Abstract
The p53 tumor suppressor protein, a key regulator of cellular responses to genotoxic stress, is stabilized and activated after DNA damage. The rapid activation of p53 by ionizing radiation and radiomimetic agents is largely dependent on the ATM kinase. p53 is phosphorylated by ATM shortly after DNA damage, resulting in enhanced stability and activity of p53. The Mdm2 oncoprotein is a pivotal negative regulator of p53. In response to ionizing radiation and radiomimetic drugs, Mdm2 undergoes rapid ATM-dependent phosphorylation prior to p53 accumulation. This results in a decrease in its reactivity with the 2A10 monoclonal antibody. Phage display analysis identified a consensus 2A10 recognition sequence, possessing the core motif DYS. Unexpectedly, this motif appears twice within the human Mdm2 molecule, at positions corresponding to residues 258–260 and 393–395. Both putative 2A10 epitopes are highly conserved and encompass potential phosphorylation sites. Serine 395, residing within the carboxy-terminal 2A10 epitope, is the major target on Mdm2 for phosphorylation by ATM in vitro. Mutational analysis supports the conclusion that Mdm2 undergoes ATM-dependent phosphorylation on serine 395 in vivo in response to DNA damage. The data further suggests that phosphorylated Mdm2 may be less capable of promoting the nucleo-cytoplasmic shuttling of p53 and its subsequent degradation, thereby enabling p53 accumulation. Our findings imply that activation of p53 by DNA damage is achieved, in part, through attenuation of the p53-inhibitory potential of Mdm2.
Keywords: Radiation, proteolysis, ubiquitination, nucleo-cytoplasmic shuttling
The p53 tumor suppressor plays a major role in the cellular response to various stress signals. The cellular levels of the p53 protein significantly increase in response to stress such as DNA damage, activation of oncogenes, and hypoxia (for review, see Giaccia and Kastan 1998; Oren 1999; Prives and Hall 1999; Vousden 2000). The wild-type p53 protein normally has a very short half-life, owing to degradation by the ubiquitin-proteasome machinery (Maki et al. 1996). Central to the regulation of p53 is the Mdm2 protein. Transcription of the mdm2 gene is strongly activated by p53 (Barak et al. 1993; Wu et al. 1993). The Mdm2 protein, in turn, binds p53 and conceals its transactivation domain (Momand et al. 1992; Oliner et al. 1993). Moreover, Mdm2 promotes the rapid degradation of p53 (Bottger et al. 1997; Haupt et al. 1997; Kubbutat et al. 1997). This negative auto-regulatory feedback loop enables the tight regulation of p53 activity and stability (Freedman and Levine 1999; Juven-Gershon and Oren 1999; Momand et al. 2000).
The ability of Mdm2 to promote p53 degradation is attributable to its E3 ubiquitin ligase activity (Honda and Yasuda 1997, 1999). Within Mdm2, the RING finger domain is of particular importance for this E3 activity, in vitro as well as in vivo (Kubbutat et al. 1997; Honda and Yasuda 1999; Fang et al. 2000). In addition, the degradation and stability of p53 are strongly dependent on the subcellular localization of both p53 and Mdm2. p53 can shuttle between the nucleus and the cytoplasm (Middeler et al. 1997; Weber et al. 1999). Mdm2, a predominantly nuclear protein, also shuttles constantly between the cytoplasm and the nucleus (Roth et al. 1998). The export of p53 into the cytoplasm is essential for its effective Mdm2-mediated degradation (Roth et al. 1998; Lain et al. 1999; Tao and Levine 1999a,b). Cytoplasmic export of p53 requires its prior ubiquitination by Mdm2 (Boyd et al. 2000; Geyer et al. 2000). There remains some uncertainty as to whether Mdm2 also contributes in additional ways to p53 shuttling (Roth et al. 1998; Lain et al. 1999; Stommel et al. 1999).
Another p53 regulator is the ARF tumor suppressor protein. ARF directly blocks the E3 activity of Mdm2, and in addition sequesters Mdm2 in the nucleolus, away from p53 (Honda and Yasuda 1999; Tao and Levine 1999a,b; Weber et al. 1999; Zhang and Xiong 1999); a nucleolar localization signal within the RING domain of Mdm2 is also required for its nucleolar sequestration by ARF (Lohrum et al. 2000; Weber et al. 2000). Several cellular proteins, including E2F1, Ras, Myc, β-catenin, pRb, and c-Abl, can stabilize p53; in many cases, stabilization is achieved through induction of ARF (for review, see Sharpless and DePinho 1999; Sherr and Weber 2000).
