Abstract
To perceive red and far-red light, plants have evolved specific photoreceptors called phytochromes. Even though the spectral properties of all phytochromes are very similar, they show a distinct mode of action. Here we describe EID1, a negatively acting component of the signaling cascade that shifts the responsiveness of the phytochrome A (phyA) signaling system associated with hypocotyl elongation from red to far-red wavelengths. EID1 is a novel nuclear F-box protein that contains a leucine zipper whose integrity is necessary for its biological function. EID1 most probably acts by targeting activated components of the phyA signaling pathway to ubiquitin-dependent proteolysis.
Keywords: Light signaling, phytochrome, F-box protein, SCF complex, Arabidopsis
Plants are sessile organisms that are forced to adapt to a given environment. Therefore, compared with animals, plants have achieved an enormous developmental plasticity that makes possible very flexible responses to diverse exogenic factors. Among these exogenic factors, light is probably one of the dominating signals that controls gene expression and development throughout all phases of the plant life cycle (Neff et al. 2000). To perceive red and far-red light, plants use the phytochrome family of photoreceptors that comprises five members [phytochromes A–E (phyA–E)] in Arabidopsis. All phytochromes can exist in two different conformers: the red light-absorbing, biologically inactive Pr form and the far-red light-absorbing, biologically active Pfr form. Because both forms can be photoconverted, phytochromes can function as red/far-red photoreversible light switches when exposed to pulses of red and far-red light (Mancinelli 1994; Neff et al. 2000).
Even though the spectral properties of all phytochromes are very similar (Eichenberg et al. 2000), they show a distinct mode of action. The products of the PHYB, PHYC, PHYD, and PHYE genes remain light stable, with the PHYB gene showing the most expression. As expected, photoresponses triggered by the light-stable phytochromes show their highest sensitivity in red light and follow the classical red/far-red photoreversibility (Neff et al. 2000). In contrast, phyA accumulates to a high level in the dark and its Pfr form is rapidly degraded. PhyA is responsible for the so-called very low fluence responses and exhibits its highest effects under strong continuous far-red light that induces the so-called high-irradiance responses (HIRs). The HIR effect is very important for the regulation of plant development in canopy shade, but it shows many aspects that are difficult to explain by the spectral properties of a photoreversible phytochrome alone. First, the action spectrum of HIR reveals a maximum at ∼720 nm, at which wavelength the responses of the light-stable phytochromes become completely inhibited. Furthermore, the extent of the HIR depends on photon fluence rates instead of photon fluences. Thus, the respective signaling cascade has not only to sense the total amount of photons that reach the tissue, but must also be able to detect the amount of photons that reaches the plant in a given time (Mancinelli 1994; Neff et al. 2000).
Several approaches have been undertaken to identify components of the phytochrome signaling cascade. Pharmacological and microinjection studies indicate an involvement of heterotrimeric G proteins, cGMP, and calcium/calmodulin in phytochrome signaling (Neuhaus et al. 1993; Bowler et al. 1994). Yeast two-hybrid screenings revealed several components, including PIF3, PKS1, and NDPK2 (Neff et al. 2000), that interact with both phyA and phyB. PIF3 is a basic helix-loop-helix (bHLH) protein that can bind to G-box DNA motifs present in many light-regulated promoters (Martinez-Garcia et al. 2000).
Genetic approaches have resulted in the isolation of Arabidopsis mutants, which exhibit different light-dependent phenotypes (Hardtke and Deng 2000; Neff et al. 2000). Until now, at least eight independent mutants have been identified that are defective in phyA-dependent responses. So far, FAR1, FIN219, PAT1, HFR1, REP1, and RSF1 have been characterized at the molecular level (Hudson et al. 1999; Bolle et al. 2000; Fairchild et al. 2000; Hsieh et al. 2000; Soh et al. 2000; Spiegelman et al. 2000). HFR1, REP1, and RSF1 are allelic and code for a bHLH protein similar to PIF3. The biochemical function of the other proteins is not defined. In contrast to the large number of loss-of-function mutants, only two mutants, spa1 and eid1, show enhanced phyA-dependent responses (Hoecker et al. 1999; Büche et al. 2000) and thus most probably encode for negatively acting components of phyA signaling. SPA1 contains WD40 repeats and exhibits some weak homology with protein kinases, but its biochemical function also remains to be elucidated.
