Abstract
The chromosome 8p11–12 Werner syndrome (WRN ) locus encodes a RecQ helicase protein of unknown function that possesses both 3′ → 5′ helicase and 3′ → 5′ exonuclease activities. We show that WRN cell lines display a marked reduction in cell proliferation following mitotic recombination, and generate few viable gene conversion-type recombinants. These findings indicate that WRN plays a role in mitotic recombination, and that a loss of WRN function may promote genetic instability and disease via recombination-initiated mitotic arrest, cell death, or gene rearrangement.
Keywords: Werner syndrome, RecQ, helicase, mitotic recombination, genetic instability, cancer
Werner syndrome (WRN; MIM #277700) is an uncommon autosomal recessive disease in which features of premature aging are associated with genetic instability and an elevated risk of cancer. The Werner syndrome clinical phenotype mimics premature aging: following puberty, patients rapidly develop premature graying and loss of hair, scleroderma-like skin changes, osteoporosis, atherosclerosis, bilateral cataract formation, diabetes mellitus, and hypogonadism (Epstein et al. 1966; Goto 2000). Spontaneous genetic instability has been identified in multiple cell lineages in WRN patients, and may be an important determinant of the increased risk of cancer (for review, see Goto et al. 1996; Moser et al. 2000a; van Brabant et al. 2000). Most Werner patients die prematurely, of either cancer or cardiovascular disease, with an average age at death of 47 yr (Epstein et al. 1966; Goto 2000).
The chromosome 8p WRN gene encodes a 162 kD member of the human RecQ helicase family that is unique in possessing an N-terminal 3′ → 5′ exonuclease domain in addition to the central, conserved 3′ → 5′ RecQ helicase consensus domain (for review, see Shen and Loeb 2000b). A majority of WRN patients have mutations in the WRN gene that truncate the WRN protein with loss of a C-terminal nuclear localization signal. Cells containing these mutant alleles produce little or no detectable protein (Goto et al. 1999; Moser et al. 1999, 2000b). There is growing evidence that WRN, like most other helicases, functions as part of a multiprotein complex. Other proteins that may associate or act with WRN include replication protein A (RPA), DNA polymerase δ, PCNA, DNA topoisomerase I, p53 and Ku (for review, see Shen and Loeb 2000b). Mutations in genes encoding these or other WRN-interacting proteins may be responsible for the ∼10% of patients who fulfill clinical and diagnostic criteria for WRN, but lack WRN mutations (Goto et al. 1999; J. Oshima, pers. comm.). One example is a well-documented 29-year-old Japanese female who had clinical and cellular phenotypes that were indistinguishable from WRN patients with truncating WRN mutations, but lacked inactivating WRN mutations (Matsumura et al. 1985; Fukuchi et al. 1989; Prince et al. 1999; see below).
RecQ helicases play a role in recombination or genome stability assurance in many organisms (for review, see Chakraverty and Hickson 1999; Shen and Loeb 2000b; van Brabant et al. 2000). Thus we reasoned that WRN might play a comparable role in human somatic cells, and we explored this possibility by using two different chromosomally integrated mitotic recombination reporter plasmids to measure spontaneous mitotic recombination rates, infer potential recombination pathways and determine the outcome of mitotic recombination events in WRN and control fibroblast cell lines.
Results
The mitotic recombination reporter plasmids pNeoA and pLrec contain direct repeats of genetically inactive neomycin phosphotransferase (neo) or β-galactosidase (lacZ) genes (Figs. 1A, 2A) that can give rise to neo+ or lacZ+ recombinants by gene conversion, by unequal sister chromatid exchange or by intrachromosomal recombination (Meyn 1993). pNeoA-derived mitotic recombinants that are neo+ can be identified by cell growth and colony formation in the presence of G418, whereas pLrec-derived lacZ+ recombinants can be identified in the absence of further cell division by staining for β-galactosidase activity.
