Abstract
In Saccharomyces cerevisiae, gene silencing at the HMR and HML loci is normally dependent on Sir2p, Sir3p, and Sir4p, which are structural components of silenced chromatin. Sir2p is a NAD+-dependent histone deacetylase required for silencing. Silencing can be restored in cells lacking Sir proteins by a dominant mutation in SUM1, which normally acts as a mitotic repressor of meiotic genes. This study found that mutant Sum1-1p, but not wild-type Sum1p, associated directly with HM loci. The origin recognition complex (ORC) was required for Sum1-1p-mediated silencing, and mutations in ORC genes reduced association of Sum1-1p with the HM loci. Sum1-1p-mediated silencing also depended on HST1, a paralog of SIR2. Both Sum1-1p and wild-type Sum1p interacted with Hst1p in coimmunoprecipitation experiments. Therefore, the SUM1-1 mutation did not change the affinity of Sum1p for Hst1p, but rather relocalized Sum1p to the HM loci. Sum1-1–Hst1p action led to hypoacetylation of the nucleosomes at HM loci. Thus, Sum1-1p and Hst1p could substitute for Sir proteins to achieve silencing through formation of a compositionally distinct type of heterochromatin.
Keywords: Position effect, epigenetic, histone deacetylase, meiosis, chromatin immunoprecipitation
Transcriptional repression is as important as activation in determining the expression profile of a genome, yet it is less well understood. Repression mechanisms are often categorized as being either locus specific or regional. In yeast, locus-specific repression is mediated by regulatory sites, typically located in the vicinity of promoters, which are bound by site-specific DNA-binding proteins. These proteins recruit other protein complexes, or corepressors that mediate repression of adjacent genes.
In yeast, the two best-studied corepressors are the Ssn6–Tup1 and the Sin3–Rpd3 complexes. The Sin3–Rpd3 complex (Zhang et al. 1998; Dorland et al. 2000) is recruited to target loci by site-specific DNA-binding proteins, such as Ume6p (Kadosh and Struhl 1997). The binding of Rpd3p results in highly localized deacetylation of histones H3 and H4 (Kadosh and Struhl 1998; Rundlett et al. 1998). Rpd3p homologs in mammals, HDAC1 and HDAC2, are also in large complexes and are recruited to specific sites of repression (for review, see Ayer 1999; Ahringer 2000). Like Sin3–Rpd3, the Ssn6–Tup1 complex is recruited to chromatin by site-specific DNA-binding proteins, such as a1/α2. Tup1p binds to the tails of histones H3 and H4 in vitro (Edmondson et al. 1996) and forms a scaffold along nucleosomes in vivo, thereby stabilizing the positions of nucleosomes in an ordered, repressive conformation (Ducker and Simpson 2000). The Ssn6–Tup1 complex coimmunoprecipitates with Rpd3p and Hos2p, histone deacetylases (Watson et al. 2000), and may also inhibit transcription by direct interaction with the transcription machinery (Lee et al. 2000). For both the Ssn6–Tup1 and Sin3–Rpd3 repressor complexes, repression is a local phenomenon, restricted to individual genes adjacent to each target site (DeRisi et al. 1997).
Silencing has been distinguished operationally from repression by its gene nonspecific nature and its ability to block or reduce gene expression throughout a chromosomal domain, such as at telomeres or at the two HM loci of Saccharomyces. Silencing at the HM loci is mediated by regulatory sites known as the E and I silencers that flank both HMR and HML. These silencers provide binding sites for ORC, Rap1p, and Abf1p, which individually bind to a variety of other sites in the genome that are not subject to silencing. The silencer-binding proteins interact with Sir1p, Sir3p, and Sir4p (Moretti et al. 1994; Triolo and Sternglanz 1996), and it is presumed that a complex containing all four Sir proteins assembles at the silencer. Sir3p and Sir4p interact with histone H3 and H4 tails (Hecht et al. 1995), and this interaction is thought to allow them to spread throughout HMR and HML. Sir2p is presumed to spread with Sir3p and Sir4p, and its histone deacetylase activity results in hypoacetylation of nucleosomes throughout HMR and HML (Braunstein et al. 1993), thereby creating heterochromatin.
Similar silencing mechanisms are used at other loci. Sir2p is also found at telomeres and the rDNA array (Hecht et al. 1996; Gotta et al. 1997), at which its deacetylase activity is required for silencing (Imai et al. 2000). Silencing at telomeres requires SIR3 and SIR4, but silencing at rDNA does not. Whether other proteins serve the roles of Sir3p and Sir4p in rDNA silencing is unknown. The Sir2p family of NAD+-dependent histone deacetylases, including Hst1p–Hst4p, may also mediate other silencing or silencing-like phenomena (Brachmann et al. 1995).
This study describes a repressor protein, Sum1p, which bridges the gap between repression and regional silencing. SUM1 encodes a sequence-specific DNA-binding protein that binds near the promoters of genes required at mid-sporulation (Xie et al. 1999). Binding of Sum1p to these genes results in their repression in mitotic cells, but, as with other repressors, does not repress the neighboring genes (Xie et al. 1999). The SUM1 sequence does not suggest any function, except for a potential AT-hook domain that could mediate DNA binding.
SUM1 was discovered by virtue of the SUM1-1 mutation, which restores transcriptional repression of HMR and HML, but not of MAT, in the absence of Sir proteins (Klar et al 1985; Livi et al. 1990; Laurenson and Rine 1991). Repression is complete at HMR, allowing MATα sir cells to mate, but it is only partial at HML, and MATa sir cells do not mate. SUM1-1 is a dominant mutation (Laurenson and Rine 1991) that creates a protein with an altered function, since neither a null mutation nor overexpression of the wild-type protein results in the SUM1-1 phenotype (Chi and Shore 1996). The SUM1-1 phenotype results from a single amino acid change near the C terminus of the protein (Chi and Shore 1996). Thus, a single mutation in a transcriptional regulator dramatically alters which genes it regulates.
To understand how SUM1-1 achieves silencing in the absence of the Sir proteins, which have been considered essential structural components of heterochromatin, and to understand how a single mutation in a repressor can so dramatically alter its targets, we studied the mechanism of Sum1-1p-mediated silencing.
Results
Sum1-1p acted directly at HM loci
In principle, Sum1-1p could either directly repress transcription at HMR and HML, or it could alter the expression of another protein, which, in turn, leads to Sir-independent silencing. To determine whether Sum1-1p acts directly at HM loci, a chromatin immunoprecipitation assay was developed using an N-terminal myc-tagged allele of Sum1-1p expressed from the SUM1 promoter at the SUM1 locus. To assess the function of the tagged Sum1-1p, its ability to restore silencing at HMR was evaluated in MATα sir2Δ cells. Loss of silencing leads to simultaneous expression of a and α information, thereby disrupting mating ability. Strains containing the tagged and untagged versions of Sum1-1p both mated well (data not shown), indicating that HMR was silenced. In a more sensitive test of repression, both SUM1-1 and myc-SUM1-1 repressed transcription of a1 mRNA to an undetectable level in sir2Δ cells (data not shown). Therefore, the tagged Sum1-1p was fully functional.
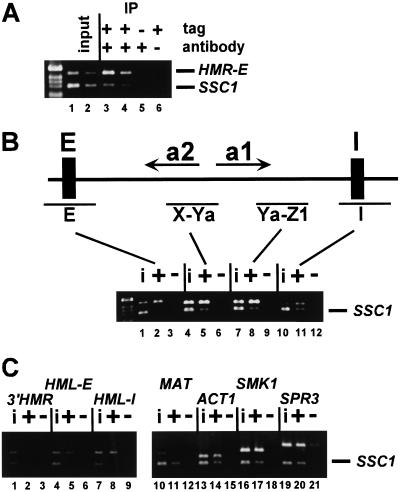
To determine whether Sum1-1p was associated with HM loci, myc-Sum1-1p and associated DNA were immunoprecipitated from crude extracts of formaldehyde cross-linked cells. This DNA was analyzed by simultaneous PCR amplification of the HMR-E silencer region and the promoter region of SSC1, whose transcription is not affected by the SUM1-1 mutation (data not shown) and served as a negative control. To monitor whether the PCR reaction was sensitive to the amount of starting DNA, a second set of PCR reactions was performed on a twofold dilution of the starting material. Sum1-1p was preferentially associated with HMR-E relative to the SSC1 negative control (Fig. 1A, cf. lanes 1,2 and 3,4). Neither HMR nor SSC1 was precipitated from cells lacking the myc tag (lane 5) or in the absence of antibody (lane 6).
Figure 1.
