Abstract
The GAL4 dimerization domain (GAL4-dd) is a powerful transcriptional activator when tethered to DNA in a cell bearing a mutant of the GAL11 protein, named GAL11P. GAL11P (like GAL11) is a component of the RNA–polymerase II holoenzyme. Nuclear magnetic resonance (NMR) studies of GAL4-dd revealed an elongated dimer structure with C2 symmetry containing three helices that mediate dimerization via coiled-coil contacts. The two loops between the three coiled coils form mobile bulges causing a variation of twist angles between the helix pairs. Chemical shift perturbation analysis mapped the GAL11P-binding site to the C-terminal helix α3 and the loop between α1 and α2. One GAL11P monomer binds to one GAL4-dd dimer rendering the dimer asymmetric and implying an extreme negative cooperativity mechanism. Alanine-scanning mutagenesis of GAL4-dd showed that the NMR-derived GAL11P-binding face is crucial for the novel transcriptional activating function of the GAL4-dd on GAL11P interaction. The binding of GAL4 to GAL11P, although an artificial interaction, represents a unique structural motif for an activating region capable of binding to a single target to effect gene expression.
Keywords: GAL4, dimerization domain, GAL11P, NMR structure
The GAL4 transcriptional activator is required for the regulation of genes involved in galactose and melibiose metabolism in the yeast Saccharomyces cerevisiae (Johnston 1987). GAL4 has a modular structure in which distinct regions of the molecule mediate recognition and binding to DNA, activation of transcription, and dimerization (Fig. 1A; Laughon 1984; Laughon et al. 1984; Keegan et al. 1986; Ma and Ptashne 1987; Lin et al. 1988; Carey et al. 1990). Only the structure of the DNA-binding domain (residues 1–66) has been solved alone in solution (Baleja et al. 1992; Kraulis et al. 1992) and in complex with DNA (Marmorstein et al. 1992). It contains an N-terminal DNA-recognition domain (residues 7–40), in which six cysteines bind two Zn+2 ions. In the free protein in solution, the region from residue 41–66 is unstructured. In the crystal structure of the complex with DNA, residues 50–63 form a coiled-coil dimerization element that extends perpendicularly away from the DNA helix (Marmorstein et al. 1992). Nevertheless, GAL4 1–65 is a monomer in the absence of DNA. Functional characterization of the 881 residue GAL4 protein has shown that a DNA-independent dimerization domain extends beyond residue 63 comprising the sequence from 65 to 94 (Carey et al. 1989); however, no structure of a construct containing the complete dimerization domain has been reported yet. Transcriptional activation function in yeast has been assigned to two regions: the primary activating region comprising residues 768–881 and a weaker one that lies between residues 148–196 (Keegan et al. 1986; Ma and Ptashne 1987; see Fig. 1).
Figure 1.
Functional domains of GAL4 and primary sequence of the GAL4 dimerization domain (GAL450–106). (A) GAL4 contains several separable functional regions, including a DNA-binding domain, a dimerization domain, and three activating regions (the activating region between 94–106 is nonfunctional in yeast; Simkovich 1996). The C-terminal region of GAL4 (851–881) recognizes the GAL80 inhibitory protein (not shown). (B) Partial amino acid sequences of the GAL4-dd and LAC9. Conserved residues are shaded in yellow. The double mutation on GAL4-dd (87 and 90) are colored red (bold). The secondary structure elements, α1, α2, and α3 are indicated by blue cylinders. (C) Temperature dependence of the unfolding of GAL4-dd. The denaturation and renaturation curves are shown in pink and blue, respectively. The CD spectra were taken in a sample 0.5 mg/mL in 50 mM phosphate buffer at pH 7.2 in an AVIV instrument, model 62DS (AVIV Associates) at 222 nm.
GAL450–97 ordinarily mediates dimerization (Carey et al. 1989). However, the same fragment manifests a novel transcriptional activating capability in yeast cells carrying a single point mutation in the GAL11 protein. This mutant GAL11P (P standing for transcriptional potentiator) and wild-type GAL11 are located in an ∼2-MD complex with RNA polymerase and 50 or more polypeptides in what has been called the RNA–polymerase II holoenzyme (Kim et al. 1994; Barberis et al. 1995; Hengartner et al. 1995).
The P mutation replaces the N342 residue with one of any number of hydrophobic amino acids and confers to GAL11 the ability to bind to the GAL4 dimerization domain (GAL4-dd). On the contrary, GAL4-dd does not interact in vitro with wild-type GAL11 as detected by several methods (see below). Furthermore, the strength of the interaction between the GAL4-dd and each GAL11P allele as measured in vitro correlates directly with the level of gene activation observed in vivo. The interacting regions of GAL4 and GAL11P can be exchanged such that the P region of GAL11P is tethered to DNA, and the GAL4-dd is attached to the holoenzyme without loss of transcriptional activation. Taken together, these results strongly suggest that recruitment of the holoenzyme to the promoter, whether by the fortuitous GAL4–GAL11P interaction or by a classic activator, results in transcriptional activation (Barberis et al. 1995; Simkovich 1996; Farrell 1996; Farrell et al. 1996; Ptashne and Gann 1997; Gaudreau et al. 1998; Keaveney and Struhl 1998).
To elucidate the molecular basis of transcriptional activation mediated by the GAL4-dd–GAL11P interaction, we studied the solution structure of the entire GAL4 dimerization domain (residues 50–106) containing the double mutation (Q87R, K90E) and characterized its interaction with GAL11P N342V, the strongest GAL11P allele. GAL4-dd (Q87R, K90E) shows a higher in vitro affinity for GAL11P than does the wild-type GAL4-dd and activates transcription more strongly in GAL11P yeast cells (Simkovich 1996). This particular GAL4 and GAL11P pair interacts in vitro with an estimated affinity constant of 3.3 × 106/M or higher (Farrell et al. 1996; Simkovich 1996). We show that the GAL4-dd consists of three helical segments connected by flexible loops. Dimerization occurs via coiled-coil interactions of all three helices. We also analyze the interaction of the GAL4-dd with GAL11P261–352 by nuclear magnetic resonance (NMR) titration experiments and test the transcriptional activation in vivo by alanine scanning mutagenesis of GAL4-dd.
Results
GAL4-dd adopts a dynamic state lacking a sharp thermal unfolding transition
To find the best conditions for a structure determination, we recorded 1H–15N heteronuclear single quantum coherence (HSQC) spectra of GAL4-dd over a range of pH and temperature. The spectra were best in a narrow range of pH near 7.5 and at 35°C. Variation of the temperature between 10°C and 50°C results in large spectral changes that are different from the kind usually observed for cooperative thermal unfolding. Although several sharp signals that were later assigned to the C-terminal tail stayed almost invariant, most other signals changed line shapes with temperature, disappeared, or newly appeared in an uncorrelated manner. This indicates temperature-dependent noncooperative conformational changes. Consistently, the temperature dependence of the circular dichroism (CD) spectrum shown in Figure 1C lacks a sharp cooperative unfolding transition and resembles the flat melting curves of helix–coil transitions in short helical polypeptides (Zimm and Bragg 1959). Aware of the dynamic state of the protein, we pursued a characterization of GAL4-dd at 35°C and pH 7.4.
