Abstract
Ten novel small nucleolar RNA (snoRNA) gene clusters, consisting of two or three snoRNA genes, respectively, were identified from Arabidopsis thaliana. Twelve of the 25 snoRNA genes in these clusters are homologous to those of yeast and mammals according to the conserved antisense sequences that guide 2′-O-ribose methylation of rRNA. The remaining 13 snoRNA genes, including two 5.8S rRNA methylation guides, are new genes identified from A.thaliana. Interestingly, seven methylated nucleotides, predicted by novel snoRNAs Z41a–Z46, are methylated neither in yeast nor in vertebrates. Using primer extension at low dNTP concentration the six methylation sites were determined as expected. These snoRNAs were recognized as specific guides for 2′-O-ribose methylation of plant rRNAs. Z42, however, did not guide the expected methylation of 25S rRNA in our assay. Thus, its function remains to be elucidated. The intergenic spacers of the gene clusters are rich in uridine (up to 40%) and most of them range in size from 35 to 100 nt. Lack of a conserved promoter element in each spacer and the determination of polycistronic transcription from a cluster by RT–PCR assay suggest that the snoRNAs encoded in the clusters are transcribed as a polycistron under an upstream promoter, and individual snoRNAs are released after processing of the precursor. Numerous snoRNA gene clusters identified from A.thaliana and other organisms suggest that the snoRNA gene cluster is an ancient gene organization existing abundantly in plants.
INTRODUCTION
Small nucleolar RNAs (snoRNAs) play an important role in ribosome biogenesis (1). Since the 1990s, a myriad of novel snoRNAs have been identified and characterized in the nucleolus of eukaryotic cells and many snoRNA homologs were recently found in some prokaryotes, such as Archaea (2–4). On the basis of structural features they can be divided into three main families: C/D box snoRNAs, H/ACA box snoRNAs and RNase MRP (5,6). Diverse functions of snoRNAs have been determined. Six snoRNAs (e.g. snoRNAs U3, U8, U14 and U22) have been shown to be an essential component of the rRNA processing machinery (1). On the other hand, a large number of snoRNAs are involved in post-transcriptional modification of rRNA. Most C/D box snoRNAs act as guides for site-specific 2′-O-methylation of pre-rRNA, while the H/ACA box snoRNA family is responsible for pseudouridylation of pre-rRNA (7–10). Recent findings have shown that some snoRNAs can guide the methylation of U6 snRNA and cellular RNAs other than rRNA (11–13), demonstrating a critical role of snoRNAs in methylation of nuclear components. Moreover, snoRNA genes have been shown to have their own particular genomic organization and expression. Two kinds of snoRNA gene organization have been found. One is an independently transcribed gene, while the other is nested in introns of protein-encoding genes (1). The expression of intronic snoRNA depends on transcription of its host gene and processing of pre-mRNA. Intron-encoded snoRNAs are widespread in various organisms, especially in mammals. Nevertheless, the majority of snoRNA genes are independently transcribed in the yeast Saccharomyces cerevisiae.
Up to now more than 100 snoRNA genes have been identified from various eukaryotic organisms. Most of them come from vertebrates and yeast, whereas only a dozen snoRNAs have been characterized from plants. Although most new snoRNAs identified from plants appear to be homologs of yeast or animal counterparts, the gene organization in plants has shown characteristics different from yeast and animal snoRNAs. U14 snoRNA gene clusters and their expression as a polycistronic transcript were reported by Leader et al. (14,15). Six snoRNAs clustered in the first intron of the rice Hsp70 gene have also been found (16; EMBL accession nos Z70693, Z69633, Z69786, Z70229–Z70231, Z96805–Z96808). It is important to identify novel snoRNAs from plants for a better understanding of the diversity of RNA gene structure and function, as well as the mechanisms involved in evolution of the snoRNA gene family. Arabidopsis thaliana is a favorite model organism in plant molecular biology with a great part of its DNA sequence documented owing to the Arabidopsis thaliana Genome Project. Now a major task is to identify novel genes from the DNA database and reveal their biological function. Although a large number of protein-encoding genes have been identified, only a few snoRNA genes have been characterized from A.thaliana (17).
In this paper we report the identification of 25 snoRNA genes, organized in 10 gene clusters, from A.thaliana. The structure of novel snoRNAs specific to plant rRNA modification and expression of the gene clusters are analyzed and discussed.
MATERIALS AND METHODS
Search of the A.thaliana genomic database
The A.thaliana DNA database in the Stanford Genomic Resource was searched for C/D box snoRNAs that exhibit structural features of guides for rRNA ribose methylation (18). Referring to the methylated nucleotide data from yeast and vertebrates, a search for perfect 12 nt complementarity to a rRNA ribose-methylated sequence immediately followed by the sequence NCUGA was performed with the programs BLAST (19) and FASTA (20). The DNA sequences of snoRNA gene candidates obtained from the A.thaliana genome were further analyzed using the PC gene 6.0 package.