In an attempt to screen for new phyA-specific mutants, we have isolated the recessive eid1 alleles that show a strictly phyA-dependent phenotype and an alteration in all analyzed HIRs (Büche et al. 2000). The eid1 mutation results in an extremely enhanced light sensitivity and an increased temporal persistence of the HIR, which indicates an important role in photon fluence rate measurement. Thus, EID1 seems to play a key role as a repressor in HIR signaling. Here we describe the cloning and the functional characterization of EID1. EID1 is a nuclear localized F-box protein that shifts the responsiveness of the phyA signaling system associated with hypocotyl elongation from red to far-red wavelengths. It most probably acts by targeting activated components of the phyA signaling cascade to ubiquitin-dependent proteolysis.
Results
Cloning of the EID1 gene
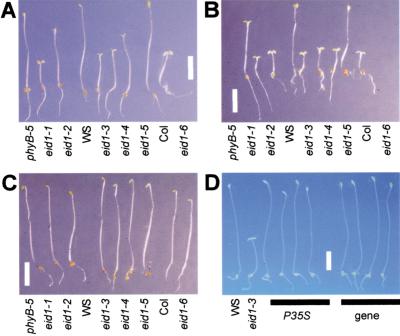
To clone the EID1 gene, we followed two different strategies: screening for new eid1 alleles in T-DNA lines and positional cloning. The increased light sensitivity observed in eid1 has been used to screen for new alleles. Under the applied low fluence rates of continuous red and far-red light, eid1 mutants exhibited a strong photomorphogenic development with open cotyledons and a reduced hypocotyl growth, whereas wild-type seedlings remained nearly etiolated (Fig. 1A,B). This phenotype was strictly light dependent because all of the mutant lines exhibited an etiolated phenotype in darkness, with elongated hypocotyls and closed cotyledons on an apical hook (Fig. 1C). One T-DNA tagged line (eid1-4) belonging to the eid1 complementation group was used to clone flanking genomic sequences by inverse PCR. The genomic fragments belong to a gene located at about 55 kbp from the luminidependens gene on chromosome 2. This close proximity to the luminidependens gene fits well with the data of the fine mapping because no recombinants with the respective marker were detectable after the analyses of 732 chromatides. Each of the six eid1 alleles isolated so far showed a mutation in an intronless open reading frame (ORF) having a length of 1008 bp (Fig. 2A). Northern blots revealed one band of ∼1350 nucleotides, corresponding to the size of the ORF of the intronless EID1 gene and the longest isolated EID1 EST (data not shown).
Figure 1.
Phenotype of 4-d-old wild type, eid1 mutant, and transgenic seedlings under different light conditions. (WS) Wassilewskija wild type; (Col) Columbia wild type. eid1-1 and eid1-2 were isolated from the phyB-5 mutant in the Landsberg erecta (Ler) background. The eid1-3, eid1-4, and eid1-5 alleles were derived from a WS background and the eid1-6 from a Col background. Scale bars, 3 mm. (A) Seedlings grown under the weak red-light field used for screening. (B) Seedlings grown under the weak far-red light field used for screening. (C) Etiolated seedlings grown in darkness. (D) Phenotype of wild-type, eid1-3, and rescued eid1-3 lines under the weak red light used for screening. The phenotype of independent transgenic lines transformed with a 35S-promoter–EID1-ORF–nos-terminator construct (P35S) or a genomic EID1 fragment (gene) is shown.
Figure 2.
