Abstract
Without the RAD51 strand exchange protein, Saccharomyces cerevisiae cannot repair a double-strand break (DSB) by gene conversion. However, cells can repair DSBs by recombination-dependent, break-induced replication (BIR). RAD51-independent BIR is initiated more than 13 kb from the DSB. Repair depends on a 200-bp sequence adjacent to ARS310, located ∼34 kb centromere-proximal to the DSB, but does not depend on the origin activity of ARS310. We conclude that the ability of a recombination-induced replication fork to copy >130 kb to the end of the chromosome depends on a special site that enhances assembly of a processive repair replication fork.
Keywords: Break-induced replication, RAD51, DNA repair, Saccharomyces cerevisiae
Break-induced DNA replication (BIR) plays a key role in the repair of double-strand breaks (DSBs) in eukaryotic chromosomes. BIR is likely to be important in restarting DNA replication after the collapse of a replication fork (for reviews, see Haber 1999; Michel 2000), but it also functions to repair DSBs created in other ways. In Saccharomyces cerevisiae, when there is homology with another chromosomal template only centromere-proximal to the DSB, repair may occur by recombination-dependent DNA replication, forming a nonreciprocal translocation (Bosco and Haber 1998). Similar types of events have been documented when linearized plasmids are introduced into yeast cells and initiate DNA replication from a template chromosome, travelling as much as several hundred kilobases to the end of the chromosome (Dunn et al. 1984; Morrow et al. 1997). BIR also may account for the analogous ALT mechanism of telomere maintenance of human tumor cells in the absence of telomerase, as well as similar types of telomere maintenance in budding yeast (Lundblad and Blackburn 1993; Le et al. 1999; Teng and Zakian 1999; Dunham et al. 2000; Teng et al. 2000).
BIR is a recombination-dependent mechanism to initiate DNA synthesis. By creating a DSB in G1 cells, we showed that this replicative process can occur before the initiation of normal DNA replication, at least in wild-type cells (Bosco and Haber 1998). BIR may be closely related to the mechanism of gene conversion induced by a DSB, based on gene conversion that appears to involve both leading- and lagging-strand DNA synthesis (Holmes and Haber 1999). Both processes may initiate in the same way, by the invasion of a single-stranded DNA end that is produced by 5′ to 3′ resection of the DSB, to establish a modified replication fork. In gene conversion, this process would terminate when the second end of the DSB engaged the replication structure, whereas in BIR, the replication process would continue to the end of the chromosome. In some circumstances, BIR and gene conversion appear to be alternative, competing outcomes of DSB-initiated recombination (Esposito 1978; Voelkel-Meiman and Roeder 1990; Malkova et al. 2000).
Nevertheless, there are significant differences between BIR and gene conversion (Malkova et al. 1996). Surprisingly, BIR can occur in the absence of the Rad51p strand exchange protein, whereas gene conversion is completely abolished (Fig. 1). However, BIR still requires the RAD52 protein (Malkova et al. 1996) and thus is apparently a recombination-dependent process. Similarly, in strains in which Rad54p, Rad55p, or Rad57p is deleted, BIR still occurs, but gene conversion is eliminated (Signon et al. 2001). In the absence of these recombination proteins, BIR is a relatively inefficient process, occurring in ∼10–15% of cell divisions. In colonies derived from single cells experiencing a DSB, ∼80% of them are sectored; some cells have lost the broken chromosome entirely whereas others retained it, by BIR ( see Fig. 1). The RAD51- and RAD54-independent pathway of BIR depends on RAD59, TID1(RDH54), MRE11, RAD50, and XRS2 (Signon et al. 2001). Double mutant combinations rad51Δ rad50Δ, rad51Δ rad59Δ, and rad54Δ tid1Δ all severely impaired in BIR, although not as strongly as in rad52Δ strains (Signon et al. 2001).
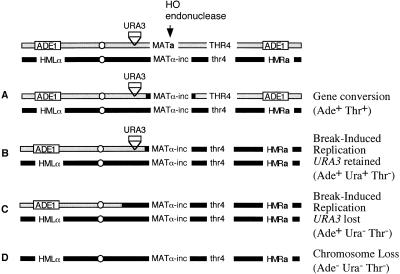
Figure 1.
