Abstract
E4BP4, a basic leucine zipper transcription factor, contains a DNA-binding domain closely related to DBP, HLF, and TEF, which are PAR proteins. Here, we show that the phase of e4bp4 mRNA rhythm is opposite to that of the dbp, hlf, and tef rhythms in the suprachiasmatic nucleus (SCN), the mammalian circadian center, and the liver. The protein levels of E4BP4 and DBP also fluctuate in almost the opposite phase. Moreover, all PAR proteins activate, whereas E4BP4 suppresses, the transcriptional activity of the reporter gene containing a common binding sequence in transcriptional assays in vitro. An electrophoretic mobility shift assay demonstrated that E4BP4 is not able to dimerize with the PAR proteins, but is able to compete for the same binding sites with them. Furthermore, we showed sustained low e4bp4 and high dbp mRNA levels in mCry-deficient mice. These results indicate that the E4BP4 and PAR proteins are paired components of a reciprocating mechanism wherein E4BP4 suppresses the transcription of target genes during the time of day when E4BP4 is abundant, and the PAR proteins activate them at another time of day. E4BP4 and the PAR proteins may switch back and forth between the on-off conditions of the target genes.
Keywords: E4BP4, PAR domain, circadian rhythm, suprachiasmatic nucleus, basic leucine zipper transcription factor, active repressor
Circadian (∼24-h) rhythms are endogenous rhythms that are observed in a wide range of life forms. The molecular dissection of these endogenous oscillatory mechanisms has advanced using cyanobacteria (Iwasaki and Kondo 2000), Neurospora (Dunlap 1999), Drosophila (Young 2000), and mice (King and Takahashi 2000) as models. In all four species, the core oscillator consists of a self-sustaining transcriptional and translational feedback loop in which cyclically expressed clock gene products negatively regulate their own expression (for review, see Dunlap 1999).
In mice, the core feedback loop is thought to be composed of a positive element, the CLOCK/BMAL1 heterodimer, and many negative elements including mPER1, mPER2, mPER3, mCRY1, and mCRY2 (Gekakis et al. 1998; Jin et al. 1999; Kume et al. 1999; Okamura et al. 1999). The transcription of the mPer1 gene, of which regulation has been well analyzed, was shown to be activated by the binding of the CLOCK-BMAL1 complex to E-boxes (CACGTG) in the promoter region of the mPer1 gene (Gekakis et al. 1998; Yamaguchi et al. 2000a), and this activation was specifically inhibited by mPER1, mPER2, mPER3, mCRY1, and mCRY2 (Jin et al. 1999; Kume et al. 1999).
In this system, all proteins, except mCRY1 and mCRY2, are called PAS proteins, because they all possess PAS (Per-Arnt-Sim) domains that are protein–protein interaction domains and function in various biological pathways (Huang et al. 1993; Lindebro et al. 1995). The importance of this domain in the circadian feedback loop is suggested by the fact that homozygous mPer2 mutant mice, of which the mPer2 gene encodes the protein lacking the PAS domain, show significant differences in circadian locomotor activity, and altered expression levels of mPer1 and mPer2 in the suprachiasmatic nucleus (SCN), the mammalian circadian center (Zheng et al. 1999).
Subsequently, the light-harvesting cryptochrome/photolyase family proteins, mCRY1 and mCRY2, also were shown to be essential components of the circadian clock feedback loop. Mice lacking mCry1 and mCry2 are behaviorally arrhythmic in constant dark (DD) conditions (van der Horst et al. 1999), and their mPer1 and mPer2 mRNA levels are constitutively high in the SCN (Okamura et al. 1999). mCRY1 and mCRY2 have structural similarities to DNA repair enzymes called photolyases, but they lack DNA repair activity (Todo et al. 1996; Kobayashi et al. 1998). Moreover, although they also contain a region similar to plant blue-light receptors (cryptochromes), they were shown to act as light-independent inhibitors of CLOCK/BMAL1 (Griffin et al. 1999).
We reported recently that DBP (named for the albumin gene D-site binding protein), a member of another transcription factor family, is closely linked to the core circadian clockwork in mammals (Yamaguchi et al. 2000b). DBP belongs to the PAR (proline and acidic amino acid–rich) basic leucine zipper transcription factor family, along with hepatic leukemia factor (HLF) and thyrotroph embryonic factor (TEF). The transcript levels of dbp are highly rhythmic in the SCN, even in DD conditions, with an amplitude comparable to that of mPer1 (Lopez-Molina et al. 1997; Yan et al. 2000). It has been proposed that this circadian dbp transcription is regulated directly by CLOCK/BMAL1-mediated activation and mPERs- and mCRYs-mediated suppression through E-box motifs located in the introns of the dbp gene (Ripperger et al. 2000; Yamaguchi et al. 2000b). Moreover, DBP can activate the mPer1 promoter in transcriptional assays in vitro, and is expressed at the right time to activate mPer1 transcription in the SCN in vivo (Yamaguchi et al. 2000b). Thus, the cyclical activity of DBP may feed back onto the central clock mechanism.
Interestingly, in the Drosophila clock, a transcription factor VRI, which is structurally related to mammalian DBP, is required for a functional Drosophila clock (Blau and Young 1999). VRI is expressed in pacemaker cells of Drosophila and its RNA levels oscillate with a circadian rhythm via the regulation of the core-clock components, including dCLOCK and dBMAL1. The gene dosage of vri affects the period of circadian locomotor activity (Blau and Young 1999). Thus, it resembles mammalian dbp in many aspects. However, VRI apparently lacks a PAR domain, although it shows strong homology in its DNA-binding domain to DBP (George and Terracol 1997). The PAR domain is a well-conserved region of DBP and two other mammalian transcription factors, HLF and TEF, and has been shown to act as an activation domain at least in the case of DBP (Lamprecht and Mueller 1999).
In mammals, one basic leucine zipper transcription factor, which has a DNA-binding domain closely related to that of DBP, HLF, or TEF, but lacks a PAR domain, has been found. In fact, this transcription factor, E4BP4 (also called NFIL3), contains the most homologous basic leucine zipper domain to VRI. E4BP4 was isolated as an adenovirus E4 promoter ATF site binding protein (Cowell et al. 1992) and a human interleukin-3 promoter binding protein (Zhang et al. 1995), and has been studied mainly in relation to the hematopoietic system (Ikushima et al. 1997; Kuribara et al. 1999).
