Abstract
Activation of the Raf/MAP kinase pathway is a critical event in tumorigenesis induced by RAS and other oncogenes, a major role of this signaling system being the regulation of cellular transcription factors. To address the contribution of MAP kinase mediated transcriptional changes to the transformed phenotype, we used an inducible form of Raf to analyze early changes in the transcription of some 6000 genes following activation of the kinase in a normal human breast epithelial cell line. Of the more than 120 significant changes in mRNA level detected, genes promoting cell proliferation, invasiveness, and angiogenesis featured prominently. Some of the most strongly induced genes encoded growth factors of the EGF family: Autocrine activation of the EGF receptor was shown to be responsible for the ability of Raf activation to protect these cells from apoptosis induced by detachment of cells from extracellular matrix (anoikis), which is a critical component of the transformed phenotype.
Keywords: Ras, Raf, transcription, apoptosis, matrix, microarray
Tumorigenesis is a multistep process during which cells acquire a characteristic set of properties that make up the malignant phenotype. The dominantly acting oncogene that is implicated most frequently in human cancer is RAS, which is activated by mutation in about 25% of malignancies (Bos 1989). Ras proteins exert their transforming influence through a number of effector enzymes, including the serine/threonine kinase Raf, which activates the ERK/MAP kinase pathway; the lipid kinase phosphoinositide 3-kinase (PI 3-kinase), which activates the serine/threonine kinase PKB/Akt and the small GTPase Rac; and the exchange factor Ral-GDS, which activates the Ras-related protein Ral (Marshall 1996; Downward 1998). PI 3-kinase and PKB/Akt have been previously implicated in mediating antiapoptotic signals generated by growth factors, extracellular matrix, and activated Ras (Downward 1998; Datta et al. 1999), whereas the Raf/MAP kinase pathway has been found to play a critical role in the control of cell cycle progression (Marshall 1999). More recently, it has become clear that Raf/MAP kinase can also induce protection against certain apoptotic stimuli (Bonni et al. 1999; Erhardt et al. 1999; Kazama and Yonehara 2000; Le Gall et al. 2000) and that PI 3-kinase and PKB/Akt can contribute to cell cycle progression (Diehl et al. 1998; Klippel et al. 1998).
The transformed phenotype induced by oncogenic RAS is caused by both the short-term consequences of activating the Ras effector pathways and the longer-term effects of changes in the gene expression program of the cell that this causes. Much emphasis has been placed on the immediate cytoplasmic signaling induced by Ras activation, but each of the major Ras effector pathways also has profound effects on transcription. In particular, activation of the MAP kinase pathway by Raf induces phosphorylation and stimulation of several transcription factors, including Elk1, SRF, ATF2, and Jun (Treisman 1996), whereas PKB/Akt influences the activity of Forkhead transcription factors and possibly NF-κB (Datta et al. 1999). In this study, we have used oligonucleotide-based DNA-microarrays to assess the contribution of transcriptional changes resulting from activation of just the Raf/MAP kinase pathway alone to the Ras-transformed phenotype. The use of an inducibly activatable Raf protein allows early transcriptional events under direct control of this pathway to be studied, which is important in the establishment of the transformed phenotype. To reflect the major cell type involved in human cancer, a normal, spontaneously immortalized human epithelial cell line derived from breast tissue has been used. With this approach we were able to identify over 120 genes that were rapidly modulated in their expression levels by threefold or more in response to Raf activation. Among these, a significant number of genes encoded key regulators of proliferation, survival, angiogenesis, and invasiveness. Furthermore, the ability of Raf activation to very strongly induce autocrine expression of the EGF-like growth factors HB-EGF, TGFα, and amphiregulin was directly implicated in the ability of sustained Raf/MAP kinase pathway stimulation to protect these cells from matrix detachment-induced apoptosis, a major feature of the transformed phenotype.
Results
Activation of Raf-ER in MCF-10A cells induces sustained activation of ERK/MAP kinase and changes in cell morphology but not growth factor independence
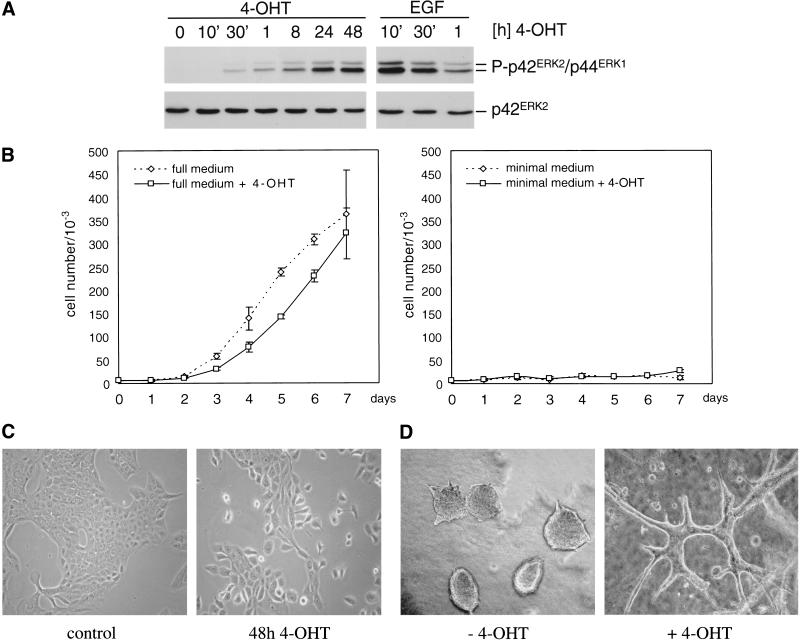
To investigate the effect of activation of Raf on epithelial cell biology, we used MCF-10A cells, a normal, spontaneously immortalized human lumenal mammary epithelial cell line (Soule et al. 1990). These cells were infected with retroviruses encoding a Raf/estrogen receptor/green fluorescent protein fusion, the kinase activity of which is stimulatable by 4-hydroxy tamoxifen (4-OHT) but not natural estrogens (Bosch et al. 1997). After drug selection and repeated fluorescence-activated cell sorting, a population highly enriched for cells expressing the fusion protein was obtained (MCF-10A ΔRaf-ER). The addition of 4-OHT to MCF-10A ΔRaf-ER cells that had been grown in growth factor–poor medium for 24 h induced rapid and sustained stimulation of ERK/MAPK phosphorylation (Fig. 1A), indicating activation of the fusion protein.
Figure 1.
Activation of ΔRaf-ER and changes in cell morphology and growth properties in response to 4-OHT. (A) Detection of MAPK-phosphorylation in MCF-10A human breast epithelial cells stably expressing the ΔRaf-ER fusion protein in response to treatment with 100 nM 4-OHT or 20 ng/mL EGF for the indicated times. Cells were starved in medium containing 5% horse serum for 24 h before treatment. Phosphorylation of p42ERK2 and p44ERK1 was detected by immunoblotting with a phosphospecific antibody (upper panel). Expression of p42ERK2 was monitored by immunoblotting with a pan-ERK antibody (lower panel). (B) Growth curves of MCF-10A ΔRaf-ER cells in medium containing different supplements in the presence (solid line, squares) or absence (dashed line, diamonds) of 4-OHT. (left) cells grown in medium containing 5% horse serum, 20 ng/mL EGF, 10 μg/mL insulin, 5 μg/mL hydrocortisone and 100 ng/mL cholera toxin (full medium); (right) cells grown in medium containing 5% horse serum without supplements (minimal medium). Values shown represent the mean and standard deviation of three replicate experiments. (C) MCF-10A ΔRaf-ER cells after treatment with solvent (0.2 μL/mL ethanol) or 100 nM 4-OHT for 48 h. The morphology of ethanol treated cell is indistinguishable from control or parental cells. (D) MCF-10A ΔRaf-ER grown in collagen gels for 10 d in the presence or absence of 100 nM 4-OHT. Cystic structures in the control image are similar to those described previously for different epithelial cell lines.