Sum1-1p associated with HM loci. (A) Lysates were prepared from formaldehyde cross-linked MATα haploids cells of the genotypes sir2Δ 7myc–SUM1-1 (lanes 1–4,6; JRY7153) and sir2Δ SUM1-1 (lane 5; JRY7152). DNA immunoprecipitated with anti-myc antibodies was analyzed by simultaneous PCR amplification of the HMR-E silencer region (top) and the SSC1 promoter (bottom). A total of 1/28,000 (lane 1) or 1/56,000 (lane 2) of the input DNA or 1/38 (lanes 3,5,6) or 1/76 (lane 4) of the immunoprecipitated DNA was analyzed. Negative control lanes had samples prepared with an untagged version of Sum1-1p (lane 5) or without antibody in the immunoprecipitation (lane 6). DNA molecular weight marker is 1-kB DNA ladder (GIBCO BRL). (B,C) Chromatin immunoprecipitation was performed as above using MATα haploids of the genotypes sir2Δ 7myc–SUM1-1 (lanes i, +; JRY7153) and sir2Δ SUM1-1 (lanes i; JRY7152) strains. A total of 1/28,000 input DNA (lanes i) or 1/50 of the immunoprecipitated DNA (lanes + and −) was amplified using primers to the indicated regions of HMR (B, top) or the indicated primer pair (C, top) and to the SSC1 promoter region (bottom). Negative control lanes had samples prepared with an untagged version of Sum1-1p (lanes −).
Chromatin silenced by Sir proteins is widely assumed to contain Sir proteins throughout the silenced domain, although they have only been shown to be at one site (Hecht et al. 1996). To determine whether Sum1-1-mediated silencing behaved similarly, the distribution of Sum1-1p across HMR was examined (Fig. 1B). In addition to HMR-E, Sum1-1p also associated with the HMR-I silencer (Fig. 1B, lanes 10–12; cf. ratios of HMR products with SSC1 products in input [i] and immunoprecipitate [+]), and with two regions overlapping the coding sequence of the a1 and a2 genes (X-Ya, lanes 4–6; Ya-Z1, lanes 7–9). Therefore, Sum1-1p was clearly present at multiple sites across the HMR locus.
The size of the region repressed by Sir-mediated silencing is constrained by boundary elements (Bi et al. 1999; Donze et al. 1999), and the Sir proteins are thought to associate only with sequences between these boundaries. Similarly, Sum1-1p was not associated with a region just beyond a boundary element at the right side of HMR-I (Fig. 1C, lanes 1–3). Therefore, Sum1-1p appeared to associate with the HMR locus, much as the Sir proteins are thought to.
Sum1-1p mediates transcriptional repression at HML as well as HMR, but not at MAT (Livi et al. 1990). This phenotype predicted that Sum1-1p should also be found at HML but not at MAT. Sum1-1p associated with both silencers at HML (Fig. 1C, lanes 4–9), and was not associated with MAT (lanes 10–12). Therefore, Sum1-1p was localized to the genes that it silenced, ruling out indirect models in which Sum1-1p altered expression of some other gene(s) that, in turn, caused silencing.
SUM1-1 mutation relocalized Sum1p
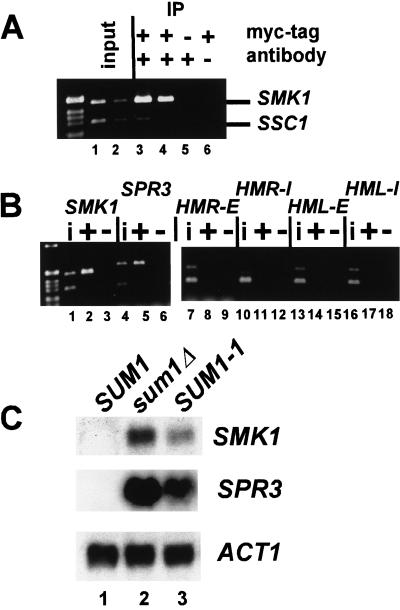
In principle, the SUM1-1 mutation could restore silencing to Sir− strains by relocalizing the Sum1p repressor to HM loci. Alternatively, wild-type Sum1p may normally reside at HM loci, but be unable to cause silencing. To distinguish between these models, the association of wild-type Sum1p with HM loci was examined by chromatin immunoprecipitation by use of a myc-tagged allele of wild-type Sum1p. The function of this tagged Sum1p was assessed by measuring expression of mRNA from SPR3, a gene that is normally repressed by Sum1p (Xie et al. 1999). SPR3 mRNA was derepressed and easily detected in a sum1Δ strain, but was undetectable in both a wild-type strain and the myc-SUM1 strain (data not shown), indicating that the tagged allele of SUM1 was functional.
The SMK1 gene served as a positive control for immunoprecipitation because Sum1p binds to the promoter of SMK1 in mobility shift assays (Xie et al. 1999). Consistent with this result, Sum1p associated with the SMK1 promoter in chromatin immunoprecipitation assays (Fig. 2A, cf. lanes 1,2 and 3,4). Sum1p also associated with the SPR3 promoter, as expected (Fig. 2B, lanes 4–6). In contrast, in the same samples, wild-type Sum1p was not associated with any of the silencers (lanes 7–18). Therefore, the SUM1-1 mutation restored silencing to sir mutant strains by redirecting Sum1p to HM loci.
Figure 2.
Wild-type Sum1p did not associate with HM loci. (A) Chromatin immunoprecipitation was performed as in Fig. 1, using MATα haploids of the genotypes sir2Δ 3myc–SUM1 (lanes 1–4,6; JRY7172) and sir2Δ SUM1 (lane 5; JRY4565). Immunoprecipitated DNA was analyzed by simultaneous PCR amplification of the SMK1 promoter (top) and the SSC1 promoter (bottom). A total of 1/28,000 (lane 1) or 1/56,000 (lane 2) of the input DNA or 1/38 (lanes 3,5,6) or 1/76 (lane 4) of the immunoprecipitated DNA was analyzed. (B) Chromatin immunoprecipitation was performed using sir2Δ 3myc–SUM1 (lanes i, +; JRY7172) and sir2Δ SUM1 (lanes −; JRY4565). A total of 1/19,000 input DNA (lanes i) or 1/50 of the immunoprecipitated DNA (lanes + and −) was amplified using primers to the indicated loci (top) and to the SSC1 promoter region (bottom). (C) RNA from MATα haploids of the genotypes SUM1 (lane 1, W303), sum1Δ (lane 2, JRY7170), and SUM1-1 (lane 3, MC89) was analyzed for SMK1 (top), SPR3 (middle), or ACT1 (bottom) mRNA.
To determine whether the SUM1-1 mutation reduced the association of the mutant protein with wild-type binding sites, or simply expanded the range of sites to which it bound, the association of Sum1-1p with SMK1 and SPR3 was compared with wild-type Sum1p's association. The amount of these loci immunoprecipitated with myc–Sum1-1p was three to sixfold less than was obtained with wild-type myc–Sum1p (Fig. 1C, lanes 16–21 for Sum1-1p and Fig. 2B, lanes 1–6 for Sum1p; cf. fraction of input SMK1 or SPR3 recovered). Therefore, Sum1-1p was only slightly associated with the promoter regions of SMK1 and SPR3, indicating that the mutant protein lost most of its ability to associate with its original site of action.The SUM1-1 mutation resulted in significant derepression of both SMK1 and SPR3 (Fig. 2C, cf. lanes 1 and 3), although these genes were still somewhat repressed compared with a sum1Δ strain (lane 2).
ORC was required for Sum1-1p-mediated repression of HMR
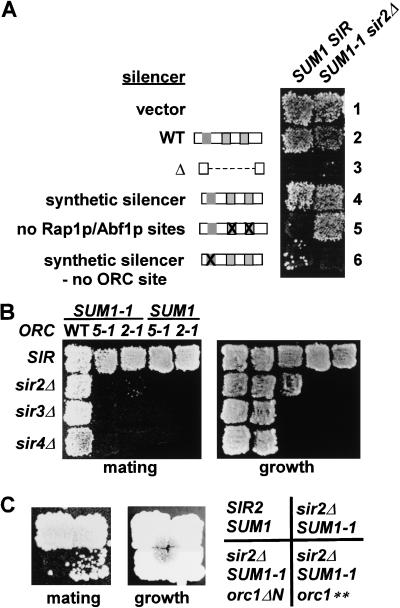
To identify elements required for the association of Sum1-1p with HM loci, sequences required for Sum1-1p-mediated silencing were identified. Plasmids containing the HMRa locus flanked by an intact I silencer and wild-type or mutant E silencers were transformed into MATα strains. Silencing mediated by Sir proteins (Fig. 3A, left) or Sum1-1p (right) was detected by a mating assay that led to growth. Sum1-1p could silence HMR containing a wild-type E silencer (Fig. 3A, row 2), but could not silence it when the E silencer was deleted (Fig. 3A, row 3). Therefore, sequences within the E silencer were required for Sum1-1p-mediated silencing.