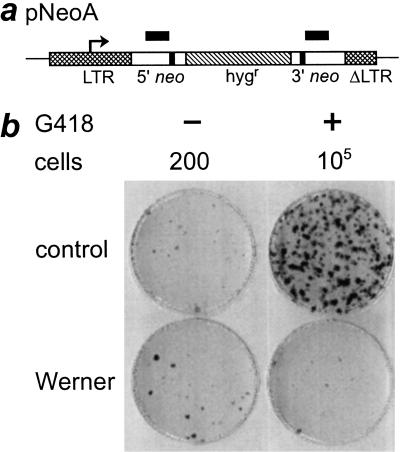
Figure 1.
Reduced recovery of viable pNeoA recombinants in WRN cells. (a) pNeoA contains a direct repeat of bacterial neomycin phosphotransferase alleles (neo; open boxes) where each allele has been inactivated with an 8-bp BamHI linker (filled boxes), separated by a hygromycin resistance gene (hygr; hatched box). neo expression is controlled by a retroviral long terminal repeat (LTR, cross-hatched box; transcription start site, arrow). ΔLTR (cross-hatched box) is a partial LTR copy containing a 299-bp deletion (Meyn 1993). The location and extent of the 592-bp region of perfect sequence identity between the 5′ and 3′ neo alleles is shown by the black bars over the two alleles. (b) Reduced recombination-dependent G418-resistant colony formation in WRN cell lines. Dishes seeded with either 200 or 105 pNeoA-containing cells were grown in the absence or presence of G418, then crystal violet-stained to determine colony forming efficiency (left column) or pNeoA recombinant frequency (right column). Replicate cultures were grown from pools that contained 47 to 486 independent pNeoA clones/cell line. Results shown are from replicate cultures of GM639-500.1 (control) and AG11395-500.2 (Werner). Comparable results were obtained using replicate cultures derived from a second control line (GM-847) and from a WRN cell line (WV1) that lacks WRN protein (additional results not shown).
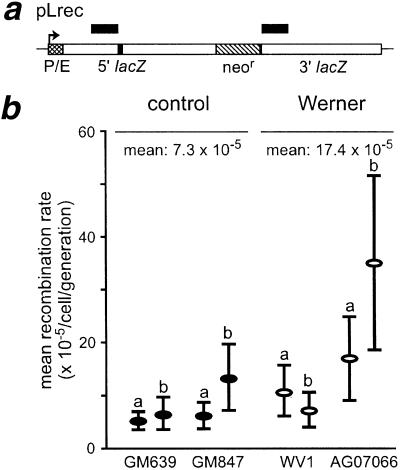
Figure 2.
Mitotic recombination rate of control and WRN pLrec sublines. (a) pLrec contains a direct repeat of bacterial lacZ alleles (open boxes) that have been inactivated by an 8-bp XhoI linker (5′ allele, filled box) or by a 727-bp deletion (3′ allele), separated by the neomycin resistance gene (neor; hatched box), lacZ expression is under SV40 early promoter control (P/E; cross-hatched box). The location and extent of the 693-bp region of perfect sequence identity between the 5′ and 3′ lacZ alleles are shown by the black bars over the two alleles. (b) Mitotic recombination rates for independent control and WRN pLrec sublines that lack (WV1-derived) or retain (AG07066-derived) WRN protein. A total of 273 independent G418-resistant colonies were screened by Southern blot hybridization to identify seven sublines that contained intact single copies of pLrec: control sublines 639L22 (a), 639rec75 (b) and 847L22 (a); and WRN sublines WVP45 (a), WVP46 (b), PSVL30 (a) and 811L45 (b). One additional GM847-derived pLrec subline, LNL1 (GM847b) was kindly provided by M.S. Meyn (Yale University, CT and Hospital for Sick Children, Toronto). Recombination rates were calculated from the fraction of colonies that contained no Lac+ cells (the P0 fraction) using data from 860 colonies/pLrec subline (range 226–1559) where mean colony size was 374 cells (range 229–530). The difference in recombination rate between WRN and control cells was not significant: P = 0.22, with 95% confidence intervals of −9.8, 30.2.