RNA extraction and northern hybridization
Fresh A.thaliana seedlings for the isolation and preparation of RNA were kindly provided by Dr Li Jayang (Genetics Institute, Chinese Academy of Science). Total cellular RNA was isolated and purified according to the method described by Chomczynski et al. (21). An aliquot of 10 µg total RNA was analyzed by electrophoresis on 8% acrylamide/7 M urea gels. Electrotransfer onto nylon membrane (Hybond-N+; Amersham) was followed by UV irradiation for 5 min. Hybridization with 5′-labeled probes was performed as previously described (22).
Reverse transcription analyses and mapping of ribose methylation
Reverse transcription was carried out in a 20 µl reaction mixture containing 10 µg total RNA and 20 ng 5′-labeled primer in the presence of 250 µM dNTPs. After denaturation at 65°C for 5 min the mixture was cooled to 42°C for l0 min, then 200 U of MMLV reverse transcriptase (Promega) were added and incubated at 42°C for 60 min. The cDNA synthesized by reverse transcription was analyzed by electrophoresis on 8% acrylamide/7 M urea gels. Sequence analysis of cDNA was performed after purification of full size primer extension products on denaturing polyacrylamide gels as described previously (18).
Ribose-methylated nucleotides of A.thaliana rRNAs were determined by primer extension at low dNTP concentrations as follows. Two reverse transcription reactions, containing 5 µg total cellular RNA and 0.1 pmol oligodeoxynucleotides labeled at the 5′-end with 32P, were carried out in the presence of either 4 µM or 1 mM dNTPs. In order to map the methylation position precisely, a rDNA sequence ladder was prepared and used as a molecular weight marker. The rDNA fragments surrounding the methylated nucleotide sequences of A.thaliana 5.8S, 18S and 25S rRNA were amplified by PCR with the primer pairs R58/LH2, 18N5/18N9R and At28Sc4F/Z44M2116, respectively, then cloned into the SmaI site of plasmid pTZ18. The plasmid DNA insert was directly sequenced with the same primer as used for rRNA methylation mapping and run in parallel with the reverse transcription reaction as a molecular weight marker.
RT–PCR experiment
Total cellular RNA was treated with DNase before reverse transcription with the primers. About 50 µg RNA in 100 µl of DNase I buffer was incubated with 10 U of RQ1 RNase-free DNase (Promega) and subsequently submitted to phenol/chloroform/isoamyl alcohol (50:49:1) extraction. After reverse transcription with a reverse primer, PCR was carried out by adding the corresponding forward primer labeled at the 5′-end with 32P.
Oligodeoxynucleotides
Oligonucleotides were synthesized and purified by Sangon Co. (Shanghai, China). The sequences of oligonucleotide probes and primers used for northern blotting and reverse transcription were as follows: 5′-ACTCAGTCCCTTAGATGTTCA-3′ (Pu25), 5′-GATCAGAATATGATGTCATTCT-3′ (Pz27), 5′-CCTCAGAATTGCAGAAGTATCA-3′ (Pz37), 5′-GAATCAGAAAGAACGTAGCGAG-3′ (Pz41), 5′-CCATCAGCAACCAAAAGTTATC-3′ (Pz42), 5′-GACTCAGAATATGGGTGGTGA-3′ (Pz43), 5′-GAGTCAGAAGTAGCTGATTATA-3′ (Pz44) and 5′-CCTCAGCAGTCCTAGTCAAGA-3′ (Pz45). These oligonucleotides were complementary to the 3′-end of the U25a, Z27a, Z37a, Z41a, Z42a, Z43a, Z44a and Z45 snoRNA genes, respectively. Arabidopsis thaliana rRNA ribose-methylated nucleotides were assayed by reverse transcription with the following primers: 5′-CGGGATTCTGCAATTCACACCA-3′ (R58), 5′-CATTGTTCCATCGACCAGAGGC-3′ (Z42M1840), 5′-CGCTCCACCAACTAAGAACGGC-3′ (Z43M1263), 5′-CCGCAGGGACCATCGCAATGC-3′ (Z44M2116), 5′-CAGGCTCCAGCTATCCTGAGG-3′ (Z44M943) and 5′-CTGATCCCTGGTCGGCATCG-3′ (Z45M1011). The following oligonucleotides were used for amplification of A.thaliana 5.8S, 18S and 25S rRNA gene fragments: 5′-CGGGATTCTGCAATTCACACCA-3′ (R58), 5′-GTCGAATTCGTAGGTGAACCTGCGGAAGGATCATTG-3′ (LH2), 5′-TGGTGCCAGCAGCCGCGGTA-3′ (18N5), 5′-CTAAGGGCATCACAGACCTG-3′ (18N9R), 5′-TATAGGGGCGAAAGACTAATCG-3′ (At-28Sc4F) and 5′-CCGCAGGGACCATCGCAATGC-3′ (Z44 M2116). RT–PCR experiments were performed with the following primers: 5′-CCATCAGCAACCAAAAGTTATC-3′ (Z42at) and 5′-CCAATGAGGACATCAGATTATA-3′ (Z37aF). The probes and primers used in northern hybridization and reverse transcription were 5′-labeled with [γ-32P]ATP (Yahui Co.) as described (23).