MCF-10A cells are dependent on the presence of a mixture of supplements, including EGF and insulin, for proliferation in culture. Figure 1B shows growth curves of MCF-10A ΔRaf-ER cells in the presence or absence of 4-OHT in medium supplemented with different growth factors. In medium containing 5% horse serum and all supplements (full medium, left panel), activation of Raf led to an initial but transient decrease in proliferation, consistent with the previous findings that Raf can induce growth inhibitory pathways (Sewing et al. 1997). In a growth factor–poor medium, 5% horse serum without supplements (minimal medium), these cells show no increase in numbers regardless of whether 4-OHT is present (right panel), indicating that Raf activation does not induce complete growth factor independence.
Parental MCF-10A cells and uninduced MCF-10A ΔRaf-ER cells displayed typical morphological characteristics of epithelial cells, growing in monolayer culture as islands of cells in close contact. Induction of Raf kinase activity by addition of 4-OHT led to disruption of cell–cell contacts, cell scattering, and establishment of a more fibroblastoid morphology (Fig. 1C). When cultured in collagen gels, MCF-10A cells grow as lumen-filled cysts (Soule et al. 1990); on Raf activation, the cells start to form branched structures (Fig. 1D) very similar to those reported to be associated with an invasive phenotype in other epithelial cell lines (Montesano et al. 1999). Finally, although expression of V12 Ras led to induction of colony growth in soft-agar, activation of Raf alone was not sufficient to induce transformation of MCF-10A cells in this assay (data not shown), indicating that Ras uses pathways in addition to Raf to transform cells (for review, see Shields et al. 2000).
Activation of Raf leads to protection from detachment-induced apoptosis
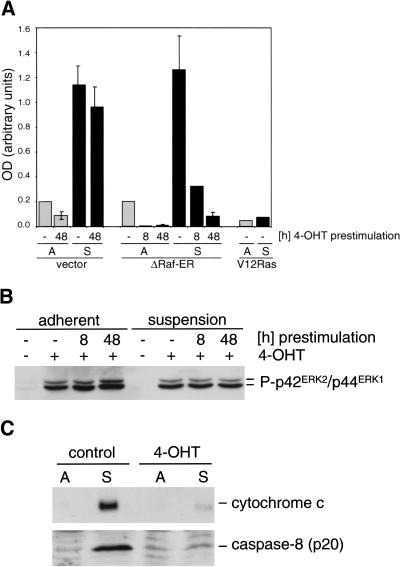
Although Raf is not able to transform MCF-10A cells by itself, we asked whether it could provide an important component of the Ras-transformed phenotype, resistance to detachment-induced apoptosis (anoikis; Frisch and Francis 1994). Although a major effector pathway mediating Ras-induced survival is thought to involve activation of protein kinase B (PKB/Akt) in a PI 3-kinase–dependent manner (Khwaja et al. 1997), we asked whether the sustained activation of Raf that was observed in MCF-10A ΔRaf-ER cells after 4-OHT treatment could be sufficient to induce survival from detachment-induced apoptosis. When attachment of the cells to substrate was prevented by plating cells on poly-HEMA coated dishes in minimal medium, parental MCF-10A cells infected with empty vector and uninduced MCF-10A ΔRaf-ER cells showed a six- to sevenfold increase in apoptosis within 24 h, measured by DNA fragmentation (Fig. 2A). When Raf was activated by treating the cells with 4-OHT for 8 or 48 h before culture in suspension, induction of apoptosis was abolished (Fig. 2A). In addition, MCF-10A cells expressing V12 Ras were also protected from detachment-induced apoptosis (Fig. 2A), as has been reported previously (Rytömaa et al. 1999).
Figure 2.
Activation of Raf induces survival from detachment-induced apoptosis and prevents release of cytochrome c from mitochondria. (A) MCF-10A cells infected with empty vector or stably expressing ΔRaf-ER or V12 Ras were pretreated with 100 nm 4-OHT for 8 or 48 h as indicated, detached by trypsin treatment, and plated on normal (light bars) or poly-HEMA coated dishes (dark bars) in minimal medium in the presence or absence of 4-OHT. After 24 h, DNA fragmentation was measured by Cell Death ELISA. Values represent the mean and standard deviation of three independent experiments. (B) Phosphorylation of p42ERK2/p44ERK1 detected with a phosphospecific antibody using cytoplasmic lysates from cells prepared in parallel to the experiment shown in A. (C) Raf activation prevents cytochrome c release in detached cells. MCF-10A ΔRaf-ER cells were pretreated with either 100 nM 4-OHT or solvent for 48 h, detached, and cultivated on normal (A) or poly-HEMA–coated dishes (S) for 24 h. (upper panel) cytochrome c detected in cytoplasmic lysates obtained by disrupting the plasma membrane with digitonin; (lower panel) detachment induced activation of caspase-8 in the presence or absence of 4-OHT. Detection of the cleaved p20 subunit of caspase-8 indicates activation of the protease.
As shown in Figure 2B, the ΔRaf-ER protein is capable of activating p42/p44ERK phosphorylation in response to 4-OHT, both in attached cells and in cells grown in suspension. Although physiological signaling from Raf to ERK has been found to be attenuated following loss of adhesion in fibroblasts (Renshaw et al. 1997), the artificial Raf construct used here is potent enough to overcome this level of dependence on matrix, as has also been found for oncogenic mutant Ras proteins (Khwaja et al. 1997).
The mechanisms by which apoptosis is triggered in response to loss of adhesion to the extracellular matrix are not completely understood. We investigated whether detachment leads to cytochrome c release from mitochondria into the cytoplasm, a key event in many forms of apoptosis, and whether Raf activation effects this. Using differential lysis of the plasma membrane by digitonin and subsequent detection of cytoplasmic cytochrome c by immunoblotting, we could show that preactivation of Raf by 4-OHT treatment for 48 h was sufficient to protect cells from detachment-induced cytochrome c release as well as activation of caspase-8 (Fig. 2C).
Analysis of differential mRNA abundance in response to Raf activation
To investigate changes in gene expression resulting from activation of Raf that might account for the changes in cell behavior reported in Figures 1 and 2, we made use of Affymetrix GeneChip oligonucleotide microarrays, which allow the parallel study of mRNA abundance of over 6000 human genes. To avoid changes reflecting the difference between proliferating and quiescent cells, we chose to use conditions in which the cells were exposed to a minimum of other stimuli, and activation of Raf was not sufficient to cause cell proliferation, as shown in Figure 1B (right panel). In addition, two different times of 4-OHT treatment were used to be able to discriminate between early and late events induced by Raf activation: a relatively short induction for 8 h as well as long-term treatment for 72 h. Because long-term culture of MCF-10A cells in minimal medium induces apoptosis (data not shown), cells were pretreated with 4-OHT in full medium for 48 h and then starved in minimal medium in the presence of 4-OHT for 24 h before lysis (total of 72 h of 4-OHT treatment). In parallel, another set of MCF-10 ΔRaf-ER cells was cultured in minimal medium for 16 h, and 4-OHT was added 8 h before lysis (total of 8 h of 4-OHT treatment). Cells treated with solvent (0.2 μL/mL of ethanol) for 48 h and cultured in minimal medium containing solvent for 24 h were used as control. To account for potential changes in gene expression induced by 4-OHT alone, MCF-10A cells infected with the empty vector were treated and analyzed in the same manner.