GAL4-dd contains three helices that dimerize via coiled-coil interactions
Figure 1B shows the amino acid sequence of the GAL4-dd (50–106) aligned with the primary sequence of Lac9, the transcriptional activator that regulates the expression of galactose and lactose genes in Kluveromyces lactis (Das and Hollenberg 1982, Salmeron and Johnston 1986; Wray et al. 1987). Resonance assignments of GAL4-dd were pursued with NMR triple-resonance and nuclear Overhauser enhancement (NOE) methods. Of the 57 residues, 43 were assigned unambiguously. Five residues at the N terminus (50–54) and four residues at the C terminus (101 and 103–105) could not be assigned. Flexible tails often remain unassigned in NMR analyses. Furthermore, nine residues from interior sites, subsequently showed to be located in connecting loop regions (67, 69, 73–77, 84, and 86), could not be identified. Either they were absent or the few unassigned signals showed no spectral properties enabling assignment. The absence of peaks is consistent with the dynamic state of GAL4-dd mentioned above.
Observation of characteristic NOE patterns and analysis of chemical shifts revealed that there are three short helices at positions 55–66, 78–84, and 87–97 (Fig. 1B). However, there are no long-range contacts between the three helices indicating that the structure is extended. The first helix α1 coincides with the terminal helix in the crystal structure of the complex of GAL41–65 with DNA (Marmorstein et al. 1992).
To characterize the mode of dimerization, we followed a previously described procedure (Walters et al. 1997a) in which a uniformly 15N- and 2H-labeled sample was mixed with unlabeled protein. In H2O solution, all amide groups are protonated, and all cross peaks between 15N-bound amide protons and aliphatic protons in a three-dimensional 15N-dispersed NOE spectroscopy (NOESY) experiment must be across the dimerization interface. In this way, 40 intermonomer NOE contacts were found (involving 16 residues of each monomer), 24 for α1, two for the loop between α1 and α2, six for α2, six for α3, and two for the region beyond α3.
Structure of the GAL4 dimerization domain
Structures of GAL4-dd were calculated with a simulated annealing protocol within the CNS software package (Brünger et al. 1998). The constraints used for the structure calculations are listed in Table 1. They consist of short and medium-range intermonomer constraints that establish the secondary structure of the three helices and the 40 intermonomer constraints that determine the parallel dimer structure. Because only one resonance signal per residue was observed, the dimer must be symmetric, and C2 symmetry was enforced in the structure calculations. Figure 2A shows a ribbon diagram of a representative structure. The overall dimensions are ∼13 Å × 62 Å × 5 Å. Figure 2B shows the backbone atoms of the 17 lowest energy structures with the C, Cα, and N atoms, of α1, α2, and α3 simultaneously superimposed. The average root mean-square deviation (rmsd) from the mean structure is 3.8 Å. However, the rmsd values decrease considerably when the backbone atoms of the helices are separately superimposed and become 0.45 Å, 1.7 Å, and 0.75 Å for α1 (residues 55–66), α2 (residues 78–84), and α3 (residues 87–97), respectively (Fig. 2C). α1 and α3 helices show the classic coiled-coil conformation. The α3 helices are less super-coiled than the α1 pair, however, and their interactions seems to be weaker as fewer interhelix contacts were observed. The short α2 helices pack onto each other with angles >90° in an almost antiparallel orientation (see Fig. 2A). A relatively long loop connects α1 and α2, whereas α2 and α3 are linked by only a couple of residues. The longest loop bears a strictly hydrophobic segment from F68 to F72. In contrast, the subsequent segment from P73 to L77 is predominately hydrophilic. Because there was no observable signal for any of the latter residues, we conclude that this segment might undergo slow conformational exchange.
Table 1.
Structural statistics for the NMR structure calculations
| Restraint type
|
Number of restraints
|
|---|---|
| NOE distance restraints per monomer (TOTAL)a | 436 |
| Intra-residue | 230 |
| Inter-residue | 206 |
| Sequential | 113 |
| Medium | 93 |
| i,i + 2 | 44 |
| i,i + 3 | 40 |
| i,i + 4 | 9 |
| Hydrogen Bonds | 38 |
| Inter-monomer | 40 |
| Ramachandran plot for all assigned residues | |
| Most favorable region (%) | 71.0 |
| Additionally allowed region (%) | 20.7 |
| Generously allowed region (%) | 5.1 |
| Disallowed region (%) | 3.2 |
| Average RMSD from idealize covalent geometry | |
| Bonds (Å) | 0.002 ± 0.0001 |
| Angles (°) | 0.420 ± 0.0140 |
| Impropers | 0.292 ± 0.0234 |
| Average RMSD (Å) of C, Ca, N backbone atoms to the average structure | |
| α1, α2, α3 | 3.8 |
| α1 (residues 55–66) | 0.45 |
| α2 (residues 78–84) | 1.70 |
| α3 (residues 87–97) | 0.75 |
No violation greater than 0.3 Å was found. The 18 residues with unassigned amide group including proline 73 are: 50–54, 67, 69, 74–77, 84, 86, 101, and 103–105.
Figure 2.
Three-dimensional structure of the GAL4-dd. (A) A stereoview of the ribbon diagram of a representative structure of the GAL4-dd showing residues H53 to D100. The N and C termini are labeled N and C, respectively. The two monomers are colored differently. Unassigned residues P73 to L77 within the loop connecting α1 and α2 are colored orange. This figure was generated with the program MOLMOL (Koradi et al. 1996). (B) A stereoview of the backbone atoms from residues L54 to D100 of the 17 lowest energy structures with helices α1, α2, and α3 (blue) simultaneously superimposed. The average backbone RMSD of α1, α2, and α3 to the mean structure is 3.8 Å. (C) A stereoview of the 17 lowest energy structures whereby the backbone atoms from residues H53 to D100 of each of the three helices were separately superimposed. For clarity, the loops regions are displayed for only one structure. The backbone atoms of the unassigned P73 to L77 segment are colored in orange as in A. The average backbone RMSD of α1, α2, and α3 to the mean structure is 0.45, 1.7, and 0.75 Å, respectively. (D) Ensemble of the 17 structures aligned by superimposing the backbone heavy atoms of helix α1.