Nucleotide and amino acid sequences of EID1. (A) Nucleotide and predicted amino acid sequences of wild-type and mutant alleles. The sequences of the Ler and WS gene are given as a standard. Deviations of the published Col sequence (accession no. AL161494) and the eid1 mutant alleles are shown. The start of the longest EST clone (161I3T7) is marked by a number sign and its polyadenylation site is indicated by a slash (/). Stop codons are indicated by asterisks (*). Amino acid sequences with similarities to an F-box are underlined with a bold line and the leucine zipper domain is double underlined. The acidic domain is single underlined and the basic domain is underlined with a wave line. (B) Alignment of F-box domains identified from F-box proteins known to be expressed in Arabidopsis. Unusual flower organs (UFO), accession no. X89224; transport inhibitor response 1 (TIR1), accession no. AF005048; Zeitlupe (ZTL), accession no. AF254413. (1) S or T; (2) large hydrophobic amino acids; (3) basic amino acids. Highly conserved amino acids are capitalized. Amino acids identical to the consensus are boxed in black. Gray shadings with white letters indicate amino acids similar to the consensus and gray shadings with black letters mark similarities between the Arabidopsis F-box proteins.
For complementation analysis, the strong eid1-3 mutant was transformed with a 35S-promotor–EID1-ORF–nos-terminator construct and a genomic fragment that includes the putative promoter and the terminator of EID1. Transgenic eid1-3 lines exhibited a clearly reduced light sensitivity and they appear like wild-type seedlings under selective light conditions (Fig. 1D). Thus, the EID1-ORF and the genomic fragment were sufficient to repress the hypersensitive phenotype in the eid1-3 mutant. Therefore, we conclude that the respective DNA sequence codes for the EID1 gene product.
EID1 is a novel type of F-box protein
The deduced EID1 protein can be subdivided into at least five different domains (Fig. 2A). The most N-terminal domain shows homology with F-box proteins (Fig. 2B). SCF (Skp1, Cdc53, and F-box) proteins are the major components of the so-called SCF complexes that function as ubiquitin ligases. SCF complexes are involved in the proteasome-dependent degradation of many regulatory proteins in yeast, animals, and plants (Craig and Tyers 1999).
A leucine zipper pattern is adjacent to the F-box motif (Fig. 2A). Leucine zipper domains facilitate the homo- or heterodimerization of polypeptides (Busch and Sassone-Corsi 1990), and they are present in different families of transcription factors that also play an important role in the light signaling of plants (Hardtke and Deng 2000; Neff et al. 2000). The functional significance of the respective domain in EID1 is underlined by the mutation in the weak eid1-2 allele, which results in a glycine to cysteine transition immediately in front of the second leucine residue of the zipper pattern. To our knowledge, EID1 is the first F-box protein reported to contain a leucine zipper that seems to be necessary for its function.
The C-terminal part of EID1 has no significant homology with known proteins. It contains a highly acidic domain followed by a basic domain. The functional importance of the C-terminal part is emphasized by the phenotype of the eid1-3, eid1-5, and eid1-6 alleles that show structural alterations or deletions in this area (Fig. 2A).
EID1 interacts with ASK1 and ASK2
The F-box protein function of EID1 is further underlined by the results of a yeast two-hybrid screening by using the full-length polypeptide fused to the GAL4 binding domain as bait. From 534 positive clones, ∼95% were identical to ASK1 and one clone was identified as being ASK2. ASK1 and ASK2 are two Arabidopsis homologs (Arabidopsis SKP1-like) of the yeast Skp1 protein (Gray et al. 1999).
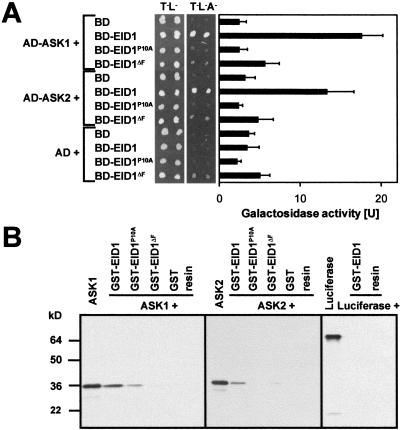
To demonstrate that the F-box domain of EID1 mediates interaction with ASK1 and ASK2, two different mutated constructs were tested. For the EID1ΔF construct, the first 54 amino acids containing the F-box domain were deleted, and for the EID1P10A mutant, the highly conserved proline amino acid at position 10 was changed to an alanine residue. Both mutations abolished the interaction of EID1 with ASK1 and ASK2 in a two-hybrid assay (Fig. 3A).
Figure 3.