Repair of a DSB in a MATa/MATα-inc diploid. In the absence of Rad51p, gene conversion (A) is virtually eliminated. Repair occurs by break-induced replication, producing Ade+Thr− cells that may either retain a URA3 marker (B) or else occur such that URA3 is lost (C). If no repair occurs, the entire broken chromosome is lost, and cells become Ade− Thr− (D). Many colonies repair the DSB after one or more cell divisions, so that the colonies often are sectored Ade+ Thr−/Ade− Thr−.
Essentially all DSB-induced mitotic recombination requires the Rad52 protein (reviewed in Pâques and Haber 1999), which has strand-annealing activity in vitro (Mortensen et al. 1996). Rad52p physically and genetically interacts with Rad51p and with the single-strand DNA-binding protein complex, RPA. However, several types of DSB-induced recombination can occur in the absence of Rad51p, Rad54p, Rad55p, and Rad57p. These events include single-strand annealing (SSA) of homologous sequences flanking a DSB (Ivanov et al. 1996) and maintenance of chromosome ends in the absence of telomerase (Le et al. 1999). On centromeric plasmids, but not on chromosomes, even gene conversion between inverted repeats can occur without Rad51p (Ivanov et al. 1996). It has been suggested that such recombination could occur by two Rad51p-independent processes, BIR coupled to SSA (Kang and Symington 2000). In recombining plasmids, the requirement for Rad51p depends on a favorable chromatin structure of the recombining sequences (Sugawara et al. 1995). When donor sequences are heterochromatic, Rad51p is required, whereas when the sequences are in a less constrained chromatin state, Rad51p is not needed. Rad52p is needed in every situation. These experiments suggest that successful BIR might only be able to be initiated at special locations along the chromosome, at sites that are sufficiently accessible to allow strand invasion in the absence of Rad51p.
We asked if BIR initiated only at a limited number of locations. Surprisingly, repair did not initiate anywhere in the first 13 kb proximal to the DSB. In the absence of Rad51p, most of the BIR events on chromosome III depend on sequences adjacent to an origin of DNA replication, but the sequences needed for origin function are not required. Moreover, we show that this sequence does not provide an especially open site for strand invasion of the template, because it is only needed on the broken chromosome. This cis-acting site may enable the BIR process to perform the extensive replication that occurs from this point to the end of the chromosome, >130 kb away.
Results
RAD51-independent BIR initiates nonrandomly along the right arm of chromosome III
To determine where BIR occurs, we constructed a set of diploids homozygous for rad51Δ in which a URA3 marker was inserted centromere-proximal to the MATa locus, the site of the DSB induced by HO endonuclease (Fig. 1). Cleavage takes place ∼202 kb from the left end of chromosome III (Kostriken and Heffron 1984). The MATα-inc locus on the homologous chromosome is not cleaved by HO, which is expressed from a galactose-inducible promoter. Loss of the broken chromosome yields Ade− Thr− cells, but BIR produces Ade+ Thr− cells (Malkova et al. 1996). Many colonies are, in fact, multiply sectored, having sectors that have lost the broken chromosome and one or more sectors that have retained the left arm of the broken chromosome, by BIR. A few Ade+ Thr+ cells appear to be the result of nonhomologous end-joining of the HO-cleaved MATa locus rather than authentic gene conversion events (data not shown). A few Ade+ Thr− cells also could occur because of a combination of two different Rad51p-independent processes, BIR and SSA (Kang and Symington 2000), but if this occurs, it is quite rare.
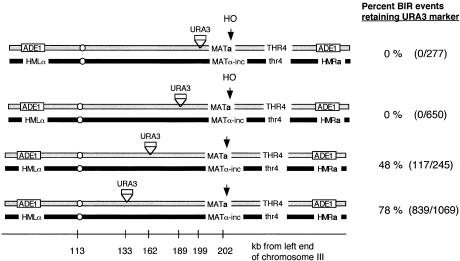
When the URA3 gene was inserted into the top chromosome, either 3 or 13 kb centromere-proximal to the DSB, none of the BIR events retained the URA3 marker (Fig. 2). For example, when URA3 was 13 kb proximal to the DSB at MAT, there were no Ura+ colonies or sectors among 650 colonies that had undergone BIR. This argues that repair of the DSB had occurred more centromere-proximally, far from the site of the DSB. However, as seen in Figure 2, when URA3 was inserted 162 kb from the left end of the chromosome (40 kb proximal to the cleavage site, respectively), ∼48% of the colonies yielded BIR events that retained the URA3 marker (Ade+ Ura+ Thr−), either as an entire colony or as one or more sectors against a background of cells that had lost the broken chromosome. When the URA3 marker was placed more centromere-proximal, at kilobase 133, 78% of the colonies showing evidence of BIR events retained URA3. In some cases, the colonies were sectored Ura3+/ Ura3−. These data indicate that RAD51-independent recombination is successful only when it is initiated at nonrandom sites along the chromosome, >13 kb proximal to the site of the DSB.