polyA+ RNA prepared from these cells was used to generate biotin-labeled cRNA, which was then hybridized onto GeneChip HuGeneFL microarrays and detected by fluorescent staining. Signal intensities of 20 perfect match oligonucleotides, as well as 20 corresponding mismatch oligonucleotides, for each gene were used to calculate relative mRNA abundance (average difference between signal intensity of perfect match and mismatch oligonucleotide) and relative changes in expression level (fold change) at 8 and 72 h of 4-OHT treatment compared with the ethanol-treated control. For a detailed description of the GeneChip software, see Affymetrix documentation or Lockhart et al. (1996) and Lipshutz et al. (1999). Results obtained with RNA from empty vector and ΔRaf-ER cells after 8 and 72 h of 4-OHT treatment were subjected to a filter query, allowing only those data sets to pass that were called increased or decreased by the GeneChip software and showed a fold change of three or more in both replicates of the ΔRaf-ER expressing cells but no significant change in cells transfected with empty vector after 4-OHT treatment (see also Material and Methods). The resulting 132 probe sets representing 124 genes and expressed sequence tags (ESTs) differentially expressed in response to Raf activation were classed into functional groups, which are shown in Table 1. Overall, the number of genes induced by Raf activation is much larger than the number of genes that show down-regulation (105 vs. 18 at 72 h), which is consistent with MAPK being primarily an inducer of transcription rather than an inhibitor. After 72 h of Raf activation, a significant amount of differential gene expression could be the result of secondary events that are not directly the result of transcription factors activated by Raf, but induced by accumulation of their target gene products. Thirty-two of the 132 probe sets represented genes only differentially expressed after prolonged 4-OHT treatment (72 h), whereas 17 were found to be regulated in a transient fashion, peaking after 8 h of 4-OHT treatment. It is noted that expression of a large number of transcriptional regulators is induced by Raf activation, including the well-characterized Jun subunit of AP-1 (Cook et al. 1999). mRNA levels of c-Fos, the other subunit of AP-1, were found to be transiently up-regulated 2.9-fold after 8 h of tamoxifen treatment (data not shown) but were removed from the analysis because the data did not meet the filter criteria. However, discrimination of primary and secondary target genes could not be addressed experimentally by inhibition of protein synthesis, as low concentrations of cycloheximide are sufficient to induce apoptosis in MCF-10A cells (data not shown).
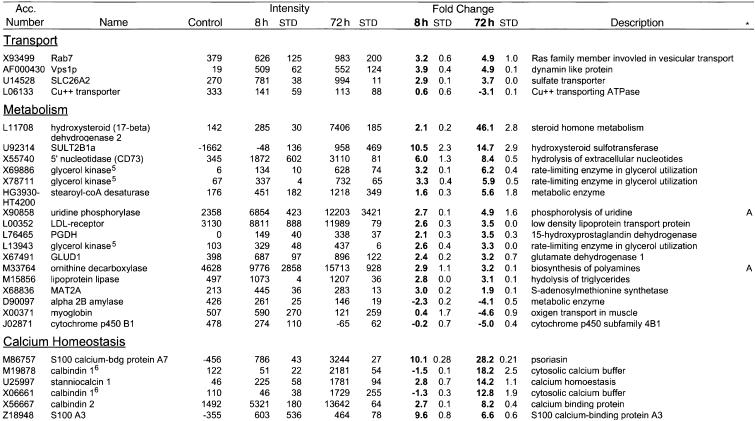
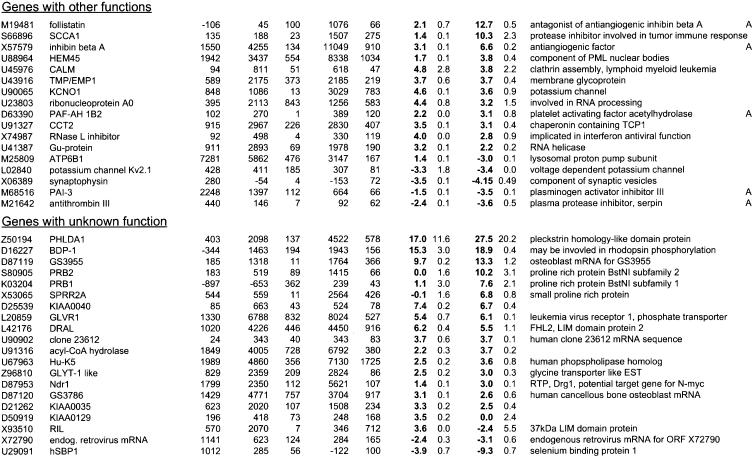
Table 1.
Differential gene expression induced by Raf activation in MCF-10A cells
Parental MCF-10A or MCF-10A ΔRaf-ER cells were pre-treated with 100nM 4-OHT or solvent (ethanol) for 48 h in full medium and subsequently grown in minimal medium for 24 h in the presence of 100 nM 4-OHT (72 h) or solvent (control) prior to lysis. In parallel samples, cells were grown in minimal medium for 16 h followed by induction with 100 nm 4-OHT in minimal medium for 8 h (8 h) prior to lysis. Samples after 8 h or 72 h of 4-OHT stimulation were prepared in duplicates. Relative mRNA abundance was measured by hybridisation to Affymetrix HuGeneFL GeneChip microassays. Values shown are the mean and standard deviation of the “Average Difference” and “Fold Change” calculated by the GeneChip (software of genes that meet the filter criteria (see Materials and Methods). The complete data set is available on http://www.icnet.uk/labs/downward/data.html.
Accession numbers are from GenBank or the TIGR gene index. Functional categories were assigned according to information retrieved from GeneCards (Weizmann Institute of Science) or Medline.
Genes represented by more than one probe set on the array. (1) PTHLH (M17183 an M24351), (2) interleukin 8 (M28130 and Y00787), (3) c-Myc (HG3523-HT4899, L00058 and M13929), (4) CD66a (HG2850-HT4814 and X16354), (5) glycerol kinase (X69886, X78711 and L13943), and (6) calbindin 1(M19878 and X06661).
A, Genes implicated in angiogenesis or blood coagulation. S, Genes implicated in protection from detachment induced apoptosis.
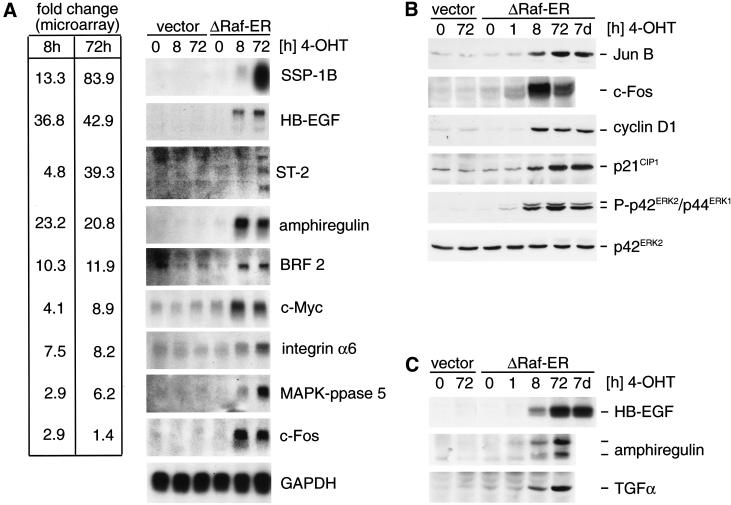
To verify changes in mRNA abundance measured using microarrays, a subset of the identified Raf target genes was detected using Northern blot analysis. As shown in Figure 3A, the analyzed genes show increases in mRNA levels on Raf activation that are comparable or higher than the fold change measured using microarrays.
Figure 3.
Differential expression of Raf target genes. (A) Northern blot detection of Raf target genes identified by microarray analysis. RNA from empty vector or ΔRaf-ER MCF-10A cells treated with 4-OHT for the indicated times while cultured in minimal medium for 24 h before lysis (in the same way as described for the experiment shown in Table 1) was separated by denaturing electrophoresis, blotted onto nylon membranes, and probed with radioactively labeled cDNA probes for the indicated genes. For comparison, fold change values measured in the microarray experiment (Table 1) are indicated on the left. (B) Whole cells lysates from cells prepared in parallel to samples used for RNA preparation in 3A were used to detect JunB, c-Fos, cyclin D1, and p21CIP1 by immunoblotting. In addition, whole cell lysates from MCF-10A ΔRaf-ER cells treated with 4-OHT for 7 d were used (lane 7). Activation of ΔRaf-ER by 4-OHT was monitored by detection of p42ERK2/p44ERK1 MAPK phosphorylation using a phosphospecific antibody. (C) Same cell lysates as in B were used to detect HB-EGF, TGFα, and amphiregulin by immunoblotting. Amphiregulin was detected as two bands representing the 17-kD precursor and a smaller processed form.
