Abstract
Abasic (AP) sites are one of the most frequently formed lesions in DNA, and they present a strong block to continued synthesis by the replicative DNA machinery. Here we show efficient bypass of an AP site by the combined action of yeast DNA polymerases δ and ζ. In this reaction, Polδ inserts an A nucleotide opposite the AP site, and Polζ subsequently extends from the inserted nucleotide. Consistent with these observations, sequence analyses of mutations in the yeast CAN1s gene indicate that A is the nucleotide inserted most often opposite AP sites. The nucleotides C, G, and T are also incorporated, but much less frequently. Enzymes such as Rev1 and Polη may contribute to the insertion of these other nucleotides; the predominant role of Rev1 in AP bypass, however, is likely to be structural. Steady-state kinetic analyses show that Polζ is highly inefficient in incorporating nucleotides opposite the AP site, but it efficiently extends from nucleotides, particularly an A, inserted opposite this lesion. Thus, in eukaryotes, bypass of an AP site requires the sequential action of two DNA polymerases, wherein the extension step depends solely upon Polζ, but the insertion step can be quite varied, involving not only the predominant action of the replicative DNA polymerase, Polδ, but also the less prominent role of various translesion synthesis polymerases.
Keywords: Abasic sites, mutagenic bypass, yeast, DNA polymerase δ, DNA polymerase ζ
Abasic (AP) sites represent one of the most frequently formed DNA lesions in eukaryotes, and it has been estimated that a human cell loses as many as 104 purines per day from its genome (Lindahl and Nyberg 1972). In Saccharomyces cerevisiae, AP sites are efficiently repaired by the AP endonucleases encoded by the APN1 and APN2 genes. APN1 and APN2 provide alternate pathways for the removal of AP sites, and consequently, simultaneous inactivation of both the genes results in a dramatic decline in the efficiency to repair AP sites (Johnson et al. 1998).
If the AP sites are not removed by excision repair processes, they present a block to the replication machinery. During replication, AP sites can be bypassed either by a specialized mutagenic DNA polymerase, or by error-free mechanisms such as recombination or a copy-choice type of DNA synthesis. In S. cerevisiae, genes in the RAD6 epistasis group promote replication through DNA lesions (Prakash 1981). The REV1, REV3, and REV7 genes of this group are essential for UV-induced mutagenesis (Lawrence and Hinkle 1996), and these genes are also indispensable for mutagenesis induced by AP sites (Johnson et al. 1998). REV1 encodes a deoxycytidyl transferase activity (Nelson et al. 1996a), and the REV3- and REV7-encoded proteins together form DNA polymerase ζ (Nelson et al. 1996b).
Although Polζ is absolutely required for damage-induced mutagenesis, and therefore for the mutagenic bypass of a variety of DNA lesions, our recent studies have indicated that on its own, Polζ bypasses UV lesions very inefficiently (Johnson et al. 2000a). This is because Polζ is very inefficient in inserting nucleotides opposite the 3′ T of a cis–syn thymine–thymine (T–T) dimer or a (6–4) T–T photoproduct. Polζ, however, efficiently extends from nucleotides placed opposite the 3′ T of these lesions by another DNA polymerase (Johnson et al. 2000a). Because Rev1 can insert a C opposite the AP site (Nelson et al. 1996a), and because Rev1 is essential for mutagenesis induced by AP sites (Johnson et al. 1998), in principle, mutagenic bypass of AP sites could be subsumed by the combined action of Rev1 and Polζ. However, we find that mutational inactivation of the Rev1 C-transferase activity has little effect on AP mutagenesis, and sequence analyses of mutations resulting from AP bypass indicate that in vivo, C is inserted rather infrequently opposite AP sites. These observations suggest that the C-transferase activity of Rev1 is not likely to have a predominant role in AP bypass.
In addition to the requirement of Rev1 and Polζ, genetic studies in yeast have indicated a requirement of Polδ for the mutagenic bypass of DNA lesions. A mutation, pol3-13, in the catalytic subunit of Polδ confers a deficiency in UV mutagenesis (Giot et al. 1997), and deletion of POL32, which encodes a nonessential subunit of Polδ, also lowers UV mutagenesis (Gerik et al. 1998). Because Polδ is essential for the replication of both DNA strands and will be the first polymerase to encounter the DNA lesion, it could promote damage bypass by inserting a nucleotide opposite the lesion, which could then be extended by Polζ. Here we provide evidence supporting such a role for Polδ in AP bypass. From the pattern of mutations induced by AP sites in yeast, we infer that in vivo, A is the residue inserted most frequently opposite AP sites, and our biochemical studies indicate that Polδ primarily inserts an A opposite the AP site. Efficient bypass of this lesion occurs when Polδ is combined with Polζ.
Results
Spectrum of CAN1s to can1r mutations resulting from AP bypass
To determine the mutational specificity of AP sites in vivo, we sequenced mutations resulting from their bypass in yeast cells treated with the alkylating agent methyl methanesulfonate (MMS). MMS alkylates the bases in DNA, particularly adenine at the N3 position (3MeA) and guanine at the N7 position (7MeG). The removal of alkylated bases by an N-methyl purine DNA glycosylase (Roy et al. 1994; Bjoras et al. 1995) results in an AP site that can be acted upon by the AP endonuclease activity of Apn1 or Apn2 proteins. We examined MMS-induced CAN1s to can1r forward mutations in the apn1Δ apn2Δ strain because the repair of AP sites is severely inhibited in this strain. Sequence analysis of a number of independent can1r mutations formed in the apn1Δ apn2Δ strain after MMS treatment indicated that ∼70% of these were base substitutions and ∼30% were +1 or −1 frameshift mutations (Table 1). The most frequent base substitutions were G : C to T : A transversions (40%). Considering that the majority of AP sites arise from the loss of A or G residues, we calculate that A, C, G, and T nucleotides were inserted opposite AP sites with relative frequencies of 64%, 14%, 11%, and 11%, respectively. These observations indicate that, in vivo, incorporation of an A residue opposite the AP site is the major base substitution caused by this DNA lesion.
Table 1.
MMS induced can1rmutations in the apn1Δ apn2Δ strain
| Mutations
|
Number (frequency)
|
|---|---|
| A:T → T:A | 2 (5%) |
| G:C → T:A | 16 (40%) |
| G:C → C:G | 3 (7.5%) |
| A:T → G:C | 4 (10%) |
| G:C → A:T | 3 (7.5%) |
| +1 frameshifts | 7 (17.5%) |
| −1 frameshifts | 5 (12.5%) |
Yeast cells were suspended in 0.05 M KPO4 (pH 7) buffer, and treated with 0.08% MMS for 20 min at 30°C. Following termination of the reaction by the addition of an equal volume of 10% sodium thiosulfate, cells were spun down, resuspended in water, and plated on synthetic complete medium lacking arginine and supplemented with canavanine.