Reduction in viable mitotic recombinants in WRN cells
Experiments performed with replicate cultures from pools of independent, pNeoA-containing WRN or control cells revealed a ≥22.8-fold reduction in the rate of recombination-dependent G418-resistant colony formation in WRN cells (1.6 vs. 36 × 10−5/generation; P = 0.02; Fig. 1). This reduction was observed in two independent WRN cell lines with different WRN-inactivating mutations (WV1 and AG11395; Fig.1 and additional data not shown). Control experiments demonstrated that this reduction in colony formation was recombination-dependent: comparable numbers of G418-resistant colonies could be generated by transfecting WRN or control cell lines with pSV2neo or with a pNeoA variant (pNeoC) having an intact neo+ allele; and G418-resistant WRN and control clones had comparable colony-forming efficiencies when grown in the presence of G418. Blot hybridization analyses indicated that WRN and control clones contained comparable small numbers of integrated pNeoA plasmid DNA molecules (1–3 per cell; additional results not shown).
Recombination initiation appears normal in WRN cells
WRN and control cells containing single integrated copies of pLrec (Fig. 2A) were used to further characterize this defect by determining whether reduced numbers of WRN recombinants reflected a reduced rate of recombination or a failure of recombinant progeny to grow. pLrec allows this distinction to be made because Lac+ recombinants, in contrast to neo+ recombinants, can be unambiguously detected in the absence of cell division. Independent WRN and control pLrec sublines had comparable mitotic recombination rates (17.4 vs. 7.3 × 10−5/cell/generation) that were not significantly different (P = 0.22; Fig. 2B). pLrec clones from the WRN variant cell line AG07066 that expresses WRN protein had higher recombination rates than WRN sublines that lacked protein, although this difference was not statistically significant (P = 0.29).
Limited growth with loss of conversion-type recombinants in WRN
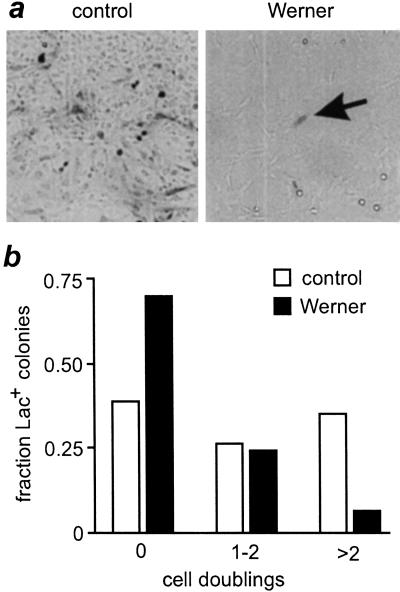
Few WRN-derived Lac+ recombinants in the above experiment went on to divide, in contrast to a majority of the control Lac+ recombinants. This difference was quantified by determining the number of Lac+ cells in 278 Lac+ WRN or control colonies: 70% of Lac+ WRN colonies (89/127) contained single Lac+ cells. Moreover, only 5.5% of WRN Lac+ colonies (7/127) had greater than four Lac+ cells, in contrast to 35% (53/151) of control Lac+ colonies (Fig. 3). This difference in the distribution of Lac+ cell numbers in WRN and control Lac+ colonies, corrected for colony size, was significant (P < 0.001). Control experiments ruled out the possibility that WRN cells were being selectively growth-arrested or killed upon β-galactosidase expression, or that we were observing rare non-recombinant cells that were expressing senescence-associated β-galactosidase activity (additional results not shown).
Figure 3.
Reduced cell division of Lac+ WRN mitotic recombinants. (a) Control and WRN colonies were fixed and stained with X-gal to reveal Lac+ (dark, arrow) cells. A majority of WRN colonies had single Lac+ cells (arrow), in contrast to control Lac+ colonies. (b) Frequency distribution of the number of Lac+ WRN (filled bars) and control (open bars) colonies in which Lac+ recombinant cells had undergone no, 1–2 or >2 cell divisions. Lac+ cells were present in 127/2200 WRN and 151/4684 control colonies.