To correlate differences observed in mRNA abundance with changes in protein expression, whole cell lysates prepared in parallel to mRNA samples were used for detection of a subset of proteins whose mRNAs showed significant up-regulation in response to Raf activation (Fig. 3B). JunB, c-Fos, p21CIP1, and cyclin D1, all of which have been shown previously to be transcriptionally induced by activation of the MAPK-pathway (Hipskind et al. 1994; Albanese et al. 1995), show strong up-regulation of protein levels on Raf activation.
Transcripts coding for three different peptides of the EGF-family of growth factors, HB-EGF, amphiregulin, and TGFα were strongly up-regulated in response to Raf activation (42.9-fold, 20.8-fold, and 3.3-fold, respectively, at 72 h in Table 1). Interestingly, HB-ECF and amphiregulin are the most strongly induced of all of the genes on the microarray following 8 h of Raf activation (36.8-fold and 23.2-fold, respectively), which is also reflected by a significant induction of transcript levels measured by Northern blotting (Fig. 3A). Detection of HB-EGF, TGFα, and amphiregulin protein revealed that synthesis of all three peptides is induced on Raf activation (Fig. 3C).
Interestingly, phosphorylation of ERK/MAPK and up-regulation of Jun B, cyclin D1, p21CIP1, and HB-ECF could still be detected in lysates from ΔRaf-ER cells in which Raf was activated by 4-OHT treatment for 7 d (Fig. 3B and C).
Raf activation induces expression of soluble growth factors that can activate p42/p44–MAPK and PKB/Akt
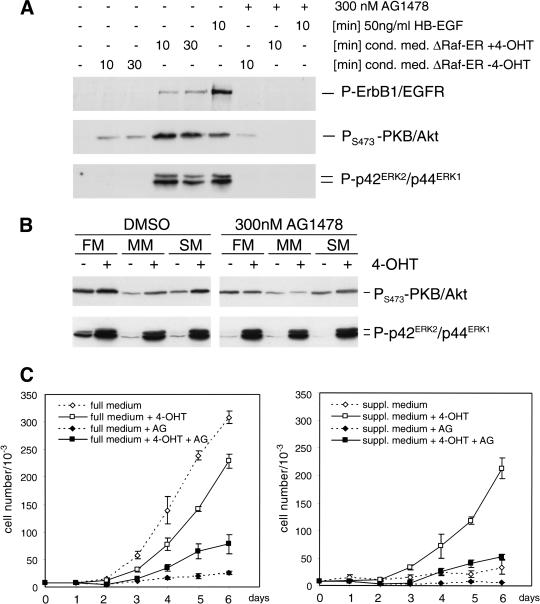
We investigated whether soluble factors produced by cells expressing activated Raf were able to stimulate EGF receptor-induced signaling pathways; supplement-free minimal medium that had been conditioned by 4-OHT treated MCF-10A ΔRaf-ER cells for 24 h induced rapid phosphorylation of ErbB1/EGFR, p42/p44ERK, and PKB/Akt in parental MCF-10A cells. Phosphorylation of all three proteins could be completely abolished by preincubation with AG1478, a selective inhibitor of EGFR tyrosine kinase activity (Fig. 4A). The minor activation of PKB/Akt phosphorylation induced by conditioned medium from ΔRaf-ER cells in the absence of 4-OHT was independent of EGFR function because it was insensitive to AG1478 treatment (Fig. 4A, lanes 2,3,7).
Figure 4.
Raf-induced synthesis of soluble factors leads to activation of EGF-receptor signaling and reduced growth factor dependence in MCF-10A cells. (A) MCF-10A ΔRaf-ER cells were treated with 100 nM 4-OHT or solvent for 48 h and grown in minimal medium for 24 h. Medium was collected and used to stimulate parental MCF-10A cells, which had been cultured in minimal medium for 24 h. To specifically inhibit EGF-receptor tyrosine kinase activity, 300 nM AG1478 was added 30 min before addition of conditioned medium where indicated. Phosphorylation of ErbB1/EGFR, PKB/Akt, and p42ERK2/p44ERK1 MAPK was detected by immunoblotting of whole cell lysates using phosphospecific antibodies. (B) MCF-10A ΔRaf-ER cells were treated with 100 nM 4-OHT or solvent for 24 h while being cultured in full medium (FM), minimal medium (MM), or medium supplemented with 10 μg/mL insulin, 5 μg/mL hydrocortisone, and 100 ng/mL cholera toxin (SM) in the presence or absence of 300 nM AG1478. Whole cell lysates were used to detect phosphorylation of PKB/Akt and p42ERK2/p44ERK1 MAPK. (C) Growth curves of MCF-10A ΔRaf-ER cells cultured in the presence (solid line, squares) or absence (dashed line, diamonds) of 4-OHT. (Left) Cells grown in full medium; (right) cells grown in medium supplemented with 10 μg/mL insulin, 5 μg/mL hydrocortisone, and 100 ng/mL cholera toxin (suppl. medium). Where indicated, 300 nM AG1478 was added to the culture (solid squares and diamonds).
To investigate whether autocrine activation of EGF receptor signaling would also induce activation of endogenous PKB/Akt we analyzed lysates from ΔRaf-ER cells grown under different conditions in the presence or absence of 4-OHT. As shown in Figure 4B, activation of Raf induces an detectable increase in phosphorylation of endogenous PKB/Akt. This increase was dependent on EGF receptor function because it could be completely abolished by AG1478, whereas tamoxifen-dependent activation of ERK/MAPK phosphorylation was not affected by inhibition of EGF receptor function (Fig. 4B).
Synthesis of autocrine factors induced by Raf activation also altered growth factor dependency of MCF-10A cells. Figure 4C shows growth curves of MCF-10A ΔRaf-ER cells grown in the presence or absence of 4-OHT in full medium (left panel) or minimal medium supplemented with insulin, hydrocortisone, and cholera toxin, which by itself was not sufficient to support proliferation of MCF-10A cells (right panel). Raf activation in full medium resulted in a transient decrease in proliferation (see also Fig. 1B). This could be because of the induction of inhibitors of proliferation such as p21CIP1, as has been shown previously (Fig. 3B). However, in supplemented medium, activation of Raf was sufficient to induce a similar proliferation rate to that seen in full medium (Fig. 4C, right panel). Inhibition of EGF receptor function by addition of AG1478 blocked proliferation of MCF-10A ΔRaf-ER cells in full medium, which was only slightly relieved by Raf activation (Fig. 4C, left panel). Proliferation in supplemented medium induced by Raf activation was dependent on EGF receptor function because it was completely abolished in the presence of AG1478 (Fig. 4C, right panel). Although basal levels of ERK/MAPK phosphorylation were higher in full medium compared to minimal or supplemented medium, activation of Raf by addition of 4-OHT resulted in similar levels of ERK/MAPK phosphorylation under all growth conditions (Fig. 4B, lower panel). Thus, at least a component of the Raf-induced growth response in these cells relies on autocrine EGF-like factor signaling.
These results indicate that Raf activation induces synthesis of soluble growth factors that activate EGF receptor signaling. Although Raf clearly induces certain growth inhibitory pathways, it can compensate for EGF dependency in supporting proliferation of MCF-10A cells.
Survival in suspension induced by Raf is mediated by autocrine activation of the EGF receptor
The strong induction of expression of EGF-like growth factors observed in response to Raf activation led us to investigate whether autocrine signaling could be involved in protection from detachment-induced apoptosis. For autocrine production of EGF-like factors to be able to protect MCF-10A cells from apoptosis in suspension, it is necessary that they produce the factors when detached as well as during adherent growth. Figure 5A shows that MCF-10A ΔRaf-ER cells prestimulated with 4-OHT for 24 h and then placed in suspension culture for 24 h show as strong induction of HB-EGF, amphiregulin, and TGFα proteins as cells that are continually adherent.