Figure 3.
ORC was required for Sum1-1p-mediated silencing. (A) MATα haploids of the genotypes SIR SUM1 (W303) and sir2Δ SUM1-1 (JRY7152) were transformed with a plasmid vector (pRS316, line 1) or the vector containing HMRa (pJR759, line 2), HMRa-eΔ (pJR760, line 3), HMRa-syntetic silencer (pJR900, line 4), HMRa -no Rap1p/Abf1p binding sites (pJR891, line 5), or HMRa-synthteic silencer/no ORC binding site (pJR901, line 6). Representative transformants were tested for their mating ability by replica plating onto an a mating-type tester lawn (TD4) on minimal medium requiring plasmid retention for growth. Mating was indicated by growth of prototrophic diploids. (B) MATα haploids of the genotypes SIR SUM1-1 (MC89), sir2Δ SUM1-1 (JRY7152), sir3Δ SUM1-1 (JRY7161), sir4Δ SUM1-1 (JRY7166), SIR orc5-1 SUM1-1 (JRY7149), sir2Δ orc5-1 SUM1-1 (JRY7157), sir3Δ orc5-1 SUM1-1 (JRY7164), sir4Δ orc5-1 SUM1-1 (JRY7169), SIR orc2-1 SUM1-1 (JRY7150), sir2Δ orc2-1 SUM1-1 (JRY7159), orc5-1 (JRY7146), and orc2-1 (JRY7147) were tested for their mating ability as in A using mating tester JRY2726. (C) MATα haploids of the genotypes SIR SUM1 (W303), sir2Δ myc-SUM1-1 (JRY7153), sir2Δ myc-SUM1-1 orc1Δ1-235 (orc1ΔN, JRY7176), and sir2Δ myc-SUM1-1 ORC1 (Q236H, I237M) (orc1**, JRY7177) were tested for their mating ability as in B.
To determine whether one or more of the three known protein binding sites were important for Sum1-1p silencing, a synthetic silencer, which resembles the wild-type silencer only in the presence and spacing of the ORC, Rap1p, and Abf1p-binding sites (McNally and Rine 1991), was tested. The synthetic silencer supported Sum1-1p-mediated silencing (Fig. 3A, row 4). Therefore, Sum1-1p-mediated silencing depended on one or more of the known binding sites, and not on other sequences within the natural E silencer. Mutation of both the Rap1p and Abf1p-binding sites together had no effect on Sum1-1p-mediated silencing (Fig. 3A, row 5). In contrast, mutation of the ORC-binding site eliminated Sum1-1p-mediated silencing (Fig. 3A, row 6). Therefore, the binding site for ORC, but not for Rap1p or Abf1p, was important for Sum1-1p-mediated silencing.
The requirement for an ORC-binding site for Sum1-1p-mediated silencing implied that ORC itself was required. SUM1-1 suppressed sir silencing defects in the presence of wild-type ORC (Fig. 3B, first column), but silencing was completely abolished by the orc5-1 (second column) or orc2-1 (third column) temperature-sensitive mutations in cells grown at the permissive temperature.
The only known role of ORC in Sir-mediated silencing at HM loci is binding to Sir1p (Fox et al. 1997; Gardner et al. 1999). The N-terminal domain of Orc1p is required for this interaction (Triolo and Sternglanz 1996). Deletion of this domain disrupts Sir-mediated silencing but not the replication function of ORC (Bell et al. 1995). Similarly, the N-terminal domain of Orc1p was also required for Sum1-1p-mediated silencing. The orc1Δ1-235 allele completely eliminated Sum1-1p-mediated silencing (Fig. 3C). In fact, a Q236H I237M mutation, which has no effect on Sir-mediated silencing (Bell et al. 1995), significantly reduced Sum1-1p-mediated silencing (Fig. 3C). Thus, specific residues in the N-terminal portion of Orc1p were required for Sum1-1p-mediated silencing.
Sum1-1p-mediated silencing was more dependent on ORC than was Sir-mediated silencing. In SIR strains bearing wild-type silencers, the orc5-1 and orc2-1 mutations did not noticeably disrupt silencing (Fig. 3B, fourth and fifth columns), presumably due in part to the abilities of Sir3p and Sir4p to bind directly to Rap1p. The greater dependence of Sum1-1p on ORC was consistent with Sum1-1p-mediated silencing being independent of Rap1p and Abf1p-binding sites.
Given the dependence of Sum1-1p on ORC, we tested whether wild-type Sum1p also required ORC for its function. RNA from orc mutants was examined for the expression of SMK1 and SPR3. No derepression was observed under any condition tested (data not shown). Therefore, wild-type Sum1p-mediated repression did not require ORC. The dependence of mutant Sum1-1p on ORC, and the requirement for an ORC-binding site at the HMR locus for repression of that locus, indicated that the SUM1-1 mutation either created or enhanced an affinity of Sum1p for ORC.
The role of ORC in Sum1-1p-mediated silencing
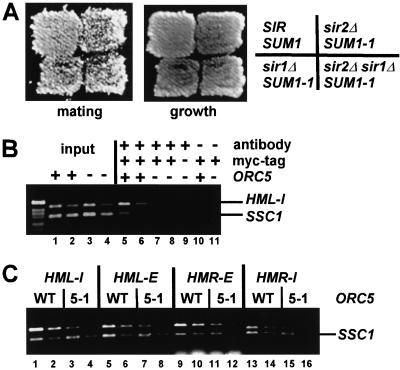
In Sir-mediated silencing, ORC recruits Sir1p (Fox et al. 1997; Gardner et al. 1999), which in turn is required for the establishment of silencing. If ORC plays the same role in Sum1-1p-mediated silencing, then Sir1p should be required for Sum1-1p-mediated silencing. However, mating was not significantly reduced in a MATα SUM1-1 sir1Δ sir2Δ strain (Fig. 4A). Therefore, the role of ORC in Sum1-1p-mediated silencing was not to recruit Sir1p.
Figure 4.
ORC was required for Sum1-1p association with HM loci. (A) MATα haploids of the genotypes SIR SUM1 (W303), sir2Δ SUM1-1 (JRY7152), sir1Δ SUM1-1 (JRY7173), and sir1Δ sir2Δ SUM1-1 (JRY7174) were tested for their mating ability, as in Fig. 3B. (B) Chromatin immunoprecipitation was performed as in Fig. 1, using MATα haploids of the genotypes sir2Δ ORC5 7myc–SUM1-1 (JRY7153), sir2Δ orc5-1 7myc–SUM1-1 (JRY7158), and sir2Δ orc5-1 SUM1-1 (JRY7157). Immunoprecipitated DNA was analyzed by simultaneous PCR amplification of the HML-I silencer (top) and the SSC1 promoter (bottom). A total of 1/23,000 (lanes 1,3) or 1/46,000 (lanes 2,4) of the input DNA or 1/25 (lanes 5,7,9–11) or 1/50 (lanes 6,8) of the immunoprecipitated DNA was analyzed. (C) Chromatin immunoprecipitation was performed using the same strains described in B. A total of 1/25 or 1/50 of the immunoprecipitated DNA was amplified using primers to the indicated loci (top) or to SSC1 (bottom).
ORC might recruit Sum1-1p to HM loci, either through direct contact or indirectly through another protein. Alternatively, ORC may be required to activate Sum1-1p's silencing function after it is recruited to HM loci by some other means. These models were distinguished by testing whether the association of Sum1-1p with HM loci was ORC dependent. Sum1-1p association with the HML-I silencer was reduced, but not totally eliminated, in an orc5-1 strain (Fig. 4B, cf. lanes 5,6 and lanes 7,8). Similar results were observed at the other three silencers (Fig. 4C, cf. ratios of HM product to SSC1 product in ORC5 and orc5-1 samples), and in four other separate immunoprecipitation experiments. The absence of a complete dependence on ORC for Sum1-1p's HM association was likely due, at least in part, to the necessity to use hypomorphic alleles of ORC.