Figure 2.
Position of BIR as determined by the retention of a URA3 marker placed centromere-proximal to the DSB at MAT. The retention of URA3 markers inserted 3, 10, 40, and 69 kb proximal to the DSB was determined among colonies in which BIR had occurred. (BIR) break-induced replication; (DSB) double-strand break.
Most BIR events depend on a sequence near ARS310
Inspection of the DNA sequences between 162 and 189 kb revealed one possibly interesting cis-acting element that might account for the success of BIR in that region: the origin of DNA replication, ARS310, at kilobase 166. Although there are two other sequences closer to MAT that function as origins of DNA replication on plasmids, ARS310 is the nearest centromere-proximal origin that is functional on an intact chromosome III (Newlon et al. 1993). To determine if the region containing ARS310 was indeed important for the initiation of BIR, we constructed a diploid homozygous for a 845-bp deletion that includes ARS310 (ars310–Δ1, Fig. 3A). The frequency of colonies that lacked any BIR sectors increased from 19% to 50% of the colonies, and equally strikingly, it dramatically reduced the proportion of BIR events that retained URA3 at kilobase 133 from 78% to 36% (Fig. 4A). Thus, in ars310–Δ1, BIR occurs less often, and, when it occurs, it usually is initiated even further from the site of the DSB. However, as shown below, the sequences that facilitate BIR are actually adjacent to ARS310 and not dependent on its activity.
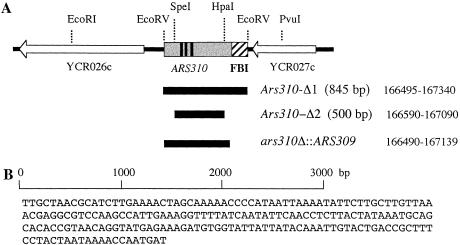
Figure 3.
Deletion analysis of the ARS310 region. (A) Map of the region containing ARS310 and the locations of three deletion/modifications created in this region. (B) Two hundred base pair sequence of FBI, the facilitator of rad51-independent BIR. (FBI) facilitator of BIR; (BIR) break-induced replication.
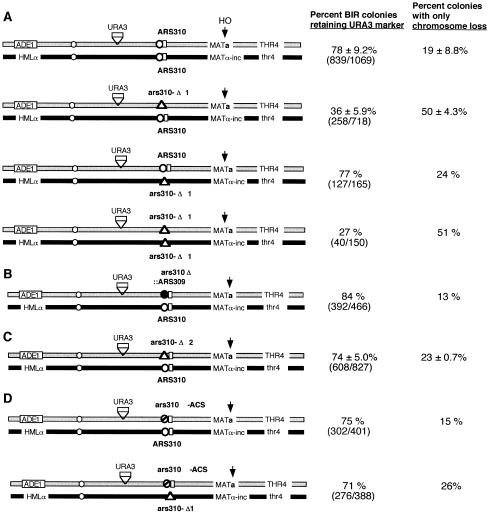
Figure 4.
Role of the ARS310 region in facilitating rad51Δ-independent BIR. (A) Effect of homozygous and heterozygous ars310–Δ1 deletions. (B) Effect of the ars310Δ :: ARS309 replacement of part of the ARS310 region. (C) Effect of the ars310–Δ2 deletion, which leaves the FBI region (indicated by rectangle) intact. (D) Effect of 3-bp substitutions in each of the three ACS sequences within ARS310. (BIR) break-induced replication.