Regulation of p53 protein levels and activity is achieved primarily through posttranslational mechanisms, with phosphorylation playing a major role (Ashcroft et al. 1999, 2000; Oren 1999). The p53 protein is a target for phosphorylation by a plethora of protein kinases (for review, see Fuchs et al. 1998; Giaccia and Kastan 1998; Jayaraman and Prives 1999; Meek 1999; Prives and Hall 1999). Stress-induced phosphorylation of serines and threonines within the amino-terminal region of p53 contributes to activation and stabilization of p53, by attenuating its binding to Mdm2 as well as augmenting its interaction with components of the transcriptional machinery.
A major activator of p53 in response to ionizing radiation is the ATM kinase. ATM belongs to a family of protein kinases that possess a phosphoinositide 3-kinase-related domain at their carboxyl termini; these enzymes are involved in controlling genome stability, cell cycle progression, and responses to DNA damage in various organisms (for review, see Kastan and Lim 2000; Shiloh 2001). The p53 protein is phosphorylated by ATM on serine 15 (Siliciano et al. 1997; Banin et al. 1998; Canman et al. 1998; Khanna et al. 1998), which may render p53 more resistant to the inhibitory effects of Mdm2 (Shieh et al. 1997), as well as enhance its transcriptional activity (Lambert et al. 1998; Dumaz and Meek 1999).
It appears reasonable that the interplay between p53 and Mdm2 is modulated by modification not only of p53, but also of Mdm2. In fact, Mdm2 can be phosphorylated on multiple sites (Henning et al. 1997; Mayo et al. 1997; Gotz et al. 1999), as well as be modified by covalent attachment of Sumo (Buschmann et al. 2000). We described previously the rapid ATM-dependent phosphorylation of Mdm2 on exposure of cells to ionizing radiation (IR) and radiomimetic drugs (Khosravi et al. 1999). This is reflected in a faster electrophoretic mobility of Mdm2, with a concomitant decrease in its apparent reactivity with the monoclonal antibody 2A10 (Khosravi et al. 1999; Maya and Oren 2000). Moreover, Mdm2 can be phosphorylated directly by ATM in vitro (Khosravi et al. 1999).
We now report that microinjection of the 2A10 antibody increases substantially the steady state levels of endogenous wild-type p53. Human Mdm2 carries two putative 2A10 epitopes, spanning positions 258–260 and 393–395, respectively. Both epitopes are highly conserved in evolution and contain critical serines, at positions 260 and 395, whose phosphorylation may affect 2A10 reactivity. We show that serine 395 (S395) is a major target for phosphorylation by ATM in vitro. Moreover, mutational analysis supports the conclusion that S395 undergoes ATM-dependent phosphorylation in vivo following DNA damage. The data further suggests that S395 phosphorylation attenuates the ability of Mdm2 to promote the nuclear exit of p53 and to target p53 for degradation. These results support the existence of a dual mechanism for p53 accumulation and activation following DNA damage, where the simultaneous phosphorylation of both p53 and Mdm2 ensures an ef fective release of p53 from the inhibitory action of Mdm2.