RESULTS
Detection of 10 C/D box snoRNA gene clusters
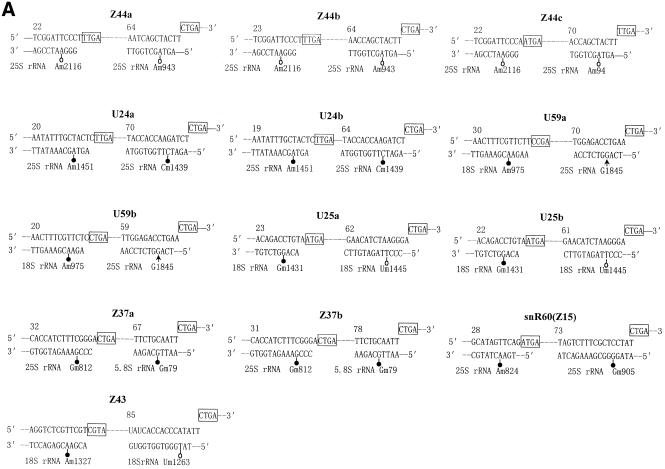
We have searched the A.thaliana genome database in the Stanford Genomic Resource for antisense snoRNA genes that possess a long sequence complementarity to rRNA. A number of DNA sequences coding for snoRNA were obtained. When the flanking sequences of these snoRNA genes were further analyzed it was found, unexpectedly, that in most cases one or two snoRNA genes, located upstream or downstream, were linked with a previously identified snoRNA gene to form a snoRNA gene cluster. Here we focus our attention on 10 C/D box snoRNA gene clusters, termed A.thaliana clusters II, III, IV, V, VI, VII, VIII, IX, X and XI, respectively, with reference to the four Z2 snoRNA genes previously named A.thaliana snoRNA gene cluster I (24). The sequences of the 10 snoRNA gene clusters are showed in Figure 1.
Figure 1.
The sequences of the snoRNA gene cluster from A.thaliana. snoRNA genes are in capital letters; the C/D and C′/D′ boxes are boxed by solid and dashed lines, respectively. A bar is drawn over sequence complementary to rRNA and arrows indicate nucleotides involved in the terminal stem.
Ten snoRNA gene clusters containing 25 snoRNAs are scattered over all chromosomes of A.thaliana, especially on chromosome 5 (see Table 1). All the snoRNA coding sequences in the clusters exhibit the hallmark structure of C/D box antisense snoRNA. This includes the presence of two short conserved motifs, box C (5′-UGAUGA-3′) and box D (5′-CUGA-3′), which are immediately flanked by a 4–20 bp stem involving both termini of the RNA molecule. These features are the key signals for both exonucleolytic processing and nucleolar localization (25–27). Most of the snoRNA genes had clear C′ and D′ boxes in the central region of the gene. Each snoRNA gene had at least a 10–16 nt long complementarity to rRNA, which is known as the functional antisense element involved in guiding 2′-O-ribose methylation of rRNA (28).
Table 1. Localization of 10 snoRNA gene clusters.