The disorder between the helices, which is apparent in Figure 2B, is primarily a variation of the amount of twist around the C2 symmetry axis between the α1–α1‘, α2–α2‘, and α3–α3′ coiled coils. This is shown in Figure 2D in which the structures were aligned by superimposing only the backbone atoms of the α1 helices. The figure shows that there is variability of orientation around the C2 symmetry axis, of the large loop, the α2 and α3 helices relative to the α1–α1‘. The tilt angle between the α1–α1‘ and α3–α3′ coiled coils varies rather little in the ensemble of calculated structures. This is reasonable because any permanent tilt would make the dimer asymmetric, which would contradict the symmetric nature of the observed NMR spectra. From these considerations, we conclude that the average solution structure of GAL4-dd is extended as shown in Figure 2A, and the disorder consists of the amount of twist and presumably a transient variation of the lengths of the helices.
Dimerization interface
The GAL4-dd dimerizes in solution because of multiple coiled-coil type interactions between symmetry related atoms situated along each monomer. The identification of the intermonomers contacts forming the dimer interface was essential for obtaining the structure of the GAL4 dimer. Intermonomer NOEs were obtained using a previously described asymmetric deuteration strategy (Walters et al. 1997a).
Figure 3A and B shows the superposition of the 17 lowest energy structures with the side chains of residues forming the dimer interface within α1 and α3 displayed. A projection of the helices onto a wheel diagram is shown in Figure 3C and D. Annotating the heptad repeat sequence characteristic of coiled coils (Lupas 1996) by letters from a to g, α1 contains the sequence a1b1c1d1e1f1g1a2b2c2d2 in which the hydrophobic residues (in boldface) at positions a1, d1, a2, and d2 corresponding to the residues L54, V57, L61, and L64, respectively, form the core of the dimer interface. The same arrangement was reported for α1 in the crystal structure of GAL4 complexed with DNA (Marmorstein et al. 1992). The disruption of the coiled-coil conformation occurs at the second half of the second heptad (e2f2g2) where position g2 is occupied by a hydrophobic residue (L67) instead of a hydrophilic one presumably causing the interruption of the coiled coil.
Figure 3.
Dimerization interface within α1 and α3 of GAL4 (50–106). (A) A stereoview of the superimposed backbone atoms within α1 for the 17 lowest energy structures. The side chains of V57, L61, and L64 forming intermonomer contacts are displayed in different colors for each monomer. (B) A stereoview of the superimposed backbone atoms within α3 for the 17 lowest energy structures. The side chains of I89, L93 forming intermonomer contacts and F97 are shown in different colors for each monomer. (C) Helical wheel representation of α1 with hydrophobic, polar, and charged residues displayed in red, green. and blue, respectively. (D) Helical wheel representation of α3. Same color code for the residues as C but glycine in black.
Among the residues located in the loop between α1 and α2, I71 shows intermonomer contacts. In the short helix α2, I80, L81, and K82 also show dimer contacts. Finally, in α3, we observe intermonomers contacts for I89, E90, and L93, but the packing of the α3 helices appears to be less tight than in α1. Considering that the number of observed intermonomer and intramonomer NOE distance restraints was higher for α1 coiled coil, the differences may reflect variations in the dynamic behavior of the molecule. Therefore, we assayed this possibility by studying the mobility of the GAL4-dd. The dynamic properties of the GAL4-dd may be relevant for the transcriptional activation function as will be discussed below.
Protein mobility
To understand whether the apparent disorder in some parts of the GAL4-dd structure is due to mobility rather than lack of constraints, and to provide a basis for a possible correlation between mobility and activation function, we studied the motional properties of the GAL4-dd by using 15N relaxation experiments. Three relaxation parameters were measured, the nitrogen longitudinal relaxation rate RN(Nz), the nitrogen transverse relaxation rate RN(Nx,y), and the heteronuclear NOE (XNOE) (Fig. 4; Peng and Wagner 1992). The 15N relaxation parameters are related to the rotational diffusion of the N–H bond due to overall tumbling and internal motion, and so they contain information on the molecular mobility of the protein. Increased protein mobility is manifested in faster RN(Nz) rates, slower RN(Nx,y) rates and augmented XNOE magnitudes. The GAL4-dd (55–102) displays average values for these relaxation parameters comparable to those measured for related proteins of similar size, such as PUT3 (Walters et al. 1997b). The average values are 2.43/sec for RN(Nz) with a range of 1.66 to 2.88/sec, 9.48/sec for RN(Nx,y) with a range of 4.48 to 12.9/sec, and 0.48 for the XNOE with a range of 0.22 to 1.94. It is readily apparent from Figure 4 that the relaxation behavior of the GAL4-dd is not uniform throughout its amino acid sequence. The N-terminal half bearing α1 displays a lower average mobility than the C-terminal half composed of α2 and α3 with an overall average RN(Nx,y) of 10.6/sec and 9.3/sec and an overall XNOE average value of 0.32 and 0.42, respectively. Leu 61 and Leu 64, which form the core of the dimer interface in α1, show the lowest XNOE values. Of note, the mobility of α3 increases continuously toward the end and continues to grow toward the unstructured end (Fig. 4B,C). This is consistent with the picture of a fraying helix with a poorly defined helix boundary.
Figure 4.
15N relaxation rates and heteronuclear NOE enhancements for GAL4-dd. (A) Relaxation rates for the nitrogen longitudinal magnetization, RN(Nz). (B) Relaxation rate for the nitrogen transverse magnetization, RN(Nx,y). (C) Heteronuclear NOE enhancements, XNOE = (Isat-Ieq)/Ieq, where Isat and Ieq represent the steady state heteronuclear NOE and equilibrium value, respectively. All relaxation parameters where measured at 400 MHz with a 1-mM 15N isotopically labeled sample at pH 7.4 and at 35°C. Relaxation rates and XNOE values for residues with unassigned amide resonances and P73 are represented as zero values in all graphs. The signal of residue F68 in the XNOE experiment was too weak to allow analysis. The helices are indicated below the residue number by blue rectangles. L61 and L64 within the α1 are indicated by arrows. The numbers above each data point of the XNOE plot indicate the number of intermonomers NOEs determined for each residue as described (Walters et al. 1997a).
The different dynamics properties between the α1 and α2–α3 regions can explain, partly, the relatively reduced number of NOEs observed for the C-terminal region of the GAL4-dd and consequently, the better structural definition for α1 than α2–α3. The number of intermonomer contacts observed for each residue is indicated in Figure 4C.
Alanine-scanning mutagenesis to probe the GAL4-dd–GAL11P261–352 interaction
Activation in vivo
Previous truncation analyses identified the GAL4 residues 58–97 as the minimal segment necessary for transcriptional activation in GAL11P cells (Barberis et al. 1995). To identify individual residues within this segment of GAL4 that play a role in transcriptional activation in vivo, we performed site-directed alanine substitution mutagenesis of each of the 40 residues (Fig. 5). The mutations were made in the context of a chimeric protein in which residues 50–97 of GAL4 were fused to LexA, a dimeric bacterial DNA-binding protein that functions efficiently in yeast. In GAL11P cells, the LexA–GAL4 (50–97) fusion protein stimulates the expression of a reporter gene bearing LexA-binding sites upstream of the minimal GAL1 promoter (Barberis et al. 1995; Farrell et al. 1996). Thus, it allowed us to circumvent any effects of alanine substitutions on dimerization and subsequent DNA binding of GAL4 to its cognate sites and identified those residues that specifically effect the ability of GAL4-dd to activate transcription. Figure 5A shows a scheme of the experimental design and a summary of the activity of each mutant normalized to the wild-type LexA–GAL4 (58–97) fusion protein. The results shown in Fig. 5A identify a cluster of hydrophobic residues from 69–72 (LLIF), and residues L64, E75, D76, S85, L86, I89, L92, L93, L96, and F97 as essential for activation in vivo.