Molecular analyses of independent Lac+ mitotic recombinants isolated by viable fluorescein-di-β-D-galactopyranoside (FDG) flow sorting revealed a marked reduction in gene conversion-type WRN recombinants: 2 of 9 (22%) WRN-derived Lac+ recombinants were conversion-type recombinants that retained both 5′ and 3′ lacZ alleles, in contrast to the 11 of 16 (69%) Lac+ control recombinants that were conversion-type events. All of the remaining recombinants in both WRN and control cultures (7 of 9 WRN and 5 of 16 control recombinants) contained lacZ+ recombinants that were generated by unequal sister chromatid exchange or by an intrachromosomal ‘popout’ event (additional data not shown). The reduction in WRN conversion-type recombinants was statistically significant (P = 0.03). In contrast, our control data were virtually indistinguishable from comparable, independently generated pLrec data using an overlapping set of control cell lines (Meyn 1993).
Discussion
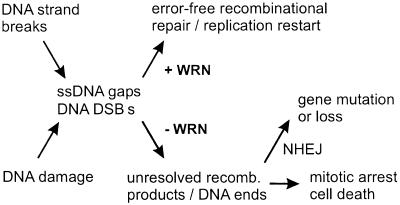
The most direct interpretation of our results is that WRN and control cells initiate mitotic recombination at comparable rates, but WRN cells often fail to resolve recombinant products to yield viable neo+ or Lac+ progeny. Molecular analysis of the subset of WRN mitotic recombinants that retained growth potential (∼10%, Figs. 1 and 3) allowed us to further identify a decrease in WRN conversion-type recombinants. These findings identify a role for WRN in mitotic recombination in human somatic cells, and suggest that WRN recombinants that have lost growth potential are likely to be arrested or dead cells that contain unresolved or disrupted gene conversion products (Fig. 4).
Figure 4.
Model of WRN function. The replication of DNA containing strand breaks or damage can result in stalled replication/synthesis forks, ssDNA gaps or DNA double strand breaks (DSBs) that are substrates for homology- and/or recombination-dependent repair. WRN acts to promote error-free homology-dependent repair or restart to generate viable recombinant products while inhibiting illegitimate recombination or end-joining. In the absence of WRN, unresolved recombination products and DNA ends may lead to checkpoint activation and mitotic arrest or cell death, or to gene rearrangement or loss mediated by nonhomologous DNA end joining (NHEJ).
A second conclusion from our results is that a direct or indirect loss of WRN function confers the same aberrant mitotic recombination phenotype. This finding is consistent with previous work that demonstrated comparable abnormalities in cell growth, genetic instability and drug sensitivity in WRN mutant and WRN variant cell lines that express WRN protein (Matsumura et al. 1985; Fukuchi et al. 1989; Prince et al. 1999). WRN is thus a genetically heterogeneous disease that, in a majority of instances, results from WRN mutations (Moser et al. 1999).
A plausible model for WRN function can be constructed from these results in which WRN functions in vivo as part of a protein complex that branch-migrates or resolves 3- or 4-stranded DNA junctions generated during DNA replication, recombination or DNA repair (Fig. 4). WRN is thus likely to participate in the conservative, recombination-mediated repair of DNA damage during or after S-phase that uses complementary genetic information on the same chromosome or a sister chromatid. WRN could also branch-migrate or unwind—and thus disrupt—illegitimate recombination intermediates in cycling or noncycling cells, to prevent mutations or gene rearrangement.