| Gene cluster |
Chromosome |
snoRNA |
Accession no. |
| II | 4 | Z44a | AJ240077 |
| Z44b | AJ440078 | ||
| U24a | AJ240063 | ||
| III | 5 | Z44c | AJ440079 |
| U24b | AJ224036 | ||
| IV | 1 | U59a | AJ240064 |
| U59b | AJ240065 | ||
| V | 5 | U25a | AJ240066 |
| SnR38a | AJ240067 | ||
| Z45 | AJ276572 | ||
| VI | 2 | Z37a | AJ240068 |
| SnR40a | AJ240069 | ||
| Z42 | AJ276571 | ||
| VII | 1 | Z41a | AJ240071 |
| Z27a | AJ010675 | ||
| VIII | 5 | Z41b | AJ240072 |
| Z27b | AJ240070 | ||
| IX | 3 | Z43 | AJ240080 |
| SnR74 | AJ240073 | ||
| SnR60 | AJ240074 | ||
| X | 2 | Z37b | AJ242537 |
| SnR40b | AJ242536 | ||
| XI | 3 | U25b | AJ276573 |
| SnR38b | AJ276575 | ||
| Z46 | AJ276574 |
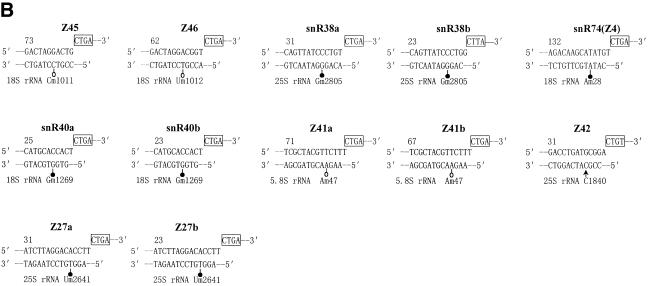
According to the relationship of antisense snoRNA structure and function (7,8,29), rRNA/snoRNA duplexes and rRNA methylation sites predicted by these new RNA guides were analyzed and are described in Figure 2. About half of the snoRNA genes from A.thaliana have two sequence complementarities to rRNA. A comparison with known snoRNA genes from various organisms showed that 12 of 25 snoRNA genes in the clusters were homologs of yeast or animal snoRNAs according to the antisense element conserved among organisms. Therefore, these snoRNA genes were named as their counterparts, i.e. U24a, U24b, U25a, U25b, U59a, U59b, snR38a, snR38b, snR40a, snR40b, snR60 and snR74, implying the same function conserved in the course of evolution. Among the 25 snoRNA genes, however, 13 snoRNA genes, named Z27a, Z27b, Z37a, Z37b, Z41a, Z41b, Z42, Z43, Z44a, Z44b, Z44c, Z45 and Z46, respectively, were identified for the first time from A.thaliana. Interestingly, six novel antisense snoRNA species (Z41a–Z46) can form an 11–16 bp long duplex with rRNA, but they do not match any known ribose-methylated nucleotide of rRNA in yeast and vertebrates. Am47 in 5.8S rRNA, predicted by Z41a and Z41b, is a unique 2′-O-ribose methylation in plants (30), however, six other potential methylation sites have never been mapped in any organism. It is possible, therefore, that the six novel snoRNA species (Z41a–Z46) may represent the specific snoRNAs for plant rRNA methylation.
Figure 2.
Methylation guide duplexes between snoRNAs and rRNAs. 2′-O-ribose methylation sites homologous to those of mammals and yeast are shown by filled circles and novel methylation sites determined in this work are depicted by open circles. Arrows indicate unmethylated nucleotides. Box D and D′ motifs (boxes) are indicated. (A) snoRNAs with two complementarities to rRNAs. (B) snoRNAs with one complementarity to rRNAs.
Positive identification of specific snoRNAs from the clusters
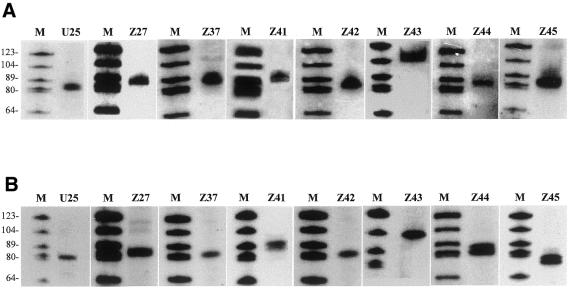
Eight oligonucleotide probes and primers were designed and synthesized according to the coding region of eight of the novel snoRNA genes, i.e. U25, Z27a, Z37a, Z41, Z42, Z43, Z44a and Z45, which were selected from clusters II, V, VI, VII and IX, respectively (see Fig. 1). Northern analyses were performed with A.thaliana total cellular RNA and the labeled oligonucleotide probes to confirm the expected snoRNAs transcribed from the gene clusters. As shown in Figure 3A, all the target snoRNAs were positively detected by northern blotting. In each case a strong band was revealed under stringent conditions of hybridization. These RNA molecules, ranging from 66 to 109 nt, were detected as expected because the 5′- and 3′-ends of most antisense snoRNAs were processed within 10 nt upstream of box C and downstream of box D, respectively. Although the snoRNAs were from different clusters, their cellular abundance was of the same order of magnitude according to the intensity of each band. In addition, northern analysis of RNA isolated from partially purified nucleoli of A.thaliana cells showed that the target snoRNAs were enriched in the nucleus (data not shown).
Figure 3.
snoRNA analyses. (A) Northern blot analyses. Aliquots of 10 µg total cellular RNA were separated in each lane and hybridized with the labeled oligonucleotide probes described in Materials and Methods. Lane M, molecular weight markers (pBR322 digested with HaeIII and 5′-end-labeled with [γ-32P]ATP). (B) Reverse transcription analyses were carried out with the 5′-end-labeled primers described in Materials and Methods.