The deoxycytidyl transferase activity of Rev1 is dispensable for mutagenesis induced by AP sites
To assess the in vivo role of the deoxycytidyl transferase activity of Rev1 in mutagenic bypass of AP sites, we examined the effect of inactivation of this activity on mutagenesis induced by AP sites. For this purpose, we altered the Asp 467 and Glu 468 residues present in the highly conserved motif III consisting of serine, isoleucine, aspartate, and glutamate residues (SIDE) in Rev1 to alanines (Johnson et al. 1999). The analogous mutation in Rad30 inactivates the DNA polymerase activity (Johnson et al. 1999), and similarly, the rev1 Ala467–Ala468 mutation completely inactivated the deoxycytidyl transferase activity of Rev1 (data not shown). To determine whether Rev1 deoxycytidyl transferase activity was required for AP mutagenesis, the ability of the rev1 Ala467–Ala468 mutation to complement the rev1Δ mutation was examined. The wild-type or the mutant gene was expressed in yeast from the native REV1 promoter on a low-copy CEN plasmid. Yeast cells were treated with various concentrations of MMS, and the rates of forward mutations at the CAN1s locus were examined. Whereas MMS-induced can1r mutations are completely abolished in the apn1Δ apn2Δ rev1Δ strain (Johnson et al. 1998), introduction of the rev1 Ala467–Ala468 mutant gene into this strain restored MMS-induced can1r mutations to the level seen in the apn1Δ apn2Δ strain (data not shown). Thus, although the Rev1 protein is absolutely required for damage-induced mutagenesis, its deoxycytidyl transferase activity is dispensable for mutagenesis induced by AP sites.
Requirement of Polδ for mutagenic bypass of AP sites
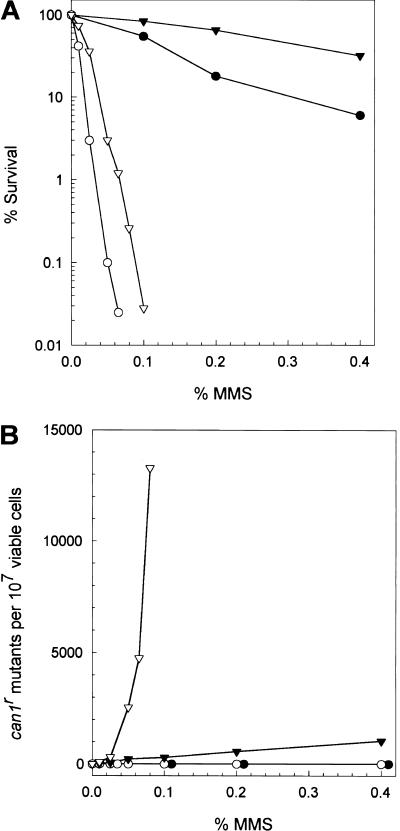
Yeast Polδ is comprised of three subunits of 125, 58, and 55 kD, which are encoded by the POL3, POL31, and POL32 genes, respectively (Gerik et al. 1998). The 125-kD catalytic subunit and the 58-kD subunit are essential for viability, but the 55-kD subunit encoded by the POL32 gene is not essential (Gerik et al. 1998). A mutation in the catalytic subunit of Polδ, pol3-13, confers UV sensitivity and a deficiency in UV mutagenesis (Giot et al. 1997). The pol32Δ mutant is also UV-sensitive and deficient in UV mutagenesis (Gerik et al. 1998). To determine the role of Polδ in mutagenesis induced by AP sites, we examined the effect of the pol32Δ mutation on MMS-induced CAN1s to can1r mutations in the apn1Δ apn2Δ strain. The pol32Δ mutant exhibits somewhat enhanced sensitivity to MMS, and MMS-induced mutations do not occur in this strain (Fig. 1; Gerik et al. 1998). For example, treatment with 0.4% MMS produced ∼1000 can1r mutants per 107 viable cells in the wild-type strain, whereas MMS-induced mutagenesis was abolished in the pol32Δ single mutant (Fig. 1B). These observations indicate a role for the Pol32 subunit of Polδ in the mutagenic bypass of MMS-induced base damage. The apn1Δ apn2Δ strain displays a very high incidence of MMS-induced mutations, ∼13,000 per 107 cells at 0.08% MMS, owing to the presence of unrepaired AP sites. By contrast, MMS-induced mutagenesis was abolished in the apn1Δ apn2Δ pol32Δ strain (Fig. 1B). The apn1Δ apn2Δ pol32Δ strain also exhibits a greater sensitivity to MMS than the apn1Δ apn2Δ strain does (Fig. 1A). These results indicate a requirement of the Pol32 subunit of Polδ for the mutagenic bypass of AP sites.
Figure 1.
Effect of the pol32Δ mutation on AP site induced mutagenesis. (A) MMS sensitivity of pol32Δ strains. Cells were grown overnight in YPD and treated with MMS at the indicated doses for 20 min. Appropriate dilutions were plated on YPD. All data points represent the average of at least three experiments. (Filled inverted triangles) EMY74.7, wild type; (filled circles) YPO69, pol32Δ; (open inverted triangles) YRP269, apn1Δ apn2Δ; (open circles) YPO71, apn1Δ apn2Δ pol32Δ. (B) MMS-induced mutagenesis in pol32Δ strains. Cells were grown overnight and treated with MMS at the indicated doses for 20 min. Appropriate dilutions were plated on either YPD for viability or on synthetic medium lacking arginine but containing canavanine for measuring the frequency of can1r induced mutations. All data points represent of the average of at least three experiments. (Filled inverted triangles) EMY74.7, wild type; (filled circles) YPO69, pol32Δ; (open inverted triangles) YRP269, apn1Δ apn2Δ; (open circles) YPO71, apn1Δ apn2Δ pol32Δ.