Figure 5.

Raf-induced survival from detachment induced apoptosis is sensitive to inhibition of EGF receptor tyrosine kinase activity and PI 3-kinase activity. (A) Induction of EGF-like growth factor expression upon Raf activation. MCF-10A ΔRaf-ER cells pretreated with 100 nM 4-OHT for 48 h were detached and plated on normal (A) or poly-HEMA coated dishes (S) in minimal medium in the presence (+) or absence (−) of 100 nM 4-OHT. After 24 h, expression of HB-EGF, amphiregulin, and TGFα and phosphorylation of p42ERK2/p44ERK1 MAPK was monitored by immunoblotting of whole cell lysates. Amphiregulin was detected as two bands representing the 17-kD precursor and a smaller processed form. (B) Down-regulation of Bcl-XL in suspension is prevented by Raf activation. Same lysates as in A were probed for Bcl-XL and Bcl-2 expression by immunoblotting. (C) EGF-like growth factors provide a survival signal against detachment induced apoptosis. Parental MCF-10A cells were detached and plated on normal (adherent, light bars) or poly-HEMA coated dishes for 24 h (suspension, dark bars) in minimal medium supplemented with EGF, HB-EGF, TGFα, or amphiregulin as indicated. DNA fragmentation was determined by Cell Death ELISA. Values represent the mean and variation of a typical experiment performed in duplicates. (D) MCF-10A ΔRaf-ER cells were detached and plated on normal (A, light bars) or poly-HEMA–coated dishes (S, dark bars) in minimal medium for 24 h in the presence (+) or absence (−) of 100 nM 4-OHT. Selective inhibitors of MEK (PD98059), PI3-kinase (LY294002), EGFR (AG1478, PD168393), or HB-EGF (Glu52-Diphtheria toxin, CRM197) were added as indicated. DNA-fragmentation was determined by Cell Death ELISA. Values represent the mean and variation of a typical experiment performed in triplicates. (E) Whole cell lysates from MCF-10A ΔRaf-ER cells treated in parallel to the experiment shown in D were analyzed for p42ERK2/p44ERK1 MAPK phosphorylation by immunoblotting. (F) MCF-10A infected with empty vector or stably expressing V12Ras were cultured in minimal medium with or without 20 ng/mL EGF for 24 h. Whole cell lysates were assayed for PKB/Akt and p42ERK2/p44ERK1 MAPK phosphorylation by immunoblotting. (G) MCF-10A infected with empty vector or stably expressing V12Ras were detached and plated on normal (A, light bars) or poly-HEMA–coated dishes (S, dark bars) in minimal medium for 24 h in the presence of 300 nM AG1478, 30 μM PD98059, or 50 μM LY294002 as indicated. DNA-fragmentation was determined by Cell Death ELISA. Values represent the mean and variation of a typical experiment performed in duplicates.
It has been shown previously that in keratinocytes inhibition of EGF receptor function and detachment from matrix induces down-regulation of Bcl-XL mRNA and protein level. We did not observe significant changes in Bcl-XL mRNA levels judged from the microarray results, but Bcl-XL protein levels were down-regulated after 24 h in suspension in MCF-10A ΔRaf-ER cells; this was completely inhibited by pretreatment with 4-OHT (Fig. 5B, upper panel). Bcl-2 protein levels were also down-regulated after 24 h in suspension but were not altered on activation of Raf by 4-OHT (Fig. 5B, lower panel).
When recombinant EGF, HB-EGF, or TGFα were added to suspension cultures of parental MCF-10A cells, they were sufficient to induce significant protection from detachment-induced apoptosis (Fig. 5C). However, addition of amphiregulin induced less survival than addition of HB-EGF or TGFα, reflecting its lower binding affinity for the EGF-receptor.
Because a single dose of these growth factors clearly can support survival of MCF-10A cells in suspension, we investigated whether inhibition of their function might compromise the ability of Raf to block detachment-induced apoptosis. We targeted one of the growth factors selectively, using a mutant form of diphtheria toxin (CRM197), which lacks enzymatic activity but still binds to its receptor, HB-EGF, and prevents it interacting with the EGF receptor. CRM197 caused a significant, albeit partial, reduction in survival in suspension induced by Raf activation (Fig. 5D). Because HB-EGF is only one of several EGF-family growth factors produced by these cells, we also assessed the effect of inhibiting the EGF receptor itself using pharmacological inhibitors. Survival in suspension induced by activation of Raf could be completely blocked when cells were treated with two selective EGF receptor tyrosine kinase inhibitors, AG1478 or PD168393 (Fig. 5D). Two other inhibitors also blocked Raf protection from anoikis: PD98059 presumably inhibits signaling by ΔRaf-ER immediately downstream at MEK, whereas LY294002 acts to inhibit PI 3-kinase and is likely to reverse EGF receptor-mediated activation of PI 3-kinase downstream from autocrine EGF-like factors. Inhibition of EGF receptor function or PI 3-kinase activity did not alter hormone-dependent activation of the ΔRaf–ER fusion protein, as shown by detection of ERK/MAPK phosphorylation in the presence of LY294002, AG1478, or PD168393 (Fig. 5E). In contrast, PD98059, which inhibits MEK activation downstream from Raf, completely blocked ERK/MAPK phosphorylation in response to 4-OHT treatment (Fig. 5E).
Having shown that Raf-dependent survival in suspension is dependent on autocrine activation of EGF receptor signaling and can be blocked by inhibition of PI 3 kinase, we asked whether EGF receptor signaling is also involved in protection from anoikis by activated Ras. MCF-10A cells stably expressing activated Ras (MCF-10A V12Ras) maintain detectable levels of PKB/Akt and ERK/MAPK phosphorylation in minimal medium, as well as showing an increased response to EGF treatment (Fig. 5F). MCF-10A V12Ras cells are protected from detachment-induced apoptosis (Figs. 2A, 5G), which was independent on EGF receptor function because addition of AG1478 did not abrogate survival. Although inhibition of MEK by PD98059 had only marginal effects, inhibition of PI 3-kinase reduced survival in suspension significantly (Fig. 5G). Inhibition of both PKB/Akt and MEK led to an even more pronounced increase in apoptosis in suspension. This supports the conclusion that, although PI 3-kinase activation seems to be more important, both Ras effector pathways contribute to survival in suspension induced by oncogenic Ras.
Taken together these results indicate that autocrine activation of PI 3-kinase through induction of expression of EGF-like growth factors is crucial for the ability of the Raf/MAP-kinase pathway to protect MCF-10A cells from detachment-induced apoptosis. In cells expressing oncogenic Ras, however, the PI 3-kinase pathway gets activated directly, rendering survival in suspension independent of EGF receptor function.
Discussion
Using a hormone-inducible version of Raf, we have studied the consequences of selective activation of one effector of the Ras oncoprotein in epithelial cells. A recent paper has reported the use of a PCR-based cDNA subtraction technique to identify 244 known genes that are differentially expressed by twofold or more between a normal rat embryo fibroblast cell line and a derivative that has been constitutively transformed by Ras (Zuber et al. 2000). Very few changes were found in common between these two sets of data: Only five genes were found to be regulated by twofold or more in the same direction in both reports, despite the fact that most of the genes identified as changed by Zuber et al. were present on the chip used in our study. Possible reasons for this include the fact that the Raf/MAP kinase pathway is only one of a number of signaling pathways directly activated by Ras. Because only a small subset of the Ras-regulated genes identified by Zuber et al. was sensitive to inhibition of MEK by PD98059, many of the changes induced may have been caused by effectors of Ras other than Raf or caused by synergistic interactions between different effector pathways.