To further confirm the identity of these RNA species and to map the 5′-ends of the snoRNAs, reverse transcription analyses of A.thaliana total cellular RNA with the same oligonucleotide primers as above were carried out. A major cDNA product was obtained for each snoRNA (Fig. 3B). Since the primers are located on the 3′-ends of the RNA species, the cDNA products have nearly the same size as the RNA molecules detected by northern hybridization. For some snoRNAs, such as Z44 and Z45, cDNA mini-heterogeneity (1–2 nt) could be distinguished. This may reflect nibbling by exonucleases during processing of the terminal nucleotide of snoRNA. The sequences of the cDNA products from each snoRNA were determined by cloning of purified cDNA bands. The sequences determined perfectly matched the corresponding snoRNA genes in the clusters, further confirming the identity of the snoRNAs.
Determination of six rRNA methylation sites predicted by the novel snoRNAs
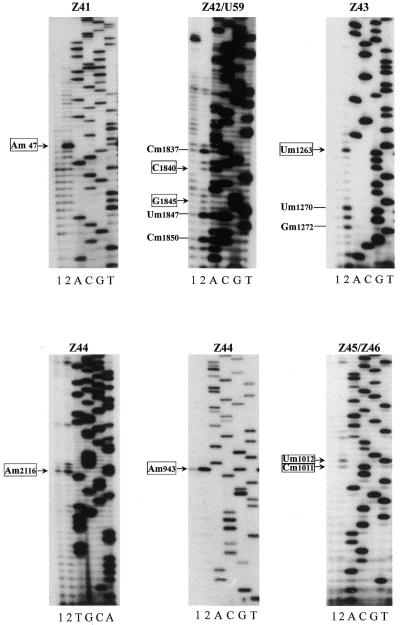
Because the ribose methylation sites of rRNA in A.thaliana have not yet been completely mapped, the methylated nucleotides predicted by the six novel snoRNAs were verified experimentally in this study. Using primer extension at low dNTP concentrations, which causes pausing at ribose-methylated nucleotides in the RNA template, we determined, as shown in Figure 4, six new methylated nucleotides in A.thaliana rRNAs, i.e. Am47 in 5.8S rRNA, Um1010, Cm1011 and Um1263 in 18S rRNA and Am943 and Am2116 in 25S rRNA. These methylated nucleotides were predicted by Z41a, Z41b, Z43, Z44a, Z44b, Z45 and Z46, respectively. However, C1840 in 25S rRNA, which was predicted by Z42, did not produce an evident pause due to methylation, as compared with three known methylated nucleotides nearby, in our primer extension assay. Likewise, G1845, indicated by additional complementarity of U59, was found to be unmethylated in the same assay.
Figure 4.
Determination of rRNA methylation sites predicted by the novel snoRNAs. Lane 1, control reaction at 1.5 mM dNTP; lane 2, primer extension at 4 µM dNTP; lanes A, C, G and T, the rDNA sequence ladder. The sites of ribose methylation were revealed by RT pauses at low dNTP concentrations. Arrows indicate potential methylation sites predicted by the novel snoRNAs.
Analyses of intergenic sequences of gene clusters
All the snoRNA genes in the clusters are arranged in a head-to-tail fashion and are closely linked. Sequence analyses showed that the intergenic spacers which separate snoRNA genes in the 10 clusters were rich in uridine (up to 40%). Most of them were of small size, ranging from 35 to 115 bp, with the exception of U25b–Z46 from cluster XI, which lie 330 bp apart (Fig. 1). No conserved promoter elements of plant snoRNA genes were found within the short sequences of the intergenic spacers. This suggests that the snoRNA genes in clusters might be transcribed together as a polycistronic precursor from an upstream promoter. Individual snoRNAs would be released after processing by a nuclease(s), as reported by Leader et al. (15).
The presence of a polycistronic transcript from cluster VI (Z37a–snR40a–Z42) of A.thaliana was determined by RT–PCR with pairs of primers designed to amplify regions that encompass the three snoRNA gene encoding sequences and their spacers. As shown in Figure 5, a precursor representing an unprocessed polycistronic transcript was detected, with the band produced by RT–PCR corresponding precisely to the size of the product, i.e. a band of 334 bp, as expected. No product was observed in any of the RT–PCR controls performed in the absence of reverse transcriptase.
Figure 5.