AP bypass by the concerted action of Polδ and Polζ
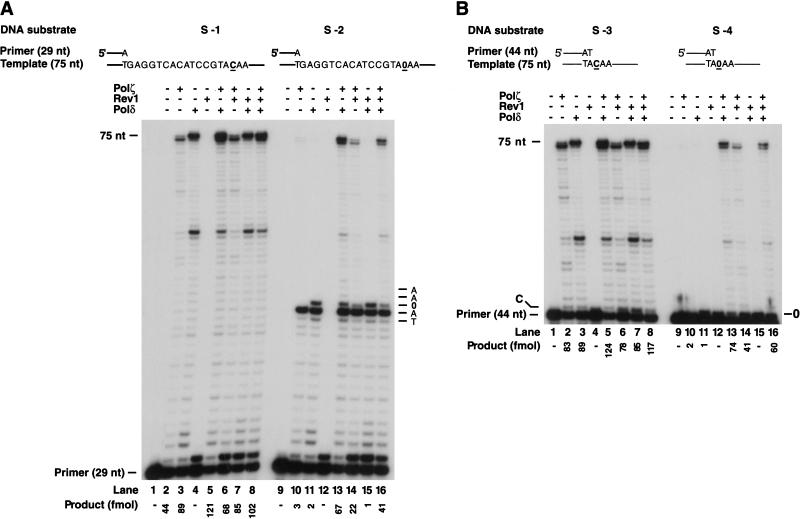
AP bypass was examined with a running-start and a standing-start DNA substrate that were constructed using a linear 75-nt template DNA containing a single AP site, or, as a control, containing a C residue instead of the AP site, and annealed to a 5′-32P-labeled 29-nt or a 44-nt primer, respectively. DNA synthesis reactions with these substrates were carried out in the presence of Polδ and Polζ. We also included Rev1 in these experiments (Fig. 2). With the running-start DNA substrate, none of the individual Polδ, Polζ, or Rev1 enzymes were able to bypass the AP site (Fig. 2A, lanes 10–12). The AP site is a strong block to synthesis by Polζ, and Polζ was unable to insert a nucleotide opposite this lesion (Fig. 2A, lane 10). By contrast, Polδ catalyzed nucleotide incorporation opposite the AP site but did not extend the resulting primer end (Fig. 2A, lane 11). In the control reactions, both polymerases carried out efficient synthesis to the end of the unmodified template (Fig. 2A, lanes 2,3). Efficient bypass of the AP site, however, could be achieved when Polδ was combined with Polζ (Fig. 2A, lane 13). Compared to the synthesis on undamaged DNA (Fig. 2A, lane 5), Polδ and Polζ together replicated through 55% of the AP sites (Fig. 2A, lane 13). In agreement with the previously published ability of Rev1 to promote AP bypass in combination with Polζ (Nelson et al. 1996a), Rev1 and Polζ together replicated through 32% of the AP sites (Fig. 2A, cf. lanes 6 and 14). However, Rev1 did not stimulate AP bypass when it was combined with Polδ (Fig. 2A, lane 15), nor did it increase the bypass activity of the Polδ/Polζ combination (Fig. 2A, cf. lanes 13 and 16). The key observation here is that the two DNA polymerases, Polδ and Polζ, together carry out efficient AP bypass, and in this reaction, Polδ inserts the nucleotide opposite the AP site and Polζ then extends from that nucleotide.
Figure 2.
Translesion synthesis on DNA template containing a single AP site using Polδ, Polζ, and Rev1. Sequences adjacent to the primer–template junction are shown for the 5′- 32P-labeled primers and 75-nt template. The position corresponding to the model abasic site on the template is indicated by 0. (A) Running-start DNA synthesis on undamaged (lanes 1–8) or single-abasic-site-containing (lanes 9–16) 75-nt template annealed with the 29-nt primer. Polζ (10 nM), PolΔ (10 nM), and Rev1 (10nM) or the indicated combinations of these proteins were incubated with the DNA substrate (20 nM) for 5 min at 30°C in the presence of 100 μM of each of four dNTPs. The reaction products were resolved on 10% denaturing polyacrylamide gel and visualized by autoradiography. The gel was analyzed by PhosphorImager and the fraction of total radioactivity present as 46- to 75-nt products was used to calculate femtomoles of product. (B) Standing-start DNA synthesis on an undamaged (lanes 1–8) or a single-AP-site-containing (lanes 9–16) 75-nt template annealed with the 44-nt primer. The reactions and the analysis were carried out as in A.
With the standing-start substrate, essentially similar results were observed (Fig. 2B). Thus, the combination of Polδ and Polζ bypassed the AP site about 60% as efficiently as the replication of undamaged DNA (Fig. 2B, cf. lanes 5 and 13).
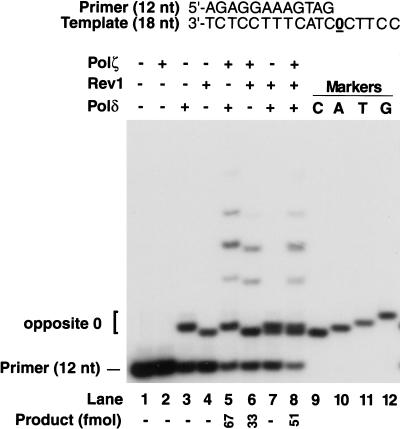
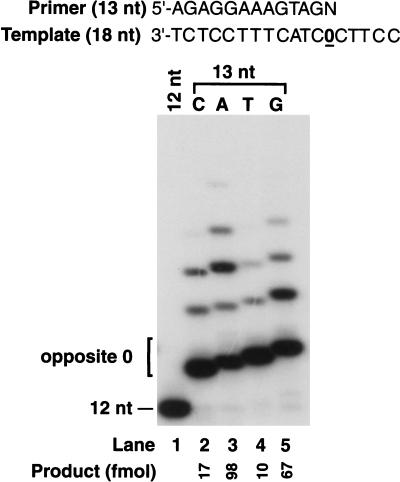
Nucleotide incorporated opposite the AP site by Polδ
To identify the deoxynucleotide inserted by Polδ opposite the AP site, we used an 18-nt template having an AP site at position 13 from the 3′ end and primed with a standing-start 12-nt primer (Fig. 3). As markers we used 13-nt oligonucleotides containing the 12-nt primer with an additional C, A, T, or G residue at position 13; these were distinguished by their relative electrophoretic mobility on a 20% polyacrylamide gel (Fig. 3, lanes 9–12). We found that Polδ mostly inserts an A (∼95%) and some G (∼5%) opposite the AP site (Fig. 3, lane 3), and Polζ extends from the inserted nucleotide (Fig. 3, lane 5). We also examined the ability of Rev1 and Polζ to insert nucleotides opposite the AP site and to extend therefrom on this template. As expected, Rev1 incorporates only a C across from the lesion (Fig. 3, lane 4), and Polζ extends from this nucleotide (Fig. 3, lane 6). When Polδ and Rev1 are combined, both A and C are inserted opposite the AP site, but there is no extension (Fig. 3, lane 7); extension, however, occurs upon the addition of Polζ (Fig. 3, lane 8).
Figure 3.
Nucleotide incorporation opposite AP site and extension from the ensuing primer end. Nucleotide incorporation opposite an AP site in reactions catalyzed by different combinations of Polζ, Polδ, and Rev1. Standing-start primer extension reactions were carried out on an AP-site-containing 18-nt template primed with the 5′-32P-labeled 12-nt oligonucleotide. The position of the AP site on the template is indicated by 0. The following enzyme concentrations were used: Polζ (20 nM), Polδ (20 nM), and Rev1 (20 nM). Electrophoretic mobilities of reaction products were compared with those of 13-nt oligonucleotide standards containing a C (lane 9), an A (lane 10), a T (lane 11), or a G (lane 12) residue opposite the AP site at position 13.