In addition, inducible activation of a signaling protein, in contrast to long-term over-expression of an activated mutant, may reflect more closely the delivery of a signal by growth factor treatment. Furthermore, genetic alterations or attenuation of signaling pathways observed after constitutive expression of activated oncogenes are largely avoided. The data presented here represent the earlier transcriptional changes induced by Raf that contribute to the establishment of the transformed state, probably avoiding more downstream events characteristic of the maintenance and consequences of that state that may have been more strongly represented in the work of Zuber et al. (2000). Other differences between the two studies could reflect the different cell types used; we chose epithelial cells, because most human tumors derive from this lineage, but only a small fraction from fibroblasts. Another recent study used subtractive hybridization of mRNAs to identify some 20 Raf-induced genes in Rat-1 fibroblasts. Only one of these (ornithine decarboxylase) was also seen to be increased in our study, possibly lending weight to the likelihood that the spectrum of genes induced in response to Raf activation differs markedly between different cell types. Another difference between these two studies was that Heinrich et al. (2000) looked at cells proliferating in 10% fetal calf serum, whereas in our study the cells were quiescent in mitogen-poor medium.
Two other studies have analyzed the results of stimulating growth signaling pathways on gene expression using microarrays. One looked at the effects of serum; the other, platelet-derived growth factor receptor activation, both in fibroblasts. Approximately 5% of the genes up-regulated following activation of Raf in epithelial cells were also induced by serum in primary human fibroblasts, whereas close to 15% were induced by PDGF-receptor activation in a murine fibroblast cell line. The immediate early gene expression response in fibroblasts to serum or PDGF is thus likely to differ very significantly from the gene expression changes seen here following Raf activation in epithelial cells.
Among the genes most strongly up-regulated by Raf activation are HB-EGF and amphiregulin. Induction of autocrine factors of the EGF family by Ras has long been recognized to contribute to the reduced requirement for growth factors observed in transformed cells. In the case of MCF-10A cells, autocrine production of these factors does not render the cells growth factor independent for proliferation, but it does provide protection from apoptosis following detachment from the extracellular matrix (see below).
In addition, we observed up-regulation of some genes involved in cell proliferation, including cyclin D1, which has been reported previously to be induced by the Raf/MAP kinase pathway (Hipskind et al. 1994). Although Raf activation did not induce growth factor independent proliferation (see Fig. 1B), it was sufficient to induce a transient increase in S phase cells (data not shown), which could contribute to the observed induction of genes such as cyclin D1.
As well as induction of genes involved in proliferation, some inhibitors of growth signaling were also up-regulated, such as MAP kinase phosphatases, thought to be part of a negative feedback loop. Although the cell cycle inhibitor protein p21CIP1, which has previously been shown to induce cell cycle arrest in response to ERK/MAPK activation, was found to be up-regulated by Raf activation in MCF10A cells, we did not observe differences in p21CIP1 mRNA levels. This may indicate that posttranscriptional mechanisms are responsible for increased p21CIP1 protein levels but could also be caused by failure in detection of p21CIP1 mRNA by the microarray.
The large number of transcriptional regulators that are induced also indicates the likely importance of progressive changes in gene expression through secondary alterations in transcription factor abundance.
Another major category of Raf-regulated genes were those involved in the control of cell adhesion and invasion. Up-regulation of α6β4 integrin and laminin expression is characteristic of many invasive carcinomas. In addition, matrix metalloproteinase 1 is important in tumor invasion and in processing of growth factor precursors. CD66a, a cell adhesion molecule that is required for morphogenesis and is implicated in tumorigenesis, was found to be up-regulated, indicating that even in the absence of transformation, Raf activation initiates changes in gene expression characteristic of malignancy.
One other characteristic of transformed cells that could be seen to be induced rapidly at the transcriptional level by Raf was the production of proangiogenic peptide factors, such as VEGF, IL-8, FGF2, and PTHLH (Hanahan and Folkman 1996). Induction of ornithine decarboxylase and uridine phosphorylase, which catalyze production of small molecule inducers of angiogenesis, was also seen. Furthermore, the inhibitors of blood coagulation anti-thrombin 3 and protein C inhibitor (PAI-3) were down-regulated; the shift in favor of blood coagulation is known to promote angiogenesis.
We have been particularly interested in the mechanism whereby Ras activation can protect epithelial cells from detachment-induced apoptosis. This is a very important part of the transformed phenotype: Normal epithelial cells are unable to grow or survive when they lose contact with the appropriate extracellular matrix proteins. However, transformed cells gain the ability to move away from their original site without arresting or dying, leading to the formation of distant metastases. Understanding the mechanisms by which commonly activated oncogenes such as Ras allow cells to survive apoptosis, especially that induced by loss of adhesion, may therefore provide important insights into how malignancies develop and progress.
Our previous work showed that the principal mechanism whereby the activated Ras oncogene protects epithelial cells from detachment-induced apoptosis involved direct activation of PI 3-kinase and Akt (Khwaja et al. 1997). However, the Raf/ERK pathway can also provide some protection from this form of cell death (Le Gall et al. 2000), although the mechanism involved has not been defined. Several possibilities exist, including both direct mechanisms and changes in the transcriptional program of the cell. An example of direct signaling is that the ERK target Rsk2 can phosphorylate and inhibit the pro-apoptotic protein BAD, a member of the Bcl-2 family (Bonni et al. 1999; Tan et al. 1999; Shimamura et al. 2000), although BAD may not be expressed in the cells used here (Downward 1999). In addition, Raf may interact directly with Bcl-2 at the mitochondria (Wang et al. 1994). However, from the data presented here, it is clear that Raf protection from detachment-induced apoptosis is dependent on the function of an autocrine loop involving transcriptional induction of EGF-like growth factors. Raf-induced protection is blocked by various different inhibitors of EGF receptor and of PI 3-kinase activities, suggesting that the autrocrine factors are stimulating PI 3-kinase activity in the cells in suspension, and that this is required for survival. Expression of activated forms of PKB/Akt is sufficient to cause survival of MDCK and MCF-10A cells in suspension (Khwaja et al. 1997; A. Schulze and J. Downward, unpubl.). Raf could therefore support MCF-10A cell survival in suspension through transcriptional induction of an EGF-like autocrine loop leading to EGFR receptor, PI 3-kinase and PKB/Akt activity. We cannot exclude the possibility that there is a more direct contribution of the Raf/MAP kinase pathway to survival, but clearly Raf activation is not sufficient to induce survival in the absence of EGF receptor function, indicating that autocrine signaling is crucial for protection from apoptosis in this system.
Looking for other explanations of the protective effect of Raf at the level of transcriptional changes, we saw only a few changes in expression of apoptosis regulatory genes. Most of these would be expected to increase cell sensitivity to apoptosis rather than decrease it (e.g., increases in the level of Bak and Fas). The only one that could be obviously protective was up-regulation of osteoprotegerin, a soluble decoy receptor for ligands for Fas-related death receptors. However, we have previously been unable to inhibit anoikis by the addition of decoy death receptors (Rytömaa et al. 1999). No regulation of expression of apoptosis regulating genes controlled by the PKB/Akt pathways NF-κB (IAPs, A1) or Forkhead (Fas ligand, Bim) was seen, suggesting that survival signaling downstream from autocrine activation of the EGF receptor and PKB/Akt is through direct phosphorylation of apoptosis regulators and not through transcriptional control. Bcl-XL protein levels drop on detachment (Rodeck et al. 1997), and Raf activation prevents this. However, EGF receptor inhibitors failed to reverse this, although removing protection from anoikis, suggesting that this up-regulation of Bcl-XL was not sufficient to maintain cell survival (A. Schulze and J. Downward, unpubl.).