To investigate the possible role of E4BP4 in the circadian oscillatory mechanism in mammals, and the possible interactions between the E4BP4 and PAR proteins (DBP, HLF, and TEF), we examined the expression profiles of their mRNAs and proteins. Among the four structurally related transcription factors, dbp, hlf, and tef mRNAs showed similar peak and trough times, but only e4bp4 mRNA fluctuated in almost the opposite phase in both the SCN and liver. The protein levels of E4BP4 and DBP also fluctuated in almost the opposite phase. Moreover, here, we demonstrate that all PAR proteins activate, whereas E4BP4 suppresses, the transcriptional activity of the reporter gene containing a common binding sequence in transcriptional assays in vitro. These observations suggest that the E4BP4 and PAR proteins complement each other well in regulating the circadian oscillatory mechanism.
Results
Transcripts of the E4BP4 and PAR transcription factor family oscillate in almost the opposite phase in the SCN
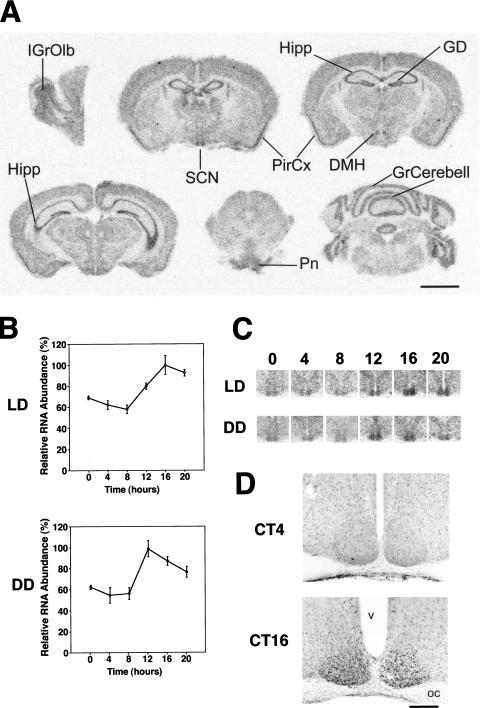
We first examined the distribution of e4bp4 mRNA in the mouse brain using in situ hybridization. The expression of e4bp4 was high in the SCN, hippocampus, gyrus dentatus, and piriform cortex, and moderate in the internal granular layer of olfactory bulb, dorsomedial hypothalamic nucleus, pontine nuclei, and the granular layer of cerebellum (Fig. 1A). Expression of e4bp4 was low in other brain regions. These hybridization signals were specific, because hybridization with a sense (control) probe showed no signals in brain sections (data not shown).
Figure 1.
Spatial and temporal expression profiles of the e4bp4 mRNA and E4BP4 proteins. (A) Distribution of e4bp4 mRNA in the mouse brain. IGrOlb, internal granular layer of the olfactory bulb; SCN, suprachiasmatic nucleus; Hipp, hippocampus; GD, gyrus dentatus; PirCx, piriform cortex; DMH, dorsomedial hypothalamic nucleus; Pn, pontine nuclei; GrCerebell, granular layer of the cerebellar cortex. Scale bar, 3 mm. (B) Rhythmic expression of e4bp4 mRNA in the SCN. Quantitative analysis of e4bp4 mRNA expressed in the SCN in LD and DD conditions. Relative e4bp4 mRNA abundance was determined by quantitative in-situ hybridization using isotope-labeled probes, with the mean peak values adjusted to 100 (n = 5, means ± SEM). (C) Representative in-situ hybridization autoradiographs showing e4bp4 mRNA in the SCN at various time points under LD and DD conditions. Numbers on the top indicate Zeitgeber (LD) or circadian (DD) times. (D) Circadian expression of E4BP4 immunoreactivity in the SCN at CT4 and CT16. OC, optic chiasma; v, third ventricle. Scale bar, 400 μm.
Next, we examined the temporal expression profile of e4bp4 mRNA in the SCN, the mammalian circadian center, with quantitative in-situ hybridization (Fig. 1B,C). In 12-h light:12h dark (LD) cycles, e4bp4 mRNA displayed a clear diurnal rhythm in the SCN showing a 1.7-fold amplitude with a peak at ZT16 (ZT, Zeitgeber time in LD-conditions; ZT0 is light-on and ZT12 is light-off) and a trough at ZT8 (P < 0.001). Under DD conditions, the rhythm of e4bp4 mRNA was clear, with a peak at circadian time (CT) 12 (where CT0 is subjective dawn and CT12 is subjective dusk) and a trough at CT4 (P < 0.001) (Fig. 1B,C).
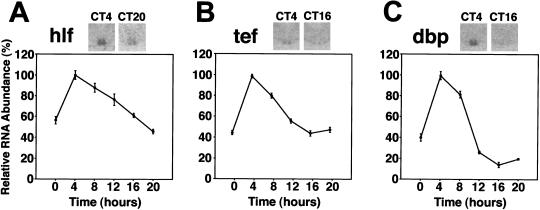
To compare the expression profile of e4bp4 with that of structurally related genes, we performed quantitative in-situ hybridization of the PAR transcription factors (hlf, tef, and dbp) in the SCN in DD conditions. The peak expression level of dbp mRNA was the highest and that of tef mRNA was the lowest of the three (see inserted pictures in Fig. 2). Interestingly, all three mRNAs showed the same expression profile, with a peak at CT4 and a trough at CT16–20 (P < 0.001, respectively; Fig. 2). The peak-trough ratio of dbp was the highest of three (for hlf, tef, and dbp, 2.3-, 2.3-, and 7.0-fold, respectively). In conclusion, e4bp4 mRNA fluctuates in almost the opposite phase to the structurally related PAR gene mRNAs (hlf, tef, and dbp) in the SCN.
Figure 2.
Circadian expression of hlf, tef, and dbp mRNAs in the SCN. Quantitative in-situ hybridization analyses of hlf, tef, and dbp mRNAs in the SCN in DD conditions are shown. Values are means ± SEM (n = 5). The mean peak values are adjusted to 100. Representative photomicrographs are shown at the top.