HST1 was required for Sum1-1p-mediated repression
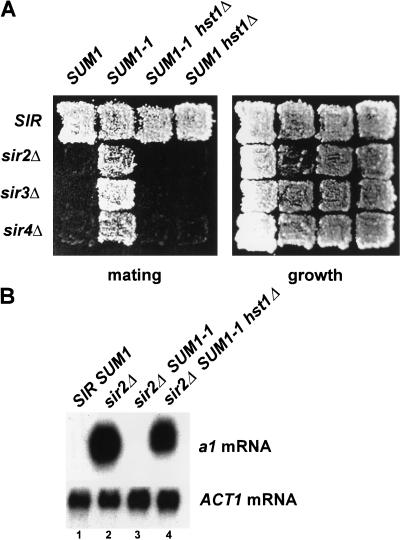
One model for the mechanism of Sum1-1p-mediated silencing is that Sum1-1p brings Sir-like proteins that, with Sum1-1p, substitute for Sir2p, Sir3p, and Sir4p to create a novel form of heterochromatin. The four HST genes are paralogs of SIR2, and thus were candidates for being Sir2p substitutes in Sum1-1p-meditated silencing. Hst1p was the strongest candidate because it is required with SUM1 for repressing a promoter fusion construct regulated by the SMK1 Sum1p-binding site (Xie et al. 1999). Deletion of HST1 eliminated SUM1-1 function (Fig. 5A, cf. second and third columns). Importantly, hst1Δ alone did not affect Sir-mediated silencing or the process of mating itself (fourth column), indicating that the hst1Δ mutation specifically affected Sum1-1p-mediated silencing. Additionally, SUM1-1 silencing measured by transcription of a1 at HMR was also HST1 dependent (Fig. 5B). Therefore, Sum1-1p-mediated silencing required HST1. HST1 was also required for SUM1-1 to suppress sir3Δ and sir4Δ-silencing defects in strains expressing Sir2p (Fig. 5A, third column, third and fourth row). Deletion of HST2, HST3, or HST4 had no effect on Sum1-1p-mediated silencing (data not shown).
Figure 5.
HST1 was required for Sum1-1p-mediated silencing. (A) MATα haploids of the genotypes SIR SUM1 (W303), sir2Δ (JRY4565), sir3Δ (JRY7160), sir4Δ (JRY7165), SIR SUM1-1 (MC89), sir2Δ SUM1-1 (JRY7152), sir3Δ SUM1-1 (JRY7161), sir4Δ SUM1-1 (JRY7166), SIR SUM1-1 hst1Δ (JRY7148), sir2Δ SUM1-1 hst1Δ (JRY7154), sir3Δ SUM1-1 hst1Δ (JRY7162), sir4Δ SUM1-1 hst1Δ (JRY7167), SIR hst1Δ (JRY7145), sir2Δ hst1Δ (JRY7156), sir3Δ hst1Δ (JRY7163), and sir4Δ hst1Δ (JRY7168) were tested for their mating ability as in Fig. 3B. (B) RNA from MATα haploids of the genotypes SIR SUM1 (lane 1, W303), sir2Δ (lane 2, JRY4565), sir2Δ SUM1-1 (lane 3, JRY7152), and sir2Δ SUM1-1 hst1Δ (lane 4, JRY7154) was analyzed for a1 (top) or ACT1 (bottom) mRNA.
Sum1-1p and Hst1p coimmunoprecipitate
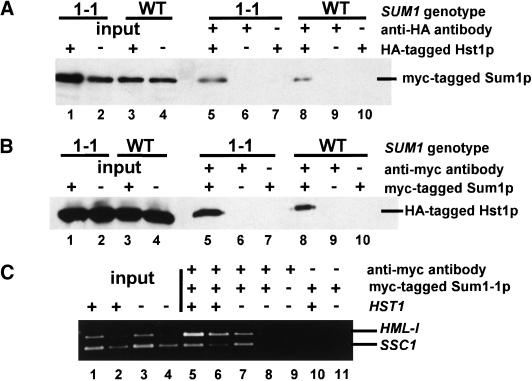
Because Sum1-1p and Hst1p were both required for Sum1-1p-mediated silencing, it was possible that they interacted physically. To test for such an interaction, coimmunoprecipitation experiments were performed by use of epitope-tagged versions of the two proteins. An integrated HA-tagged Hst1p, expressed from the HST1 promoter, was only partially functional, judged by its ability to restore silencing in a sir2Δ SUM1-1 strain compared with wild-type Hst1p (data not shown). However, this HA-tagged Hst1p expressed from a CEN plasmid restored complete silencing (data not shown), indicating that the tagged Hst1p must still interact with any partner necessary for silencing function. Tagged Hst1p was immunoprecipitated from strains containing myc-tagged Sum1-1p, and the immunoprecipitates were evaluated for the presence of Sum1-1p (Fig. 6A). Sum1-1p coprecipitated with HA-tagged Hst1p (Fig. 6A, lane 5). Conversely, Hst1p coprecipitated with myc-tagged Sum1-1p (Fig. 6B, lane 5). Therefore, Sum1-1p and Hst1p were physically associated.
Figure 6.
Hst1p was in complexes with both Sum1-1p and wild-type Sum1p. (A) A MATα haploid of the genotype sir2Δ sum1Δ hst1Δ (JRY7151) was transformed with a plasmid expressing 3myc–Sum1-1p (pJR2291, lanes 1,2,5–7) or 3myc–Sum1p (pJR2292, lanes 3,4,8–10) and a second plasmid expressing Hst1–5HAp (pJR2289, lanes 1,3,5,7,8,10) or Hst1p (pJR2288, lanes 2,4,6,9). Transformed cells were grown in minimal medium requiring maintenance of both plasmids. A total of 1/30 of input (lanes 1–4) or 1/15 of immunoprecipitated samples (lanes 5–10) was analyzed for the presence of myc-Sum1-1p or myc-Sum1p by immunoblotting. Negative control lanes had samples prepared with an untagged version of Hst1p (lanes 6,9) or without antibody in the immunoprecipitation (lanes 7,10). (B) A MATα haploid of the genotype sir2Δ sum1Δ hst1Δ (JRY7151) was transformed with a plasmid expressing Hst1–5HAp (pJR2289, all lanes) and a second plasmid expressing 3myc–Sum1-1p (pJR2291, lanes 1,5,7), Sum1-1p (pJR2293, lanes 2,6), 3myc–Sum1p (pJR2292, lanes 3,8,10) or Sum1p (pJR2294, lanes 4,9). A total of 1/30 of input (lanes 1–4) or 1/15 of immunoprecipitated samples (lanes 5–10) was analyzed for the presence of Hst1–5HAp by immunoblotting. (C) Chromatin immunoprecipitations used MATα haploids of the genotypes sir2Δ HST1 7myc–SUM1-1 (JRY7153), sir2Δ hst1Δ 7myc–SUM1-1 (JRY7178), and sir2Δ hst1Δ SUM1-1 (JRY7157). A total of 1/23,000 (lanes 1,3) or 1/46,000 (lanes 2,4) of the input DNA or 1/25 (lanes 5,7,9–11) or 1/50 (lanes 6,8) of the immunoprecipitated DNA was analyzed.
To determine whether the SUM1-1 mutation changed the affinity of Sum1p for Hst1p, coimmunoprecipitation experiments were also conducted by use of a tagged version of wild-type Sum1p. Sum1p coimmunoprecipitated with tagged Hst1p (Fig. 6A, lane 8), and Hst1p coimmunoprecipitated with tagged Sum1p (Fig. 6B, lane 8). In both cases, the amount of material precipitated was comparable with the amount precipitated with mutant Sum1-1p. Therefore, the change in function resulting from the SUM1-1 mutation was probably not due to a change in affinity for the Hst1p corepressor.
Because Sum1-1p was bound to Hst1p and also associated with HM loci, it was likely that Hst1p was also associated with HM loci. Additionally, HST1 was required with SUM-1 for hypoacetylation of HM loci in sir2Δ strains, as described below, again suggesting that Hst1p was associated with HM loci. However, we were unable to preferentially precipitate any DNA known to associate with either Sum1-1p or Sum1p with antibodies against HA-tagged Hst1p (data not shown). Perhaps Hst1p does not cross-link well, or is too indirectly associated with the DNA to be detected by this assay. Alternatively, Hst1p may be only transiently associated with DNA.
The presence of Sum1-1p, but not Sum1p, at HM loci implied that Sum1-1p was responsible for bringing Hst1p to HM loci, and not the other way around. However, it was still possible that Hst1p itself contributed to the presence Sum1-1p at HM loci. In fact, Sum1-1p association with HMR was slightly reduced in an hst1Δ strain, although to a lesser extent than in orc mutants (Fig. 6C). Hst1p may stabilize the association of Sum1-1p with HMR, perhaps by providing another binding surface.