EID1 interacts with the Skp1-like proteins ASK1 and ASK2. (A) Analysis of the interaction in the yeast two-hybrid system. Growth on control plates lacking tryptophan and leucine (T− L−) and on selective plates lacking both amino acids and adenine (T− L− A−) is shown. The plates were photographed after 4 d. Galactosidase activity is given as the mean ± SD of five independent yeast lines for each combination of constructs. (AD) GAL4 activation domain; (BD) GAL4 binding domain. (B) ASK1 and ASK2 interaction with EID1 in GST pull-down experiments. The SDS-PAGE of 35S-labeled ASK1, ASK2, and the Luciferase control are shown. An aliquot (0.5 μL) of the labeled proteins was loaded as a reference. The proteins used in the pull-down experiments are indicated on the top. (GST) Glutathione-S-Transferase; (resin) control with empty Glutathione-S-Sepharose.
The results of the two-hybrid assays could be verified by pull-down experiments by using fusion proteins of GST to wild-type EID1 or its mutated forms. The GST fusion proteins were expressed in Escherichia coli and bound to Glutathione-S-Sepharose. [35S]methionine-labeled ASK1 and ASK2 polypeptides were added and binding was analyzed by SDS-PAGE and autoradiography. ASK1 and ASK2 showed a strong interaction with the chimeric GST–EID1 polypeptide (Fig. 3B). The interaction was clearly reduced or abolished with the mutated GST–EID1ΔF and GST–EID1P10A fusion proteins. No interactions were detected with GST, the empty Glutathion-S-Sepharose, and the labeled Luciferase as negative controls.
EID1 is localized in the nucleus
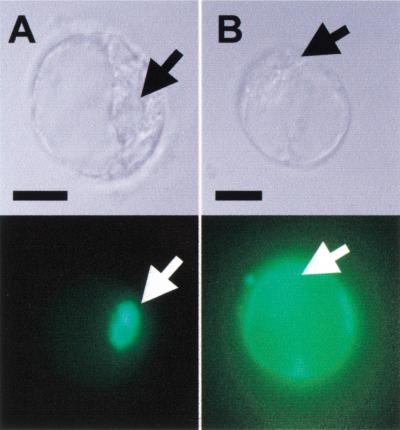
Translational fusions to green fluorescent protein (GFP) were used to investigate the subcellular localization of EID1. In transiently transformed parsley protoplasts, EID1–GFP appeared homogeneously distributed in the nucleus in dark- and light-treated protoplasts (Fig. 4A). In contrast, cytoplasmic localization became detectable with the EID1mNLS–GFP protein for which the first two basic amino acids in the basic domain were replaced by serine and glutamate residues (Fig. 4B). This result clearly demonstrates that the basic domain in EID1 is a functional nuclear localization sequence (NLS).
Figure 4.
Cellular distribution of EID1–GFP in transiently transformed parsley protoplasts. (A) Protoplasts were transfected with a plasmid directing expression of EID1–GFP. (B) Protoplasts were transfected with a plasmid directing expression of GFP linked to the Cterminus of EIDmNLS, which contains mutations in the putative NLS. After transformation cells were kept in darkness for 24 h before analysis. The top panels show bright light field images and the lower panels show fluorescence images. The position of the nucleus is indicated by an arrow. Scale bars, 10 μm.