E4BP4 and DBP proteins also fluctuate in almost the opposite phase in the SCN
Next, we examined E4BP4 protein expression in the SCN with immunocytochemistry using anti-E4BP4 serum (Fig. 1D). Strong E4BP4 immunoreactivity was observed in the nuclei in the majority of the SCN cells (>80%) sampled at night (ZT14, ZT16, and ZT20) in LD conditions and at subjective night (CT12, CT16, and CT20) in DD conditions. This immunoreactivity was specific, because preincubation with an affinity-purified E4BP4 antigen completely abolished the signals (data not shown). In contrast, the SCN sampled at daytime (ZT4, ZT6) in LD conditions, and at subjective daytime (CT4), contained only a few and weakly stained E4BP4-immunoreactive nuclei (Fig. 1D). The similarity between the temporal expression profile of the E4BP4 protein and the e4bp4 mRNA suggests the rapid translation and nuclear accumulation of E4BP4, compared with the mPER1 protein, which was delayed by about 6 h relative to mPer1 mRNA (Field et al. 2000). Moreover, the expression profiles of E4BP4 proteins obviously are different from those of DBP proteins in the SCN we reported previously (Yamaguchi et al. 2000b). Because DBP proteins accumulate in the subjective morning peaking at CT6 in the SCN, the E4BP4 proteins were found to oscillate in almost the opposite phase, as in the case of their transcripts.
An opposite phase relationship also is observed in the liver
Because non-SCN tissues also rhythmically express various genes for which transcription oscillates in the SCN (Falvey et al. 1995; Fonjallaz et al. 1996; Zylka et al. 1998), we examined the temporal expression profile of e4bp4 mRNA in the liver of DD-housed mice.
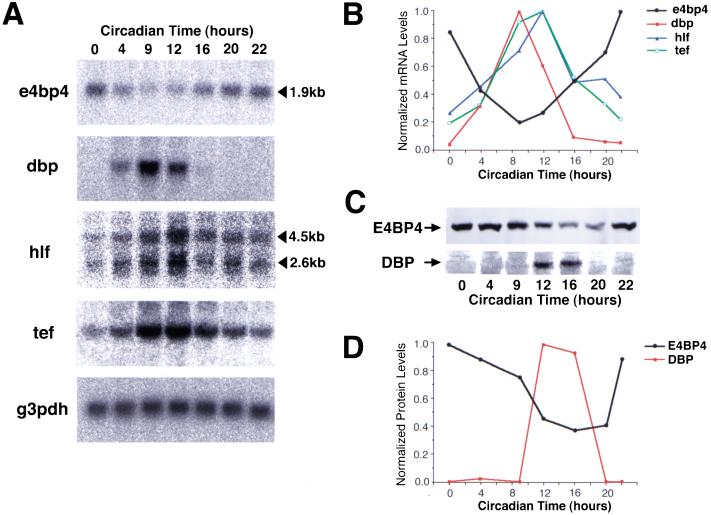
Northern blot analysis showed that a 1.9-kb transcript was detectable at all times examined, but its abundance displayed a clear rhythm with a maximum at CT22 and a minimum at CT9 (Fig. 3A,B). In contrast, dbp, hlf (both 2.6-kb and 4.5-kb transcripts), and tef mRNAs cycle in almost opposite phases peaking at the end of subjective day, although the peak time of dbp mRNA was slightly earlier than the others (Fig. 3A,B). Therefore, the phase difference between e4bp4 and PAR family mRNAs in the SCN is conserved in the liver, although the phase of each gene expression in the liver is 5–10 h later than that in the SCN.
Figure 3.
Circadian accumulation of e4bp4, dbp, hlf, and tef mRNAs in the mouse liver. (A) Northern blot analysis of the total RNAs (10 μg) isolated from the liver of DD-housed mice at different time points. The sizes of e4bp4 and hlf mRNAs are shown on the right. No significant variation in the amounts of loaded mRNA was detected by hybridization with a glyceraldehyde-3-phosphate dehydrogenase (g3pdh) cDNA probe. (B) Rhythmic expression of e4bp4 (black) is plotted in comparison with dbp (red), hlf (blue), and tef (green). The hlf mRNA levels represent the sum of the quantitated radioactivities of 4.5-kb and 2.6-kb transcripts. Normalized mRNA levels are shown with each peak value adjusted to 1.0. (C) Western blot analysis of liver nuclear extracts prepared from DD-housed mice, with anti-E4BP4 antiserum (upper panel) and anti-DBP antiserum (lower panel). (D) Rhythmic expression of E4BP4 (black) is plotted in comparison with DBP (red). Protein levels were determined by Western blot analysis in (C). The peak values are adjusted to 1.0.
We next examined the temporal expression profiles of the E4BP4 and DBP proteins in the liver. Nuclear extracts were prepared from the livers of DD-housed mice and fractionated by SDS-polyacrylamide gel electrophoresis. Immunoblot analysis with anti-E4BP4 serum revealed a single band of ∼55 kD, in accordance with the predicted mass of 53 kD (Fig. 3C). The abundance of E4BP4 protein varied, with high levels between CT22 and CT4, and low levels between CT12 and CT20 (Fig. 3C,D). Therefore, the E4BP4 protein was abundant in the liver at about subjective dawn, the same as e4bp4 mRNA.
When the liver samples were probed using an antiserum to DBP, the highest protein levels were observed only between CT12 and CT16 (Fig 3C,D), consistent with previous reports by Schibler and coworkers (Fonjallaz et al. 1996; Lopez-Molina et al. 1997). Thus, E4BP4 and DBP proteins also fluctuate in almost the opposite phase in the liver, as in the SCN.
All PAR proteins activate, whereas E4BP4 suppresses the mPer1 promoter through the same sequence
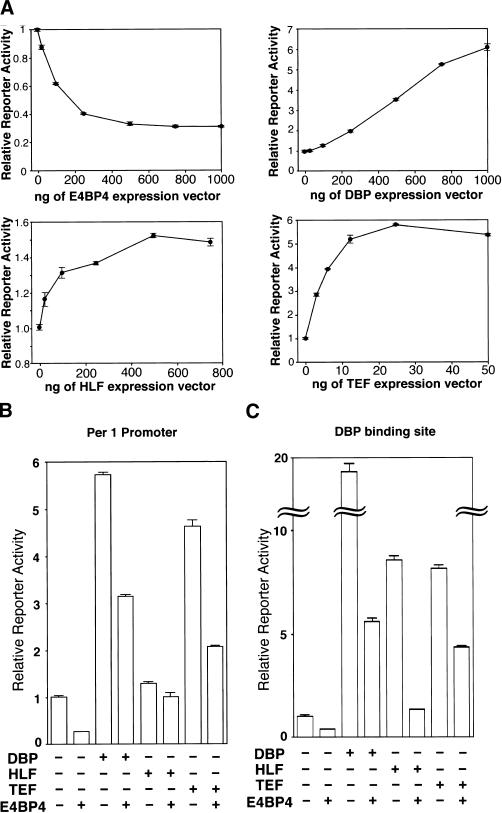
The optimal binding motif for E4BP4, RT(G/T)AYG TAAY (where R is a purine and Y is a pyrimidine), and those for the three PAR proteins are almost identical (Drolet et al. 1991; Cowell et al. 1992; Falvey et al. 1996; Hunger et al. 1996). We showed previously that the mPer1 promoter contains a DBP-binding sequence, AT TATGCAAC (9 of 10 bases are identical to the optimal binding motif), through which DBP is able to activate the mPer1 promoter, at least in transient transfection analysis (Yamaguchi et al. 2000b). In this study, we examined the effects of E4BP4, HLF, and TEF on the mPer1 promoter. A 1.3-kbp fragment of the 5′-flanking region of the mPer1 gene containing an endogenous promoter and the DBP-binding site was subcloned into a promoterless reporter vector for use in transcriptional analysis in HepG2 cells.