Figure 5.
Alanine scanning mutagenesis experiments identify GAL4 residues crucial for the activation in vivo and electrophoretic mobility shift assays identify GAL4 mutants that bind GAL11P263–352. (A) In vivo activation assay. (Top) Design of the experiment: Two LexA-binding sites are located 50 bp upstream of the GAL1 promoter TATA box. The expression of the reporter gene (lacZ) is regulated by the ability of LexA derivatives to interact with GAL11P and recruit the transcriptional machinery to this promoter. (Bottom) Summary of the alanine scanning mutagenesis of GAL4, which identify residues that are essential for activation by GAL11P in vivo. The levels of activation of each mutant were normalized against the wild-type LexA(1–202) + Gal4(50–97) derivative. Ala 91 in the wild-type GAL4 was replaced with a leucine. (B) Control for the gel-shift experiments. GAL4 (1–97) binds to its cognate site whereas neither GST–GAL11 nor GST–GAL11P binds this oligonucleotide. As shown in lanes 11–16, GST–GAL11P but not GST–GAL11 binds the GAL4–DNA complex, causing a further decrease in the electrophoretic mobility of the olignucleotide–protein complex (supershift). GST–GAL11263–352 was at the final concentration of 1.6, 3.2, and 4.8 μM in lanes 2–4 and 11–13, respectively and so was GST–GAL11P263–352 in lanes 5–7 and 14–16. GAL4 (30 nM) was present in lanes 9 and 11–16. GAL4 (15 nM) was added to lane 8, and 45 nM GAL4 was added to lane 10. (C–G) Each of the alanine substituted derivatives of GAL4 (1–97) binds to the cognate DNA-binding site, and their GST–GAL11P263–352 binding properties are approximated by the intensity of the supershift. In this experiment, 3 μM GST–GAL11P263–352 was added to each GAL4–DNA complex. GAL4 mutants were at ∼30-nM concentrations; at this concentration, each derivative saturably binds the labeled DNA probe.
To ascertain if the loss of activation occurs because of a loss in interaction, we purified wild-type GAL4 (1–97) and 39 mutant forms each of which bears an alanine mutation at a single position scanning the residues 58–97, except for position 91, which is an alanine in the wild type and substituted by leucine. The mutant GAL4, D76A, was insoluble and thus refractory to further analysis. The in vitro interaction of these GAL4 derivatives with a fragment of GAL11 and GAL11P (residues 263–352) expressed as a GST fusion protein was measured in a gel mobility shift assay (Fig. 5B–G). In this assay, neither GAL11 nor GAL11P bind to DNA bearing a GAL4 DNA-binding site (Fig. 5B, lanes 1–7), but, as expected, GAL4 (1–97) does (Fig. 5B, lanes 8–10). Conversely, Figure 5B shows that GAL11P (lanes 14–16), but not GAL11 wild type (lanes 11–13), binds to the DNA-tethered GAL4 as indicated by the supershift of the protein–DNA complex. Figure 5(C–G) shows that all 39 GAL4 mutants bound the GAL4 DNA site, and 26 of these interacted with GAL11P as shown by the supershift.
There is a strong correlation between the ability of each GAL4 mutant to stimulate activation in vivo as measured in Figure 5A and the ability to interact with GAL11P in vitro as measured in Figure 5C–G. Thus, of the 39 mutants, those that interacted with GAL11P in vitro, all activated in vivo. Moreover, all of the 13 derivatives that failed to interact in vitro (positions 64, 69–72, 75, 85, 86, 89, 92, 93, 96, and 97) also failed to activate in vivo. K90A bound Gal11P very weakly in vitro but activated well in vivo. Conversely, E58A bound well in vitro but did not activate well in vivo; however, it was particularly difficult to purify, and thus its inability to activate transcription might be a consequence of instability in vivo.
The results from the alanine scanning are elucidated further by visualizing the crucial mutations (Fig. 7B, below) on the GAL4-dd structure (Fig. 1B). Leu 69 to Phe 72 map on the hydrophobic part of the loop connecting α1 and α2 whereas E75 and D76 are on the hydrophilic section of the same loop. M83 to L86 constitute the loop between α2 and α3, and I89, L92, L93, L96, and F97 are in helix α3. Leu 64 form the core of α1 dimerization interface, and the effect of its mutation is probably purely structural.
Figure 7.
GAL4 residues critical for binding GAL11P and transcriptional activation in vivo map to the hydrophobic α1–α2 loop, α3 and beyond. (A) Molecular surface representation of one representative structure of GAL4-dd. Residues experiencing large and small or no changes in their chemical shifts on addition of GAL11P are displayed in red and blue, respectively. Unassigned residues are displayed in gray. The mutations in the double mutant version of GAL4-dd, Q87R, and K90E, are labeled. (B) Residues that critically affect transcriptional activation in vivo when mutated to alanine are displayed in yellow onto the same molecular structure as in Fig. 7A. A ribbon diagram of the GAL4-dd, with the same color code, with the side chains atoms of the residues sensitive to alanine mutagenesis within α1, α2, and α3 is displayed in the right panel. This figure was generated with the program MOLMOL (Koradi et al. 1996).
NMR titration of GAL4-dd with GAL11P261–352
To identify the GAL11P-binding site, we titrated 15N-labeled GAL4-dd with unlabeled GAL11P261–352 and monitored the effect with 1H–15N HSQC spectra. Three of five spectra recorded in this series are shown in Figure 6B (0, 0.5, and 1 mM GAL11P). Increasing concentrations of GAL11P from 0 to 2 mM, in 0.25-mM increments, were added to 1 mM (dimer) 15N-GAL4-dd while measuring 1H–15N HSQC spectra. Because only the signals of protons covalently attached to 15N nuclei are detected, all resonances observed arise from the 15N-labeled GAL4-dd while the GAL11P peptide is invisible. We obtained the following results.