Results
Microinjection of the 2A10 antibody enables p53 accumulation
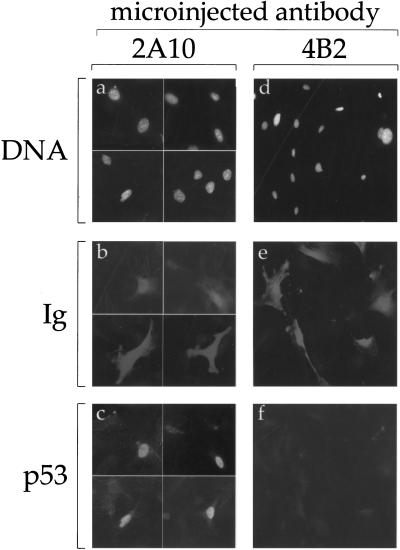
IR and radiomimetic agents elicit rapid ATM-dependent phosphorylation of Mdm2 (Khosravi et al. 1999). The timing of this process is compatible with a role in the subsequent accumulation of p53; however, a causal relationship was not established. To better evaluate the relationship between Mdm2 phosphorylation and p53 accumulation, we took advantage of the fact that the ATM-dependent phosphorylation of Mdm2 decreases its reactivity with the monoclonal antibody (MoAb) 2A10 (Khosravi et al. 1999; Maya and Oren 2000). This suggested that the region of Mdm2 recognized by 2A10 may contribute to the regulation of Mdm2 activity, and that interference with the proper performance of this region might thus compromise the biochemical activity of Mdm2. To test this prediction, primary mouse embryo fibroblasts (MEFs) were microinjected with 2A10. Steady state levels of p53 in the injected cells were monitored by indirect immunofluorescence. As shown in Figure 1, the level of endogenous p53 in noninjected MEFs was practically below detection, apparently because in the absence of stress, p53 is constitutively targeted for rapid Mdm2-mediated degradation. However, cells microinjected with 2A10 (identified by positive immunoglobulin staining, Fig. 1b) exhibited prominent nuclear accumulation of endogenous p53 (Fig. 1c), suggesting that binding of 2A10 to Mdm2 disrupts the ability of Mdm2 to target p53 for degradation. The accumulation of p53 was not caused by a nonspecific effect of the microinjection, because it was not induced by a different Mdm2-specific MoAb, 4B2 (Fig. 1e,f), directed against the amino-terminal part of Mdm2 (Chen et al. 1993).
Figure 1.
Microinjection of 2A10 induces p53 accumulation. Primary MEFs were microinjected with ascites fluid of either the anti-Mdm2 monoclonal antibody 2A10 (panels a–c), or the anti-Mdm2 monoclonal antibody 4B2 (panels d–f). Twenty-nine h later, cells were fixed and stained with DAPI (panels a,d), Cy3-conjugated goat anti-mouse antibody to visualize the microinjected mouse monoclonal antibodies (panels b,e), and anti-p53 polyclonal serum followed by FITC-conjugated donkey anti-rabbit antibody, to visualize endogenous p53 (panels c,f). Panels a–c) represent the same microscopic fields, photographed at three different wavelengths, as is also the case for panels d–f. Ig = immunoglobulin.
The selective ability of 2A10 to abrogate the p53-destabilizing effect of Mdm2 supports the conjecture that the corresponding epitope resides within a region of Mdm2 critical for its proper activity. Similar observations were reported recently by Lane and coworkers (Midgley et al. 2000).
Mapping of the 2A10 epitopes on Mdm2
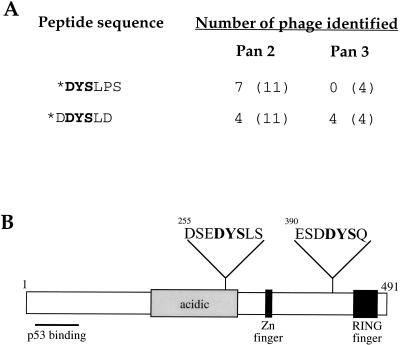
As a first step towards identifying the residue(s) on Mdm2 that are phosphorylated by ATM, we attempted to map the 2A10 epitope through phage display epitope library analysis. Screening of a 6-mer phage peptide library resulted in the selection of 2A10-reactive peptides sharing the core sequence 1-DYS-5, with a preference for D at position 1 and a strong preference for L at position 5 (Fig. 2A and data not shown). Unexpectedly, inspection of the sequence of human Mdm2 revealed two putative 2A10 epitopes (Fig. 2B). One of these (EDYSL) is located at positions 257–261 of Mdm2, whereas the other (DDYSQ) resides at positions 392–396. Of note, the preceding residues are also very similar between both putative epitopes. Importantly, both putative 2A10 epitopes reside within sequences that are highly conserved through evolution (Momand et al. 2000), suggesting that both sites are parts of functionally important domains.
Figure 2.
Mdm2 encompasses two putative 2A10 epitopes. (A) A 6-mer phage display peptide library was screened with the 2A10 antibody as described in Materials and Methods. Eleven positive clones obtained after the second panning, as well as four positive clones obtained after the third panning, were subjected to DNA sequencing. All clones were found to fall into only two sequence classes. The deduced amino acid sequences of the corresponding 6-mer peptides, present in the positive phage, are shown. Based on this data and on the screening of another independent library (data not shown), the consensus 2A10 epitope was deduced as 1-DYS-5, where 1 is preferentially D and 5 is preferentially L; the core DYS sequence is indicated in bold. Also shown is the number of positive phage found to carry the indicated sequence, out the total number of positive phage (in parentheses) sequenced after each panning. (B) Positions of the two putative 2A10 epitopes on Mdm2 are indicated. The positions of the main identified structural domains of the protein (Momand et al. 2000) are also indicated. Zn finger = zinc finger.