The transcriptional activity of the mPer1 promoter was suppressed in a dose-dependent manner by increasing amounts of the transfected-E4BP4 expression vector (Fig. 4A). In contrast, the HLF- and TEF- expression vectors, as well as the DBP-expression vector, activated the mPer1 promoter (Fig. 4A).
Figure 4.
Transcriptional regulation by E4BP4 and PAR proteins. (A) Transcriptional regulation of the mPer1 promoter by E4BP4 and each PAR protein alone. A reporter plasmid (10 ng) containing a 1.3-kbp mPer1 promoter including the DBP-binding site, and increasing doses of each expression plasmid, were cotransfected. Each value is the mean ± SEM of triplicate results for a single assay. A similar pattern of activation or suppression was reproduced in another experiment. (B) Transcriptional regulation of the mPer1 promoter by the coexpression of E4BP4 and each PAR protein. Presence (+) or absence (−) of DBP-, HLF-, TEF-, and E4BP4-expression plasmids (1000, 500, 25, and 1000 ng, respectively) is noted. (C) Transcriptional regulation of an HSV-TK-driven reporter plasmid containing a DBP-binding site derived from the mPer1 promoter. The presence (+) or absence (−) of DBP-, HLF-, TEF-, and E4BP4-expression plasmids (500, 100, 5, and 500 ng, respectively) is noted. The total amount (1 μg) of expression plasmids per well was adjusted by adding a pcDNA3 plasmid. (B,C) Each value is the mean ± SEM of three replicates for a single assay. The results shown are representative of at least three independent experiments.
We next tested for possible interactions between the E4BP4 and PAR proteins. The 1.3-kbp mPer1 promoter was cotransfected with E4BP4 expression plasmids and each of the PAR proteins expression plasmids. E4BP4 not only suppressed basal promoter activity (74.2% [P < 0.001]), but also significantly reduced the DBP-, HLF-, and TEF-mediated transcriptions (45.2% [P < 0.001], 22.0% [P < 0.005], and 55.4% [P < 0.001], respectively) (Fig. 4B).
To test whether the DBP-binding site actually is responsible for these transcriptional regulations, we examined the transcriptional activity of a construct in which three copies of a 20-bp sequence centered on the DBP-binding site (ATTATGCAAC) were linked in tandem and subcloned into a reporter vector containing the herpes simplex virus thymidine kinase (HSV-TK) minimal promoter. Again, DBP, HLF, and TEF produced a substantial increase in transcriptional activity (19.3-fold [P < 0.001], 8.5-fold [P < 0.001], and 8.1-fold [P < 0.001], respectively) and E4BP4 reduced the basal transcriptional activity (61.8% [P < 0.001]; Fig. 4C). Each of the PAR protein- (DBP, HLF, and TEF) induced transcriptions also was reduced (71.3% [P < 0.001], 84.6% [P < 0.001], and 46.4% [P < 0.001], respectively) by the coexpression of E4BP4 (Fig. 4C). Similar results were obtained when an optimal binding motif for all four proteins, ATTACGTAAC, was used as the reporter construct (data not shown). Moreover, when the three DBP-binding sites were mutated, the transcriptional activity of the reporter construct was not affected significantly by the expression of DBP, HLF, TEF, or E4BP4 (data not shown).
Therefore, we conclude that E4BP4 and all PAR proteins are able to suppress and activate transcriptional activity, respectively, through the same sequence and that E4BP4 is able to reduce the PAR protein-induced transcriptions in HepG2 cells. Taken together with the inverse expression profiles of E4BP4 and DBP in vivo, this indicates that E4BP4 suppresses transcription of the target genes during the time of day when E4BP4 is abundant, and DBP activates them, possibly in cooperation with HLF and TEF, at another time of day. These regulations may increase the oscillation amplitudes of the target genes.
E4BP4 competes for the same binding site with the PAR proteins, but does not heterodimerize with them
Although it is known that different PAR family members are able to homodimerize and heterodimerize (Fonjallaz et al. 1996), and that E4BP4 is able to homodimerize (Cowell et al. 1992), the interactions between E4BP4 and PAR proteins are unknown. Moreover, it is uncertain whether the affinities of the four transcription factors for a common recognition site are close enough to compete for the same binding site with each other. To elucidate the mechanism of the antagonistic regulation of PAR proteins and E4BP4 on the same sequence, we performed electrophoretic mobility shift assays (EMSA) with a 32P-radiolabeled probe encompassing a sequence responsible for such transcriptional regulation. As expected, each of the in-vitro translated E4BP4, DBP, HLF, and TEF proteins generated single strong bands resulting from complex formation with the probe (Fig. 5). To compare the binding affinities of E4BP4 and PAR proteins for the probe quantitatively, we performed EMSAs, titrating the amount of a radiolabeled probe against a constant amount of protein (Fig. 5). After the radioactivity of the bound and free forms was measured, the dissociation constants (Kd) were determined by Scatchard plots (Fig. 5). The Kds for binding of E4BP4, DBP, HLF, and TEF to the probe were 2.1, 1.3, 4.0, and 2.8 nM, respectively.
Figure 5.
Relative affinities of E4BP4 and the three PAR proteins for an optimal binding site (ATTACGTAAC). A constant amount of protein was incubated with increasing amounts of a radiolabeled probe. After gel electrophoresis and autoradiography (top panels), the radioactive bands corresponding to the bound and free forms were quantitated. The concentration of the bound probe was plotted against the total input probe to show saturation curves (middle panels). These data also were plotted by the method of Scatchard to determine the Kd values (bottom panels). The slope of the best-fit line is equal to −1/Kd. The correlation coefficients (R2) are indicated in the bottom panels.