The enhancer sequences are only required on the broken chromosome
If the function of the region including ARS310 were to provide a site that was particularly accessible for strand invasion or strand annealing without RAD51, it might only be required on the recipient (bottom) chromosome. We created additional rad51Δ diploids in which the 845-bp region was deleted only on the top or bottom chromosome (Fig. 4A). The results (Fig. 4A) indicate that the region removed in ars310–Δ1 is important only on the broken chromosome. Deletion of this region from the recipient chromosome gave results indistinguishable from the wild-type diploid, whereas deleting it from the top chromosome gave results indistinguishable from the homozygous deletion.
Most BIR events initiate distal to the enhancer region
To determine more precisely where BIR events initiated, we used the diploid that retained ARS310 on the top chromosome but carried the ars310–Δ1 deletion on the unbroken chromosome (Fig. 4A). We then examined cells that had retained URA3 at kilobase 162 (which is 4 kb to the left of ARS310) to determine if ARS310 on the top chromosome had been retained or if the region had become homozygous for ars310–Δ1 (data not shown). In 14 of 15 cases, the region remained heterozygous for the presence of ARS310, indicating that BIR must have initiated to the right of ARS310.
ARS310 is not required to stimulate RAD51-indepenent BIR
To establish whether ARS310 and its surrounding sequences were unique, we created two different alterations of the region. For these experiments, we used diploids in which only the top (broken) chromosome was modified. First, we inactivated ARS310 by introducing pairs of base pair substitutions in each of the three ACS sequences that contribute to the activity of ARS310 (Theis and Newlon 2001). These mutations eliminated origin firing as determined by two-dimensional gel analysis (data not shown). In two independent strains, BIR occurred to the same extent as in a wild-type strain (Fig. 4D). Thus, origin activity per se is not required.
We then created a second, smaller deletion of ARS310 (ars310–Δ2), a HpaI-SpeI deletion that removes 500 bp of ARS310 but leaves the adjacent 200 bp intact (Fig. 4C). The results show that these ARS-adjacent sequences have most or all of the activity needed to promote efficient BIR. Compared with a deletion of ars310–Δ1, in which 50% of the cells lost the broken chromosome and only 36% of the BIR events retained the URA3 marker located at kilobase 133, the presence of the 200-bp segment reduced chromosome loss to 23% and increased URA3 retention to 74%. Thus, neither ARS activity nor the 500-bp fragment containing ARS310 is needed to stimulate BIR.
We also replaced 649 bp of ARS310 with ARS309 (see Fig. 3A), by using a construct that retains the same 200-bp region as in the ars310–Δ2 deletion. We confirmed that ARS309 was a functional origin of DNA replication in this new location (data not shown). The ars310Δ :: ARS309 replacement restored BIR events retaining the URA3 marker at kilobase 133 to nearly the same level as with the intact ARS310 and its surrounding sequences (Fig. 4B). From these experiments, we concluded that the ability to promote BIR resided in the 200 bp distal to ARS310 (and retained in the ars310Δ :: ARS309 construct).
Discussion
The key question that we have attempted to address in this study is how recombination-mediated DNA replication can occur in the absence of Rad51p. Rad51p is the only protein expressed in mitotic cells that is homologous with the RecA strand exchange protein of bacteria, the only known family of strand exchange proteins (Shinohara et al. 1992). Accordingly, gene conversion between an HO-cleaved MAT locus and an intact template on a homologous chromosome is abolished in rad51Δ diploids. Nevertheless, these cells are able to repair a DSB by a recombination-dependent (i.e., RAD52 dependent) process that we have termed break-induced replication, analogous to recombination-dependent processes that function in bacteriophage T4 replication initiation and that can accomplish recombination in phage λ (for reviews, see Kowalczykowski 2000; Kreuzer 2000). BIR also has been invoked to explain how cells with a broken replication fork in both prokaryotes and eukaryotes can reestablish replication (Kogoma 1996; Haber 1999; Michel 2000) and also how the stability of chromosome ends can be maintained in the absence of telomerase (Walmsley et al. 1984; Lundblad and Blackburn 1993; Bosco and Haber 1998; Le et al. 1999; Teng and Zakian 1999; Dunham et al. 2000; Teng et al. 2000). As with the BIR system described here, maintenance of S. cerevisiae cells in the absence of telomerase also requires RAD52 but is independent of RAD51. Both the single BIR event that we have studied and telomere maintenance without telomerase occur by two RAD52-dependent pathways, one of which requires RAD51, RAD54, RAD55, and RAD57 and the other of which involves RAD50, MRE11, XRS2, RAD59, and TID1 (Lundblad and Blackburn 1993; Le et al. 1999; Teng and Zakian 1999; Teng et al. 2000; Signon et al. 2001).