The WRN protein may perform several of these activities: it has been shown to unwind and branch-migrate model Holliday junctions and related DNA substrates, and can efficiently bind and unwind or degrade a range of non-B DNA structures in vitro (Constantinou et al. 2000; Shen and Loeb 2000a). A more likely scenario is that activities intrinsic to WRN are selectively targeted, augmented or regulated by the interaction of WRN with other proteins form a functional complex (for review, see Shen and Loeb 2000b). The mild clinical phenotype of WRN argues that WRN function may be partially redundant, one of several activities that process or resolve mammalian recombination products (Constantinou et al. 2001). WRN may be required, for example, to deal with only certain types of DNA damage or specific sequence contexts (e.g., tetrahelical DNA or DNA palindromes; Fry and Loeb 1999).
There are several intriguing parallels between our results and work in bacterial and yeast recombination or RecQ mutants. lac recombination substrates generate comparable amounts of β-galactosidase activity from lac recombination substrates, but ∼100-fold fewer viable Lac+ recombinants in recombination-deficient recB− or recC− than in rec+ E.coli. Viable Lac+ recombinants can be recovered in recBCsbcBC mutants that activate the alternate RecFOR pathway that includes bacterial RecQ (Birge and Low 1974). E. coli strains mutant in the RuvABC or RecG Holliday junction resolvases display reduced viability, reduced recombination and enhanced sensitivity to DNA damaging agents. The dependence of these cellular phenotypes upon recombination has been demonstrated by showing, for example, that recA mutations that abolish recombination partially suppress ruv mutant phenotypes (Kowalczykowski et al. 1994; West 1997; Sharples et al. 1999).
Comparable results have been observed in budding and fission yeast that lack RecQ helicases. Budding yeast deficient in the Sgs1p RecQ helicase are hyper-recombinant, DNA damage repair-deficient and display reduced viability after induced recombination (Gangloff et al. 1994; Watt et al. 1996). These phenotypes can be partially suppressed by rad51 mutations that inactivate homologous recombination (Gangloff et al. 2000). Fission yeast that lack the rqhi RecQ helicase are DNA damage-sensitive and display aberrant mitoses (Murray et al. 1997; Stewart et al. 1997). Both mutant phenotypes can be partially suppressed by heterologous expression of a bacterial resolvase protein (Doe et al. 2000).
A defect in mitotic recombination or recombinational repair is an attractive mechanistic explanation for the origin of WRN cellular phenotypes. These include S-phase prolongation, an elevated S-phase arrest fraction, reduced cell proliferation, sensitivity to DNA damaging agents such as 4NQO and camptothecin, cytogenetic abnormalities, and a deletion mutator phenotype (for review, see Fukuchi et al. 1989; Moser et al. 1999; Shen and Loeb 2000b). Failed or inefficient recombinational repair of DNA crosslinks, DNA double strand breaks (DSBs) or stalled or inactivated replication forks could lead to chromosome breakage or loss, gene rearrangement and mutation or cell death (Fig. 4; Thompson and Schild 1999; Michel 2000). Cell viability in the absence of WRN may depend on DNA end joining, which itself may promote additional gene rearrangement or mutation (Kanaar et al. 1998; Karran 2000). A better understanding of WRN function should help to delineate roles for WRN in mitotic recombination and to indicate how a loss of WRN function generates the WRN cellular and clinical phenotype that includes an elevated risk of neoplasia and non-neoplastic disease.
Materials and methods
Cell lines and culture
Werner syndrome patient-derived SV40 fibroblast cell lines included WV1, AG11395 (WS780) and AG07066 (PSV811). WV1 and AG11395 contain WRN mutations and lack detectable WRN protein (Huschtscha et al. 1986; Saito and Moses 1991; Prince et al. 1999; J. Oshima, pers. comm.). AG07066 (PSV811) was derived from a 29-year-old female Japanese WRN patient (Goto et al. 1981; Fujiwara et al. 1985) and displays WRN cellular phenotypes but lacks inactivating WRN mutations (Fukuchi et al. 1989; Marciniak et al. 1998; Prince et al. 1999). Control SV40 fibroblast cell lines GM639 and GM847 were obtained from the NIGMS Human Genetic Mutant Cell Repository (Camden, NJ). All lines were grown in Dulbecco-modified Eagle's medium (DMEM) containing 4500 mg/L glucose (Irvine Scientific, Santa Ana, CA) supplemented with 10% (v/v) fetal bovine serum (FBS; Hyclone Laboratories, Logan, UT), penicillin G sodium (100 units/mL), and streptomycin sulfate (100 μg/mL) in a humidified 37°C, 7% CO2 incubator. A modified CaCl2 transfection protocol was used to introduce plasmid DNA into cell lines (Chen and Okayama 1987).