Because the four examined proteins showed similar relative affinities to the probe, we reasoned that the PAR proteins bound to the probe may be displaced by E4BP4 in a competitive fashion. To confirm this possibility, we synthesized each protein in a separate reaction and combined a constant amount of each PAR protein, and increasing amounts of E4BP4, prior to a mobility shift assay. Because the mobility of the full-length E4BP4-probe complex was similar to those of the complexes generated by DBP, HLF, and TEF, we used a truncated E4BP4 protein, which yielded a more rapidly migrating complex. This protein retained the basic and leucine zipper domains (amino acids 65–250) and had an affinity for the probe (Kd = 2.8 nM) comparable to that of full-length E4BP4.
The complexes yielded by DBP, HLF, and TEF diminished clearly with increasing amounts of the complexes generated by the truncated E4BP4 (Fig. 6A). No additional band with intermediate mobility representing the heterodimer formation of the PAR proteins and the truncated E4BP4 was detectable (Fig. 6A). This also was the case when each PAR protein and E4BP4 was synthesized simultaneously in the same reaction mixture by combining the DNA templates prior to the transcription and translation in vitro (Fig. 6B). Conversely, the cotranslation of long- and short-E4BP4 resulted in an additional band demonstrating their dimerization as expected (Fig. 6B). These data suggest that E4BP4 is not able to dimerize with DBP, HLF, and TEF, but is able to compete for the same binding site with them. Therefore, the antagonistic regulation of the PAR and E4BP4 proteins in transcriptional assays may be explained by this competitive mechanism.
Figure 6.
E4BP4 competes for the same binding sequence with PAR proteins. (A) E4BP4 and PAR proteins were synthesized in separate reactions prior to combining them. A constant amount of each PAR protein, increasing amounts of E4BP4, and the probe (1 ng) were incubated for 15 min at 30°C before loading the mixture on to the gel. The concentration of input protein (nM) in each reaction was determined by Scatchard plots, as shown in Figure 5 and indicated in parentheses. Each arrow indicates the position of the complex generated by each transcription factor. (B) E4BP4 and PAR proteins were synthesized simultaneously in the same reaction mixture before incubation with 1 ng of the probe. Excess amounts of each PAR-protein or long-E4BP4 (65–324) expression plasmids (shown in the left-hand part of the figure), and excess amounts of short-E4BP4 (65–250) expression plasmid (right-hand part of the figure) were combined prior to the transcription and translation in vitro. The amounts of the used expression vectors (ng) are indicated in parentheses. The positions of the complex generated by each transcription factor alone, and the heterodimeric complex formed between long and short E4BP4 are indicated by the arrows.
e4bp4 and dbp mRNA levels are inversely regulated in mCry-deficient mice
We demonstrated that the transcript levels of the e4bp4 and PAR transcription factor family, and the protein levels of E4BP4 and at least DBP, oscillate in almost the opposite phase in the SCN and liver. Moreover, E4BP4 and PAR proteins were shown to have suppressing and activating functions on the same sequence, respectively. These findings indicate that the E4BP4 and PAR proteins complement each other well in regulating target genes, and that they are the paired components of a reciprocating mechanism.
To investigate relationships with the putative core oscillation mechanism, we examined the temporal expression profiles of e4bp4 and dbp in the SCN and liver of mice lacking both the mCry1 and mCry2 genes. These mutant mice completely lack free-running rhythmicity in constant darkness (van der Horst et al. 1999). In wild-type control animals, the typical circadian variation in e4bp4 mRNA levels in the SCN was observed with low levels at CT4 (40 h after animals were placed in DD) and high levels at CT12 (48 h in DD) (1.7-fold, P < 0.001) (Fig. 7A). In contrast, e4bp4 mRNA levels were low at both times in mCry-deficient mice (Fig. 7A).
Figure 7.
Expression of e4bp4 and dbp in wild-type and mCry1−/−mCry2−/− mice. (A) Expression levels of e4bp4 and dbp in the SCN of wild-type (open bars) and mCry-deficient (solid bars) mice in constant darkness were determined by quantitative in-situ hybridization. The mean peak values of the wild-type mice were adjusted to 100%. Values are expressed as means ± SEM (n = 3). Representative autoradiographs are shown at the top. Because mCry-deficient mice are arrhythmic, we used an environmental time scale (hours in DD) rather than a circadian time scale. *, significant differences in wild-type mice, P < 0.001; there was no significant difference in mCry1−/−mCry2−/− mice. (B) Temporal expression profiles of e4bp4 and dbp in the liver of wild-type and mCry1−/−mCry2−/− mice. Northern blots of the total RNAs (10 μg) prepared from the livers of DD-housed mice are shown. g3pdh was used as a control.
We then examined dbp mRNA levels in the SCN of mCry double-mutant mice. In marked contrast, mCry double-mutant mice showed high dbp mRNA levels in the SCN (Fig. 7A). This finding is consistent with the proposed regulatory mechanism for the transcription of the dbp gene: The CLOCK/BMAL1 heterodimer activates, whereas mCRYs mainly suppresses, the transcription of the dbp gene through the E-box motifs (Ripperger et al. 2000; Yamaguchi et al. 2000b).
Sustained low e4bp4 and high dbp mRNA levels also were observed in the liver of mCry-deficient mice (Fig. 7B). Therefore, these results indicate that both e4bp4 and dbp are regulated directly or indirectly by the core oscillatory mechanism including CLOCK/BMAL1, mPERs, and mCRYs, maintaining an inverse relationship.
Discussion
On the basis of our results, we propose two working models that can explain the relationship between the e4bp4 gene and the putative core feedback loop including CLOCK/BMAL1, mPERs, and mCRYs (Fig. 8A,B).
Figure 8.
Models of the role of E4BP4 and the PAR proteins, and the regulation of the e4bp4 gene. (A) The e4bp4 gene is regulated by an unidentified transcriptional repressor (X) that is activated (+) by CLOCK/BMAL1 and suppressed (−) by mPERs and mCRYs, as is the case with the dbp, hlf, and tef genes. DBP, HLF, and TEF activate, whereas E4BP4 suppresses the transcriptions of target genes at different times of day. This regulation increases the amplitude of the oscillation of the target genes. (B) The e4bp4 gene is regulated by an unidentified repressor (Y) that is regulated by DBP, HLF, TEF, and E4BP4 itself.
In the model shown in Figure 8A, the e4bp4 gene is regulated by an unidentified transcriptional repressor (X) that is regulated by CLOCK/BMAL1 and the negative elements, mPERs and mCRYs, as in the case of the dbp, hlf, and tef genes. When X mRNA is translated rapidly and the produced X protein accumulates in the nuclei of the SCN cells with little delay and depresses expression of e4bp4, the phase of the e4bp4 rhythm thus is expected to be opposite to that of the dbp, hlf, and tef rhythms. Rapidly translated E4BP4 suppresses the transcriptions of target genes during the time of day when E4BP4 is abundant, and the PAR proteins activate them at another time of day. Thereby, the E4BP4 and PAR proteins increase the amplitude of the rhythmically expressed transcript levels of the target genes (Fig. 8A).