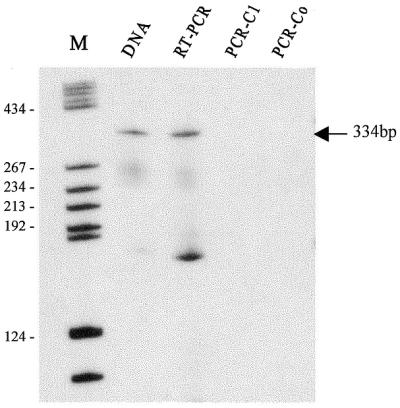
Detection of a polycistronic snoRNA transcript by RT–PCR with primer pair Z37aF/Z42at. Lane DNA, control PCR performed on the total DNA; lane RT–PCR, PCR amplification after reverse transcription of total RNA; lane PCR-C1, control PCR performed on total RNA without reverse transcription; lane PCR-Co, control PCR carried out with water as template; lane M, molecular weight marker (pBR322 digested with HaeIII).
It is worth noting that, besides being U-rich, the sequences of the intergenic spacers of each cluster are very different from each other. A hairpin-like structure, predicted by the RNA folding program, was found for most of the intergenic spacers of the A.thaliana snoRNA gene clusters, which is reminiscent of the processing signal recognized by nuclease(s) in yeast snoRNA polycistronic precursors (18).
DISCUSSION
Novel snoRNA identification through comparative analyses
It has been shown that all sites of rRNA ribose methylation are specified through the same snoRNA-guided process (28,31). The number of these modifications varies considerably between organisms, from 55 for yeast to 107 for human rRNA (32), showing an increasing number of methylated sites in multicellular organisms. Interestingly, all of the methylated sites have been found to be located in the phylogenetically conserved regions of rRNA and about two-thirds of rRNA ribose-methylation sites in yeast are conserved in human. The pattern of rRNA methylation in diverse organisms and the mechanism(s) that produces it remain to be elucidated. Recently, computer searches of DNA databases relying on the presence of sequences and structural features has represented a powerful means for detecting novel snoRNAs in some vertebrates and the yeast S.cerevisiae (8,18,22,31). Nevertheless, the application of this method to plant genomic sequences has been limited or has involved complicated computations, because the sites of rRNA ribose methylation have not been completely mapped in any plant. Thus, we have tried to search the A.thaliana genomic sequence, based on the known pattern of rRNA ribose methylation of yeast and human, to identify cognate snoRNA genes in A.thaliana. By this means we have identified 10 novel snoRNA gene clusters containing 25 snoRNAs and a series of snoRNA gene candidates (L.H.Qu and H.Zhou., unpublished results). An interesting phenomenon observed in A.thaliana concerning snoRNA gene organization is that many snoRNA genes were in clusters consisting of two or more snoRNA genes. By analysis of the clusters we were able to discover six novel snoRNA species that did not match any known methylated nucleotide in yeast and human. Together with mapping of the predicted methylated sites, five novel snoRNA species were suggested to be specific guides of rRNA modification in plants. However, Z42 is a typical antisense snoRNA with a 12 nt complementarity to 25S rRNA, but the target nucleotide predicted by Z42 was not methylated in A.thaliana. Like yeast snoRNA snR190 (31), the function of Z42 as a rRNA methylation guide remains to be determined. Identification and characterization of cognate snoRNAs through comparative analyses will contribute to a more comprehensive pattern of 2′-O-methylation of rRNA from A.thaliana and make it possible to find novel snoRNA genes characteristic of plants.
SnoRNA homologs among distant organisms
Although a large number of rRNA methylation sites have been highly conserved during evolution, identification of snoRNA homologs among distant eukaryotes might be problematic in some cases. The fact that some snoRNAs share partial antisense elements with their counterparts in other organisms or possess two antisense elements homologous to two different snoRNAs with a single antisense element suggests a more complex definition of snoRNA homologs from different organisms. Arabidopsis thaliana snR40, for instance, shares only one functional element, instead of two, with its yeast counterpart. Conversely, A.thaliana U25 and U59 have an additional sequence complementarity to rRNA compared with their mammalian counterparts. Yeast U24 is an example that differs from vertebrate U24 by one of two antisense elements at the 3′-end of the molecule (8). Arabidopsis thaliana U24, with two sequence complementaries to 25S rRNA, is a yeast rather than vertebrate homolog, implying an ancestor of the U24 gene shared by these two organisms. However, A.thaliana Z27a and Z27b, two copies of the same snoRNA species, represent unique cognate homologs of animal snoRNA because the methylated nucleotide Um2641 in 18S rRNA predicted by Z27a and Z27b is not methylated in yeast. This methylated site is conserved in vertebrates (32), but the corresponding snoRNA guide has not yet been identified. More interestingly, Z37a and Z37b represent particular snoRNA species in plants, because they have two special sequence complementarities, one of which is responsible for Gm79 in 5.8S rRNA, while the other is the same as snR39b. Gm79 in 5.8S rRNA is a conserved methylation site between vertebrates and plants, but not yeast (32). In contrast, snR39b has been identified as a methylation guide snoRNA with a single antisense element in both yeast (32) and mouse (L.H.Qu and H.Zhou, unpublished results). Since most members of the C/D box antisense snoRNA family have been shown to possess a bimodular structure with two functionally independent, but structurally related, halves, our observations from analyses of snoRNA homologs implies that recombination between elementary modules in different snoRNAs might have occurred during evolution. The results also reveal a complicated pattern of rRNA modification in plants.