EID1 is necessary for a far-red light shift in the phyA-dependent regulation of hypocotyl elongation
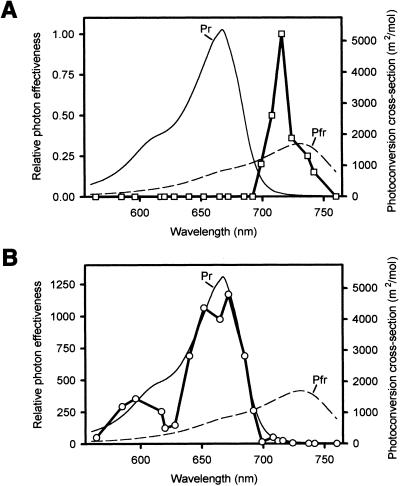
The eid1 mutants exhibited an extremely enhanced sensitivity in a broad-banded red light field as long as phyA was not completely degraded at higher fluence rates. Analysis of eid1 phyA double mutants further confirmed that the observed hypersensitivity in red and far-red light is strictly phyA-dependent (Büche et al. 2000). To test for the spectral sensitivity in greater detail, we determined action spectra for hypocotyl elongation in phyB-5 and phyB-5 eid1-1 mutants under low light fluence rates. phyB-5 mutants were used to exclude responses in the red-light region of the electromagnetic spectrum that are predominantly regulated by phyB in Arabidopsis. A typical HIR action spectrum with a maximum at 716 nm was obtained for phyB-5 seedlings (Fig. 5A). In contrast, the action spectrum with the phyB-5 eid1-1 mutant was completely altered (Fig. 5B). Even though the eid1 mutant exhibited a 23-fold increase in light sensitivity at 716 nm, the maximum light response was detectable around 670 nm, where it exhibited a 1170-fold higher value for relative photon effectiveness. The shape of the phyB-5 eid1-1 action spectrum is very similar to the absorption cross-section of the inactive Pr form of the photoreceptor and thus, to the cross-section expected for the formation of the physiologically active Pfr form (Fig. 5B).
Figure 5.
Action spectra for hypocotyl elongation in phyB-5 and phyB-5 eid1-1 seedlings. Fluence rate response curves were measured at different wavelengths and the fluence rate that led to 40% inhibition compared with dark controls was determined. The value obtained for phyB-5 at 716 nm (0.12 μmole/m2sec) was set to 1 and the relative photon effectiveness for phyB-5 and phyB-5 eid1-1 at different wavelengths was calculated. The determined values are symbolized by squares or circles connected with a bold line. Photoconversion cross-sections are plotted as thin lines by using published data for purified oat phytochrome (Mancinelli 1994). (A) Action spectra for phyB-5. (B) Action spectra for phyB-5 eid1-1.
Discussion
This report describes the identification of the EID1 gene, a negatively acting component that plays an important role in phyA-specific regulation of plant photomorphogenesis. The intronless gene was isolated by fine mapping and T-DNA tagging. The identity of the gene was verified by the presence of mutations in the coding region of all six independent eid1 alleles and by complementation of the Eid1 phenotype. The apparent length of the EID1 transcripts in Northern blots fitted to the expected size of the longest EST.
Because the spectral properties of the different types of phytochromes are almost identical (Eichenberg et al. 2000), they should exhibit their highest physiological responses in red light that leads to the formation of a high Pfr level. However, phyA clearly differs from all other phytochrome types because it is most effective under strong far-red light and loses its activity under continuous red light. We could demonstrate that, in the absence of EID1, the light-labile phyA can function as a red-light photoreceptor under continuous irradiation similar to the light-stable types of phytochrome. To our knowledge, this is the first example that a component of a signaling cascade leads to a spectral shift in the sensitivity of a photoreceptor. Usually in the eye and all known other systems the spectral diversity of photoreceptor molecules is obtained by structural modifications that result in an altered absorption spectra. To allow efficient perception of far-red light, plants followed an alternative strategy. They suppress the signal following irradiation with high fluence rates of red light by phyA degradation (Büche et al. 2000) and use EID1 to suppress phyA responses at low fluence rates.
In this context it is highly interesting that EID1 encodes an F-box protein that interacts with the Arabidopsis Skp1-homologs ASK1 and ASK2. Thus, EID1 is most probably a component of an SCF complex. SCF complexes function as ubiquitin ligases in different signaling pathways, whereby the F-box component has been proposed to provide substrate specificity (Craig and Tyers 1999). Because the level and the degradation of phyA was not altered in eid1 mutants, a direct function in the proteolysis of the photoreceptor can be excluded (Büche et al. 2000). EID1 most probably acts by targeting other positively acting components of the phyA signaling pathway to degradation. Good candidates would be positively acting, nuclear localized components of the phyA signaling pathway such as FAR1 and the bHLH transcription factor HFR1 (Hudson et al. 1999; Fairchild et al. 2000). In most systems, the F-box proteins exclusively interact with the phosphorylated forms of their target proteins. Because protein phosphorylation most probably also plays an important role in phytochrome signaling (Fankhauser et al. 1999; Hardtke and Deng 2000; Watson 2000), it is worthwhile to speculate that EID1 only interacts with its targets after a Pfr-dependent modification. If this modification also leads to a functional activation, this mechanism would result in an antagonism between degradation and activity. EID1-dependent proteolysis might eliminate all activated signaling intermediates at low Pfr amounts formed under weak light. Under strong far-red light, the level of the Pfr form of phyA remains relatively high (Mancinelli 1994), which would result in a higher level of activated intermediates that can no longer be completely removed by EID1 function.
