg-1(cc545ts) unc-54(r293) I. axIs1101 is an integrated array containing pDP#MM016b [unc-119 rescuing fragment (Maduro and Pilgrim 1995)] and pMM280 [(med-1∷GFP fusion kindly provided by Joel Rothman, Maduro et al. 2001)].
ama-1 RNA interference
Double-stranded RNA interference was used to knock out ama-1 gene function by use of the bacterial feeding method (Timmons and Fire 1998). A ∼2-kb SacI–KpnI genomic fragment of ama-1 (Bird and Riddle 1989) was subcloned into L4440 and transformed into Escherichia coli HT115. Ampicillin-resistant transformants were grown in LB with 75 ug/ml ampicillin overnight. IPTG was added to the bacterial culture (final concentration of 5 mM) prior to spreading on NGM plates containing 75 ug/ml ampicillin and 0.3 mM IPTG. L4 hermaphrodites were placed on the bacterial lawn to feed on the ama-1 dsRNA for 19–22 h at 25°C. Most embryos produced after that time failed to express med-1:GFP and pes-10:GFP fusions (Seydoux and Fire 1994; Maduro et al. 2001) and arrested around the 100-cell stage, confirming loss of ama-1 function (Powell-Coffman et al. 1996).
Whole mount in situ hybridization
In situ hybridization was performed by use of single-stranded DNA probes labeled with digoxigenin (Roche Molecular Biochemicals) as described in Seydoux and Fire 1995.
Immunofluorescence microscopy
Embryos were permeabilized by freeze cracking (Epstein and Shakes 1995) and fixed following one of four protocols as follows: 5 min in −20°C MeOH and 20 min in formaldehyde fix [1X PBS, 0.08 M HEPES (pH 6.9), 1.6 mM MgS04, 0.8 mM EDTA, 3.7% formaldehyde] for visualizing GFP and NOS-2; 10–60 sec in −20°C MeOH and 30 min in formaldehyde fix for visualizing PIE-1 and RNAPII; 15 min in −20°C MeOH and 15 min in formaldehyde fix for visualizing P granules and ELT-2; 15 min in −20°C MeOH and 10 min in −20°C acetone for visualizing P granules only.
Primary antibodies used were anti-NOS-2 (Subramaniam and Seydoux 1999), anti-PIE-1 (Tenenhaus et al. 1998), mAb H5 (Warren et al. 1992; Kim et al. 1997; Patturajan et al. 1998), K76 (Strome and Wood 1982) and anti-ELT-2 (Fukushige et al. 1998). Secondary antibodies used were as follows: Cy3 or Rhodamine-conjugated goat anti-rabbit IgG, Cy3 or Rhodamine-conjugated goat anti-mouse IgG, and FITC-conjugated goat anti-mouse IgM (Jackson Immuno Research). DAPI (1 ug/ml) was added to secondary antibody dilutions to visualize DNA. Samples were mounted in Vectashield (Vector laboratories) and examined with a Zeiss-Axioplan2 equipped with a Photometrics Coolsnap digital camera.
Transgenic rescue assay
Wild-type and mutant transgenes were engineered and expressed essentially as described (Batchelder et al. 1999) with the exception that the rescue assay was performed in the strain dpy-18(e364) pie-1(zu127)/qC1; axIs1101 [med-1:GFP]. Mutations in pie-1 were created by recombinant PCR and confirmed by DNA sequencing. F2 Dpy Rol hermaphrodites (pie-1 mutant homozygotes containing the transgene) were placed individually on plates and allowed to lay eggs for 1–2 days at 25°C. The hermaphrodites were removed from the plates and the embryos inside their uteri were processed for staining. Eight to twenty-seven cell embryos were scored as rescued for transcriptional repression if MED-1:GFP fluorescence was observed only in the E and MS lineage (as in wild type). Twenty-eight to one hundred embryos were scored as rescued for NOS-2 expression if anti-NOS-2 staining was observed in the cytoplasm of one or two adjacent cell(s). Embryos laid on the plate were scored 3 days later for viability and fertility. Only hermaphrodites that laid 5+ eggs were scored. A hermaphrodite was scored as giving rise to viable progeny if at least one of its eggs developed into a viable adult. A hermaphrodite was scored as giving rise to fertile progeny if a majority of its adult progeny were fertile (embryos in uterus). Data presented in Figure 3 were compiled from at least three independent transgenic lines for each transgene. Because transgenes in complex arrays become progressively silenced with each generation, all of the lines were scored in the F2 generation after injection and were not maintained further.