Sum1-1p-mediated repression resulted in histone deacetylation
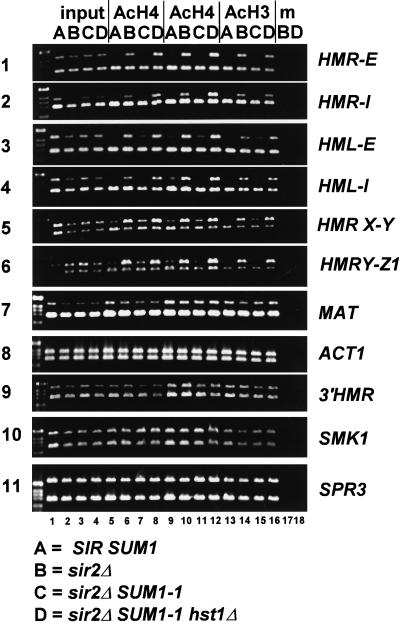
Sir2 family members, including Hst1p, are NAD+-dependent histone deacetylases (Imai et al. 2000; Landry et al. 2000; Smith et al. 2000). Nucleosomes at silenced HM loci are hypoacetylated, particularly in response to Sir2p overexpression (Braunstein et al. 1993). The role of Hst1p in Sum1-1p-mediated repression (Fig. 5A,B) implied that histones H3 and H4 at HM loci would be hypoacetylated in a SUM1-1 mutant. Therefore, chromatin immunoprecipitation assays using antibodies against acetylated histones were performed to test this hypothesis. As expected, all four silencers (Fig. 7, panels 1–4) and the a1–a2 coding region (Fig. 7, panels 5,6) were under-represented in the immunoprecipitated material from the SIR strain (Fig. 7, lanes A), but not from the sir2Δ strain (Fig. 7, lanes B). In contrast, MAT and ACT1 (Fig. 7, panels 7,8), nonsilenced loci, were nearly equally abundant in both samples. Therefore, the HM loci were underacetylated in the SIR strain, as expected. In a parallel experiment using a sir2Δ SUM1-1 strain (Fig. 7, lanes C), all four silencers (Fig. 7, panels 1–4) and the a1–a2 coding region (Fig. 7, panels 5,6) were hypoacetylated, whereas MAT and ACT1 (Fig. 7, panels 7,8) were not. Therefore, Sum1-1p-mediated silencing resulted in hypoacetylation of the histone tails at both HM loci.
Figure 7.
Sum1-1p-mediated silencing was associated with histone deacetylation. Chromatin immunoprecipitations used MATα haploids of the genotypes SIR SUM1 (lanes A, W303), sir2Δ (lanes B, JRY4565), sir2Δ SUM1-1 (lanes C, JRY7152), and sir2Δ SUM1-1 hst1Δ (lanes D, JRY7154). DNA immunoprecipitated with one of two different anti-acetyl-histone H4 antibodies (AcH4, lanes 5–8,9–12) or anti-acetyl-histone H3 antibody (AcH3, lanes 13–16) was analyzed by simultaneous PCR amplification of the indicated region (top) and the SSC1 promoter (bottom). A total of 1/25,000 of the input DNA (lanes 1–4) and 1/50 (lanes 5–8,17,18) or 1/250 (lanes 9–16) of the immunoprecipitated DNA was analyzed. Negative control lanes had samples prepared without antibody in the immunoprecipitation (lanes 17,18).
To determine whether this deacetylation was dependent on HST1, a sir2Δ SUM1-1 hst1Δ strain (Fig. 7,lanes D) was examined. In contrast to the sir2Δ SUM1-1 strain, all four silencers and the a1–a2 coding region were acetylated to an extent comparable with the control loci, indicating that HST1 was required for the Sum1-1p-mediated deacetylation of HM loci. This result was consistent with Hst1p being the responsible histone deacetylase. Thus, both Sir2p and Hst1p acted through a common mechanism to generate heterochromatin.
If nucleosome deacetylation were dependent on association of Sum1-1p and Hst1p with chromatin, then deactylated nucleosomes, like Sum1-1p itself, should not extend beyond a boundary element at HMR. The chromatin distal to the boundary on the I-silencer side was equally acetylated in all strains (Fig. 7, panel 9). Therefore, within the resolution of the experiment, nucleosome deacetylation correlated with the presence of Sum1-1p.
Wild-type Sum1p also interacted with Hst1p (Fig. 6; Xie et al. 1999). Therefore, it was possible that nucleosomes at the promoters of genes repressed by wild-type Sum1p were also deacetylated. To test this hypothesis, the immunoprecipitates were examined for SMK1 or SPR3 promoter regions. If Sum1p–Hst1p-dependent deacetylation occurred, it would be greatest in SUM1 strains (Fig. 7, lanes A,B), reduced in a mutant SUM1-1 strain (Fig. 7, lanes C), and not seen when the Hst1p deacetylase is absent (Fig. 7, lanes D). However, SMK1 and SPR3 promoters (Fig. 7, panels 10,11) were equally well represented in all four strains, indicating that wide-spread histone deacetylation did not occur at these promoters.
SUM1-1 slow growth was dependent on HST1 and ORC
Consistent with previous reports (Chi and Shore 1996), we found that SUM1-1 cells divided slowly, with a doubling time between 3 and 3.5 h at 23°, compared with wild-type cells, which doubled approximately every 2 h. This slow growth was not due to silencing of the HM loci, which are also silenced in wild-type strains, or to the loss of wild-type SUM1 function, because sum1Δ strains doubled every 2 h. Sum1-1p may affect the growth rate by partially silencing one or more critical genes. Alternatively, Sum1-1p might interfere with DNA replication, through interaction with ORC. Interestingly, HST1 and ORC, which are both required for Sum1-1p-mediated silencing at HM loci, were required for this slow growth. SUM1-1 orc and SUM1-1 hst1 strains doubled every 2 to 2.5 h. Thus, Sum1-1p, Hst1p, and ORC may act together at other loci.
Discussion
The mechanism of Sum1-1p-mediated silencing
This study has established the mechanism of action of the enigmatic suppressor mutation, SUM1-1. The paradox of the SUM1-1 mutation has been its ability to restore silencing in the absence of Sir proteins, which are key structural components of heterochromatin. We discovered that the SUM1-1 mutation altered Sum1p in a manner that led to the formation of a compositionally distinct form of heterochromatin, in which Sum1-1p, Hst1p, and perhaps other proteins substitute for Sir proteins. Sum1-1p was associated with multiple sites within HMR and HML (Fig. 1B,C), and therefore, acted directly at those loci, where wild-type Sum1p does not act. Sum1-1p required ORC (Fig. 3) and HST1 (Fig. 5) for silencing. ORC was required for association of Sum1-1p with HM loci (Fig. 4B,C). Hst1p was physically associated with Sum1-1p (Fig. 6) and was required for Sum1-1p-associated deacetylation of histones at HM loci (Fig. 7). These data inspired a model for how Sum1-1p causes silencing. In this model, Sum1-1p, in partnership with Hst1p and perhaps other proteins, associates with ORC at silencers. The deacetylase activity of Hst1p then leads to hypoacetylation of nucleosomes at HM loci.
This study also explored how the SUM1-1 mutation changes the action of Sum1p. The mutation altered the localization of the protein, thereby reducing repression of wild-type Sum1p targets and causing new repression at HM loci (Figs. 1 and 2). It did not change the affinity of Sum1p for Hst1p (Fig. 6), but did result in a new requirement for ORC (Fig. 3; data not shown). Perhaps the mutation, a threonine to isoleucine change in a locally hydrophobic region (Chi and Shore 1996), creates an especially hydrophobic sticky patch, analogous to the sickle cell anemia allele, and this hydrophobic patch creates an affinity for ORC. Alternatively, the mutation could reduce site-specific DNA binding, thereby permitting the protein to associate with other, lower-affinity sites. Thus, the SUM1-1 mutation either created or exposed an affinity for ORC, which in turn directed the protein to new loci.
Although wild-type Sum1p binds DNA directly (Xie et al. 1999), several lines of evidence favored models in which ORC was required for Sum1-1p association with HM loci through a physical link between the two proteins. First, Sum1-1p did not associate with HM loci through the wild-type Sum1p-binding site, because Sum1p was not found at HM loci (Fig. 2B). Moreover, Sum1-1p had reduced association with promoters containing Sum1p-binding sites (Fig. 1C; M. Pierce and A. Vershon, pers. comm.). Second, ORC was required for Sum1-1p-mediated silencing (Fig. 3) and contributed to association of Sum1-1p with HM loci (Fig. 4B,C). Therefore, Sum1-1p was likely targeted to HM loci through ORC.