Activation of Raf is able to induce the production of autocrine EGF-like factors, as reported previously for HB-EGF (McCarthy et al. 1995), through a mechanism involving Ets-2 phosphorylation and AP-1 (McCarthy et al. 1997). Through this autocrine loop, Raf activation can thus also stimulate PI 3-kinase and PKB/Akt and hence protect cells from detachment-induced death. A threshold of Raf activity may be required because Raf-CAAX, an activated form of Raf that is less potent than ΔRaf-ER, is unable to protect cells from anoikis (Khwaja et al. 1997). It should be noted that protection from anoikis in Ras-transformed MCF-10A cells is resistant to EGFR receptor tyrosine kinase inhibitors, presumably because of the ability of Ras to activate the PI 3-kinase pathway directly. It has been shown, however, that full transformation of MCF-10A cells by oncogenic Ras requires autocrine activation of the EGF receptor (Ciardiello et al. 1990).
The data described here provide a description of the early transcriptional changes induced by activation of the Raf/MAP kinase pathway in normal epithelial cells. Many characteristics of the transformed cells can be seen in the gene expression patterns of these cells after just a few hours, even though several more days of Ras activation would be required to induce the full transformed phenotype. The preponderance of changes in the expression of transcriptional regulators and the production of autocrine growth factors that control other intracellular signaling pathways begins to provide a picture of how the network of progressive alterations in cell function is established that can eventually promote malignancy.
Material and methods
Cell culture and retroviral infection
MCF-10A cells were grown in Ham's nutrient mixture F12/DMEM (1:1) containing 5% horse serum and 10 μg/mL insulin, 20 ng/mL EGF, 5 μg/mL hydrocortisone, and 100 ng/mL cholera toxin (full medium). Minimal medium consisted of Ham's nutrient mixture F12/DMEM (1:1) containing 5% horse serum, whereas for supplemented medium 10 μg/mL insulin, 5 μg/mL hydrocortisone, and 100 ng/mL cholera toxin were added to minimal medium. 4-hydroxytamoxifen (4-OHT) was purchased from Sigma and dissolved in ethanol to obtain a 0.5 mM stock solution.
Retroviral vectors pBabe-puro-EGFP-ΔRaf:hbER (Bosch et al. 1997), pLXSN-V12Hras (Rodriguez-Viciana et al. 1997), or corresponding empty vectors were packaged in GP +E cells and used to infect MCF-10A cells expressing the ecotropic retrovirus receptor. Cells were selected with 2.5 μg/mL puromycin or 0.5 mg/mL G418 for 2 wk, and pBabepuro-EGFP-ΔRaf:hbER cells were sorted twice for EGFP-expression by FACS.
Culture in collagen gels was performed as described in Khwaja et al. (1998).
Antibodies and immunoblotting
Anti-phospho-p42/p44 Erk1/2 (T202, Y204) was purchased from New England Biolabs; anti-panERK and anti-Bcl-XL from Transduction Laboratories; anti-cytochrome c and anti-cyclin D1 (DCS-6) from Neomarkers; anti-TGFα, anti-HB-EGF, and anti-amphiregulin from R & D Systems; anti-phospho-EGFR from Calbiochem; anti-Bcl-2 from Pharmingen; and anti-estrogen receptor, anti-p21CIP1, anti-JunB, and anti-c-Fos from Santa Cruz. Anti-phospho-S473-PKB/Akt has been described previously (Khwaja et al. 1997). Cells were lyzed in buffer containing 1% Triton X-100 or in SDS sample buffer. Proteins were separated by SDS-PAGE and blotted onto PVDF membrane, incubated with antibody solutions, and detected using ECL.
Detachment-induced apoptosis and detection of cytochrome c release
MCF-10A cells were detached by incubation with trypsin, washed, and plated at a density of 105 cells/mL in 1.5 mL medium containing 5% horse serum onto normal or poly-HEMA coated plates. After 24 h, cells were harvested and DNA fragmentation was quantified using a Cell Death Detection ELISA kit (Roche Pharmaceuticals) according to manufacturers instructions. PD98059, LY294002, AG1478, and PD168393 were purchased from Calbiochem. Glu52-Diphtheriatoxin (CRM197) was obtained from Sigma. Cytochrome c was detected in cytoplasmic fractions as described in (Rytömaa et al. 2000).
RNA preparation and array hybridisation
Total RNA was prepared using Trizol Reagent (GIBCO BRL), following the manufacturers protocol. Poly A +-RNA was purified from total RNA with Oligotex latex beads (QIAGEN). Biotin-labeled cRNA was prepared following Affymetrix protocols (see Lockhart et al. 1996; Wodicka et al. 1997), and 17.5 μg cRNA were hybridized onto Affymetrix GeneChip HuGeneFL probe arrays, representing over 6000 known human genes and ESTs.
Data analysis
Raw data were analyzed using Affymetrix GeneChip software v 3.01 (for explanation of quantitative analysis, see Lockhart et al. 1996). The probe set intensity (average difference) is proportional to the abundance of the specific mRNA it represents and is calculated by comparing hybridization signal of the perfect match oligonucleotide to that of the mismatch oligonucleotide and averaged over a set of 20 specific oligonucleotide pairs for each gene. Total signal intensity of different probe arrays was scaled to the same value before comparison. Fold change was calculated using the Affymetrix GeneChip software by pairwise comparison of corresponding probe pairs from experiment (8 h or 72 h of 4-OHT treatment) and baseline (control) probe arrays. Genes with changes in mRNA abundance in response to Raf activation were selected by defining a filter query (Microsoft Access): Only those probe sets were allowed to pass which were called different by the GeneChip software and showed increases or decreases equal or larger than threefold in both replicate experiments. To account for high noise at low signal intensities, genes for which the average difference values were below 100 throughout the experiment (in both baseline and experiment) were eliminated from the analysis. To avoid saturation effects at high signal intensities, genes with an average differences larger than 10,000 throughout the experiment were also eliminated. Finally, the filter query disregarded changes induced by 4-OHT alone that were detected in 4-OHT treated empty vector infected cells.
Northern blotting
Ten micrograms of total RNA was separated by denaturing agarose/formaldehyde gel electrophoresis, blotted onto charged Nylon membranes, and immobilized by UV cross-linking. Fragments of the human cDNAs for integrin α6, small proline rich protein 1B (SSP-1B), c-Myc, HB-EGF, amphiregulin, c-Fos, ST-2, MAP-kinase phosphatase 5 (MAPK-ppase 5), and BRF 2 were isolated from corresponding IMAGE clones and labeled with [α-P32]dCTP using a random prime labeling kit (Stratagene). Hybridizations were performed using Quickhyb hybridization solution (Stratagene). Signals were detected by autoradiography and/or PhosphorImaging. GAPDH was detected using a labeled fragment of the rat GAPDH cDNA.
Acknowledgments
We wish to thank Affymetrix for the opportunity to use the GeneChip technology and for supplying the microarrays within the Academic User Center program (supported in part by NIH grant PO1HGO1323) and especially G. Tanimoto (Affymetrix, Santa Clara) for his continuous help with the microarray experiments and data analysis. We also thank J. Sgouros (ICRF, London) for help with data analysis and A. Harris for helpful comments. A.S. was supported by an EMBO long-term fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL 44-20-7269-3533; FAX 44-20-7269-3094.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.191101.