Analysis of ΔC
The pie-1 ORF was PCR amplified from pie-1(zu154) homozygotes. Sequence analysis revealed the presence of a premature stop codon at codon 243. Embryos derived from pie-1(zu154) unc-25(e156) mothers do not express detectable PIE-1 (Tenenhaus et al 1998). In contrast, embryos derived from smg-1(cc545ts) unc-54(r293); pie-1(zu154) unc-25(e156) mothers express a truncated form of PIE-1 (ΔC, Fig. 6), indicating that this allele can be suppressed by mutations that eliminate nonsense-mediated RNA decay. Transcriptional repression was assayed in embryos derived from smg-1(cc545ts) unc-54(r293) I; pie-1(zu154) unc-25(e156) mothers grown at 25°C by immunofluorescence with anti-RNAPII–H5 [which stains transcriptionally active nuclei (Seydoux and Dunn 1997)] and by in situ hybridization with a probe to the zygotically expressed vet-5 mRNA (data not shown; Seydoux et al. 1996). NOS-2 expression was assayed by anti-NOS-2 immunofluorescence. The fate of P3 descendents was examined by staining 550-cell embryos with antibodies against P granules [marker of germ cells; (Strome and Wood 1982)] and ELT-2 [marker of intestinal cells; (Fukushige et al. 1998)]. Cells positive for both ELT-2 and P granule staining were seen in 22/26 smg-1(cc545ts); pie-1(zu154) embryos [full genotype: smg-1(cc545ts) unc-54(r293); pie-1(zu154) unc-25(e156)] in 13/13 pie-1(zu154) embryos[full genotype: dpy-18(e364) pie-1(zu154) unc-25(e156)], and in 0/5 smg-1(cc545ts) embryos [full genotype: smg-1(cc545ts) unc-54(r293)] and in 0/8 pie-1(zu154)/+ control embryos [full genotype: dpy-18(e364) pie-1(zu154) unc-25(e156); eDp6]. These findings confirm that P2 descendents adopt EMS-like fates in embryos expressing ΔC as they do in embryos that lack PIE-1 entirely.
Immunoblotting
Protein extraction and Western analysis were performed as described in Tenenhaus et al. (1998) with the exception that the blot was incubated with anti-PIE-1 antibody at a dilution of 1:20. Protein extracts were derived from mixed stage populations derived from the following strains: N2, smg-1(cc545ts) unc-54(r293); pie-1(zu154) unc-25(e151)/qC1, smg-1(cc545ts) unc-54(r293), and pie-1(zu154) unc-25(e156)/qC1.
GFP:PIE-1 transgenes and confocal microscopy
Wild-type and mutant pie-1 transgenes were tagged with GFP as described (Reese et al. 2000). Subcellular localization of GFP transgenes within the germ-line blastomere P2 was documented by use of a confocal laser scanning microscope (Noran Oz). A Krypton-Argon laser (Omnichrome, series 43) was used to generate an excitation wavelength of 488 nm and a barrier filter was used to detect 500+ nm emissions. Z-axis images were collected at 0.5-um intervals through the cell (P2). Figure 5 shows the most central focal plane. Localization of the GFP:PIE (ZF2) mutant was similar to wild type, although occasional embryos appeared to have reduced levels of the fusion on P granules (M. Dunn and G. Seydoux, unpubl.). In addition to reduced levels in interphase nuclei shown in B, the GFP:PIE-1(AEAA) mutant also failed to associate with centrosomes during mitosis (K. Reese, M. Dunn, and G. Seydoux, unpubl.).
Acknowledgments
We thank Matt Wallenfang for his initial analysis of pie-1(zu154); smg-1(cc545) embryos, Kim Reese for GFP:PIE-1(AEAA), Andy Fire for smg-1(cc545), Joel Rothman for med-1∷GFP, Lisa Timmons for RNAi feeding reagents, Jim McGhee for anti-ELT-2 antibody, and Keith Blackwell and Joel Rothman for their thoughtful comments on the manuscript. This work was supported by grants from the Packard Foundation and the National Institutes of Health (R01HD37407).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gseydoux@jhmi.edu; FAX (410) 502-6718.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.876201
References
- Anderson R, Fassler R, Georges-Labouesse E, Hynes RO, Bader BL, Kreidberg JA, Schaible K, Heasman J, Wylie C. Mouse primordial germ cells lacking beta1 integrins enter the germline but fail to migrate normally to the gonads. Development. 1999;126:1655–1664. doi: 10.1242/dev.126.8.1655. [DOI] [PubMed] [Google Scholar]
- Bai C, Tolias PP. Cleavage of RNA hairpins mediated by a developmentally regulated CCCH zinc finger protein. Mol Cell Biol. 1996;16:6661–6667. doi: 10.1128/mcb.16.12.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino SM, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes & Dev. 1997;11:1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Halsell SR, Cooperstock RL, Kloc M, Karaiskakis A, Fisher WW, Fu W, Hamilton JK, Etkin LD, Lipshitz HD. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J. 1999;18:2610–2620. doi: 10.1093/emboj/18.9.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelder C, Dunn MA, Choy B, Suh Y, Cassie C, Shim EY, Shin TH, Mello C, Seydoux G, Blackwell TK. Transcriptional repression by the Caenorhabditis elegans germ-line protein PIE-1. Genes & Dev. 1999;13:202–212. doi: 10.1101/gad.13.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendel-Stenzel MR, Gomperts M, Anderson R, Heasman J, Wylie C. The role of cadherins during primordial germ cell migration and early gonad formation in the mouse. Mech Dev. 2000;91:143–152. doi: 10.1016/s0925-4773(99)00287-7. [DOI] [PubMed] [Google Scholar]