Steady-state kinetic analyses of nucleotide insertion opposite the AP site and of subsequent extension by Polζ
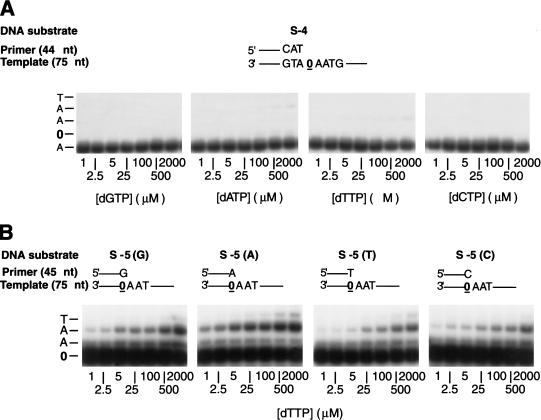
Next, we measured the kinetic parameters of nucleotide insertion opposite the AP site and of subsequent extension by Polζ. The kinetics of insertion of a single deoxynucleotide opposite the AP site, and as a control opposite an undamaged C or G residue, and the kinetics of addition of the next correct nucleotide to various 3′-primer termini situated across from the AP site or opposite undamaged C or G residues were determined as a function of deoxynucleotide concentration under steady-state conditions. The patterns of insertion of nucleotides opposite the AP site and of extension from the G, A, T, or C 3′-primer termini situated across from the AP site are shown in Figure 4. From the kinetics of deoxynucleotide incorporation, the apparent Km and Vmax values were determined, and the frequency of deoxynucleotide insertion (finc) and extension (f0ext) were calculated (Mendelman et al. 1990; Goodman et al. 1993; Creighton et al. 1995). As shown in Table 2, Polζ misincorporates nucleotides opposite a template C or G with a frequency of ∼10−4, and it inserts nucleotides opposite the AP site with a frequency of 10−4 to 10−5. However, Polζ extends from the primer end situated opposite the abasic site very efficiently. Compared to the extension from a G opposite C in the template, Polζ extended from an A, a G, a C, and a T residue opposite the AP site with frequencies of ∼3 × 10−1, 1 × 10−1, 2 × 10−2, and 7 × 10−3, respectively (Table 3). Thus, Polζ extends from an A or a G opposite the AP site about 3- and 10-fold less efficiently than it extends from a G opposite template C, whereas C opposite the AP site is extended about 50-fold less efficiently than is G opposite C. Polζ also extends from 3′-terminal mispaired ends quite readily. For example, an A opposite template C is extended about 25% as efficiently as a G opposite template C. Overall, Polζ extends from base mispairs with frequencies of 10−1 to 10−2 (Table 3).
Figure 4.
Steady-state deoxynucleotide incorporation and primer-extension reactions catalyzed by Polζ on an AP-site-containing DNA template. (A) Deoxynucleotide incorporation opposite a template AP site. Polζ (5 nM) was incubated for 1 min at 30°C with the S-4 primer–template DNA substrate (20 nM) and increasing concentrations (0–2000 μM) of a single deoxynucleotide (dGTP, dATP, dTTP, or dCTP) in the standard reaction buffer. The quenched samples were analyzed by 10% denaturing polyacrylamide gel electrophoresis. (B) Extension of primers containing a G, an A, a T, or a C residue opposite the AP site by Polζ. Primers differing only in the last nucleotide at the 3′ end were annealed separately to an AP-site-containing DNA template as shown on the top. Reactions were carried out in the presence of increasing dTTP concentration (0–2000 μM) as in A. The position of the AP site in the template is indicated by 0.
Table 2.
Kinetic parameters of nucleotide insertion reactions catalyzed by yPolζ
| Site
|
dNTP added
|
Km (μM)
|
Vmax (%/min)
|
Vmax/Km
|
finc
|
|---|---|---|---|---|---|
| Insertion opposite abasic site | |||||
| dGTP | 543 ± 42 | 1.4 ± 0.19 | 0.003 | 2.1 × 10−5 | |
| 5′----CAT | dATP | 81 ± 2.9 | 4.8 ± 0.07 | 0.059 | 4.2 × 10−4 |
| ----GTA0 AA-- | dTTP | 125 ± 8.5 | 3.3 ± 0.09 | 0.026 | 1.8 × 10−4 |
| dCTP | 113 ± 14 | 1.4 ± 0.12 | 0.012 | 8.6 × 10−5 | |
| Insertion opposite C | |||||
| dGTP | 0.11 ± 0.008 | 15.4 ± 2.0 | 140 | 1 | |
| 5′----CAT | dATP | 267 ± 18 | 2.1 ± 0.14 | 0.0078 | 5.5 × 10−5 |
| ----GTAC AA-- | dTTP | 552 ± 96 | 3.8 ± 1.1 | 0.0069 | 4.9 × 10−5 |
| dCTP | 321 ± 14 | 2.9 ± 0.32 | 0.009 | 6.4 × 10−5 | |
| Insertion opposite G | |||||
| dGTP | 344 ± 24 | 4.3 ± 0.12 | 0.0125 | 1 × 10−4 | |
| 5′----CAT | dATP | 118 ± 5 | 1.4 ± 0.33 | 0.012 | 1 × 10−4 |
| ----GTAG AA-- | dTTP | 428 ± 54 | 3.6 ± 0.28 | 0.0084 | 7 × 10−5 |
| dCTP | 0.14 ± 0.03 | 16.7 ± 1.1 | 119.3 | 1 | |
Table 3.