Sum1-1p-mediated silencing also required HST1. Hst1p was physically associated with Sum1-1p (Fig. 6A,B), which in turn was associated with HM loci (Fig. 1B,C). In addition, Hst1p, a histone deacetylase, together with Sum1-1p were required for hypoacetylation of HM loci in sir2Δ strains (Fig. 7), implying that Hst1p acted at, and therefore was associated with, HM loci. Hst1p did not recruit Sum1-1p to HM loci, because wild-type Sum1p was also physically associated with Hst1p (Fig. 6A,B) but was not found at HM loci (Fig. 2B). Therefore, Hst1p was brought to the HM loci by Sum1-1p and acted as a cosilencer through a mechanism similar to that of the Rpd3p corepressor.
The mechanism of wild-type Sum1p repression
Comparison of Sum1-1p-mediated silencing and Sum1p-mediated repression indicated that the SUM1-1 mutation changed the way Sum1p functions. For example, ORC appeared not to play a role in repression by wild-type Sum1p because genes repressed by Sum1p were not derepressed in orc mutant strains (data not shown). However, these experiments required use of hypomorphic rather than null alleles of ORC genes. Therefore, the possibility of a role for ORC in Sum1p function could not be completely excluded.
Another difference between wild-type Sum1p-mediated repression and Sum1-1p-mediated silencing was the extent of hypoacetylation of nucleosomes at repressed loci. This study did not detect hypoacetylation of the promoter regions of SMK1 and SPR3. Either Sum1p repression at these loci did not involve stable histone deacetylation, or the region of deacetylation was short or dispersed, and thus was not detected by this assay. In contrast, the hypoacetylation caused by SUM1-1 was evident over a region spanning several kilobases.
The roles of ORC and Hst1p in other types of repression
The requirement for ORC in Sum1-1p-mediated silencing may reflect a fundamental role of ORC in multiple chromatin structures. In yeast, the N-terminal portion of Orc1p is required for Sir-mediated silencing but is dispensable for replication (Bell et al. 1995). Deletion of the Orc1p N-terminal domain also disrupted Sum1-1p-mediated silencing (Fig. 3C). This domain interacts with Sir1p (Triolo and Sternglanz 1996) and is critical in recruiting Sir1p to the silencer (Gardner et al. 1999). Similarly, in Drosophila, mutations in ORC1 affect position and variegation, and the N terminus of Orc1p recruits HP1 to heterochromatin (Pak et al. 1997). Together, these results suggest a general role for ORC in heterochromatin and imply that the N-terminal portion of Orc1p has a special silencing function. Perhaps the silencing proteins recruited by Orc1p, including HP1, Sir1p, and Sum1-1p, share a common structural motif.
The role of HST1 in Sum1-1p-mediated silencing has implications for the roles of HST2-4 and other SIR2 orthologs. Much as Hst1p, a histone deacetylase, was brought to HM loci by Sum1-1p, the Sir2p family of proteins may generally act as cosilencers recruited to chromosomal loci by adapter proteins analogous, but not necessarily homologous, to Sum1-1p. Once at these loci, they would act enzymatically to deacetylate histone tails, thereby altering chromatin structure. Given that the Sir2 family has five members in yeast, at least seven members in humans, and orthologs in essentially all genomes analyzed, recruitment of these proteins by repressors may be a common theme in gene silencing.
Other sites and types of repression
There are interesting parallels and differences between the mechanisms of Sir-mediated silencing and Sum1-1p-mediated silencing. Both require the HMR-E silencer. However, Sir-mediated silencing utilizes three silencer-bound proteins, whereas Sum1-1p-mediated silencing requires ORC but not the binding of Rap1p or Abf1p. In both cases, the silencer-bound proteins appear to act by recruiting silencing proteins, either the Sir proteins or Sum1-1p and Hst1p. These complexes, containing either the Sir2p or Hst1p deacetylase, then appear to associate with multiple sites throughout the HM loci, resulting in the deacetylation of nucleosomes. The requirement for a deacetylase to act in the place of Sir2p during Sum1-1p-mediated silencing strengthens the link between hypoacetylation and repression. Whether Sum1-1p carries out the functions of both Sir3p and Sir4p or other as yet unknown analogous proteins associate with Sum1-1p and Hst1p remains to be determined. Therefore, for both Sum1-1p and Sir-mediated silencing, the requirement for silencer-binding proteins and the involvement of deacetylated nucleosomes were the same, but the proteins that create and ultimately constitute the heterochromatin differed.
Sir-mediated and Sum1-1p-mediated silencing represent two separate silencing routes that can be, but are not always, independent. The results of this and other studies indicate that, in some cases, Sum1-1p interacts with the Sir proteins. In a SIR SUM1-1 strain, a synthetic silencer bearing a mutation of the ORC-binding site is silenced (Laurenson and Rine 1991). In contrast, in this study, the same silencer was not silenced in a sir2Δ SUM1-1 strain (Fig. 3A). This lack of silencing was also observed in a sir3Δ SUM1-1 strain (data not shown). Thus, in the original experiment, the importance of the ORC-binding site for Sum1-1p-mediated silencing was masked by the presence of Sir proteins. Similarly, SIR2 is also required for SUM1-1 to suppress the silencing defect of a strain bearing both rap1-12 and a deletion of the ORC-binding site at HMR (Chi and Shore 1996). Therefore, it appears that when both Sir proteins and Sum1-1p are present, the two silencing routes can jointly silence a locus that neither could silence alone.
The interaction of Sum1-1p with Sir proteins and the requirement for ORC in Sum1-1p-mediated silencing explained the puzzling observation that SUM1-1 does not suppress a sir2Δ defect in telomeric silencing but does enhance telomeric silencing in a SIR strain (Chi and Shore 1996). Like HM silencing, telomeric silencing requires Sir2p, Sir3p, and Sir4p. However, synthetic telomeres that can initiate silencing lack ORC-binding sites, consisting instead of polymers of Rap1p-binding sites. Because Rap1p contributed little if anything to Sum1-1p silencing at HMR, whereas ORC was of central importance, it would appear that in the absence of bound ORC at telomeres, Sum1-1p would have no way of associating with the telomeres to form heterochromatin in a sir2Δ mutant. In contrast, Sum1-1p could interact with Sir proteins at telomeres to enhance silencing.
The slow growth phenotype of SUM1-1 strains and the dependence of this slow growth on ORC and HST1 suggested that Sum1-1p-mediated silencing might occur at other loci. Genes located near ORC-binding sites are good candidates to be repressed by Sum1-1p, because the presence of ORC appears to be required for association of Sum1-1p with a locus. However, only a subset of the ∼400 ORC-binding sites at origins of replication could be sites of Sum1-1p-mediated repression because the cells are viable. Therefore, some origins must have additional features that predispose them to be silenced.
Mechanistic links between repression and silencing
This study points to similarities in the mechanisms of promoter-local repression and long-range silencing. Sum1p is a repressor that acts directly at the SMK1, SPR3, and other promoters, and its effect does not spread to a nearby, divergently transcribed gene (Xie et al. 1999). On the other hand, Sum1-1p causes silencing at a distance through the formation of heterochromatin. Thus, two nearly identical proteins act through mechanisms that have sometimes been considered completely different. This narrow margin separating repression from silencing is perhaps the most striking finding of this study. Other clear similarities among repression and silencing mechanisms are emerging. For example, Ssn6–Tup1 associates with histone tails and forms a scaffold along nucleosomes (Edmondson et al. 1996; Ducker and Simpson 2000), much as the Sir proteins are thought to do (Hecht et al. 1996). Additionally, Rpd3–Sin3 repression, Ssn6–Tup1 repression, Sir-mediated silencing, and Sum1-1p-mediated silencing all use histone deacetylases recruited by DNA-binding proteins.
The chromosomal context in which the Sum1 protein finds itself may determine its range of action. For example, the presence or absence of nearby boundary elements could determine the extent of spreading. Perhaps genes like SMK1 have boundary elements that limit the extent of deacetylation. Alternatively, elements like the HMR-E silencer may have features that promote the binding and subsequent movement or spreading of Sum1-1p and Hst1p.
Materials and methods
Plasmids and epitope tagging
Plasmids pRS316, pRS412, and pRS416 (Sikorski and Hieter 1989) and pJR759, pJR760, pJR891, pJR900, and pJR901 (Laurenson and Rine 1991) were described previously. Table 1 lists plasmids generated for this study. SUM1-1 and SUM1 plasmids were derived from plasmids DMC283 and DMC326, respectively (Chi and Shore 1996). To generate myc-tagged alleles of SUM1 or SUM1-1, the first three codons of SUM1 were altered by site-specific mutagenesis to ATGCCTAGG, changing amino acids 2,3 from S,E to P,R, and creating an AvrII site at the 5′ end of the ORF. A multimerized myc tag flanked by AvrII sites was ligated into this site. To generate pJR2291–pJR2294, the coding region was excised with ApaI and SacI and ligated into these sites of pRS412. HST1 plasmids were derived from pLP316 (Brachmann et al. 1995). The SacI fragment containing HST1 was ligated into the SacI site of pRS416 or pRS406. To generate an HA-tagged allele of HST1, the last three codons of HST1 were altered by site-specific mutagenesis to CACCGGTAA, changing the last two amino acids from Q,Q to H,R, and creating an AgeI site at the 3′ end of the ORF. A multimerized HA tag flanked by AgeI sites was ligated into this site.