The snoRNA gene cluster is an ancient organization existing abundantly in plants
No snoRNA gene cluster has been found in mammals, although multiple snoRNAs may be produced simultaneously by processing a mRNA precursor transcribed from a host gene having numerous single snoRNA-containing introns (33,34). In yeast, in addition to a few intron-encoded snoRNA genes, major C/D box antisense snoRNA genes are dispersed as independently transcribed singlets and only five antisense snoRNA gene clusters have been identified from the yeast genome (31). The identification of 11 snoRNA gene clusters in A.thaliana reveals the abundance of gene clusters in plants. The finding of snoRNA gene clusters in both plants and yeast suggests that snoRNA gene clusters, which have developed more complexity in plants, are an ancient gene organization conserved through the course of evolution. This suggestion is supported by a recent study of snoRNA gene clusters from a protozoan, Trypanosoma brucei (35). SnoRNA gene clusters would represent an effective regulation of the transcriptional unit and produce snoRNAs in stoichiometric amounts. Nevertheless, maturation of snoRNAs is required by processing of the polycistronic transcript. It has been shown by Leader et al. that such processing is independent of splicing and requires endonucleolytic activity (36). Recently, RNase III, an endonuclease involved in rRNA and tRNA processing, has been shown to be a key player in the processing of polycistronic transcripts from snoRNA gene clusters in yeast (37). Yet, its role and recognition signal remain to be determined in plant snoRNA processing. More detailed analyses of the A.thaliana genomic database will provide further insights into the general genomic organization of snoRNA genes and thereby shed light on the mechanism of expression of snoRNAs in plants.
snoRNA sequences determined in this work have been deposited in the EMBL sequence database; the accession numbers are displayed in Table 1.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Micheal Epperley for revising the text of the manuscript. This work was supported by the National Natural Science Foundation of China (key projects 39730300 and 39525007) and a Fund for Distinguished Young Scholar from the Education Ministry of China.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +86 20 8411 2399; Fax: +86 20 8403 6551; Email: AJ010675, AJ224036, AJ240063–AJ240074, AJ240077, AJ240080, AJ242536, AJ242537, AJ276571–AJ276575, AJ440078, AJ440079
References
- 1.Maxwell E.S. and Fournier,M.J. (1995) The small nucleolar RNAs. Annu. Rev. Biochem., 35, 897–934. [DOI] [PubMed] [Google Scholar]
- 2.Smith C.M. and Steitz,J.A. (1997) Sno storm in the nucleolus: new roles for myriad small RNPs. Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- 3.Omer A.D., Lowe,T.W., Russell,A.G., Ebhardt,H., Eddy,S.R. and Dennis,P.P. (2000) Homologs of small nucleolar RNAs in Archaea. Science, 21, 517–522. [DOI] [PubMed] [Google Scholar]
- 4.Gaspin C., Cavaille,G., Erauso,G. and Bachellerie,J.P. (2000) Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J. Mol. Biol., 297, 895–906. [DOI] [PubMed] [Google Scholar]
- 5.Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small nucleolar RNAs defined by different box elements with related functions. Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- 6.Tollervey D. and Kiss,T. (1997) Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol., 3, 337–342. [DOI] [PubMed] [Google Scholar]
- 7.Kiss-Laszlo Z., Henry,Y., Bachellerie,J.P., Caizergues-Ferrer,M. and Kiss,T. (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 8.Nicoloso M., Qu,L.H., Michot,B. and Bachellerie,J.P. (1996) Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation rRNA. J. Mol. Biol., 260, 178–195. [DOI] [PubMed] [Google Scholar]
- 9.Ni J., Tien,A.L. and Fournier,M.J. (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell, 89, 565–573. [DOI] [PubMed] [Google Scholar]
- 10.Ganot P.H., Bortolin,M.L. and Kiss,T. (1997) Site-specific pseudouridine formation in eukaryotic pre-rRNA is guided by small nucleolar RNAs. Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- 11.Tycowski K.T., You,Z.H., Graham,P.J. and Steitz,J.A. (1998) Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell, 2, 629–638. [DOI] [PubMed] [Google Scholar]