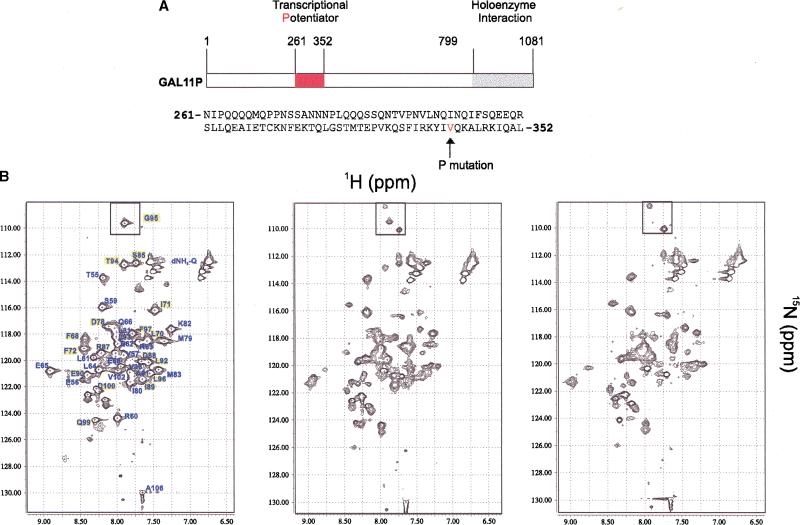
Figure 6.
GAL11P binds the GAL4-dd dimer with slow exchange kinetics, and as a monomer. (A) Functional domains and partial amino acid sequence of GAL11P261–352. The N342V mutation is labeled in red. Position 342 is surrounded by a relatively high number of hydrophobic and basic residues. (B) NMR titration of 15N-labeled GAL4-dd (50–106) with unlabeled GAL11P (261–352). (1H,15N) HSQC spectra of 1.0 mM GAL4 (50–106) dimer alone (left), in the presence of 0.5 mM GAL11P (middle), and in the presence of 1.0 mM GAL11P (right). The sample contained 50 mM phosphate buffer at pH 6.8, 100 mM NaCl. Cross peaks are labeled in the spectra of free GAL4 (left). The signal of G95 that splits in two asymmetric peaks on GAL11P binding is marked with a box at the top of the spectra. Several other signals, primarily from helix α3 disappear in the presence of GAL11P. The assignment of these signals is highlighted in the left panel. Numerous new signals of lower intensity appear in the right panel manifesting widespread asymmetry of GAL4-dd in the complex with GAL11P.
GAL11P forms a tight complex with GAL4-dd with slow exchange kinetics (koff < 5/sec) because adding GAL11P causes disappearance of a subset of the HSQC peaks and appearance of new peaks proportional to the concentration of the ligand (Fig. 6B). Addition of the wild-type GAL11 peptide under otherwise identical conditions had no effect on the GAL4-dd resonances up to a 1:3 ratio of GAL4-dd to wt-GAL11 (data not shown).
GAL11P binds as a monomer to the GAL4-dd dimer because addition of GAL11P to the 1 mM (dimer) sample of GAL4-dd causes spectral changes up to 1 mM (monomer) GAL11P concentration. No significant spectral changes happen at higher concentration of GAL11P. The relative concentrations of the GAL4-dd versus GAL11P peptide particularly for this assays were verified by amino acid analysis.
Formation of the monomer–dimer complex introduces asymmetry into the GAL4-dd dimer (Fig. 6B). Although the spectrum of the complex has not been assigned, a splitting of signals is observed for several residues, most clearly documented for G95, which is the only glycine and thus well separated from all other signals. The peak at 109.6 ppm 15N chemical shift splits in two peaks at 108.3 and 110.1 ppm 15N chemical shift. Many other peaks disappear in the titration and numerous new signals of half the intensity appear. Thus, introduction of asymmetry is manifested in many other peaks. However, the new peaks cannot easily be assigned because they are far away from those of the free GAL4-dd, and a complete assignment of the complex was not achieved. The assignments for those peaks that clearly disappear in the complex are highlighted in yellow in the left panel of Figure 6B; those that remain unchanged are not.
Examination of the chemical shift perturbation of the NMR resonances on titration shows that there are no or only very minor changes observed for the entire helix α1. All residues of the loop connecting α1 and α2 disappear with increasing GAL11P concentration. The helix α2 is only affected at its C-terminal end by the addition of GAL11P. However, all of the helix α3 and the subsequent residues 97–100 disappear or split in two components. Because the binding of GAL11P peptide to GAL4-dd undoubtedly brings G95 to the vicinity of GAL11P, we assumed that the nearby residues whose resonances also are perturbed in the titration experiments also must be in close proximity to GAL11P. However, many of these GAL4 residues that participate in the dimerization may play a role in direct interactions with GAL11P, as observed in the case of G95. These observations indicate that GAL11P binds GAL4-dd via the α1–α2 loop and the helix α3.
Figure 7 shows the residues sensitive to GAL11P identified by NMR titration (Fig. 7A) and by alanine-scanning mutagenesis (Fig. 7B) mapped onto the molecular surface of GAL4-dd. Residues that are largely perturbed on GAL11P addition in the NMR titration are colored in red (Fig. 7A). Two regions stand out: the hydrophobic α1–α2 loop and α3. Alanine substitutions that decrease activation in vivo to lower than 10% of the wild-type GAL4-dd are colored in yellow onto the same molecular surface representation of the GAL4-dd (Fig. 7B). Comparison of A and B in Figure 7 reveals a remarkably close match between the red patches identified by NMR titration (left panel) with the yellow ones identified by alanine scanning (right panel). Thus, in general the GAL4-dd residues shown to be sensitive to GAL11P in the NMR titration experiment are in close agreement with those shown to be crucial for transcriptional activation in vivo with GAL11P.
Discussion
GAL4-dd structure
The GAL4-dd structure is a C2-symmetric homodimer and contains three distinct coiled-coil helix pairs connected by mobile loops. The average structure is extended but mobile and poorly defined with respect to the twist angle around the C2 symmetry axis. Tilting motions that would bend the C2 symmetry axis are indicated by the distribution of the ensembles of calculated structures but must be fast on the NMR time scale because there is no solid evidence for asymmetry in the NMR spectra of the free GAL4-dd dimer. The roughly linear melting behavior, the absence of cooperative thermal unfolding, and the appearing and disappearing of HSQC peaks with variation of temperature indicate a unique dynamic nature of the GAL4-dd structure. This could be pictured as a transient melting and folding of the helix ends and a concomitant structural change of the connecting loops.
Complex structure
Our NMR titration data indicate that the dimeric GAL4-dd forms a tight complex with monomeric GAL11P261–352 with dissociation rates that are slow on the NMR time scale (koff < 5/sec). This is consistent with a Kd of 300 nM or lower (Farrell et al. 1996; Simkovich 1996). The monomer-to-dimer association renders the GAL4-dd asymmetric, which is clearly documented by peak doubling in the NMR spectra (Fig. 6B). This is a unique observation for the interaction of an activation domain with its target protein. Structural data on complexes between homodimers and monomers are rare but have been reported, such as the complex between human growth hormone and its dimeric receptor (de Vos et al. 1992) or the RNA bacteriophage coat protein–operator complex (Valegard et al. 1994). The symmetry-breaking aspects have been discussed extensively in Bardsley et al. (1998). As discussed for the previous cases, binding of the first GAL11P monomer changes the conformation of the GAL4-dd dimer in a way that abrogates affinity for the second binding site. This is an extreme case of negative cooperativity.