Kinetic parameters of extension reactions catalyzed by yPolζ
| Site
|
dNTP added
|
Km (μM)
|
Vmax (%/min)
|
Vmax/Km
|
f 0ext
|
|---|---|---|---|---|---|
| Extension from G, A, T, or C opposite abasic site | |||||
| 5′----CATG | dTTP | 6.5 ± 0.55 | 12 ± 2.2 | 1.85 | 9.5 × 10−2 |
| ----GTA0 AA-- | |||||
| 5′----CATA | dTTP | 2.3 ± 0.17 | 13 ± 1.5 | 5.65 | 2.9 × 10−1 |
| ----GTA0 AA-- | |||||
| 5′----CATT | dTTP | 49 ± 3.7 | 6.8 ± 0.24 | 0.14 | 7.2 × 10−3 |
| ----GTA0 AA-- | |||||
| 5′----CATC | dTTP | 21 ± 1.2 | 8.4 ± 1.3 | 0.4 | 2.1 × 10−2 |
| ----GTA0 AA-- | |||||
| Extension from G, A, T or C opposite C | |||||
| 5′----CATG | dTTP | 0.35 ± 0.02 | 6.8 ± 0.56 | 19.4 | 1 |
| -----GTAC AA-- | |||||
| 5′----CATA | dTTP | 2.3 ± 0.22 | 12 ± 0.89 | 5.22 | 2.6 × 10−1 |
| -----GTAC AA-- | |||||
| 5′----CATT | dTTP | 15 ± 0.9 | 8.9 ± 1.3 | 0.59 | 3.0 × 10−2 |
| ----GTAC AA-- | |||||
| 5′----CATC | dTTP | 26 ± 2.8 | 3.9 ± 0.08 | 0.15 | 7.7 × 10−3 |
| -----GTAC AA-- | |||||
| Extension from G, A, T or C opposite G | |||||
| 5′----CATG | dTTP | 6.9 ± 0.2 | 10.7 ± 1.8 | 1.55 | 3.6 × 10−2 |
| ----GTAG AA-- | |||||
| 5′----CATA | dTTP | 2.1 ± 0.14 | 8.4 ± 0.77 | 4.0 | 9.3 × 10−2 |
| -----GTAG AA-- | |||||
| 5′----CATT | dTTP | 28.8 ± 4.6 | 14.1 ± 1.7 | 0.48 | 1.1 × 10−2 |
| -----GTAG AA-- | |||||
| 5′----CATC | dTTP | 0.23 ± 0.018 | 9.9 ± 1.6 | 43 | 1 |
| -----GTAGAA-- | |||||
Polζ extension of primer end situated opposite the AP site
Next, we examined the ability of Polζ to extend from various 3′ termini situated across from the AP site in the 18-nt template in the presence of 100 μM of each of the four dNTPs (Fig. 5). Although Polζ extended each of the C, A, T, or G 3′-primer ends situated opposite the AP site (Fig. 5, lanes 2–5), extension from A was ∼5-fold more efficient than from the C primer end, and the order and the frequency of extension from various 3′-terminal deoxynucleotides were A : G : C : T = 9.8 : 6.7 : 1.7 : 1. Thus, even under saturating dNTP concentrations, Polζ shows a preference for extending from an A opposite the AP site.
Figure 5.
Extension of primers with various 3′-terminal primer dNMPs opposite the template AP site by Polζ. Four different 5′-32P-labeled 13-nt primers, differing only in their 3′-terminal nucleotide, were annealed to an 18-nt template containing an AP site. In the template–primer shown, N designates the position of the variable terminal primer dNMP and 0 designates the position of the AP site. Lane 1 has a 12-nt primer and lacks the 13th nucleotide indicated by N. DNA substrate (20 nM) was assayed with 20 nM Polζ (lanes 1–5) in the presence of each of the four dNTPs (100 μM).
Discussion
To determine the mutational specificity of AP sites in eukaryotes, we analyzed the sequence of can1r mutations obtained following MMS treatment of the apn1Δ apn2Δ yeast strain. Our results indicate that A is inserted most frequently opposite the AP site, and that C, G, and T together are inserted at about 50% the frequency of A. Previously, in one study done in yeast with plasmids containing an AP site, C was found to be inserted preferentially (∼80%) opposite this lesion (Gibbs and Lawrence 1995), whereas in another study with the SUP4 gene carried on a plasmid, the frequency of spontaneous A · T → C · G events increased in the apn1Δ strain (Kunz et al. 1994), suggesting an insertion of G opposite the AP site resulting from the loss of an A. Thus, the mutagenic consequences of AP sites in yeast may differ depending on whether the lesion is on a plasmid or in the genome. Also, we note that mutagenesis at the SUP4 locus differs from mutagenesis at the majority of loci (Drake 1991).
NMR structural studies have shown that A fits into the double helix opposite the AP site without conferring any distortion. Such DNA retains all aspects of B-form DNA, in which the A residue and the abasic residue lie inside the double helix, and the melting temperature of the A · AP site is the same as that of the A · T base pair (Cuniasse et al. 1987, 1990; Kalnik et al. 1988). Thus, the geometry of the A residue opposite the AP site is very similar to that of the A · T base pair. G opposite the AP site is less stable, but at low temperatures this residue is also predominantly intrahelical (Cuniasse et al. 1990). By contrast, a pyrimidine opposite the AP site is not stable, and in this case, both the pyrimidine and the abasic sugar are extrahelical (Cuniasse et al. 1990). In Escherichia coli, synthesis past an AP site is accompanied by the preferential incorporation of an A opposite this lesion, a phenomenon known as the “A-rule” (Strauss 1991). Polδ may act in a similar fashion, as it predominantly inserts an A opposite the AP site. Although the geometry of an A · AP pair resembles that of the A · T base pair, Polδ nonetheless can distinguish between these pairs, since it does not extend from the A residue of an A · AP pair.
Genetic studies in yeast have indicated the absolute requirement of Polζ for mutagenesis induced by AP sites (Johnson et al. 1998). However, on its own, Polζ bypasses this lesion very poorly. This is because Polζ is highly inefficient in inserting deoxynucleotides opposite the AP site, but it efficiently extends from the nucleotide inserted opposite the AP site by another DNA polymerase. Steady-state kinetic analyses further demonstrate that Polζ extends most efficiently from an A opposite the AP site. Relative to the extension from G opposite template C, Polζ extends from an A opposite the AP site with a frequency of 3 × 10−1, and it extends from a G opposite this lesion with a frequency of ∼10−1. Thus, the insertion of an A opposite the AP site by Polδ, followed by the efficient extension of this primer end by Polζ, explains our in vivo mutagenesis results of the preferential incorporation of an A opposite this DNA lesion.
In addition to the insertion of an A, we also observe the insertion of C, G, and T residues opposite the AP site. Although our studies on mutational inactivation of Rev1 C-transferase activity have provided no evidence for the requirement of this activity in the mutagenic bypass of AP sites, they do not rule out a minor contribution of this activity to AP bypass. Since only ∼10% of can1r mutations are expected to derive from the insertion of C opposite the AP site, it would be difficult to ascertain such a limited involvement of Rev1 C-transferase activity in AP bypass from the analyses of mutation frequencies. Furthermore, there is the possibility that enzymes other than Rev1 also insert a C opposite the AP site. Thus, although the Rev1 protein is essential for mutagenesis induced by AP sites, its C-transferase activity is not prominently involved in the bypass of this lesion. Presumably, the indispensability of the Rev1 protein for AP mutagenesis derives from its structural role during Polζ-dependent bypass of this lesion (see Fig. 6). Since Polδ would be the first polymerase to arrive at the AP site, we surmise that its ability to insert an A opposite this lesion supercedes the insertion of C by Rev1, and also, since Polζ extends from an A opposite the AP site about 15-fold more efficiently than from C (Table 3), we expect A to be incorporated much more frequently during AP bypass than is C. Unlike the efficient bypass of a cis–syn T–T dimer (Johnson et al. 2000b; Washington et al. 2000), or an 8-oxoguanine lesion (Haracska et al. 2000), Polη does not bypass an AP site; however, Polη may contribute to the insertion of a G opposite the AP site (Haracska et al. 2001). In humans, Polι, a very low fidelity enzyme, would additionally function in the insertion of nucleotides opposite the AP sites (Johnson et al. 2000a). Therefore, AP bypass in eukaryotes is mediated by the sequential action of two DNA polymerases, wherein the extension step depends solely on Polζ, but the insertion step can be quite varied, involving not only the predominant action of the replicative DNA polymerase, Polδ, but also the less prominent role of various translesion synthesis polymerases.