Here, we show that Rad51p-independent repair does not occur at random sites proximal to the DSB; rather, most of the repair events initiate between 13 and 34 kb proximal to the DSB. We identified a 200-bp segment adjacent to ARS310 that is necessary for this activity. Whether the activity is entirely contained in these 200 bp or extends distally has not been determined. We have named this Facilitator of BIR as FBI. There is probably at least one additional sequence on chromosome III with similar activity, because approximately one-third of the original BIR events occur in a strain deleted for FBI. This sequence is not near ARS309, because a deletion of this ARS does not affect BIR in the absence of FBI; moreover, ARS309 inserted 3 kb proximal to MAT does not change the location of BIR events (data not shown).
How FBI, situated 34 kb proximal to the DSB, allows BIR to occur without Rad51p is not clear. Initially, we speculated that the ARS310 region, partly denatured by the persistent binding of origin recognition complex (ORC) proteins (Lee et al. 2000), might provide an especially accessible site for strand invasion in the absence of Rad51p. However, this idea is inconsistent with the finding that FBI is needed only on the broken chromosome. Hence, the template chromosome does not have to be particularly open.
One possibility is that FBI helps to assemble a processive strand-invasion and replication complex that allows BIR to proceed to the chromosome end. We know that DNA synthesis during repair is less processive than in normal DNA replication (Pâques et al. 1998). Moreover, colonies produced by single cells of rad51Δ diploids that suffered a DSB are often small and nibbled, as if some cells died and failed to give rise to offspring that would populate that part of the growing colony (Malkova et al. 1996). Because simple chromosome loss is not a lethal event and many cells in the colony are 2n-1, these lethal sectors imply that many cells tried to perform BIR, but these attempts ended in a lethal failure. Perhaps the template chromosome and the partially replicated chromosome became trapped in mitosis, causing cell death.
FBI may provide a way for Mcm proteins or some alternative helicase to associate with the BIR replication fork (Fig. 5C) or to recruit the proteins that act in the absence of Rad51p. Most likely, the repair fork lacks the Mcm proteins that are suggested to act as the helicase at normal replication forks (Aparicio et al. 1997; Labib et al. 2000). Mcm proteins are loaded at origins by interactions with origin-binding proteins (Tanaka et al. 1997); they may not assemble at a replication fork established by recombination.
Figure 5.
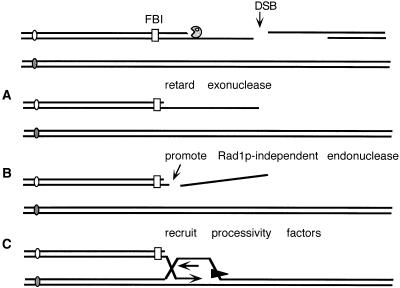
Possible roles of the FBI sequence in promoting Rad51p-independent BIR. (A) DNA is resected by 5′ to 3′ exonucleases, leaving long 3′ single-stranded DNA (ssDNA) tails. FBI may stall the exonuclease, allowing other processing of the ssDNA. (B) The 3′-ended tails must be cleaved off to provide a 3′ end that can act as a primer to initiate new DNA synthesis. FBI may participate in this Rad1p-Rad10p independent process. (C) FBI may help recruit processivity or other factors (shown as a black triangle) needed for BIR to the end of the chromosome, >130 kb away. (DSB) double-strand break; (FBI) facilitator of BIR.
Alternatively, FBI could restrain degradation of the broken chromosome (Fig. 5A) to allow the assembly of the replication–strand-invasion complex. Another step that must occur is to remove a long 3′-ended single-stranded DNA tail created by 5′ to 3′ exonuclease degradation (Fig. 5B). This step is needed to provide a 3′ end that can act as a primer to initiate new DNA synthesis. Although in some circumstances such a tail can be removed by Rad1p-Rad10p endonuclease (Pâques and Haber 1997), Rad51p-independent BIR is Rad1p independent (Signon et al. 2001). In these respects, FBI has some similarities to the properties of the Chi sequence that modifies the activities of RecBCD nuclease in Escherichia coli (for review, see Kuzminov 1999). One difference is that Chi-dependent recombination is promoted further away from the DSB, whereas 14 of 15 repair events occur upstream of ars310-Δ1, showing that FBI promotes recombination upstream of this site.