Despite the tight interaction, the complex is in a dynamic state. This is concluded from the HSQC spectrum recorded at the end point of the titration (Fig. 6B, right) and from the spectra recorded with excess of GAL11P (data not shown). These spectra contain far less than the number of signals expected for an entirely folded GAL4-dd dimer in complex with the unlabeled GAL11P. Conversely, an HSQC spectrum of free GAL11P261–352 shows only 48 weak peptide NH cross peaks (data not shown). Thus, GAL4-dd, GAL11P261–352, and their complex contain large portions that undergo apparently slow conformational exchange, which would make a more detailed structural characterization difficult.
GAL4-dd shows a novel transcriptional activation motif
The NMR titration and the alanine scanning experiments locate the GAL11P-binding site to the loop regions, the terminal helix α3, and part of the segment beyond α3. In wt-GAL4-dd, these regions bear six negative charges (E75, D76, D78, D84, D88, and D100) balanced by three basic side chains (R74, K82, and K90; Fig. 1B). The double mutant (Q87R, K90E) that introduces an extra negative charge shows higher affinity to GAL11P and activates transcription stronger. Thus, the novel activation region of GAL4-dd shows the feature of high acidity present in other classic activating domains. Intriguingly, the GAL11P-binding face on GAL4-dd coincides with part of an acidic segment (residues 75–147) that previously been has shown to activate promoters bearing GAL4-binding sites in HeLa cell extracts (Lin et al. 1988). Therefore, GAL11P may use the same activator motif.
In contrast with earlier studies of activating domains, which all revealed a single helix in the target-bound state (Kussie et al. 1996; Radhakrishnan et al. 1997; Uesugi et al. 1997) the novel activation motif of the GAL4-dd is a coiled-coil dimer supporting two acidic/hydrophobic loops. The mobility experiments revealed that the GAL4-dd (the terminal α3 helix pair) shows enhanced mobility gradually increasing toward the C terminus (Fig. 4). This may be important for activation function and is consistent with the observation that mutations stabilizing the α3 dimerization reduce the activation potential (A.Z. Ansari and M. Ptashe, unpubl.).
The apparent need for weak dimer interaction of the α3 helices may explain why the double mutant version of the GAL4-dd studied here is a better activator. Both residues R87 and E90 are placed to the same side of the helix facing away from the dimerization interface. Interaction between the two side chains may stabilize the individual helices but not the dimer interaction. This is consistent with the previously mentioned observation that stabilizing the dimer reduces the activation function.
The recruitment of GAL11P by GAL4-dd relies on polar and hydrophobic interactions
The clustering of negative charges in the GAL11P-binding site of GAL4-dd suggest an electrostatic interaction. Consistently, the segment flanking the P-mutation in GAL11P has excess positive charge with five basic residues within ±10 positions (Fig. 6A). It needs to be explained, however, why the N342V mutation in GAL11P generates a binding site for the GAL4-dd. The introduction of a hydrophobic residue at position 342 might cause folding of GAL11P, and the new binding function might be a conformational effect rather than a direct hydrophobic interaction of V342 with GAL4-dd. However, mutagenesis experiments in which residue at position 342 in GAL11 has been systematically substituted with various residues showed that only hydrophobic substitutions confer the transcriptional potentiator phenotype to GAL11. The different hydrophobic substitutions lead to different in vitro binding affinities for GAL4 and likewise different degrees of in vivo transcriptional activation (Barberis et al. 1995; Farrell 1996; Farrell et al. 1996). It is difficult to imagine that such differences in affinities reflect different degree of structural stabilization toward permissive binding conformations. The large effect of such mutations at position 342 on binding affinity suggests a direct contact between the residue at this position with GAL4-dd. Thus, the structure of GAL4-dd and the features of the binding site suggest that the hydrophobic residue V342 and other nearby hydrophobic groups (Y340, I341, F336, I337, A445, L446, and I449; see Fig. 6A) in GAL11P might interact with the hydrophobic section of the loop connecting α1 and α2 in GAL4-dd. Mutational analysis indicates that in GAL11P bearing another hydrophobic substitution at position 342 (N342I), changing the neighboring hydrophobic residue I341 to Asn, reduces transcriptional activation some 10-fold. Moreover, adding the change Q343I to this GAL11 double mutant (N342I–I341N) improves activation by twofold. This observation further support the idea that hydrophobic residues neighboring 342 participate in GAL4–GAL11P interactions.
Hydrophobic interactions are usually tighter than hydrophilic ones. This might explain the low Kd and the slow dissociation rates, which is unusual for activator–target interactions. For comparison, the VP16 activation domain is in fast exchange with the bound state, and the helical structure of the bound conformation was elucidated with transferred NOE experiment (Uesugi et al. 1997). Typical activators are capable of contacting several targets to recruit the transcriptional machinery to the promoter (Ptashne and Gann 1997; Koh et al. 1998 and references therein). Thus, rapid exchange may allow them to recruit and orient the holoenzyme, and other components of the transcriptional machinery appropriately on the promoter (Kim et al. 1994; Koleske and Young 1994; Kuras and Struhl 1999; Li et al 1999). The slow exchange kinetics observed here may be biologically relevant for the newly acquired ability of GAL4-dd to function as an activator in the presence of GAL11P. GAL4-dd can only interact with the P region of GAL11P, thereby recruiting the holoenzyme to the promoter. The thermodynamic and structural properties of GAL4-dd binding to GAL11P described in this study are consistent with the notion that GAL4-dd needs to hold on to the holoenzyme long enough to permit binding of other essential components of the transcriptional machinery to the promoter. However, the significance of a kinetic versus a thermodynamic explanation remains to be tested. Notably, this singular activator-target interaction suffices for high levels of transcriptional activation that are comparable to those elicited by strong natural activators.
Materials and methods
Sample preparation
Dr. R. Marmorstein kindly provided the purification procedure for GAL450–106. GAL450–106 was used instead of 50–97 because the latter expresses poorly in bacteria. GAL450–106 was expressed in BL21-DE(3) pLysS Escherichia coli cells from a pRSETA expression vector (Invitrogen). Cells were grown at 37°C, induced 3 h with 1 mM IPTG, and harvested by centrifugation. Purification was performed through two consecutive ammonium sulfate precipitation with 0.25 and 0.15 g/mL, followed by ion exchange chromatography on a DEAE–Sepharose column and gel filtration on a Superdex 75 (16/60) column (Amersham Pharmacia Biotech). The fractions containing GAL450–106 were analyzed by SDS-PAGE electrophoresis. 15N- and 15N–13C-labeled GAL4 were obtained by using 15NH4Cl and 15NH4Cl, 13C6-D-glucose as the sole nitrogen and nitrogen-carbon sources in M9 medium, respectively. 2H–15N–labeled protein was produce as described previously (Walters 1997a).