There is also evidence that proteins of the COP/DET/FUS class of mutants, which show a light-grown phenotype in darkness, are involved in ubiquitin-dependent protein degradation. The respective proteins seem to function as general repressors of photomorphogenesis downstream of different plant photoreceptors, including phyA. COP1 contains a RING finger motif that is present in most ubiquitin ligases. It is involved in the degradation of the transcription factor HY5 that regulates certain aspects of photomorphogenesis in Arabidopsis. Components of the COP9 complex show homology with the lid subcomplex of the 19S proteasome regulatory particle (Hardtke and Deng 2000). Thus, EID1 might link phyA-dependent light signaling to COP9 complex-dependent proteolysis.
Materials and methods
Plant material, light conditions, and characterization of mutants
Seeds of the Wassilewskija wild type were mutagenized by imbibition in ethyl methanesulfonate as described by Büche et al. (2000). T-DNA lines were obtained directly from INRA or from the Nottingham Arabidopsis Stock Center. For light experiments, seedlings were grown on paper for 4 d. For screenings, standard light fields were used and fluence rates were adjusted to 15 nmole/m2sec with red light and 140 nmole/m2sec with far-red light (Büche et al. 2000). Action spectroscopy was performed as described by Schäfer and Fukshansky (1984). To obtain monochromatic light, DIL and DEPIL interference filters (Schott, Mainz, Germany) together with modified Leitz Prado 500-W projectors (Leitz, Wetzlar, Germany) and Osram Xenophot Longlife lamps (Osram, Berlin, Germany) were used. Fluence rates were measured by using a J16 photometer and a J6512 radiant energy probe (Tektronix, Beaverton, USA).
New hypersensitive mutants were crossed with eid1-3 to test for complementation. T-DNA flanking sequences in eid1-4 were isolated by inverse PCR by using the primers 5′-CCAGTGTCCTATTACCAATAGCCGAC GGTATCG-3′ and 5′-TAAGCGTAGACAACCCTCACCTGGAAACGG3′ that anneal to the T-DNA of the INRA lines (Bouchez et al. 1993). To check for mutations in the EID1 gene, we amplified genomic DNA by PCR by using two different primer pairs: 5′-AACAGTTAAGACTATTCCCG GTTTACGCG-3′ together with 5′-GTTTTGATCGGGACCTAGCTCAC GTTCTA-3′ (EID1N-R primer); and 5′-CTCGCTGTTTGTTGTCCTG GTCTCTTCC-3′ (EID1C-F primer) together with 5′-TCCTGTCCAGAA AGTAAGAAGAAGGTGCC-3′. DNA fragments were sequenced directly. Mapping was done as described by Koniecny and Ausubel (1993). For DNA extraction we used a DNeasy plant mini kit (QIAGEN).