pNeoA recombination rate determinations
Pools of independent hygromycin-resistant pNeoA colonies were generated from each of two independent WRN or control cell lines by transfection (Fig. 1; Meyn 1993). Replicate cultures were grown from 100–500 cells/pool to ∼2 × 106 cells, then plated in triplicate in 60 mm dishes to determine colony-forming efficiency (200 cells/plate) or the frequency of G418-resistant cells (105 cells/plate in G418 medium). Plates were fed with fresh medium every 5–7 d, and stained with crystal violet after 18 to 21 d to identify and count colonies. The rate of generation of recombination-dependent G418-resistant colonies was calculated by a variant of the serial sampling method of Newcombe (Fukuchi et al. 1989). The statistical significance of pNeoA recombination rate differences was determined by applying Welch's t-test for groups with unequal variance after log transforming data.
pLrec recombination rate determinations
Single copy integrants of pLrec (Fig. 2; Herzing and Meyn 1993) in WRN or control cells were identified by Southern blot analysis of DNA from G418-resistant colonies that had been digested with EcoRV alone or with HindIII and BamHI. A 32P-radiolabeled, 2.95 kb KpnI–DraI fragment of lacZ was used as a probe as described previously (Fukuchi et al. 1989). pLrec sublines were grown in 8-well slides (Nalge Nunc, Rochester, NY) to yield 5 to 25 colonies/well with an average size of 250–500 cells/colony. Two slides were used to determine cells/slide and colonies/slide, respectively, by trypsinization or crystal violet staining and then counting. The remaining 8 slides/subline were fixed with 2% formaldehyde/0.2% glutaraldehyde in PBS, then stained with X-gal to identify Lac+ cells (Meyn 1993). Colonies containing one or more Lac+ cells, and the number of Lac+ cells per colony, were counted by microscopy. Individual colonies were treated as replicate cultures in recombination rate calculations by the P0 method, which does not require the assumption of equal growth potential of parental and recombinant cells (Luria and Delbrück 1943). The statistical significance of recombination rate differences was determined by applying Welch's t-test for equality of means between groups with unequal variances. Confidence intervals were calculated as described by Li et al. (1983). The statistical significance of differences in the distribution of numbers of Lac+ cells in Lac+ WRN and control colonies was determined by a χ2 test for equality of the distributions of Lac+ cells/colony, using quartiles of the combined data to determine the category cutpoints.
Molecular analysis of Lac+ recombinants
The structure of pLrec recombinants was determined by analysis of the 5′ and 3′ pLrec lacZ alleles of flow-sorted Lac+ WRN or control cells as described previously (Meyn 1993). Multiple replicate cultures of WRN and control pLrec sublines were grown from 100 to 5000 cells/subline to ∼107 cells, then loaded with the viable β-galactosidase substrate fluorescein-di-β-D-galacto-pyranoside (FDG; Molecular Probes, Eugene, OR). Lac+ cells were identified and sorted on a FACSstar PLUS cell sorter (Becton Dickinson, San Jose, CA) equipped with a 488 nm argon laser (Coherent, Palo Alto, CA). The most highly fluorescent 0.4% (1st sort) or 3% (2nd sort) of cells were collected and grown to generate colonies that were X-gal stained to verify their Lac+ phenotype prior to molecular analysis. The significance of differences in the number of Lac+ WRN and control cells that contained conversion-type, as opposed to unequal sister chromatid exchange (SCE)/popout-type, recombinant molecules was determined by calculating the exact probability of a 2 × 2 contingency table.