The expression of GAL11P261–352 was performed similarly from a pRSETA vector in BL21-DE(3) pLysS E. coli cells. Purification of GAL11P261–352 was performed by partial fractionation with 0.3 g/mL ammonium sulfate followed by ion exchange chromatography by using SP-Sepharose and gel filtration on a Superdex-75 column.
Verification of the identity of the purified proteins was performed by mass spectrometry and amino acid analysis. The NMR protein samples ranged from 0.5 to 1 mM concentration in 50 mM sodium phosphate buffer at pH 7.4.
Alanine scanning mutagenesis
Mutations of the GAL4 segment in the context of LexA(1–202) + Gal4(50–97) were generated by site-directed PCR mutagenesis. The PCR products were subcloned between XbaII–SalI sites in frame with the LexA coding sequence in RJR 238 bearing a chromosomal origin of replication, a centromere, and a His 3 histidine auxotrophy selection marker. The LexA derivatives are expressed by a strong yeast actin promoter and the transcriptional unit ends with a GAL11 terminator. Each of the 40 mutants was confirmed by sequencing. Expression was tested in a GAL11P strain from a chromosomally integrated β-gal reporter gene bearing two LexA-binding sites 50 bp upstream of the GAL1 promoter TATA box. β-gal assays were performed as described previously (Barberis et al. 1995; Simkovich 1996).
Electrophoretic mobility shift assay
A double stranded oligonucleotide of 30 bp containing a single GAL4-binding site (5′-TCCGGAGGACTGTCCTCCGGT-3′) was labeled with 32P at the 5′ end. The labeled DNA was incubated in a 20-μL reaction volume, with each of the GAL4 mutants and with GST–GAL11 or GST–GAL11P in EMSA buffer (20 mM Hepes at pH 7.5, 25 mM NaCl, 5 mM MgCl2, 20 mM ZnCl2, 6% glycerol, 200 μg/mL bovine serine albumin, 3 mM β-mercaptoethanol, 2 mg/mL polydIdC). The complexes were resolved on a 9% polyacrylamide gel in 0.5× Tris-borate run for 2.5 h at 100 V at 9°C. The bands were visualized by Phosphor-imaging on a Fuji-Bas PhosphorImager.
The GAL4 mutants were expressed in BL21-DE(3) pLysS bacterial strain and purified on a SP-Fast Flow Sepharose resin (Pharmacia) as described previously (Ansari et al. 1998). GST–GAL11 or GST–GAL11P (residues 263–352 of GAL11) also were expressed in this strain and purified on a glutathione–Sepharose (Pharmacia) column by using standard procedures (Smith and Johnson 1988). Both proteins were dialyzed against the EMSA buffer.
NMR spectroscopy and assignments
All NMR spectra were recorded on Bruker AMX 500, Varian UnityPlus 400, Varian Unity 500, Inova 500, and UnityPlus 750 spectrometers at 35°C. Spectra were processed using FELIX (Molecular Simulations) and analyzed using XEASY (Bartels et al. 1995) on Silicon Graphics workstations. Backbone assignments were obtained using double-resonance 15N-dispersed NOESY triple resonance three-dimensional (1H, 15N, and 13C) HNCA and HN(CO)CA spectra. Side-chain resonances were assigned using 15N- and 13C-dispersed NOESY experiments. HNHA and HNHB experiments were used to assign Hα and Hβ, respectively (Archer et al. 1991; Madsen et al. 1993; Vuister and Bax 1993). Intermonomer contacts were obtained as described previously (Walters et al. 1997a) using a 200-msec mixing time for the 15N-dispersed NOESY. Complete mixing of the two homodimers species, unlabeled and 2H–15N–labeled, was achieved by heating the sample for 10 min to 60°C. No mixing was observed at room temperature over a period of 1 wk.
NOE and distance restraints
Distance restraints were obtained using a two-dimensional NOESY spectrum with a mixing time of 80 msec and 15N- and 13C-dispersed NOESY (Talluri and Wagner 1996) spectra with mixing times of 120 and 80 msec, respectively. Cross peaks were integrated using the XEASY software package (Bartels et al. 1995). The intensities of NOE cross peaks arising from interactions between Hα and HN nuclei within helical regions were used to calibrate the 15N-dispersed NOESY spectrum (Wüthrich 1986). The 13C-dispersed NOESY spectrum was calibrated by assuming the weakest cross peak corresponding to an interaction between Hα and Hβ, within well-defined helical elements, to be 5 Å. NOE derived restraints were supplemented with 38 hydrogen bonds per monomer within regular secondary structural elements. Secondary structural elements were identified based on the Hα chemical shift index (Wishart et al. 1992) and NOE connectivity. For each hydrogen bond, two restraints were used such that the distances between the amide proton and the oxygen and between amide nitrogen and oxygen were restricted to be 1.8–2.5 Å and 2.5–3.3 Å, respectively. Distance symmetry restraints for all assigned residues were used to generated a twofold symmetric arrangement of the dimer as described previously (Brünger 1992).
Structure calculations
Structure calculations were performed using CNS version 0.5 (Brünger et al. 1998) on an R10000 IndigoII Silicon Graphics workstation. The molecular dynamics simulated annealing was performed using NMR distance restraints and distance symmetry restraints starting from an extended duplicated strand. With the exception of the NOESY cross peaks obtained using the GAL4 (50–106) heterodimer and the hydrogen bonds, all NOEs were treated as ambiguous (Nilges 1993). The InsightII program (Molecular Simulations) and MOLMOL (Koradi et al. 1996) were used for displaying and visual inspection of the three-dimensional structures. PROCHECK (Laskowoski et al. 1993) was used to further assess the quality of the structures. Residues in which the amide group was unassigned were dismissed from the statistics reported for the Ramachandran plot (Table 1). Coordinates of the structures have been deposited at the Protein Data Bank.
Relaxation experiments
All relaxation experiments were performed on a Varian UnityPlus400 spectrometer. The three relaxation parameters, 15N longitudinal and transverse relaxation rates and heteronuclear NOE values, were measured using two-dimensional heteronuclear pulse sequences with pulsed-field gradients and sensitivity enhancement (Dayie and Wagner 1994). The analysis of the relaxation data was performed as described previously (Peng and Wagner 1992). Each peak was integrated by volume by using XEASY and fitted to single exponential function by using the Levenburg–Marquardt nonlinear least squares method implement in PLOT (New Unit). Uncertainties in the relaxation rates were estimated with Monte Carlo simulations by using one repeated time point as described (Peng and Wagner 1992).
Acknowledgments
We are indebted to Dr. Kylie Walters for sharing her valuable experience in solving dimers structures throughout all this work. We are grateful to Dr. Yingxi Lin and Dr. Christoph Fahlke for many helpful discussions, to Gregory Heffron, Dr. Hiroshi Matsuo, and Dr. Walfrido Antuch for their assistance in NMR spectroscopy. We thank the Helen Hay Whitney Foundation for support to A.Z.A. and Howard Hughes Medical Institute for a predoctoral fellowship to N.S. This work was supported by grants from the NIH and the NSF.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL wagner@hms.harvard.edu; FAX (617) 432-4383.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.873901.