An inspection of the 200-bp FBI sequence shows several putative protein-binding sites, none of which is obviously implicated in DNA replication functions. However, the small size of this region should make it possible to learn what its key features are.
Materials and methods
Strains
Diploids are isogenic to those described by Signon et al. ( 2001) and are the result of crossing haploid strains EI515 and YLS23. Modifications of the ARS310 region and the insertion of a URA3 marker (described below) were performed, and then deletions of the RAD51 gene were introduced into derivatives of strains EI515 and YLS23 (Malkova et al. 1996) using the a linearized restriction fragment of pJH683 (rad51Δ :: URA3) or pJH1079 (rad51Δ :: LEU2).
A 1.1-kb HindIII URA3 fragment was introduced 3 and 13 kb centromere-proximal to MAT by using linearized fragments of plasmids pAF228 (a gift from B. Dujon, Institut Pasteur) and pJH106, respectively. The same URA3 marker was introduced at positions 133 and 162 kb from the left end of chromosome III, ∼69 and 40 kb centromere-proximal to the HO cleavage site at MAT using plasmids pLS96 and pLS98, respectively. Details are available on request.
Construction of ARS deletions and replacements
Plasmids used for deleting or moving ARS elements were constructed in the URA3 vector, pRS306, and were used for two-step gene replacements (Boeke et al. 1987). Plasmids were digested with an appropriate restriction enzyme to direct integration and were used to transform yeast strains YLS23 and EI515 (Gietz et al. 1992). Transformants were streaked on plates containing 5-fluoro-orotic acid to select strains that had lost the URA3 marker (Boeke et al. 1984) and in which the wild-type chromosome III segment was replaced by the desired construct.
The ars310–Δ1 deletion plasmid, pYND54, carried a 5.7-kb SacII-HindIII fragment of chromosome III from which the 849-bp EcoRV fragment (chromosome III base pairs 166495–167340) containing ARS310 had been deleted (A. Dershowitz and C.S. Newlon, in prep.). The ars310–Δ2 construct was created by deleting the 500-bp SpeI-HpaI fragment (chromosome III base pairs 166590–167090) containing ARS310 from plasmid ARS310BX (Theis and Newlon 2001). To insert ARS309 in place of ARS310, we amplified a 421-bp fragment containing ARS309 (chromosome III base pairs 131948–132368) and fused it to the appropriate left- and right-flanking fragments, including base pairs 165177–166490 and 167139–169198, respectively. For ARS309 inserted 3 kb proximal to MAT, the 421-bp ARS309 region was between base pairs 198042 and 198110. Plasmid ΔARS309 (Dershowitz and Newlon 1993) was used to make the ARS309 deletion.
Media and growth conditions
Rich medium (YEPD) and synthetic complete medium with bases and amino acids omitted as specified were as described (Kaiser et al. 1994). YEP-glycerol and YEP-galactose (YEP-Gal) consisted of 1% yeast extract/2% Bacto peptone media supplemented with 3% (v/v) glycerin or 2% (w/v) galactose, respectively. YEPD medium containing 0.015% (v/v) methyl methane sulfonate was used to assess Rad− phenotypes. Cultures were incubated at 30°C.
Analysis of DNA repair
Logarithmically growing cells grown in YEP-glycerol were plated on YEP-Gal and grown into colonies, as described previously (Malkova et al. 1996). Cells were grown overnight in 50 mL of YEP-glycerol to a cell density of 1–5 × 107 cells per milliliter. Appropriate dilutions of cells were plated on YEP-Gal, grown to colonies, and analyzed. The colonies then were replica-plated onto nutritional drop-out media to determine the fate of the ADE1, THR4, and URA3 markers. Colonies that either were fully Ade+ Thr− or contained Ade+ Thr− sectors against an Ade− Thr− background were counted and scored for the presence of the URA3 marker.