In addition to the serine residue within the core DYS motif, additional serines reside in close proximity to both putative 2A10 epitopes (Fig. 2B). In principle, phosphorylation of any of those serines in response to DNA damage may also affect 2A10 reactivity, as may phosphorylation of the core tyrosine residue, if Mdm2 can undergo such modification in vivo. Of particular note, S395, located within the carboxy-terminal putative 2A10 epitope, is followed by a glutamine residue and thus constitutes a potential ATM phosphorylation site (Kim et al. 1999; O'Neill et al. 2000).
S395 of Mdm2 is phosphorylated in vitro by ATM
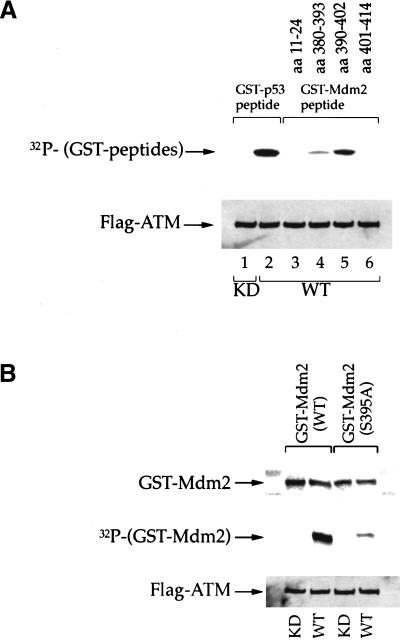
To gain definitive information on the site(s) targeted by ATM, a set of human Mdm2-derived synthetic peptides containing the ATM consensus sequence, SQ, were assayed for phosphorylation by ATM in vitro (Fig. 3A). A GST–p53 peptide, serving as a positive control (Canman et al. 1998), was efficiently phosphorylated by wild-type ATM (Fig. 3A, lane 2) but not by kinase-dead ATM (Fig. 3, lane 1). Substantial phosphorylation was also seen when GST was fused with a peptide consisting of residues 390–402 of human Mdm2 (Fig. 3A, lane 5). In contrast, two other SQ-containing peptides were not phosphorylated at all (Fig. 3, lanes 3,6), whereas a weak signal was obtained with a peptide corresponding to residues 380–393 (Fig. 3, lane 4). This suggested strongly that the major ATM phosphorylation site resides between residues 390–402. The only serine within this segment that is followed by a glutamine is S395, thus implicating it as the principal target of ATM in vitro.
Figure 3.
Serine 395 is the major site on Mdm2 for phosphorylation by ATM in vitro. (A) Immunoprecipitated wild-type (WT) or kinase dead (KD) FLAG-ATM were incubated with a fixed amount of E. coli-expressed recombinant proteins, consisting of fusions between GST and peptides derived from various regions of human Mdm2. The amino acid positions corresponding to each peptide are indicated in the upper part of the panel (GST–Mdm2 peptide). Kinase assays were performed in vitro as described in Materials and Methods. A fusion between GST and a peptide corresponding to residues 9–21 of human p53 (GST–p53 peptide) was used as a positive control. Upper panel: 32P incorporation into the various GST peptides. Lower panel: Western blot with anti-FLAG M2 monoclonal antibody, confirming the presence of similar amounts of ATM protein in all reactions. (B) Immunoprecipitated WT or KD FLAG-ATM were incubated with recombinant proteins consisting of fusions between GST and full-length Mdm2, either wild-type or S395A. Kinase assays were performed in vitro as in A. Upper panel: Western blot with an Mdm2-specific monoclonal antibody, to assess the input amount of fusion protein in each reaction. Middle panel: 32P incorporation into the GST–Mdm2 fusion proteins. Lower panel: Western blot with anti FLAG M2 monoclonal antibody.