Acknowledgments
This work was supported by grants from the NIA, the NCI and the University of Washington Werner Syndrome Program Project (1 PO1 CA77852) to R.J.M., and by R29 grant CA77607 from the NCI to M.J.E. We thank M.S. Meyn for providing the pLrec and pNeoA recombination plasmids and the LNL1 pLrec subline of GM847; G. Smith for several important practical suggestions; the UW Pathology and Immunology Flow facilities for help in establishing the viable sorting of Lac+ cells; A. Hackmann for help with figures; M. Tewson and L. Rogers for technical help; and A. Blank, B. Brewer, L. Loeb, M. Moser, J. Shen and A. Taylor for comments on the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL monnat@u.washington.edu; FAX (206) 543-3967.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.877001.
References
- Birge EA, Low KB. Detection of transcribable recombination products following conjugation in Rec+, RecB− and RecC− strains of Escherichia coli K12. J Mol Biol. 1974;83:447–457. doi: 10.1016/0022-2836(74)90506-3. [DOI] [PubMed] [Google Scholar]
- Chakraverty RK, Hickson ID. Defending genome integrity during DNA replication: A proposed role for RecQ family helicases. BioEssays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou A, Tarsounas M, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID, West SC. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Reports. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou A, Davies AA, West SC. Branch migration and Holliday junction resolution catalyzed by activities from mammalian cells. Cell. 2001;104:259–269. doi: 10.1016/s0092-8674(01)00210-0. [DOI] [PubMed] [Google Scholar]
- Doe CL, Dixon J, Osman F, Whitby MC. Partial suppression of the fission yeast rqh1− phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 2000;19:2751–2762. doi: 10.1093/emboj/19.11.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner's syndrome: A review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine. 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- Fry M, Loeb LA. Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Kano Y, Ichihashi M, Nakao Y, Matsumura T. Abnormal fibroblast aging and DNA replication in the Werner syndrome. Adv Exp Med Biol. 1985;190:459–477. doi: 10.1007/978-1-4684-7853-2_23. [DOI] [PubMed] [Google Scholar]
- Fukuchi K, Martin GM, Monnat RJ., Jr Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc Natl Acad Sci. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast Type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- Goto M. Werner's syndrome: From clinics to genetics. Clin Exp Rheumatol. 2000;18:760–766. [PubMed] [Google Scholar]
- Goto M, Tanimoto K, Horiuchi Y, Sasazuki T. Family analysis of Werner's syndrome: A survey of 42 Japanese families with a review of the literature. Clin Genet. 1981;19:8–15. doi: 10.1111/j.1399-0004.1981.tb00660.x. [DOI] [PubMed] [Google Scholar]
- Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- Goto M, Yamabe Y, Shiratori M, Okada M, Kawabe T, Matsumoto T, Sugimoto M, Furuichi Y. Immunological diagnosis of Werner syndrome by down-regulated and truncated gene products. Hum Genet. 1999;105:301–307. doi: 10.1007/s004399900151. [DOI] [PubMed] [Google Scholar]
- Herzing LBK, Meyn MS. Novel LacZ-based recombination vectors for mammalian cells. Gene. 1993;137:163–169. doi: 10.1016/0378-1119(93)90002-k. [DOI] [PubMed] [Google Scholar]
- Huschtscha LI, Thompson KVA, Holliday R. The susceptibility of Werner's syndrome and other human skin fibroblasts to SV40-induced transformation and immortalization. Proc R Soc Lond B Biol Sci. 1986;229:1–12. doi: 10.1098/rspb.1986.0070. [DOI] [PubMed] [Google Scholar]