Table 1.
Plasmids generated for this study
| Plasmid
|
Description
|
|---|---|
| pJR2288 | pRS416 HST1 |
| pJR2289 | pRS416 HST1-5HA |
| pJR2291 | pRS412 3myc-SUM1-1 |
| pJR2292 | pRS412 3myc-SUM1 |
| pJR2293 | pRS412 SUM1-1 |
| pJR2294 | pRS412 SUM1 |
| pJR2295 | pRS415 7myc-SUM1-1 |
| pJR2296 | pRS415 3myc-SUM1 |
Yeast strains and construction
Strains used in this study were all derived from W303-1a (Table 2). The hst1Δ∷KanMX null allele was generated by one-step gene conversion (Rothstein 1983), completely deleting the HST1 ORF. The KanMX gene was amplified from plasmid pFA6a–KanMX4 (Wach et al. 1994) by use of primers 5′-TACGAACACTTCTCTTCTTTTTTGTTGTTTTTG TGAGAAAACGTACGCTGCAGGTCGAC-3′ and 5′-GCGGTATACTTATTTTTACTCCCCCTTCTGTGTTTTCTTCATCGATGAATTCGAGCTCG-3′, in which the underlined portion was complementary to the plasmid and the remainder was complementary to the yeast sequence flanking HST1. The PCR product was transformed into yeast by use of the modified LiOAC method (Schiestl and Gietz 1989) and geneticin-resistant colonies were selected. Gene disruption was confirmed by use of PCR analysis and DNA blotting. The sum1Δ∷LEU2 allele was created similarly, except the LEU2 gene was amplified from plasmid pRS415 by use of primers 5′-ATCAAACGAAAAGTTTCATACATAATTAACAAAATTCGTTTGTTGCGGGGGATTGTACTGAGAGTGCACC-3′ and 5′-TTTTTCATTTTTTTATCTATTCTCGAAACTGCCCCAACGTACGGACCAGCCTGTGCGGTATTTCACACCG-3′. The 7myc–SUM1-1 and 3myc–SUM1 alleles were generated by transforming DNA containing the complete ORF with tag plus 5′ and 3′ flanking sequences (from plasmids pJR2295 and pJR2296) into a sum1Δ∷URA3 strain and selecting for 5-FOA-resistant colonies. The orc5-1 and orc2-1 alleles (Foss et al. 1993; Loo et al. 1995) and LEU2∷ORC1(Q236H,I237M) and LEU2∷orc1Δ1-235 were described previously (Bell et al. 1995). The sir1Δ∷LEU2, sir2Δ∷TRP1, sir2Δ∷HIS3, sir3Δ∷LEU2, and sir4Δ∷HIS3 alleles were complete deletions of the ORFs generated by one-step gene conversion.
Table 2.
Strains used in this study
| Strain
|
Genotype
|
Sourcea
|
|---|---|---|
| W303-1b | MATα ade2-1 can1-100 his3-11 leu2-3, 112 trpl-1 ura3-1 | R. Rothstein |
| MC89 | W303 MATα SUM1-1 | D. Shore |
| TD4 | MATacan1 his4-519 leu2-3,112 trpl ura3-52 | G. Fink |
| JRY2726 | MATahis4 | P. Schatz |
| JRY4565 | W303 MATα sir2Δ | |
| JRY7145 | W303 MATα hst1Δ::KanMX | |
| JRY7146 | W303 MATα orc5-1 | |
| JRY7147 | W303 MATα orc2-1 | |
| JRY7148 | W303 MATα SUM1-1 hst1Δ::KanMX | |
| JRY7149 | W303 MATα SUM1-1 orc5-1 | |
| JRY7150 | W303 MATα SUM1-1 orc2-1 | |
| JRY7151 | W303 MATα sir2Δ::TRP1 hstlΔ::KanMX sumlΔ::LEU2 | |
| JRY7152 | W303 MATα sir2Δ::TRP1 SUM1-1 | |
| JRY7153 | W303 MATα sir2Δ::HIS3 7myc-SUM1-1 | |
| JRY7154 | W303 MATα sir2Δ::TRP1 SUM1-1 hstlΔ::KanMX | |
| JRY7156 | W303 MATα sir2Δ::TRP1 hst1Δ::KanMX | |
| JRY7157 | W303 MATα sir2Δ::TRP1 SUM1-1 orc5-1 | |
| JRY7158 | W303 MATα sir2Δ::HIS3 7myc-SUM1-1 orc5-1 | |
| JRY7159 | W303 MATα sirΔ::TRP1 SUM1-1 orc2-1 | |
| JRY7160 | W303 MATα sir3Δ::LEU2 | |
| JRY7161 | W303 MATα sir3Δ::LEU2 SUM1-1 | |
| JRY7162 | W303 MATα sir3Δ::LEU2 SUM1-1 hst1Δ::KanMX | |
| JRY7163 | W303 MATα sir3Δ::LEU2 hstlΔ::KanMX | |
| JRY7164 | W303 MATα sir3Δ::LEU2 SUM1-1 orc5-1 | |
| JRY7165 | W303 MATα sir4Δ::HIS3 | |
| JRY7166 | W303 MATα sir4Δ::HIS3 SUM1-1 | |
| JRY7167 | W303 MATα sir4Δ::HIS3 SUM1-1 hstlΔ::KanMX | |
| JRY7168 | W303 MATα sir4Δ::HIS3 hstlΔ::KanMX | |
| JRY7169 | W303 MATα sir4Δ::HIS3 SUM1-1 orc5-1 | |
| JRY7170 | W303 MATα sum1Δ::LEU2 | |
| JRY7172 | W303 MATα sir2Δ::HIS3 3myc-SUM1 | |
| JRY7173 | W303 MATα sirlΔ::LEU2 SUM1-1 | |
| JRY7174 | W303 MATα sirlΔ::LEU2 sir2Δ::TRP1 SUM1-1 | |
| JRY7176 | W303 MATα sir2Δ::HIS3 7myc-SUM1-1 orclΔ::TRP1 LEU2::orclΔl-235 | |
| JRY7177 | W303 MATα sir2Δ::HIS3 7myc-SUM1-1 orclΔ::TRP1 LEU2:ORC1(Q236H,I237M) | |
| JRY7178 | W303 MATα sir2Δ::HIS3 7myc-SUM1-1 hstlΔ::KanMX |
Unless otherwise indicated, all strains were produced for this study.
Mating assays
For mating assays, cells were patched on solid YPD medium (or minimal medium without uracil to retain plasmids) and grown 8–16 h before replica plating to mating-type tester lawns (JRY2726 unless otherwise indicated) on minimal medium. Plates were incubated at 30°C unless otherwise indicated and photographed after 2 or 3 d.
Immunoprecipitation techniques
For protein coimmunoprecipitation, 30 OD units cells were lysed by vortexing with glass beads in 100 μL of ice-cold Co-IP lysis buffer (50 mM Tris at pH 7.5, 5 mM EDTA, 250 mM NaCl, 0.1% NP40, 1.5 mM DTT, 0.1 mM TPCK, 1 mM PMSF, 5 μg/mL chymostatin, 2 μg/mL pepstatin A, 1 mM benzamidine, and 1× complete protease inhibitors; Boehringer-Mannheim). The volume was then increased to 500 μL with Co-IP lysis buffer, and the lysate was drawn off the beads and then spun 10 min in a microfuge at 4°. The supernatant fraction was collected, and 100 μL was used in a final volume of 400 μL Co-IP lysis buffer for each immunoprecipitation reaction. The samples were incubated at 4°C with 5 μL of rabbit polyclonal IgG antibodies (Upstate Biotech) for 3 h and then 60 μL of Protein A agarose beads for 1 h. The beads were washed twice in Co-IP lysis buffer, and proteins were eluted in SDS-PAGE sample buffer. Samples were electrophoretically fractionated on 7.5% polyacrylamide–SDS gels, transferred to membranes, and probed by use of mouse monoclonal antibodies (BAbCO).