- 12.Ganot P.H., Jady,B.E., Bortolin,M., Darzcq,X. and Kiss,T. (1999) Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol., 19, 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jady B.E. and Kiss,T. (2000) Characterisation of the U83 and U84 small nucleolar RNAs: two novel 2′-O-ribose methylation guide RNAs that lack complementarities to ribosomal RNAs. Nucleic Acids Res., 28, 1348–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leader D.J., Sanders,J.F., Waugh,R., Shaw,P.J. and Brown,J.W.S. (1994) Molecular characterisation of plant U14 small nucleolar RNA genes: closely linked genes are transcribed as a polycistronic U14 transcript. Nucleic Acids Res., 22, 5196–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leader D.J., Clark,G.P., Watters,J., Beven,A.F., Shaw,P.J. and Brown,J.W.S. (1997) Clusters of multiple different small nucleolar RNA in plants are expressed as and processed from polycistronic pre-snoRNA. EMBO J., 16, 5742–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu L.H., Zhong,L., Shi,S.H., Lu,Y.J., Zhou,H., Fang,R.X. and Wang,Q. (1997) Two snoRNAs are encoded in the first intron of the rice Hsp70 gene. Prog. Natural Sci., 7, 373–377. [Google Scholar]
- 17.Barneche F., Steinmetz,F. and Echeverria,M. (2000) Fibrillarin genes encode both a conserved nucleolar protein and a novel snoRNA involved in rRNA methylation in Arabidopsis thaliana. J. Biol. Chem., 275, 27212–27220. [DOI] [PubMed] [Google Scholar]
- 18.Qu L.H., Henras,A., Lu,Y.J., Zhou,H., Zhou,W.X., Zhu,Y.Q., Zhao,J., Henry,Y., Caizergues-Ferrer,M. and Bachellerie,J.P. (1999) Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol., 19, 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 20.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem., 162, 732–735. [DOI] [PubMed] [Google Scholar]
- 22.Qu L.H., Henry,Y., Nicoloso,M., Michot,B., Azum,M., Renalier,M., Caizergues- Ferrer,M. and Bachellerie,J.P. (1995) U24, a novel intron-encoded small nucleolar RNA with two 12 nt-long, phylogenetically conserved complementarities to 28S rRNA. Nucleic Acids Res., 23, 2669–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Zhou H., Meng,Q. and Qu,L.H. (2000) Identification of Z2 snoRNA gene cluster from Arabidopsis thaliana genome. Sci. China Ser. C, 43, 449–453. [Google Scholar]
- 25.Cavaille J. and Bachellerie,J.P. (1996) Processing of fibrillarin-associated snoRNAs from pre-mRNA intron: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie, 78, 443–456. [DOI] [PubMed] [Google Scholar]
- 26.Caffarelli E., Fatica,A., Prisley,S., De Gregorio,E., Fragapane,P. and Bozzoni,I. (1996) Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing and the stability of the mature snoRNA. EMBO J., 15, 1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 27.Samarsky D.A., Fournier,M.J., Singer,R.H. and Bertrand,E. (1998) The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J., 17, 3743–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachellerie J.P. and Cavaille,J. (1997) Guiding ribose methylation of rRNA. Trends Biochem. Sci., 22, 257–261. [DOI] [PubMed] [Google Scholar]
- 29.Tollervey D. (1996) Small nucleolar RNAs guide ribosomal RNA methylation. Science, 273, 1056–1057. [DOI] [PubMed] [Google Scholar]
- 30.Melekhovets Y.F. and Troitsky,A.V. (1993) Sequence and methylation of 5.8S rRNA in fern, Osmunda regalis. Nucleic Acids Res., 21, 2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowe T.M. and Eddy,S.R. (1999) A computational screen for methylation guide snoRNAs in yeast. Science, 283, 1168–1171. [DOI] [PubMed] [Google Scholar]
- 32.Maden B.E.H. (1990) The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acids Res., 10, 7103–7113. [DOI] [PubMed] [Google Scholar]
- 33.Tycowski K.T., Shu,M.D. and Steitz,J.A. (1996) A mammalian gene with introns instead of exons generating stable RNA products. Nature, 379, 464–466. [DOI] [PubMed] [Google Scholar]
- 34.Smith C.M. and Steitz,J.A. (1998) Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol., 18, 6897–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunbar D.A., Chen,A.A., Wormsley,S. and Baserga,S.J. (2000) The genes for small nucleolar RNAs in Trypanosoma brucei are organized in clusters and are transcribed as a polycistron. RNA, 28, 2855–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leader D., Clark,G., Watters,J., Beven,A.F., Shaw,P.J. and Brown,J.W.S. (1999) Splicing-independent processing of plant box C/D and box H/ACA small nucleolar RNAs. Plant Mol. Biol., 39, 1091–1100. [DOI] [PubMed] [Google Scholar]
- 37.Chanfreau G., Legrain,P. and Jacquier,A. (1998) Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol., 284, 975–988. [DOI] [PubMed] [Google Scholar]