In the model in Figure 8B, the e4bp4 gene is regulated by an unidentified repressor (Y) that is indirectly regulated by CLOCK/BMAL1, mPERs, and mCRYs via DBP-, HLF-, or TEF-mediated regulation. Besides our two models, it also is conceivable that the rhythmic expression of the e4bp4 gene is controlled by the cycling presence of an unidentified positive element that drives the rhythmic expression of the bmal1 gene, depending on mPER2 (Shearman et al. 2000). It is noteworthy that in all three models, the existence of an unidentified activator or repressor is indicated.
In the liver, albumin, cholesterol 7α hydroxylase, and cytochrome P450 (Cyp2c6, Cyp2a4, and Cyp2a5) genes are thought to be candidates for the target genes of the PAR proteins (Falvey et al. 1996; Lavery et al. 1999). Conversely, the interleukin 3 gene is thought to be a target of E4BP4 in T lymphocytes (Zhang et al. 1995). The consensus-binding site for E4BP4 and the PAR proteins is different from the CLOCK/BMAL1 E-box binding site. Therefore, E4BP4 and the PAR proteins potentially could regulate a set of output genes that do not also possess an E-box. Moreover, as we have demonstrated, E4BP4 and the PAR proteins regulated the transcriptional activity of the mPer1 promoter in a transcriptional assay in vitro. Thus, the cyclic activities of E4BP4 and the PAR proteins may feed back onto the central clock mechanism.
E4BP4 was shown to behave as an active transcriptional repressor that directly suppressed the transcriptional activities of genes whose promoters it bound (Cowell and Hurst 1994). This active repression is mainly because of a small transferable repression domain of 65 amino acids in the C-terminal half of the protein (Cowell and Hurst 1994). In the case of DBP, on the other hand, the PAR (proline and acidic amino acid-rich) domain, which resides amino-terminal to the basic region, has been shown to act as an activation domain (Lamprecht and Mueller 1999). Some other examples of the combination of activator and active repressor that bind the same site are found in genes controlling segmentation during early Drosophila development (e.g., Fushi tarazu and Engrailed). It has been proposed that competition between an activator and repressor is not only required, but also is sufficient to establish all-or-none switches in gene expression (Rossi et al. 2000). E4BP4 and the PAR proteins may switch back and forth between the on-off conditions.
The Drosophila transcription factor VRI contains a DNA-binding domain closely related to mammalian E4BP4, but lacks a PAR domain. It therefore is comparable to E4BP4. Recently, VRI was shown to be required for a functional Drosophila clock: Reducing vri gene dosage caused period shortening and elimination of the normal vri cycle generated long-period rhythms or arrhythmicity (Blau and Young 1999). Therefore, this family of transcription factors may have an important role in circadian clocks in both Drosophila and mammals. However, as we have shown, the phase of the e4bp4 oscillation was opposite that of the mPer1, mPer2, and mPer3 rhythms (Tei et al. 1997; Takumi et al. 1998a,b), which are regulated directly by CLOCK/BMAL1 (Gekakis et al. 1998; Jin et al. 1999). On the other hand, the cycling of vri mRNA in Drosophila is regulated directly by dCLOCK/dBMAL1, and vri mRNA thereby oscillates with the same phase as per mRNA in adult heads (Blau and Young 1999). Thus, there may be a difference in the way E4BP4 and VRI are utilized in the mammalian/Drosophila clocks.
In summary, our data indicate that E4BP4 and the PAR proteins are paired components of an oscillatory mechanism. This mechanism may be found throughout much of the animal kingdom.
Materials and methods
Animals
Male BALB/c mice (Japan Animal Company, Japan) purchased 5 wk postpartum were exposed to 2 to 4 wk of complete light (fluorescent light, 300 lux)-dark (LD) cycles and then kept in complete darkness for 2 d (DD) as a continuation of the dark phase of the last LD cycle. Wild-type and mCry1−/−mCry2−/− animals (C57Bl6:Ola 129 hybrid, 8 to 12 wk old) generously provided by Dr. G.T.J. van der Horst (Erasmus University), were housed in the same way. The experimental protocol of the current research was approved by the Committee for Animal Research at Kobe University School of Medicine.
Quantitative in-situ hybridization
In-situ hybridization histochemistry using free-floating sections was performed according to a method detailed previously (Shigeyoshi et al. 1997; Yamaguchi et al. 2000b). We used 33P-radiolabeled complementary RNA probes for mouse e4bp4 (nucleotide residues 194–793; GenBank accession no. U83148), mouse dbp (595–1100; U29762), mouse hlf (corresponding to nucleotide residues 20–543 of rat hlf; S79820), and mouse tef (corresponding to nucleotide residues 4–471 of rat tef; S58745). All fragments for templates were obtained by reverse transcription-PCR and sequenced to verify their identity and orientation. No specific hybridization signals were observed when sense cRNA probes were used. The radioactivity of each SCN on BioMax film (Kodak) was analyzed using a microcomputer interfaced to an image-analyzing system after conversion into the relative optical density; the 10 sections of the SCN then were summed. For statistical analysis, one-way analysis of variance (ANOVA) followed by Scheffe's multiple comparisons or a two-sample t test was applied.
Northern blot analysis
Northern blot analysis was performed as described (Yamaguchi and Nakanishi 1998). Ten micrograms of the total RNAs extracted from the livers of the DD-housed mice sacrificed at various circadian times were analyzed. As templates for the generation of radiolabeled probes, an 812-bp fragment of the 5′-portion of the coding region of mouse dbp, a 543-bp BglII–NruI fragment of human hlf, a 622-bp fragment of the 5′-portion of the coding region of rat tef, and a 600-bp EcoRI–NcoI fragment of mouse e4bp4 cDNA were used. The radioactivity of each band was quantitated with a microcomputer-coupled image-processing system (BAS5000, Fujifilm).