Acknowledgments
We thank members of our laboratories for fruitful discussions and suggestions. C.S.N. is supported by NIH Grant NIH GM35679. J.E.H. is supported by NIH Grant GM20056 and NSF Grant MCB-9724086. C.B.S was partially supported by NIH Training Grant T32 CA09665 and by a fellowship from UMDNJ-Graduate School of Biomedical Sciences. M.L.N. was a undergraduate research scholar of a Howard Hughes Medical Institute grant to Brandeis University.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL haber@brandeis.edu; FAX (781) 736-2405.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.875901.
References
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Lacroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-Fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Bosco G, Haber JE. Chromosome break-induced DNA replication leads to non-reciprocal translocations and telomere capture. Genetics. 1998;150:1037–1047. doi: 10.1093/genetics/150.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dershowitz A, Newlon CS. The effect on chromosome stability of deleting replication origins. Mol Cell Biol. 1993;13:391–398. doi: 10.1128/mcb.13.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- Dunn B, Szauter P, Pardue ML, Szostak JW. Transfer of yeast telomeres to linear plasmids by recombination. Cell. 1984;39:191–201. doi: 10.1016/0092-8674(84)90205-8. [DOI] [PubMed] [Google Scholar]
- Esposito MS. Evidence that spontaneous mitotic recombination occurs at the two-strand stage. Proc Natl Acad Sci. 1978;75:4436–4440. doi: 10.1073/pnas.75.9.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. DNA recombination: The replication connection. Trends Biochem Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- Holmes A, Haber JE. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, Fishman LJ, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kang LE, Symington LS. Aberrant double-strand break repair in rad51 mutants of Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:9162–9172. doi: 10.1128/mcb.20.24.9162-9172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T. Recombination by replication. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- Kostriken R, Heffron F. The product of the HO gene is a nuclease: Purification and characterization of the enzyme. Cold Spring Harb Symp Quant Biol. 1984;49:89–96. doi: 10.1101/sqb.1984.049.01.012. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC. Initiation of genetic recombination and re combination-dependent replication. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- Kreuzer KN. Recombination-dependent DNA replication in phage T4. Trends Biochem Sci. 2000;25:165–173. doi: 10.1016/s0968-0004(00)01559-0. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JF. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Le S, Moore JK, Haber JE, Greider C. RAD50 and RAD51 define two different pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DG, Makhov AM, Klemm RD, Griffith JD, Bell SP. Regulation of origin recognition complex conformation and ATPase activity: Differential effects of single-stranded and double-stranded DNA binding. EMBO J. 2000;19:4774–4782. doi: 10.1093/emboj/19.17.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of RAD51 in yeast: A possible role for break-induced DNA replication. Proc Natl Acad Sci. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Klein F, Leung W-Y, Haber JE. HO endonuclease-induced recombination in yeast meiosis resembles Spo11-induced events. Proc Natl Acad Sci. 2000;97:14500–14505. doi: 10.1073/pnas.97.26.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B. Replication fork arrest and DNA recombination. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- Morrow DM, Connelly C, Hieter P. “Break copy” duplication: A model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon CS, Collins I, Dershowitz A, Deshpande AM, Greenfeder SA, Ong LY, Theis JF. Analysis of replication origin function on chromosome III of Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1993;58:415–423. doi: 10.1101/sqb.1993.058.01.048. [DOI] [PubMed] [Google Scholar]
- Pâques F, Haber JE. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F, Leung WY, Haber JE. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol Cell Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Signon L, Malkova A, Naylor M, Haber JE. Genetic requirements for RAD51,RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol Cell Biol. 2001;21:2048–2056. doi: 10.1128/MCB.21.6.2048-2056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Ivanov EL, Fishman LJ, Ray BL, Wu X, Haber JE. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature. 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Chang J, McCowan B, Zakian VA. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- Theis JF, Newlon CS. Two compound replication origins in S. cerevisiae contain redundant ORC binding sites. Mol Cell Biol. 2001;21:2790–2801. doi: 10.1128/MCB.21.8.2790-2801.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Roeder GS. Gene conversion tracts stimulated by HOT1-promoted transcription are long and continuous. Genetics. 1990;126:851–867. doi: 10.1093/genetics/126.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley RW, Chan CS, Tye BK, Petes TD. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature. 1984;310:157–160. doi: 10.1038/310157a0. [DOI] [PubMed] [Google Scholar]