This notion was explored more directly by comparing the effect of ATM on fusion proteins containing either full-length wild-type human Mdm2 or a mutant with a serine to alanine substitution at position 395 (S395A). Whereas the wild-type Mdm2 fusion protein was an excellent in vitro ATM substrate, substitution to alanine almost completely abolished phosphorylation (Fig. 3B). Hence, S395 is indeed a major site of phosphorylation by ATM in vitro. The residual phosphorylation of S395A may represent minor in vitro phosphorylation sites, possibly residing within the amino-terminal portion of Mdm2 (Khosravi et al 1999), that are recognized with varying efficiency by different preparations of ATM. The significance of such minor sites remains unclear; however, there is evidence that not all in vitro ATM sites are actually targeted in vivo (Y. Shiloh, unpubl.).
ATM-dependent phosphorylation of S395 in vivo
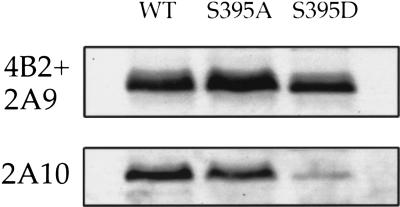
The involvement of S395 in ATM-dependent phosphorylation in vivo was explored by replacing S395 with either alanine (S395A), expected to abrogate phosphorylation, or aspartic acid (S395D), expected to mimic some aspects of phosphorylation. After transfection into H1299 cells, the 2A10 reactivity of the different in vivo-expressed Mdm2 variants was compared by Western blot analysis. Substitution of S395 by aspartic acid dramatically reduced 2A10 reactivity (Fig. 4, S395D). A mild reduction was also seen with S395A, consistent with the phage display data that implicate a requirement for serine at this position for optimal 2A10 binding.
Figure 4.
Mutation of serine 395 decreases 2A10 immunoreactivity. H1299 cells were transiently transfected with expression plasmids (600 ng each) encoding either wild-type human Mdm2 (WT), Mdm2 S395A, or Mdm2 S395D. Twenty-six h later, cells were harvested and subjected to SDS-PAGE followed by Western blot analysis. The membranes were reacted either with a mixture of the 2A9 and 4B2 Mdm2-specific monoclonal antibodies or with the 2A10 Mdm2-specific monoclonal antibody.
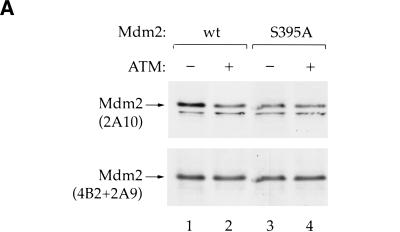
ATM-dependent Mdm2 phosphorylation can be monitored with 2A10 (Khosravi et al 1999). If S395 is phosphorylated by ATM also in vivo, it is predicted that wild-type Mdm2 but not S395A will display an ATM-dependent decrease in 2A10 reactivity. This prediction was tested by transfecting p53/mdm2 double null MEFs (174-2, gift of G. Lozano) with either wild-type or S395A Mdm2, with or without an ATM expression plasmid. Cotransfection of wild-type Mdm2 with ATM resulted in a decrease in 2A10 reactivity, consistent with ATM-dependent phosphorylation (Fig. 5A, lanes 1,2). This was not due to differences in the total amount of Mdm2, as confirmed by reprobing the same membrane with a combination of the Mdm2-specific MoAbs 4B2 and 2A9. In contrast to wild-type Mdm2, the 2A10 reactivity of S395A was completely unaffected by ATM (Fig. 5A, lanes 3,4). Hence, in transfected cells, S395A cannot undergo ATM-dependent phosphorylation within the 2A10 epitope, supporting the conclusion that S395 is also a target of ATM in vivo.
Figure 5.
Ser395 is required for ATM-dependent phosphorylation in vivo. (A) p53/mdm2 double null 174-2 cells were transiently transfected with 1.5 μg of either wild-type (lanes 1,2) or S395A (lanes 3,4) Mdm2 expression plasmid, plus 4.5 μg of either empty vector (lanes 1,3) or ATM expression plasmid (lanes 2,4). Twenty-six h later, cells were harvested and subjected to SDS-PAGE followed by Western blot analysis. The membrane was first reacted with 2A10, followed by reprobing with a mixture of the Mdm2-specific MoAbs 2A9 and 4B2. (B) p53-null AP29 cells were infected with retroviruses expressing either wild-type or S395A human Mdm2. Forty-eight h later cells were treated with 25 μM MG132 for 4 h to block further Mdm2 degradation, and then irradiated with 8Gy and harvested 30 min later. Proteins were extracted in the presence of phosphatase inhibitors. Ten percent of the extract was taken directly for SDS-PAGE (lanes 5–8), whereas the rest was subjected to immunoprecipitation with MoAb 2A9, specific for human Mdm2 (lanes 1–4). Following transfer to a nitrocellulose membrane, the membranes were reacted sequentially with 2A10 (upper row) and then with a mixture of 2A9 and 4B2 (lower row).