- Bird DM, Riddle DL. Molecular cloning and sequencing of ama-1, the gene encoding the largest subunit of Caenorhabditis elegans RNA polymerase II. Mol Cell Biol. 1989;9:4119–4130. doi: 10.1128/mcb.9.10.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Ding D, Parkhurst SM, Halsell SR, Lipshitz HD. Dynamic Hsp83 RNA localization during Drosophila oogenesis and embryogenesis. Mol Cell Biol. 1993;13:3773–3781. doi: 10.1128/mcb.13.6.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Shakes DC. C. elegans; In: Wilson L, Matsudaira P, editors. Modern biological analysis of an organism. San Diego, CA: Academic Press, Inc.; 1995. p. 655. [Google Scholar]
- Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol. 1998;198:286–302. [PubMed] [Google Scholar]
- Guedes S, Priess JR. The C. elegans MEX-1 protein is present in germline blastomeres and is a P granule component. Development. 1997;124:731–739. doi: 10.1242/dev.124.3.731. [DOI] [PubMed] [Google Scholar]
- Kim E, Du L, Bregman DB, Warren SL. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Mizuno H, Okada M. Accumulation and spatial distribution of poly-A+ RNA in oocytes and early embryos of Drosophila melanogaster. Dev Growth Differ. 1988;30:251–260. doi: 10.1111/j.1440-169X.1988.00251.x. [DOI] [PubMed] [Google Scholar]
- Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- Lamb MM, Laird CD. Increase in nuclear poly(A)-containing RNA at syncytial blastoderm in Drosophila melanogaster embryos. Dev Biol. 1976;54:31–42. doi: 10.1016/0012-1606(76)90004-x. [DOI] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro, M.F., Meneghini, M.D., Bowerman, B., Broitman-Maduro, G., and Rothman, J. H. 2001 Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3β homolog is mediated by MED-1 and -2 in C. elegans. Mol. Cell. (In press). [DOI] [PubMed]
- Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70:163–176. doi: 10.1016/0092-8674(92)90542-k. [DOI] [PubMed] [Google Scholar]
- Mello CC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 1996;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- Mickey KM, Mello CC, Montgomery MK, Fire A, Priess JR. An inductive interaction in 4-cell stage C. elegans embryos involves APX-1 expression in the signalling cell. Development. 1996;122:1791–1798. doi: 10.1242/dev.122.6.1791. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science. 1996;274:2075–2079. doi: 10.1126/science.274.5295.2075. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Pitt JN, Schisa JA, Priess JR. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol. 2000;219:315–333. doi: 10.1006/dbio.2000.9607. [DOI] [PubMed] [Google Scholar]
- Powell-Coffman JA, Knight J, Wood WB. Onset of C. elegans gastrulation is blocked by inhibition of embryonic transcription with an RNA polymerase antisense RNA. Dev Biol. 1996;178:472–483. doi: 10.1006/dbio.1996.0232. [DOI] [PubMed] [Google Scholar]
- Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes & Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Reese KJ, Dunn MA, Waddle JA, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development. 1994;120:2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- ————— Whole-mount in situ hybridization for the detection of RNA in Caenorhabditis elegans embryos. Methods Cell Biol. 1995;48:323–337. doi: 10.1016/s0091-679x(08)61394-1. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 1996;382:713–716. doi: 10.1038/382713a0. [DOI] [PubMed] [Google Scholar]
- Strome S, Wood WB. Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc Natl Acad Sci. 1982;79:1558–1562. doi: 10.1073/pnas.79.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tabara H, Hill RJ, Mello CC, Priess JR, Kohara Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development. 1999;126:1–11. doi: 10.1242/dev.126.1.1. [DOI] [PubMed] [Google Scholar]
- Tenenhaus C, Schubert C, Seydoux G. Genetic requirements for PIE-1 localization and inhibition of gene expression in the embryonic germ lineage of Caenorhabditis elegans. Dev Biol. 1998;200:212–224. doi: 10.1006/dbio.1998.8940. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Wang C, Lehmann R. Nanos is the localized posterior determinant in Drosophila. Cell. 1991;66:637–647. doi: 10.1016/0092-8674(91)90110-k. [DOI] [PubMed] [Google Scholar]
- Warren SL, Landolfi AS, Curtis C, Morrow JS. Cytostellin: A novel, highly conserved protein that undergoes continuous redistribution during the cell cycle. J Cell Sci. 1992;103:381–388. doi: 10.1242/jcs.103.2.381. [DOI] [PubMed] [Google Scholar]
- Zalokar M. Autoradiographic study of protein and RNA formation during early development of Drosophila eggs. Dev Biol. 1976;49:425–437. doi: 10.1016/0012-1606(76)90185-8. [DOI] [PubMed] [Google Scholar]