References
- Ansari AZ, Reece RJ, Ptashne M. A transcriptional activating region with two contrasting modes of protein interaction. Proc Natl Acad Sci. 1998;95:13543–13548. doi: 10.1073/pnas.95.23.13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SJ, Ikura M, Torchia DA, Bax A. An alternative 3D NMR technique for correlating backbone 15N with side chain Hβ resonances in larger proteins. J Magn Reson. 1991;95:636–641. [Google Scholar]
- Baleja JD, Marmorstein R, Harrison S, Wagner G. Solution structure of the DNA-binding domain of Cd2-GAL4 from S. cerevisiae. Nature. 1992;356:450–453. doi: 10.1038/356450a0. [DOI] [PubMed] [Google Scholar]
- Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- Bardsley B, Cho YR, Westwell MS, Williams DH. Induction of asymmetry into homodimers. Chirality. 1998;10:14–23. [PubMed] [Google Scholar]
- Bartels Ch, Xia T-H, Billeter P, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- Brünger AT. X-PLOR version 3.1. A system for x-ray crystallography and NMR. New Haven, CT: Yale University; 1992. [Google Scholar]
- Brünger AT, Adams PD, Clore MG, DeLano WL, Gros P, Groesse-Kunstleve W, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carey M, Kakidani H, Leatherwood J, Mostashari F, Ptashne M. An amino-terminal fragment of GAL4 binds as a dimer. J Mol Biol. 1989;209:423–432. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- Carey M, Lin Y, Green MR, Ptashne M. A mechanism for synergistic activation of mammalian gene by GAL4 derivatives. Nature. 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- Das S, Hollenberg CP. A high frequency transformation system for the yeast Kluveromyces lactis. Curr Genet. 1982;6:123–128. doi: 10.1007/BF00435211. [DOI] [PubMed] [Google Scholar]
- Dayie KT, Wagner G. Relaxation-rate measurements for 15N-1H groups with pulsed-field gradients and preservation of coherence pathways. J Magn Reson. 1994;111:121–126. [Google Scholar]
- de Vos AM, Ultsch M, Kossiakoff AA. Human growth hormone and extracellular domain of its receptor: Crystal structure of the complex. Science. 1992;255:306–312. doi: 10.1126/science.1549776. [DOI] [PubMed] [Google Scholar]
- Farrell S. “GAL11P and GAL4: A novel protein-protein interaction that activates transcription in yeast.” Ph.D. thesis. Cambridge, MA: Harvard University; 1996. [Google Scholar]
- Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes & Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- Gaudreau L, Adam M, Ptashne M. Activation of transcription in vitro by recruitment of the yeast RNA polymerase II holoenzyme. Mol Cell. 1998;1:913–916. doi: 10.1016/s1097-2765(00)80090-8. [DOI] [PubMed] [Google Scholar]
- Hengartner CJ, Thompson CM, Zhang J, Chao DM, Liao SM, Koleske AJ, Okamura S, Young RA. Association of an activator with an RNA polymerase II holoenzyme. Genes & Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- Johnston M. A model fungal regulatory mechanism; the gal genes of Saccharomyces cerevisiae. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaveney M, Struhl K. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- Keegan L, Gill G, Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986;231:669–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Korenberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Koh SS, Ansari AZ, Ptashne M, Young RA. An activator target in the RNA polymerase II holoenzyme. Mol Cell. 1998;1:859–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- Koleske AJ, Young RA. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ, Raine AR, Gadhavi PL, Laue ED. Structure of the DNA-binding domain of zinc GAL4. Nature. 1992;356:448–450. doi: 10.1038/356448a0. [DOI] [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires PolII holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Laskowoski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Laughon A. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984;4:260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A, Driscoll R, Wills N, Gesteland RF. Identification of two proteins encoded by the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984;4:268–275. doi: 10.1128/mcb.4.2.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-Y, Virbasius A, Zhu X, Green MR. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- Lin YS, Carey MF, Ptashne M, Green MR. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- Lupas A. Coiled-coils: New structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Madsen J C, Sørensen OW, Sørensen P, Poulsen FM. Improved pulse sequences for measuring coupling constants in 13C, 15N-labeled proteins. J Biomol NMR. 1993;3:239–244. [Google Scholar]
- Marmorstein R, Carey M, Ptashne M, Harrison S. DNA recognition by GAL4: Structure of a protein–DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Nilges M. A calculation strategy for the structural determination of symmetric dimers by 1H NMR. Proteins. 1993;17:297–309. doi: 10.1002/prot.340170307. [DOI] [PubMed] [Google Scholar]
- Peng JW, Wagner G. Mapping of spectral densities of N-H bond motions in eglin c using heteronuclear relaxation experiments. Biochemistry. 1992;31:8571–8586. doi: 10.1021/bi00151a027. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- Salmeron JM, Jr, Johnston SA. Analysis of the Kluveromyces lactis positive regulatory gene Lac9 reveals functional homology to, but sequence divergence from the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986;14:7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkovich N. “GAL4: A novel function for the dimerization domain.” Ph.D. thesis. Cambridge, MA: Harvard University; 1996. [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Talluri S, Wagner G. An optimized 3D NOESY-HSQC. J Magn Reson. 1996;112:200–205. doi: 10.1006/jmrb.1996.0132. [DOI] [PubMed] [Google Scholar]
- Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL. Induced α helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- Valegard K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L. Crystal structure of an RNA bacteriophage coat protein–operator complex. Nature. 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- Vuister GW, Bax A. Quantitative J correlation: A new approach for measuring homonuclear three bond J(HN/Hα) coupling constants in 15N-enriched proteins. J Am Chem Soc. 1993;115:7772–7777. [Google Scholar]
- Walters KL, Matsuo H, Wagner G. A simple method to distinguish intermonomers NOEs in homodimeric proteins with C2 symmetry. J Am Chem Soc. 1997a;119:5958–5959. [Google Scholar]
- Walters KL, Dayie KT, Reece RJ, Ptashne M, Wagner G. Structure and mobility of the PUT3 dimer. Nat Struct Biol. 1997b;4:744–750. doi: 10.1038/nsb0997-744. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD, Richards FM. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- Wray L, Witte M, Dickinson RC, Riley MI. Characterization of a positive regulatory gene, Lac9, that controls induction of the lactose-galactose regulon of Kluveromyces lactis. Mol Cell Biol. 1987;4:1985–1998. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich K. NMR of proteins and nucleic acids. New York: Wiley; 1986. [Google Scholar]
- Zimm BH, Bragg JK. Theory of phase transition between helix and random coil in polypeptide chains. J Chem Phys. 1959;31:526–535. [Google Scholar]