Western blot analysis
Nuclear proteins were extracted from the livers of the DD-housed mice as described previously (Yamaguchi et al. 2000b). For quantification of the nuclear proteins, a bicinchoninic acid (BCA) protein assay reagent kit (Pierce) was used. Forty micrograms of nuclear extracts were mixed with a 3× SDS sample buffer (375 mM Tris-HCl at pH 6.8, 6% SDS, 30% glycerol, 10% 2-mercaptoethanol) and boiled for 3 min. Each sample was fractionated on a 10% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Immunoblot-P membrane, ATTO, Japan). Immunoblotting was performed with a polyclonal rabbit anti-E4BP4 antiserum (No.60, 1:5000), which was kindly provided by Dr. T. Inaba (Jichi Medical School), or anti-DBP antiserum (1:2000) (Yamaguchi et al. 2000b), as a primary antibody. Horseradish peroxidase-conjugated anti-rabbit immunoglobulin (1:5000, Amersham) was used in combination with enhanced chemiluminescence (NEN) to detect proteins. The relative protein level of each band was determined with the aid of a LAS1000 analyzer (Fujifilm).
Immunocytochemistry
Under deep ether anesthesia, LD- and DD-housed male BALB/c mice were perfused with saline, followed by a fixative (2% paraformaldehyde and 0.2% picric acid in a 0.1 M phosphate buffer), and then postfixed with the same fixative for 2 h. Thirty-micrometer sections were cut in a cryostat and processed for free-floating immunohistochemistry using anti-E4BP4 antiserum (1:2000) as described previously (Yagita et al. 2000). After an application of biotinylated anti-rabbit IgG (1:1000, Vector), sections were incubated with avidin-biotin-peroxidase (1:1000, Vector) and visualized with diaminobenzidine (DAB).
Transcriptional assay
Transfection and luciferase assays were carried out as described previously (Yamaguchi et al. 2000b). Unless otherwise specified, ca. 6.0 × 105 HepG2 cells per well in six-well plates were transfected with 2 μg (total) of expression plasmids with the indicated inserts in pcDNA3 (Invitrogen), 10 ng of a reporter plasmid, and 1 ng of an internal control plasmid using LipofectAmine Plus reagent (GIBCO). The total amount of DNA per well was adjusted by adding pcDNA3. After 48 h, cells were extracted with 200 μL of passive lysis buffer (Promega), and 20 μL of the extracts were taken for assays of firefly luciferase and Renilla luciferase.
A reporter construct containing the mPer1 promoter was made by ligating a 1.3-kbp fragment of the 5′-flanking region of the mPer1 gene to the Renilla luciferase reporter gene (1212-bp HindIII–XbaI fragment of the pRL-TK vector, Promega) and the simian virus 40 polyadenylation signal. A reporter construct, in which three copies of a 20-bp sequence centered on a DBP-binding site were linked in tandem, was made by inserting annealed oligonucleotides (top strand, 5′-GATCTCTGGCAT TATGCAACCCGCCCTGGCATTATGCAACCCGCCCTGG CATTATGCAACCCGCCA-3′; bottom strand, 5′-GATCTG GCGGGTTGCATAATGCCAGGGCGGGTTGCATAATGCC AGGGCGGGTTGCATAATGCCAGA-3′ [binding sites are underlined]) into the BglII site of the pRL-TK vector (Yamaguchi et al. 2000b).
For the construction of expression plasmids, the total coding regions of mouse dbp (GenBank accession no. U29762), human hlf (M95585), rat tef (S58745), and mouse e4bp4 (U83148) were obtained by reverse transcription-PCR and subcloned into pcDNA3. All coding regions were used after confirming their sequences. For statistical analysis, a two-sample t test was applied.
Electrophoretic mobility shift assay
A double stranded oligonucleotide (top strand, 5′-GCCCGCTA CATATTACGTAACAAGCGTTCGC-3′; bottom strand, 5′-GGCGAACGCTTGTTACGTAATATGTAGCGGG-3′) containing a consensus binding sequence for the PAR and E4BP4 proteins was radiolabeled using the Klenow enzyme and [α-32P]dCTP, and used as a probe.
All proteins used in this assay were synthesized by coupled transcription-translation in vitro (TNT T7 Quick Coupled Transcription/Translation System, Promega). For construction of full-length E4BP4, DBP, HLF, and TEF expression plasmids, the total coding regions were ligated into pcDNA3 containing a C-terminal Flag tag (Yagita et al. 2000). For the long (amino acids 65–324) and short (65–250) form E4BP4, the coding regions were supplied with a nucleotide sequence, 5′-GCCACCATG-3′ (Kozak sequence and a first methionine encoding ATG), at the 5′ ends, which then was subcloned into pcDNA3 containing either a C-terminal Flag or an HA tag. All constructs were made using PCR and verified by sequence analysis. Coupled transcription/translation reactions were performed according to the manufacturer's instructions with 1 μg of template DNAs and a 90-min incubation time. To confirm that a protein of the correct size was produced, Western blot analysis with anti-Flag M2 (Sigma) or an anti-HA monoclonal (Boehringer) antibody was carried out.
Binding reactions with the indicated amounts of the labeled probe and the synthesized proteins were performed in a reaction buffer (8 mM HEPES at pH 7.9, 40 mM KCl, 0.4 mM MgCl2, 6.8% glycerol, 0.1 μg/μL bovine serum albumin, 0.8 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, and 0.1 μg/μL poly dI-dC; Amersham) at 30°C for 15 min. The resulting products were loaded on an 8% nondenaturing 1× TBE (Tris-borate/EDTA electrophoresis buffer) polyacrylamide gel, and electrophoresed at 150 V. The radioactivity of each band was quantitated as described for the Northern blotting experiments.
Acknowledgments
The authors thank Kazuhiro Yagita and Hiromi Isejima for performing immunocytochemistry and Western blotting. This work was supported in part by grants from the Special Coordination Funds of the Science and Technology Agency of Japan, the Grants-in-Aid for the Scientific Research on Priority Areas of the Ministry of Education, Science, Sports, and Culture of Japan and Mitsubishi Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL okamurah@kobe-u.ac.jp; FAX 81-78-382-5341.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.873501.