References
- Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- Bergers G, Coussens LM. Extrinsic regulators of epithelial tumor progression: Metalloproteinases. Curr OpinGenet Dev. 2000;10:120–127. doi: 10.1016/s0959-437x(99)00043-x. [DOI] [PubMed] [Google Scholar]
- Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- Bos JL. ras oncogenes in human cancer: A review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- Bosch E, Cherwinski H, Peterson D, McMahon M. Mutations of critical amino acids affect the biological and biochemical properties of oncogenic A-Raf and Raf-1. Oncogene. 1997;15:1021–1033. doi: 10.1038/sj.onc.1201270. [DOI] [PubMed] [Google Scholar]
- Browder T, Folkman J, Pirie-Shepherd S. The hemostatic system as a regulator of angiogenesis. J Biol Chem. 2000;275:1521–1524. doi: 10.1074/jbc.275.3.1521. [DOI] [PubMed] [Google Scholar]
- Brown NS, Bicknell R. Thymidine phosphorylase, 2-deoxy-D-ribose and angiogenesis. Biochem J. 1998;334:1–8. doi: 10.1042/bj3340001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciardiello F, McGeady ML, Kim N, Basolo F, Hynes N, Langton BC, Yokozaki H, Saeki T, Elliott JW, Masui H, et al. Transforming growth factor-alpha expression is enhanced in human mammary epithelial cells transformed by an activated c-Ha-ras protooncogene but not by the c-neu protooncogene, and overexpression of the transforming growth factor-alpha complementary DNA leads to transformation. Cell Growth Differ. 1990;1:407–420. [PubMed] [Google Scholar]
- Cook SJ, Aziz N, McMahon M. The repertoire of fos and jun proteins expressed during the G1 phase of the cell cycle is determined by the duration of mitogen-activated protein kinase activation. Mol Cell Biol. 1999;19:330–341. doi: 10.1128/mcb.19.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Brunet A, Greenberg ME. Cellular survival: A play in three Akts. Genes & Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- de Larco JE, Todaro GJ. Growth factors from murine sarcoma virus-transformed cells. Proc Natl Acad Sci. 1978;75:4001–4005. doi: 10.1073/pnas.75.8.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes & Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J. Ras signalling and apoptosis. Curr Opin Genet Dev. 1998;8:49–54. doi: 10.1016/s0959-437x(98)80061-0. [DOI] [PubMed] [Google Scholar]
- ————— How BAD phosphorylation is good for survival. Nat Cell Biol. 1999;1:E33–E35. doi: 10.1038/10026. [DOI] [PubMed] [Google Scholar]
- Erhardt P, Schremser EJ, Cooper GM. B-Raf inhibits programmed cell death downstream of cytochrome c release from mitochondria by activating the MEK/Erk pathway. Mol Cell Biol. 1999;19:5308–5315. doi: 10.1128/mcb.19.8.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fambrough D, McClure K, Kazlauskas A, Lander ES. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- Heinrich J, Bosse M, Eickhoff H, Nietfeld W, Reinhardt R, Lehrach H, Moelling K. Induction of putative tumor-suppressing genes in Rat-1 fibroblasts by oncogenic Raf-1 as evidenced by robot-assisted complex hybridization. J Mol Med. 2000;78:380–388. doi: 10.1007/s001090000116. [DOI] [PubMed] [Google Scholar]
- Hipskind RA, Baccarini M, Nordheim A. Transient activation of RAF-1, MEK, and ERK2 coincides kinetically with ternary complex factor phosphorylation and immediate-early gene promoter activity in vivo. Mol Cell Biol. 1994;14:6219–6231. doi: 10.1128/mcb.14.9.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Hardy JD, Sun Y, Shively JE. Essential role of biliary glycoprotein (CD66a) in morphogenesis of the human mammary epithelial cell line MCF10F. J Cell Sci. 1999;112:4193–4205. doi: 10.1242/jcs.112.23.4193. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JCF, Trent JM, Staudt LM, Hudson JJ, Boguski MS, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- Kazama H, Yonehara S. Oncogenic K-Ras and basic fibroblast growth factor prevent Fas-mediated apoptosis in fibroblasts through activation of mitogen-activated protein kinase. J Cell Biol. 2000;148:557–566. doi: 10.1083/jcb.148.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khwaja A, Lehmann K, Marte BM, Downward J. Phosphoinositide 3-kinase induces scattering and tubulogenesis in epithelial cells through a novel pathway. J Biol Chem. 1998;273:18793–18801. doi: 10.1074/jbc.273.30.18793. [DOI] [PubMed] [Google Scholar]
- Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne PH, Downward J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippel A, Escobedo MA, Wachowicz MS, Apell G, Brown TW, Giedlin MA, Kavanaugh WM, Williams LT. Activation of phosphatidylinositol 3-kinase is sufficient for cell cycle entry and promotes cellular changes characteristic of oncogenic transformation. Mol Cell Biol. 1998;18:5699–5711. doi: 10.1128/mcb.18.10.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall M, Chambard JC, Breittmayer JP, Grall D, Pouyssegur J, Van Obberghen-Schilling E. The p42/p44 MAP kinase pathway prevents apoptosis induced by anchorage and serum removal. Mol Biol Cell. 2000;11:1103–1112. doi: 10.1091/mbc.11.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Marshall CJ. Ras effectors. Curr Opin Cell Biol. 1996;8:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- McCarthy SA, Chen D, Yang BS, Garcia Ramirez JJ, Cherwinski H, Chen XR, Klagsbrun M, Hauser CA, Ostrowski MC, McMahon M. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol Cell Biol. 1997;17:2401–2412. doi: 10.1128/mcb.17.5.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SA, Samuels ML, Pritchard CA, Abraham JA, M. McMahon M. Rapid induction of heparin-binding epidermal growth factor/diphtheria toxin receptor expression by Raf and Ras oncogenes. Genes & Dev. 1995;9:1953–1964. doi: 10.1101/gad.9.16.1953. [DOI] [PubMed] [Google Scholar]
- Montesano R, Soriano JV, Hosseini G, Pepper MS, Schramek H. Constitutively active mitogen-activated protein kinase kinase MEK1 disrupts morphogenesis and induces an invasive phenotype in Madin-Darby canine kidney epithelial cells. Cell Growth Differ. 1999;10:317–332. [PubMed] [Google Scholar]
- Rabinovitz I, Mercurio AM. The integrin alpha 6 beta 4 and the biology of carcinoma. Biochem Cell Biol. 1996;74:811–821. doi: 10.1139/o96-087. [DOI] [PubMed] [Google Scholar]
- Renshaw MW, Ren XD, Schwartz MA. Growth factor activation of MAP kinase requires cell adhesion. EMBO J. 1997;16:5592–5599. doi: 10.1093/emboj/16.18.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodeck U, Jost M, DuHadaway J, Kari C, Jensen PJ, Risse B, Ewert DL. Regulation of Bcl-xL expression in human keratinocytes by cell- substratum adhesion and the epidermal growth factor receptor. Proc Natl Acad Sci. 1997;94:5067–5072. doi: 10.1073/pnas.94.10.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- Rytömaa M, Martins LM, Downward J. Involvement of FADD and caspase-8 signalling in detachment-induced apoptosis. Curr Biol. 1999;9:1043–1046. doi: 10.1016/s0960-9822(99)80454-0. [DOI] [PubMed] [Google Scholar]
- Sewing A, Wiseman B, Lloyd AC, Land H. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5588–5597. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding ras: “It ain't over ‘til it’s over.”. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- Shimamura A, Ballif BA, Richards SA, Blenis J. Rsk1 mediates a MEK-MAP kinase cell survival signal. Curr Biol. 2000;10:127–135. doi: 10.1016/s0960-9822(00)00310-9. [DOI] [PubMed] [Google Scholar]
- Soule HD, Maloney TM, Wolman SR, Peterson WD, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- Tan Y, Ruan H, Demeter MR, Comb MJ. p90(RSK) blocks bad-mediated cell death via a protein kinase C-dependent pathway. J Biol Chem. 1999;274:34859–34867. doi: 10.1074/jbc.274.49.34859. [DOI] [PubMed] [Google Scholar]
- Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- Wang HG, Miyashita T, Takayama S, Sato T, Torigoe, Krajewski S, Tanaka S, Hovey L, Troppmair J, Rapp UR, et al. Apoptosis regulation by interaction of Bcl-2 protein and Raf-1 kinase. Oncogene. 1994;9:2751–2756. [PubMed] [Google Scholar]
- Wodicka L, Dong H, Mittmann M, Ho MH, Lockhart DJ. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- Zuber J, Tchernitsa OI, Hinzmann B, Schmitz AC, Grips M, Hellriegel M, Sers C, Rosenthal A, Schafer R. A genome-wide survey of RAS transformation targets. Nat Genet. 2000;24:144–152. doi: 10.1038/72799. [DOI] [PubMed] [Google Scholar]