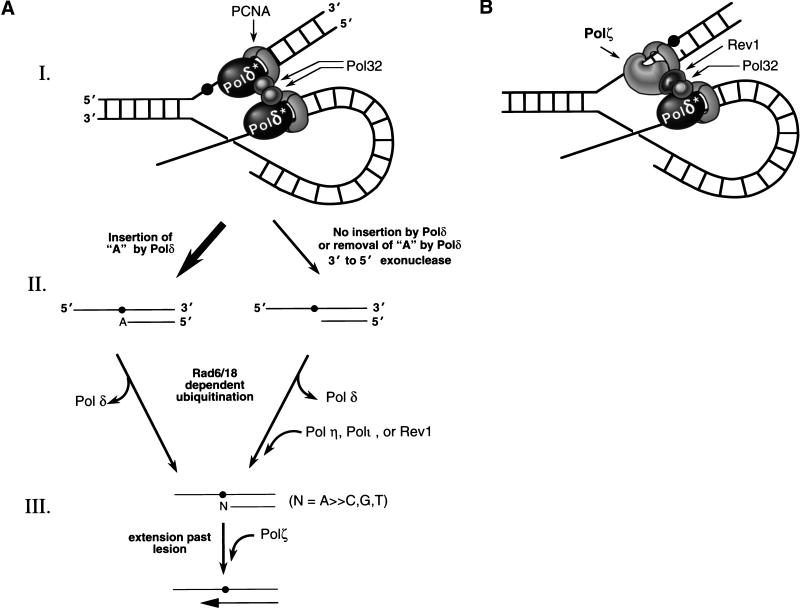
Figure 6.
Mechanisms of AP bypass. (A) The insertion and extension steps of AP bypass. (I) Polδ* represents the Pol3–Pol31 heterodimer, and the Pol32 subunit of Polδ is shown to connect with Polδ* as well as with PCNA (Burgers and Gerik 1998). An abasic site (filled circle) is present on the leading strand. (II) Following the insertion of an A or before it, the Rad6–Rad18-mediated ubiquitination leads to the displacement of Polδ stalled at the lesion site. In the absence of an A insertion (or, rarely, a G) by Polδ, Polη, Polι, or Rev1 could gain access to the lesion site and insert a nucleotide opposite the AP site. (III) Polζ extends from the nucleotide inserted opposite the AP site by Polδ or by one of the translesion synthesis DNA polymerases. (B) Roles of Pol32 and Rev1 in the mutagenic bypass of AP sites. The essential structural role of Rev1 in mutagenic bypass may be in assembling Polζ with Polδ. Rev1 may interact with Pol32, and also with PCNA/RFC, and thus mediate the assembly of Polζ with Polδ.
The absolute requirement of Polζ for the extension step explains its indispensability for AP mutagenesis. Our proposal that Polδ acts primarily in the insertion of an A, whereas Rev1 and the other translesion synthesis enzymes catalyze the insertion of C and of other nucleotides opposite the AP site, however, fails to explain the absolute requirement of the Pol32 subunit of Polδ and of Rev1 in AP mutagenesis. It is possible that the requirement of Pol32 reflects the need for this Polδ subunit in the assembly of Polζ and Rev1 at the lesion site (Fig. 6B). Because Polδ is involved in the replication of both the leading and lagging DNA strands and binds the two strands as a dimer, following the incorporation of a nucleotide opposite the AP site, Polδ may be displaced from the damage-containing template upon ubiquitin conjugation by the Rad6–Rad18 complex (Bailly et al. 1997), which is essential for damage bypass (Fig. 6A). If, however, Polδ is unable to insert a nucleotide opposite the AP site, or if the inserted nucleotide is removed by its proofreading exonuclease activity, then Rad6–Rad18-mediated displacement of Polδ may occur before the insertion step, whereupon any of the translesion synthesis enzymes such as Rev1, Polη, and additionally, Polι in humans, may gain access to the lesion site and insert a nucleotide opposite the AP site (Fig. 6A). Following the insertion of a nucleotide by Polδ or by one of the translesion synthesis polymerases, Polζ may be targeted to the lesion site via its interaction with Rev1, which in turn may bind the Pol32 subunit of Polδ remaining bound to the undamaged DNA strand (Fig. 6B). The nonenzymatic requirement of Rev1 may therefore reflect the need for this protein in the assembly of Polζ with Polδ via its Pol32 subunit. Rev1 may also facilitate interactions of Polζ with the other components of the replication machinery such as PCNA and RFC (Fig. 6B). Accordingly, the requirement of Pol32 as well as Rev1 for AP bypass may derive from their respective roles in providing access of Polζ to the lesion site.
The insertion of an A by Polδ opposite the AP site raises the possibility that Polδ is an A-rule polymerase, able to insert an A opposite various other DNA lesions as well (Strauss 1991). For example, in addition to the formation of cyclobutane dimers at TT sequences, UV induces lesions at the 5′-TC-3′ and 5′-CC-3′ sequences. The 3′-C in both sequences is highly mutagenic, and in both yeast and humans, UV-induced mutations occur predominantly by a 3′-C → T transition that results from the insertion of an A opposite the 3′ damaged C during DNA replication (Armstrong and Kunz 1990; Brash 1997; Canella and Seidman 2000). Genetic studies in yeast have shown that Polζ is responsible for the 3′-C → T mutagenesis resulting from the replicative bypass of UV lesions at TC and CC sites (Yu et al. 2001). However, because Polζ functions at the step of extending from the nucleotide incorporated opposite the 3′ residue of UV lesions by another DNA polymerase (Johnson et al. 2000a), we suggest that Polδ is the enzyme responsible for inserting an A opposite the 3′-C of UV lesions formed at TC and CC sites. Hence, by inserting an A opposite DNA lesions, Polδ could be a major contributor to DNA damage-induced mutagenesis in eukaryotes.