References
- Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- Cowell IG, Skinner A, Hurst HC. Transcriptional repression by a novel member of the bZIP family of transcription factors. Mol Cell Biol. 1992;12:3070–3077. doi: 10.1128/mcb.12.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell IG, Hurst HC. Transcriptional repression by the human bZIP factor E4BP4: Definition of a minimal repression domain. Nucleic Acids Res. 1994;22:59–65. doi: 10.1093/nar/22.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet DW, Scully KM, Simmons DM, Wegner M, Chu K, Swanson LW, Rosenfeld MG. TEF, a transcription factor expressed specifically in the anterior pituitary during embryogenesis, defines a new class of leucine zipper proteins. Genes & Dev. 1991;5:1739–1753. doi: 10.1101/gad.5.10.1739. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Falvey E, Fleury-Olela F, Schibler U. The rat hepatic leukemia factor (HLF) gene encodes two transcriptional activators with distinct circadian rhythms, tissue distributions and target preferences. EMBO J. 1995;14:4307–4317. doi: 10.1002/j.1460-2075.1995.tb00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falvey E, Marcacci L, Schibler U. DNA-binding specificity of PAR and C/EBP leucine zipper proteins: A single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol Chem. 1996;377:797–809. [PubMed] [Google Scholar]
- Field MD, Maywood ES, O'Brien JA, Weaver DR, Reppert SM, Hastings MH. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- Fonjallaz P, Ossipow V, Wanner G, Schibler U. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 1996;15:351–362. [PMC free article] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- George H, Terracol R. The vrille gene of Drosophila is a maternal enhancer of decapentaplegic and encodes a new member of the bZIP family of transcription factors. Genetics. 1997;146:1345–1363. doi: 10.1093/genetics/146.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Edery I, Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature. 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- Hunger SP, Li S, Fall MZ, Naumovski L, Cleary ML. The proto-oncogene HLF and the related basic leucine zipper protein TEF display highly similar DNA-binding and transcriptional regulatory properties. Blood. 1996;87:4607–4617. [PubMed] [Google Scholar]
- Ikushima S, Inukai T, Inaba T, Nimer SD, Cleveland JL, Look AT. Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc Natl Acad Sci. 1997;94:2609–2614. doi: 10.1073/pnas.94.6.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Kondo T. The current state and problems of circadian clock studies in cyanobacteria. Plant Cell Physiol. 2000;41:1013–1020. doi: 10.1093/pcp/pcd024. [DOI] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu Rev Neurosci. 2000;23:713–742. doi: 10.1146/annurev.neuro.23.1.713. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kanno S, Smit B, van der Horst GTJ, Takao M, Yasui A. Characterization of photolyase/blue-light receptor homologs in mouse and human cells. Nucleic Acids Res. 1998;26:5086–5092. doi: 10.1093/nar/26.22.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Kuribara R, Kinoshita T, Miyajima A, Shinjyo T, Yoshihara T, Inukai T, Ozawa K, Look AT, Inaba T. Two distinct interleukin-3-mediated signal pathways, Ras-NFIL3 (E4BP4) and Bcl-xL, regulate the survival of murine pro-B lymphocytes. Mol Cell Biol. 1999;19:2754–2762. doi: 10.1128/mcb.19.4.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht C, Mueller CR. D-site binding protein transactivation requires the proline- and acid-rich domain and involves the coactivator p300. J Biol Chem. 1999;274:17643–17648. doi: 10.1074/jbc.274.25.17643. [DOI] [PubMed] [Google Scholar]
- Lavery DJ, Lopez-Molina L, Margueron R, Fleury-Olela F, Conquet F, Schibler U, Bonfils C. Circadian expression of the steroid 15 α-hydroxylase (Cyp2a4) and coumarin 7-hydroxylase (Cyp2a5) genes in mouse liver is regulated by the PAR leucine zipper transcription factor DBP. Mol Cell Biol. 1999;19:6488–6499. doi: 10.1128/mcb.19.10.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindebro MC, Poellinger L, Whitelaw ML. Protein-protein interaction via PAS domains: Role of the PAS domain in positive and negative regulation of the bHLH/PAS dioxin receptor-Arnt transcription factor complex. EMBO J. 1995;14:3528–3539. doi: 10.1002/j.1460-2075.1995.tb07359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers JHJ, van der Horst GTJ. Photic induction of mPer1 and mPer2 in Cry-deficient mice lacking a biological clock. Science. 1999;286:2531–2534. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes & Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- Rossi FM, Kringstein AM, Spicher A, Guicherit OM, Blau HM. Transcriptional control: Rheostat converted to on/off switch. Mol Cell. 2000;6:723–728. doi: 10.1016/s1097-2765(00)00070-8. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GTJ, Hastings MH, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- Takumi T, Matsubara C, Shigeyoshi Y, Taguchi K, Yagita K, Maebayashi Y, Sakakida Y, Okumura K, Takashima N, Okamura H. A new mammalian period gene predominantly expressed in the suprachiasmatic nucleus. Genes Cells. 1998a;3:167–176. doi: 10.1046/j.1365-2443.1998.00178.x. [DOI] [PubMed] [Google Scholar]
- Takumi T, Taguchi K, Miyake S, Sakakida Y, Takashima N, Matsubara C, Maebayashi Y, Okumura K, Takekida S, Yamamoto S, et al. A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 1998b;17:4753–4759. doi: 10.1093/emboj/17.16.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- Todo T, Ryo H, Yamamoto K, Toh H, Inui T, Ayaki H, Nomura T, Ikenaga M. Similarity among the Drosophila (6–4) photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science. 1996;272:109–112. doi: 10.1126/science.272.5258.109. [DOI] [PubMed] [Google Scholar]
- van der Horst GTJ, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Yagita K, Yamaguchi S, Tamanini F, van der Horst GTJ, Hoeijmakers JHJ, Yasui A, Loros JJ, Dunlap JC, Okamura H. Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes & Dev. 2000;14:1353–1363. [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Nakanishi S. Regional expression and regulation of alternative forms of mRNAs derived from two distinct transcription initiation sites of the rat mGluR5 gene. J Neurochem. 1998;71:60–68. doi: 10.1046/j.1471-4159.1998.71010060.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Mitsui S, Miyake S, Yan L, Onishi H, Yagita K, Suzuki M, Shibata S, Kobayashi M, Okamura H. The 5′ upstream region of mPer1 gene contains two promoters and is responsible for circadian oscillation. Curr Biol. 2000a;10:873–876. doi: 10.1016/s0960-9822(00)00602-3. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Mitsui S, Yan L, Yagita K, Miyake S, Okamura H. Role of DBP in the circadian oscillatory mechanism. Mol Cell Biol. 2000b;20:4773–4781. doi: 10.1128/mcb.20.13.4773-4781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Miyake S, Okamura H. Distribution and circadian expression of dbp in SCN and extra-SCN areas in the mouse brain. J Neurosci Res. 2000;59:291–295. doi: 10.1002/(sici)1097-4547(20000115)59:2<291::aid-jnr16>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Young MW. Life's 24-hour clock: Molecular control of circadian rhythms in animal cells. Trends Biochem Sci. 2000;25:601–606. doi: 10.1016/s0968-0004(00)01695-9. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhang J, Kornuc M, Kwan K, Frank R, Nimer SD. Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Mol Cell Biol. 1995;15:6055–6063. doi: 10.1128/mcb.15.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: Differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]