- Kanaar R, Hoeijmakers JHJ, van Gent DC. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- Karran P. DNA double strand break repair in mammalian cells. Curr Opin Genet Dev. 2000;10:144–150. doi: 10.1016/s0959-437x(00)00069-1. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li I-C, Fu J, Hung Y-T, Chu EHY. Estimation of mutation rates in cultured mammalian cells. Mutat Res. 1983;111:253–262. doi: 10.1016/0027-5107(83)90068-4. [DOI] [PubMed] [Google Scholar]
- Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak RA, Lombard DB, Johnson FB, Guarente L. Nucleolar localization of the Werner syndrome protein in human cells. Proc Natl Acad Sci. 1998;95:6887–6892. doi: 10.1073/pnas.95.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura T, Nagata M, Konishi R, Goto M. Studies of SV40-infected Werner syndrome fibroblasts. Adv Exp Med Biol. 1985;190:313–330. doi: 10.1007/978-1-4684-7853-2_15. [DOI] [PubMed] [Google Scholar]
- Meyn MS. High spontaneous intrachromosomal recombination rates in ataxia-telangiectasia. Science. 1993;260:1327–1330. doi: 10.1126/science.8493577. [DOI] [PubMed] [Google Scholar]
- Michel B. Replication fork arrest and DNA recombination. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- Moser MJ, Bigbee WL, Grant SG, Emond MJ, Langlois RG, Jensen RH, Oshima J, Monnat RJ., Jr Genetic instability and hematologic disease risk in Werner syndrome patients and heterozygotes. Cancer Res. 2000a;60:2492–2496. [PubMed] [Google Scholar]
- Moser MJ, Kamath-Loeb AS, Jacob JE, Bennett SE, Oshima J, Monnat RJ., Jr WRN helicase expression in Werner syndrome cell lines. Nucleic Acids Res. 2000b;28:648–654. doi: 10.1093/nar/28.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser MJ, Oshima J, Monnat RJ., Jr WRN mutations in Werner syndrome. Hum Mutat. 1999;13:271–279. doi: 10.1002/(SICI)1098-1004(1999)13:4<271::AID-HUMU2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Murray JM, Lindsay HD, Munday CA, Carr AM. Role of the Schizosaccharomyces pombe RecQ homologue, recombination, and checkpoint genes in UV damage tolerance. Mol Cell Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince PR, Ogburn CE, Moser MJ, Emond MJ, Martin GM, Monnat RJ., Jr Cell fusion corrects the 4-nitroquinoline 1-oxide sensitivity of Werner syndrome fibroblast cell lines. Hum Genet. 1999;105:132–138. doi: 10.1007/s004399900078. [DOI] [PubMed] [Google Scholar]
- Saito H, Moses RE. Immortalization of Werner syndrome and progeria fibroblasts. Exp Cell Res. 1991;192:373–379. doi: 10.1016/0014-4827(91)90054-x. [DOI] [PubMed] [Google Scholar]
- Sharples GJ, Ingleston SM, Lloyd RG. Holliday junction processing in bacteria: Insights from the evolutionary conservation of RuvABC, RecG, and Rus A. J Bacteriol. 1999;181:5543–5550. doi: 10.1128/jb.181.18.5543-5550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J-C, Loeb LA. Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res. 2000a;28:3260–3268. doi: 10.1093/nar/28.17.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J-C, Loeb LA. The Werner syndrome gene: The molecular basis of RecQ helicase-deficiency diseases. Trends Genet. 2000b;16:213–220. doi: 10.1016/s0168-9525(99)01970-8. [DOI] [PubMed] [Google Scholar]
- Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S-phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH, Schild D. The contribution of homologous recombination in preserving genome integrity in mammalian cells. Biochimie. 1999;81:87–105. doi: 10.1016/s0300-9084(99)80042-x. [DOI] [PubMed] [Google Scholar]
- van Brabant AJ, Stan R, Ellis NA. DNA helicases, genomic instability, and human genetic disease. Annu Rev Genomics Hum Genet. 2000;1:409–459. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- Watt PM, Hickson ID, Borts RH, Louis EJ. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–944. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SC. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]