Materials and methods
Generation of yeast null mutations
The strains used in the genetic studies were EMY74.7 (MATα his3Δ-100, leu2-3, 112, trp1Δ, ura3-52) and its derivatives. To construct the pol32Δ-generating plasmid, the 1.2- and 1-kb PCR products corresponding to the 5′- and 3′-flanking regions of the POL32 gene, respectively, were directionally cloned into pUC19. The URA3 geneblaster fragment containing the yeast URA3 gene flanked by the duplicated Salmonella typhimurium hisG gene (Alani et al. 1987) was then inserted between the two PCR products. The resulting pol32Δ-generating plasmid was digested with PvuII, which releases a 6-kb fragment, introduction of which into yeast deletes nucleotides from +6 to +960 of the 1053-nt POL32 open reading frame. The presence of the pol32Δ mutation in the various yeast strains was confirmed by PCR of genomic DNA. Loss of the URA3 gene was selected for by plating strains on medium containing 5-fluoro-orotic acid.
Construction of the Rev1 C-transferase mutant
A 5.6-kb DNA fragment containing the entire yeast REV1 gene and including ∼1.1 kb of 5′-flanking DNA sequence and 1.4 kb of 3′-flanking DNA sequence, respectively, was cloned into pUC19, generating plasmid pBJ67, which was used as the starting material for constructing the Rev1 C-transferase mutant. This mutant, in which D467 and E468 are each changed to A, was generated by mutagenic PCR using the antisense oligonucleotide N4865 (5′-CACAAACAGCTGCAGCAATAGATA TAGGTAAAATC-3′) and the sense oligonucleotide N4866 (5′-TCTATTGCTGCAGCTGTTTGTGTGAGGATAATCCC-3′), resulting in a PCR fragment containing the D467, E468 → A467, A468 mutation as well as a PstI site at these positions. A 250-bp BamHI–PstI PCR fragment encompassing nucleotides +1158 to +1401 of the REV1 gene, and a 1560-bp PstI–Asp 718 PCR fragment encompassing nucleotides +1401 to the termination codon at position +2958, were directionally cloned into the BamHI–Asp 718 sites of pUC19, generating plasmid pBJ652. The presence of the A467, A468 mutation located at the PstI site was confirmed by DNA sequencing. Subsequently, the remainder of the REV1 ORF, from nucleotides +1 to +1158 along with the 5′-flanking DNA and 3′-flanking DNA, obtained from pBJ67 was cloned into pBJ652, regenerating the entire 5.6-kb REV1 DNA fragment, but now containing the rev1 D467, E468 → A467, A468 mutation. The wild-type and mutant genes were then cloned into YCplac33, a low-copy-number vector that carries the yeast URA3 gene as a selectable marker, as 5.4-kb SphI–BglII fragments, generating plasmids pBJ660 and pBJ661, respectively.
MMS sensitivity and MMS-induced mutagenesis
Cells were grown overnight in YPD medium, sonicated to disperse clumps, washed, and resuspended in 0.05 M KPO4 buffer at pH 7.0. Appropriate dilutions of MMS were added to 1-mL suspensions of cells adjusted to 1.5 × 108 cells/mL. Samples were incubated with vigorous shaking for 20 min at 30°C. Reactions were terminated by the addition of 1 mL of 10% sodium-thiosulfate. Appropiate dilutions were plated on YPD for viability determinations, and on synthetic complete medium lacking arginine but containing canavanine for determining the frequency of can1r mutations. Plates were incubated at 30°C, and colonies were counted after 3 d for viability and after 4–5 d for mutagenesis.
For determining the mutational specificity of AP sites, the apn1Δ apn2Δ strain was treated with 0.08% MMS for 20 min at 30°C. Genomic DNA was isolated from can1r mutants resulting from AP bypass and sequenced. Under these experimental conditions, relative to the spontaneous can1r mutation frequency, the increase in MMS-induced can1r mutations in the apn1Δ apn2Δ strain was >100-fold.
Purification of enzymes
Yeast Polδ was purified as described (Burgers and Gerik 1998). The purification of yeast Polζ and Rev1 was based on methods published previously, but in both cases the purification procedure included one additional chromatographic step. A GST–Rev3 fusion protein in complex with Rev7 protein (Polζ) was overexpressed in yeast strain Sc334 containing the plasmids pGST–REV3 and pREV7. The purification of GST–Rev3–Rev7 on glutathione-Sepharose 4B chromatography was carried out as described (Nelson et al. 1996b). Proteins eluted with glutathione from the matrix were dialyzed against buffer A (25 mM NaPO4 at pH 7.4, 100 mM NaCl, 10% glycerol, 0.01% NP-40, 5 mM DTT, 0.5 mM EDTA), followed by loading onto a Mini-Q column (Pharmacia). The column was washed with 20 column volumes of buffer A, and the proteins were eluted with a gradient of 10 column volumes of 100–500 mM NaCl. The GST–Rev3–Rev7 peak fractions were pooled and concentrated by dialysis in buffer A containing 200 mM NaCl and 50% glycerol. To overexpress Rev1 protein as a GST fusion protein, plasmid pBJ 392 (PKG : GST–REV1) was introduced in yeast strain LY2 (Matα gal1 reg1-501 leu2-3,-112 ura3-52 trp1Δ pep4-3 prb1-112). Cells were grown for 12 h in synthetic complete medium lacking leucine. Purification of GST–Rev1 on glutathione-Sepharose 4B was carried out as described for GST–Rev3–Rev7. The eluted protein sample was dialyzed against buffer A, followed by MiniS (Pharmacia) chromatography. GST–Rev1 protein was eluted by 100–500 mM NaCl gradient in buffer A, and pooled fractions were frozen in aliquots under liquid nitrogen and kept at −70°C.
DNA synthesis reactions
Standard DNA polymerase reactions (10 μL) contained 40 mM Tris-HCl at pH 7.5, 5 mM MgCl2, 1 mM dithiothreitol, bovine serum albumin (100 μg/mL), 10% glycerol, 100 μM dNTP, and 20 nM 5′-32P-labeled oligonucleotide primer annealed to an oligonucleotide template. Reactions were initiated by adding the enzymes Polζ, Polδ, or Rev1 in the amounts indicated in the figure legends. After incubation for 5 min at 30°C, reactions were terminated by the addition of 40 μL of loading buffer containing 20 mM EDTA, 95% formamide, 0.3% bromphenol blue, and 0.3% cyanol blue. The reaction products were subjected to electrophoresis in 10% or 20% polyacrylamide gels containing 8 M urea and visualized autoradiographically. Quantitation of reaction products was done with a Molecular Dynamics STORM phosphoimager and the ImageQuant software. DNA substrates S-2, S-4, S-5(G), S-5(A), S-5(T), S-5(C) were generated by annealing the 75-nt oligonucleotide template 5′-AGCTACC ATGCCTGCCTCAAGAGTTCGTAA0ATGCCTACACTGGA GTACCGGAGCATCGTCGTGACTGGGAAAAC-3′, which contained a model abasic site (a tetrahydrofuran moiety, purchased from Midland Co.) at the underlined position, to the 29-nt, 44-nt, and four different 45-nt 5′-32P-labeled oligonucleotide primers, N4577: 5′-GTTTTCCCAGTCACGACGATGCT CCGGTA-3′, N4309: 5′-GTTTTCCCAGTCACGACGATGCT CCGGTACTCCAGTGTAGGCAT-3′, or oligonucleotides that contain N4309 with one additional G,A,T, or C residue at its 3′ end, respectively. In the control nondamaged substrate S-1 or S-3, the 75-nt template oligonucleotide with a C residue instead of the AP site at position 45 was annealed to N4577 and N4309, respectively. The sequences of DNA substrates containing 18-nt template oligonucleotides annealed to 12-nt primer DNA are shown in the figures.