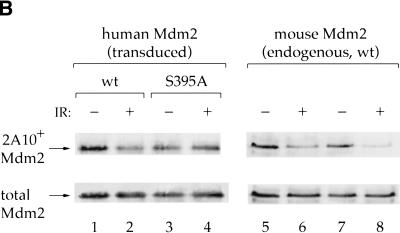
If this is indeed the case, S395A should also fail to undergo ATM-dependent phosphorylation under conditions where endogenous ATM is activated by relevant signals. Therefore, p53 null MEFs were infected with retroviruses expressing either wild-type or S395A Mdm2. Retroviral delivery was chosen because it yielded very low levels of human Mdm2, expected not to perturb the biology of the recipient MEFs. In fact, these levels were well below those of the endogenous mouse Mdm2, to the extent that they were practically undetectable by direct Western blot analysis. To allow selective visualization of the exogenous human Mdm2, it was first immunoprecipitated with the Mdm2-specific MoAb 2A9, which does not react efficiently with mouse Mdm2 (R. Maya, unpubl.). Subsequent probing of the immunoprecipitated human Mdm2 with 2A10 revealed a substantial drop in the reactivity of the wild-type protein on exposure to IR (Fig. 5B, lanes 1,2). However, the 2A10 reactivity of S395A was unaffected by IR (Fig. 5B, lanes 3,4). Of note, IR reduced the 2A10 reactivity of the endogenous murine Mdm2 in the same extracts, irrespective of whether the cells expressed wild-type or S395A human Mdm2 (Fig. 5B, lanes 5–8). The selective retention of 2A10 reactivity by S395A implies that it fails to undergo ATM-dependent phosphorylation on the 2A10 epitope in response to IR. Taken together, the findings in Figures 3 and 5 are highly consistent with the notion that cellular exposure to ionizing radiation triggers the phosphorylation of S395 by ATM.
Substitution of S395 to aspartic acid compromises the effects of Mdm2 on p53
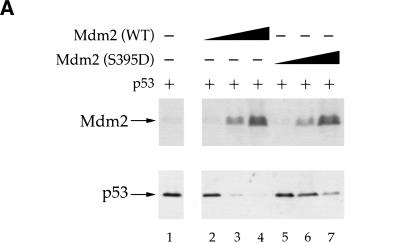
To assess the functional consequences of S395 phosphorylation, p53-null human H1299 cells were transiently transfected with a fixed amount of p53 expression plasmid, together with increasing amounts of DNA encoding either wild-type Mdm2 or S395D, the latter expected to resemble constitutively phosphorylated Mdm2. Wild-type Mdm2 elicited a pronounced decrease in steady state p53 levels (Fig. 6A, lanes 1–4), consistent with earlier reports (Haupt et al. 1997; Kubbutat et al. 1997). In contrast, despite being expressed at comparable levels (see Fig. 6A), S395D was far less capable of promoting p53 degradation (Fig. 6A, lanes 5–7). S395A was as potent as wild-type Mdm2 in some experiments, whereas in others its activity was intermediate between that of wild-type Mdm2 and S395D (data not shown), presumably owing to a mild negative effect of the substitution to alanine.
Figure 6.