Construction of mutated EID1 fragments
To create the basal construct, we amplified the EID1 coding region from genomic DNA by PCR by using the 5′ BamHI–NcoI primer (5′-TTGGATC CATGGCGGAATCTGTCTTCTCTT-3′) and the 3′ SalI primer (5′-AGAT GTCGACTTGAGACCGTGGAAGAAGAATGAAAG-3′). The BamHI–SalI fragment was cloned into the pBluescriptSK (Stratagene) vector and sequenced. All further mutations were introduced by PCR and the subcloned fragments were sequenced again. The mutated F-box constructs were obtained by using the EID1N-R primer together with the oligonucleotide 5′-ATCCATGGCGGAATCTGTCTTCTCTTGTATAGCAG AAG-3′ for the proline to alanine change in the EID1P10A construct and the olignucleotide 5′-CAGGATCCCCACCGTCATCTCCGATCTC-3′ for the deletion of the first 54 amino acid in the EID1ΔF construct. The amplified fragments were cloned into the basal construct by using an internal EcoRI site together with the original NcoI site (EID1P10A) or the introduced BamHI site (EID1ΔF). To create a GFP fusion gene, we removed the stop codon of EID1 by an SmaI restriction site by using the SmaI primer 5′-TTCCCGGGAGCGCTAGTGTAGAGAGGTAAAGCA GTCCAAGCACC-3′ together with the BamHI–NcoI primer. The amplified fragment was introduced between a 35S-promoter fragment and a GFP–nos-terminator cassette of a modified pBluescriptSK vector by using BamHI and SmaI restriction sites. The oligonucleotide 5′-CGAGATC TTCCGGTAAGCGAACGAAGGAAG-3′ spanning an internal BglII site was used together with the SmaI primer to change the amino acids in the basic domain (arginine 197 to serine, and lysine 198 to glutamate) to get the EID1mNLS construct.
Plant transformation
To create the 35S-promoter–EID1–nos-terminater construct, we cloned the basal EID1 fragment into the pPCV812 plant transformation vector by using internal BamHI and SalI restriction sites (Koncz et al. 1994). For the rescue with the EID1 genomic construct, a 4.2-kbp NsiI fragment obtained from the BAC clone T14P8 was ligated into the PstI site of the pBluescriptSK vector. The genomic fragment was isolated by using SstI and EcoRV sites, and it was subcloned into the pGPTV-bar plant transformation vector (Becker et al. 1992) that was cut with SstI and SmaI. Arabidopsis transformation was done as described by Clough and Bent (1998). Parsley protoplasts were transiently transformed by electroporation (Bowler et al. 1997).
Yeast two-hybrid screening
For two-hybrid screening, yeast strain pJ69-4A was first transformed with pGKBT7–EID1 as a bait followed by transformation with a lambda-ACT cDNA library (Lohrmann et al. 1999). The ASK1 and ASK2 coding regions were amplified by PCR as BamHI–PstI fragments and cloned into the yeast vector pGADT7 (Clontech). These constructs were used for further analysis of growth on selective media and of β-galactosidase activity (Lohrmann et al. 1999) together with EID1, EID1P10A, and EID1ΔF constructs that were subcloned into the yeast two-hybrid vector pGKBT7.
GST pull-down experiments
The EID1 constructs were subcloned into the GST gene fusion vector pGEX-4T-1 (Amersham Pharmacia). GST, GST–EID1, GST–EID1ΔF, and GST–EID1P10A were expressed in E. coli M15[pREP]. Soluble extracts were applied to 1 mL of Glutathione-S-Sepharose (Sigma) and washed two times with extraction buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, 0.14 M NaCl at pH 7.5). Equal aliquots of 35S-labeled ASK1 or ASK2 were added to 60-μL aliquots of these matrices, as well as to the empty Sepharose matrix. All samples were filled up with extraction buffer to a total volume of 200 μL and incubated for 90 min at 4°C. The matrices were washed four times with 500 μL of extraction buffer, then the bound proteins were eluted with 50 μL of elution buffer (50 mM Tris-HCl at pH 8.0, 10 mM reduced glutathione) for 10 min at 25°C. Forty microliters of the eluate were mixed with 10 μL of 5× SDS-sample buffer and incubated for 2 min at 95°C. Twenty microliters of the eluate and 0.5 μL of the 35S-labeled proteins were size-fractionated on an SDS-PAGE to visualize the labeled ASK proteins by autoradiography. 35S-labeled ASK1 or ASK2 were obtained by using their pGADT7 constructs together with a TNT Quick Coupled Transcription/Translation System (Promega) and 35S-methionine (Amersham Pharmacia).
Acknowledgments
We thank Martina Krenz and Sabine Unger for their excellent technical assistance; Klaus Harter, Kishore Panigrahi, and Tim Kunkel for critical reading of the manuscript; Michel Caboche and the people in Versailles for the gift of the T-DNA lines; the Arabidopsis Biological Resource Center in Ohio; and the Arabidopsis Stock Center in Nottingham for providing seeds, clones, and the cDNA library. This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Graduiertenkolleg “Molekulare Mechanismen pflanzlicher Differenzierung.”