Steady-state kinetic analyses
Analysis of kinetic parameters for deoxynucleotide incorporation opposite the AP site or primer extension from nucleotides opposite this lesion was done as described (Mendelman et al. 1990; Goodman et al. 1993; Creighton et al. 1995). Briefly, Polζ was incubated with increasing concentrations of a single deoxynucleotide (0–2000 μM) for 1 min under standard reaction conditions. Gel band intensities of the substrates and products were quantitated by PhosphorImager. The percentage of primer extended was plotted as a function of dNTP concentration, and the data were fit by nonlinear regression using SigmaPlot 5.0 to the Michaelis–Menten equation describing a hyperbola, v = (Vmax × [dNTP]/(Km + [dNTP]). Apparent Km and Vmax steady-state parameters were obtained from the fit and used to calculate the frequency of deoxynucleotide incorporation (finc) and the frequency of extension (fext0 using the following equation: finc or ext = (Vmax/Km)incorrect pair/(Vmax/Km)correct pair.
Acknowledgments
This work was supported by National Institutes of Health research grants GM19261 and GM58534. We thank Tom Wood for sequencing the can1rmutations, which work was performed in the Molecular Biology Core Laboratory supported by NIEHS Center Grant P30 ESO6676.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement‘ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lprakash@scms.utmb.edu; FAX (409) 747-8608.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.882301.
References
- Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted genes. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JD, Kunz BA. Site and strand specificity of UVB mutagenesis in the SUP4-o gene of yeast. Proc Nat Acad Sci. 1990;87:9005–9009. doi: 10.1073/pnas.87.22.9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- Bjoras M, Klungland A, Johansen RF, Seeberg E. Purification and properties of the alkylation repair DNA glycosylase encoded MAG gene from Saccharomyces cerevisiae. Biochem. 1995;34:4577–4582. doi: 10.1021/bi00014a010. [DOI] [PubMed] [Google Scholar]
- Brash DE. Sunlight and the onset of skin cancer. Trends Genet. 1997;13:410–414. doi: 10.1016/s0168-9525(97)01246-8. [DOI] [PubMed] [Google Scholar]
- Burgers PMJ, Gerik KJ. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase δ. J Biol Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- Canella KA, Seidman MM. Mutation spectra in supF: Approaches to elucidating sequence context effects. Mutat Res. 2000;450:61–73. doi: 10.1016/s0027-5107(00)00016-6. [DOI] [PubMed] [Google Scholar]
- Creighton S, Bloom LB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzym. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- Cuniasse P, Sowers LC, Eritja R, Kaplan B, Goodman MF, Cognet JAH, LeBret M, Guschlbauer W, Fazakerley GV. An abasic site in DNA. Solution conformation determined by proton NMR and molecular mechanics calculations. Nucl Acids Res. 1987;15:8003–8022. doi: 10.1093/nar/15.19.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuniasse P, Fazakerley GV, Guschlbauer W, Kaplan BE, Sowers LC. The abasic site as a challenge to DNA polymerase. A nuclear magnetic resonance study of G, C, and T opposite a model abasic site. J Mol Biol. 1990;213:303–314. doi: 10.1016/S0022-2836(05)80192-5. [DOI] [PubMed] [Google Scholar]
- Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Nat Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik KJ, Li X, Pautz A, Burgers PMJ. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J Biol Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- Gibbs PEM, Lawrence CW. Novel mutageneic properties of abasic sites in Saccharomyces cerevisiae. J Mol Biol. 1995;251:229–236. doi: 10.1006/jmbi.1995.0430. [DOI] [PubMed] [Google Scholar]
- Giot L, Chanet R, Simon M, Facca C, Faye G. Involvement of the yeast DNA polymerase δ in DNA repair in vivo. Genetics. 1997;146:1239–1251. doi: 10.1093/genetics/146.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF, Creighton S, Bloom LB, Petruska J. Biochemical basis of DNA replication fidelity. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- Haracska L, Yu S-L, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nature Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- Haracska, L., Washington, M.T., Prakash, S., and Prakash, L. 2001. Inefficient bypass of an abasic site by DNA polymerase η. J. Biol. Chem. (in press). [DOI] [PubMed]
- Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes & Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J Biol Chem. 1999;274:15975–15977. doi: 10.1074/jbc.274.23.15975. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000a;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J Biol Chem. 2000b;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- Kalnik MW, Chang C-N, Grollman AP, Patel DJ. NMR studies of abasic sites in DNA duplexes: Deoxyadenosine stacks into the helix opposite the cyclic analogue of 2-deoxyribose. Biochem. 1988;27:924–931. doi: 10.1021/bi00403a013. [DOI] [PubMed] [Google Scholar]
- Kunz BA, Henson ES, Roches H, Ramotor D, Nunoshiba T, Demple B. Specificity of the mutator caused by deletion of the yeast structural gene (APN1) for the major apurinic endonuclease. Proc Nat Acad Sci. 1994;91:8165–8169. doi: 10.1073/pnas.91.17.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW, Hinkle DC. DNA polymerase ζ and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surveys. 1996;28:21–31. [PubMed] [Google Scholar]
- Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochem. 1972;11:3610–3617. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- Mendelman LV, Petruska J, Goodman MF. Base mispair extension kinetics. Comparison of DNA polymerase α and reverse transcriptase. J Biol Chem. 1990;265:2338–2346. [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996a;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- ————— Thymine–thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996b;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol Gen Genet. 1981;184:471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- Roy R, Brooks C, Mitra S. Purification and biochemical characterization of recombinant N-methylpurine-DNA glycosylase of the mouse. Biochem. 1994;33:15131–15140. doi: 10.1021/bi00254a024. [DOI] [PubMed] [Google Scholar]
- Strauss BS. The ‘A-rule’ of mutagen specificity: A consequence of DNA polymerase bypass of non-instructional lesions? BioEssays. 1991;13:79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine–thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc Nat Acad Sci. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S-L, Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol Cell Biol. 2001;21:185–188. doi: 10.1128/MCB.21.1.185-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]