For chromatin immunoprecipitations, cells were cross-linked with 1% formaldehyde for 20 min; 50 OD units cells were lysed by vortexing with glass beads in 100 μL ice-cold ChIP lysis buffer (50 mM HEPES at pH 7.5, 1 mM EDTA, 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1 mM TPCK, 1 mM PMSF, 5 μg/mL chymostatin, 2 μg/mL pepstatin A, 1 mM benzamidine, and 1× complete protease inhibitors; Boehringer Mannheim). The volume was then increased to 750 μL with ChIP lysis buffer, and the lysate was drawn off the beads, sonicated four times for 10 sec each at setting 2 on a Branson Sonifier 450, and spun 10 min in a microfuge. The supernatant fraction was collected, and 150 μL was used in a final volume of 400 μL of ChIP lysis buffer for each immunoprecipitation reaction. Samples were first pre-cleared for 30 min with 30 μL of a 50% slurry of Protein A agarose beads in 0.2 mg/mL salmon sperm DNA and 0.5 mg/mL BSA (Upstate Biotech). The samples were then incubated overnight with 4 μL of antibody. Antibodies were 1 mg/mL rabbit polyclonal IgG anti-Myc tag (residues 409–420), 2–10 mg/mL ChIP grade rabbit polyclonal IgG anti-acetyl-histone H4, 1 mg/mL rabbit polyclonal IgG anti-acetyl-histone H4, or 1 mg/mL rabbit polyclonal IgG anti-acetyl-histone H3 (Upstate Biotech). Samples were incubated with 60 μL beads for 1 h, the beads were washed (wash 1: 150 mM NaCl, 20 mM Tris at pH 8.1, 2 mM EDTA, 1% Triton X-100, 0.1% SDS; wash 2: 500 mM NaCl, 20 mM Tris at pH 8.1, 2 mM EDTA, 1% Triton X-100, 0.1% SDS; wash 3: 10 mM Tris at pH 8.1, 1 mM EDTA, 0.25 M LiCl, 1% sodium deoxycholate, 1% NP40; washes 4 and 5: TE), and DNA was eluted in 500 μL of 0.1 M NaHCO3, 1% SDS. Cross-links were reversed by overnight incubation at 65°C, and the DNA was isolated by use of standard procedures.
PCR analysis of immunoprecipitated DNA
PCR reactions were performed using Amplitaq Gold enzyme (Perkin Elmer), with 3.5 mM MgCl2, 0.2 mM dNTPs, and 0.5 μM primers. After 13 min at 94°C to activate the enzyme, samples were cycled 28 times for 15 sec at 94°C, 20 sec at 55°C, and 2.5 min at 68°C. The oligonucleotides used are described in Table 3.
Table 3.
Primers for PCR amplification of ChIP DNA
| Region
|
Primer 1
|
Primer 2
|
|---|---|---|
| SSCl | gcttcggcccggttcca | cagcaagcatcttggtgcg |
| HMR-E | ctaaatcgcatttcttttcgtccac | taacaaaaaccaggagtacctgcgc |
| HMR-I | ccaacattttcgtatatggc g | cctaccacattatcaatccttgc |
| HMR-X/Ya | taccaacccatccgccg | tccgccatactacaaatatcatcc |
| HMR-Ya/Zl | gtggcattactccacttcaagtaag | caagagcaagacgatgggg |
| 3′HMR | caacatggtgttccaaagcac | gcagcttactcccaagagtgc |
| HML-E | ggatggatctagggttttatgcc | tttggcccccgaaatcg |
| HML-I | ccagctgagtaactaactctcatgg | gctgttacggagatgcaaag c |
| MAT | ggctatacgggacggatgac | gttcttagcttgtaccagaggaagc |
| ACTl | ctttttcttccacgtcctcttgc | tgggatggtgcaagcgc |
| SMKl | ggtgaccattgcgactgtgc | cataggctcccttgcccagg |
| SPR3 | gacggcgtactgtccttggg | gcttcggcccggttcca |
RNA blots
RNA was isolated from log-phase cells (Schmitt et al. 1990) and separated electrophoretically on formaldehyde agarose gels and blotted (Sambrook et al. 1989). DNA fragments for probes were generated by PCR amplification of genomic DNA (W303) by use of the primers below, and labeled with [α-32P]dCTP using the Multiprime Labeling System (Amersham). For a1 probe, GGAT GATATTTGTAGTATGGCGG and CTGTTGCGGAAAGCT GAAAC were used. For SMK1 probe, GCGACGAGGCGTG AGGGTAG and CATAGGCTCCCTTGCCCAGG were used. For SPR3 probe, CCAAGGACAGTACGCCGTC and AATGT CAACAGCGCCCC were used. For ACT1, the GenePair primer set from Research Genetics was used.
Acknowledgments
We thank Andrew Vershon and Michael Pierce for stimulating discussion and communicating unpublished results, David Shore, Lorraine Pillus, and Stephen Bell for strains and plasmids, Charity Kirk for technical assistance, Paul Kaufmann, Ann Kirchmaier, and Peter Garber for critical reading of this manuscript, and members of the laboratory for comments and suggestions. This work was supported by a postdoctoral fellowship from the Damon Runyon-Walter Winchell Cancer Research Fund (L.N.R.) and a grant from the National Institutes of Health (GM31105, J.R.). Core support was provided by an National Institute of Environmental Health Sciences Mutagenesis Center Grant.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jrine@uclink4.berkeley.edu; FAX (510) 642-6420.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.873601.
References
- Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- Ayer D. Histone deacetylases: Transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- Bell S, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- Bi X, Braunstein M, Shei G, Broach J. The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc Natl Acad Sci. 1999;96:11934–11939. doi: 10.1073/pnas.96.21.11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C, Sherman J, Devine S, Cameron E, Pillus L, Boeke J. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes & Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Rose A, Holmes S, Allis C, Broach J. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes & Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Chi M, Shore D. SUM1-1, a dominant suppressor of SIR mutations in Saccharomyces cerevisiae, increases transcriptional silencing at telomeres and HM mating-type loci and decreases chromosome stability. Mol Cell Biol. 1996;16:4281–4294. doi: 10.1128/mcb.16.8.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J, Iyer V, Brown P. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes & Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorland S, Deegenaars M, Stillman D. Roles for the Saccharomyces cerevisiae SDS3, CBK1 and HYM1 genes in transcriptional repression by SIN3. Genetics. 2000;154:573–586. doi: 10.1093/genetics/154.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker C, Simpson R. The organized chromatin domain of the repressed yeast a cell-specific gene STE6 contains two molecules of the corepressor Tup1p per nucleosome. EMBO J. 2000;19:400–409. doi: 10.1093/emboj/19.3.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson D, Smith M, Roth S. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes & Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Foss M, McNally F, Laurenson P, Rine J. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science. 1993;262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- Fox CA, Ehrenhofer-Murray AE, Loo S, Rine J. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science. 1997;276:1547–1551. doi: 10.1126/science.276.5318.1547. [DOI] [PubMed] [Google Scholar]
- Gardner K, Rine J, Fox C. A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics. 1999;151:31–44. doi: 10.1093/genetics/151.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy B, Grunstein M, Gasser S. Localization of Sir2p: The nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong C, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- ————— Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A, Kakar S, Ivy J, Hicks J, Livi G, Miglio L. SUM1, an apparent positive regulator of the cryptic mating-type loci in Saccharomyces cerevisiae. Genetics. 1985;111:745–758. doi: 10.1093/genetics/111.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov S, Heller R, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenson P, Rine J. SUM1-1: A suppressor of silencing defects in Saccharomyces cerevisiae. Genetics. 1991;129:685–696. doi: 10.1093/genetics/129.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Chatterjee S, Struhl K. Genetic analysis of the role of Pol II holoenzyme components in repression by the Cyc8–Tup1 corepressor in yeast. Genetics. 2000;155:1535–1542. doi: 10.1093/genetics/155.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livi G, Hicks J, Klar A. The sum1-1 mutation affects silent mating-type gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:409–412. doi: 10.1128/mcb.10.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S, Fox CA, Rine J, Kobayashi R, Stillman B, Bell S. The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol Biol Cell. 1995;6:741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F, Rine J. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5648–5659. doi: 10.1128/mcb.11.11.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes & Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Pak D, Pflumm M, Chesnokov I, Huang D, Kellum R, Marr J, Romanowski P, Botchan M. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- Rothstein R. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rundlett S, Carmen A, Suka N, Turner B, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schiestl R, Gietz R. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schmitt M, Brown T, Trumpower B. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Brachmann C, Celic I, Kenna M, Muhammad S, Starai V, Avalos J, Escalante-Semerena J, Grubmeyer C, Wolberger C, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Watson A, Edmondson D, Bone J, Mukai Y, Yu Y, Du W, Stillman D, Roth S. Ssn6–Tup1 interacts with class I histone deacetylases required for repression. Genes & Dev. 2000;14:2737–2744. doi: 10.1101/gad.829100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, Vershon A. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun Z, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]