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kretsch@uni-freiburg.de; FAX 49-761-203-2612.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.197201.
References
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua N-H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes & Dev. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Bouchez D, Camilleri C, Caboche M. A binary vector based on Basta resistance for in planta transformation in Arabidopsis thaliana. CR Acad Sci Paris Life Sci. 1993;316:1188–1193. [Google Scholar]
- Bowler C, Neuhaus G, Yamagata H, Chua NH. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell. 1994;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]
- Bowler C, Frohnmeyer H, Schäfer E, Neuhaus G, Chua NH. Phytochrome and UV signal transduction pathways. Acta Physiol Plant. 1997;19:475–483. [Google Scholar]
- Büche C, Poppe C, Schäfer E, Kretsch T. eid1: A new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell. 2000;12:547–558. [PMC free article] [PubMed] [Google Scholar]
- Busch SJ, Sassone-Corsi P. Dimers, leucine zippers and DNA-binding domains. Trends Genet. 1990;6:36–40. doi: 10.1016/0168-9525(90)90071-d. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Craig KL, Tyers M. The F-box: A new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog Biophys Mol Biol. 1999;72:299–328. doi: 10.1016/s0079-6107(99)00010-3. [DOI] [PubMed] [Google Scholar]
- Eichenberg K, Bäurle I, Paulo N, Sharrock RA, Rüdiger W, Schäfer E. Arabidopsis phytochromes C and E have different spectral characteristics from those of phytochromes A and B. FEBS Lett. 2000;470:107–112. doi: 10.1016/s0014-5793(00)01301-6. [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes & Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. Indentification of an SCF ubiquitin–ligase complex required for auxin response in Arabidopsis thaliana. Genes & Dev. 1999;13:1678–1691. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Deng XW. The cell biology of the COP/DET/FUS proteins. Regulating proteolysis in photomorphogenesis and beyond? Plant Physiol. 2000;124:1548–1557. doi: 10.1104/pp.124.4.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- Hsieh HL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, Deng XW. FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes & Dev. 2000;14:1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes & Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Martini N, Szabados L, Hrouda M, Bachmair A, Schell J. Specialized vectors for gene tagging and expression studies. In: Gelvin BS, Schilperoort RA, editors. Plant molecular biology manual. Dordrecht, Netherlands: Kluwer Academic Publishers; 1994. pp. 1–22. [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Lohrmann J, Buchholz G, Keitel C, Sweere U, Kircher S, Bäurle I, Kudla J, Schäfer E, Harter K. Differential expression and nuclear localization of response regulator-like proteins from Arabidopsis thaliana. Plant Biol. 1999;1:495–505. [Google Scholar]
- Mancinelli AL. The physiology of phytochrome action. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in plants. 2nd ed. Dordrecht, Netherlands: Kluwer Academic Publishers; 1994. pp. 51–59. [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J. Light: An indicator of time and place. Genes & Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- Neuhaus G, Bowler C, Kern R, Chua NH. Calcium/Calmodulin-dependent and -independent phytochrome signal transduction pathways. Cell. 1993;73:937–952. doi: 10.1016/0092-8674(93)90272-r. [DOI] [PubMed] [Google Scholar]
- Schäfer E, Fukshansky L. Action spectroscopy. In: Smith H, Holmes GM, editors. Techniques in photomorphogenesis. London: Academic Press; 1984. pp. 109–127. [Google Scholar]
- Soh MS, Kim YM, Han SJ, Song PS. REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell. 2000;12:2061–2073. doi: 10.1105/tpc.12.11.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman JI, Mindrinos MN, Fankhauser C, Richards D, Lutes J, Chory J, Oefner PJ. Cloning of the Arabidopsis RSF1 gene by using a mapping strategy based on high-density DNA arrays and denaturing high-performance liquid chromatography. Plant Cell. 2000;12:2485–2498. doi: 10.1105/tpc.12.12.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JC. Light and protein kinases. Advances Bot Res. 2000;32:149–184. [Google Scholar]