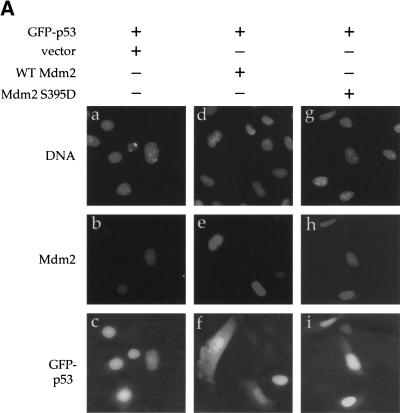
Mdm2 S395D is impaired in promoting p53 degradation. (A) H1299 cells were transiently transfected with 15 ng of human p53 expression plasmid, either alone (lane 1) or in combination with 100, 300, or 600 ng of wild-type Mdm2 or Mdm2 S395D expression plasmids (lanes 2–4 and 5–7, respectively). Twenty-six h later cells were harvested and subjected to SDS-PAGE, followed by Western blot analysis. The membranes were reacted with a mixture of the 2A9 and 4B2 Mdm2-specific monoclonal antibodies (upper panel) or with a mixture of the p53-specific monoclonal antibodies DO-1 and PAb1801 (lower panel). (B) U2-OS cells were transiently transfected with 0.5 μg of GFP-p53 expression plasmid DNA, together with 2 μg of DNA encoding either wild-type (WT) or mutant human Mdm2, as indicated above the corresponding panels. Forty-eight h later cells were fixed and stained with DAPI (panels a,d,g) and with the anti-Mdm2 monoclonal antibody 4B2 followed by Cy3-conjugated goat anti-mouse immunoglobulin serum (panels b,e,h). GFP–p53, in the same microscopic fields, was visualized by monitoring green fluorescence with an appropriate filter (panels c,f,i).
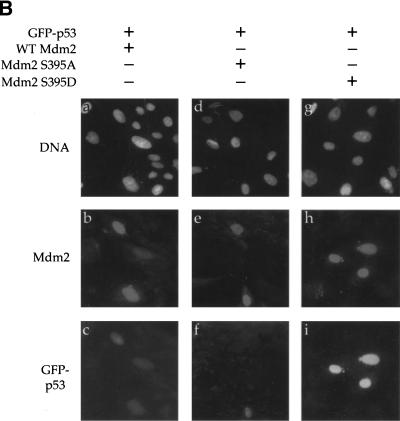
A similar picture was revealed in human osteosarcoma U2-OS cells, transiently transfected with an expression plasmid encoding a green fluorescent protein–p53 fusion protein (GFP–p53; Stommel et al. 1999), together with wild-type Mdm2, Mdm2 S395A, or Mdm2 S395D. Analysis of cultures 48 h after transfection revealed that cells transfected with a combination of GFP–p53 and wild-type Mdm2 exhibited rather faint GFP fluorescence (Fig. 6B, panels a–c), presumably owing to proteasomal degradation of the fusion protein. A similar picture was revealed with S395A (Fig. 6B, panels d–f). In both cases, close to 75% of the Mdm2 positive cells displayed only weak GFP fluorescence (data not shown). In contrast, the majority of the cells expressing S395D retained intensive fluorescence (Fig. 6B, panels g–i); weaker fluorescence was seen in only 35% of the cells. Hence in U2-OS cells, too, S395D is less effective than either wild-type Mdm2 or S395A in promoting p53 degradation. The data in Figure 6 suggests strongly that phosphorylation of Mdm2 on S395 reduces its ability to down-regulate p53.
The simplest mechanism through which S395 phosphorylation may exert its effect is by weakening the binding of Mdm2 to p53. However, analysis of H1299 cells cotransfected with p53 together with wild-type Mdm2, S395A, or S395D, failed to reveal significant differences in the ability of the various Mdm2 proteins to bring down bound p53 (data not shown). Hence, the reduced ability of S395D to promote p53 degradation is not due to a lower p53-binding capacity, and must result from interference with some other property of Mdm2.
A major outcome of p53 ubiquitination is its export from the nucleus to the cytoplasm (Boyd et al. 2000; Geyer et al. 2000), necessary for its proteasomal degradation (Roth et al. 1998; Lain et al. 1999; Stommel et al. 1999; Tao and Levine 1999a,b). We therefore compared the ability of wild-type Mdm2 and S395D to affect the subcellular localization of p53. U2-OS cells were transfected with GFP–p53, either alone or in combination with wild-type Mdm2 or S395D. Unlike in Figure 6B, cells already were analyzed 29 h after transfection, when GFP–p53 levels were still relatively high even in most cells expressing wild-type Mdm2. In the absence of cotransfected Mdm2, GFP–p53 localized primarily to the nucleus (Fig. 7A, panel c). Cotransfection with wild-type Mdm2 resulted in a significant increase in the proportion of cells containing substantial amounts of cytoplasmic GFP–p53 (Fig. 7A, panel f), consistent with the ability of Mdm2 to dictate nuclear export of p53. However, in cells cotransfected with S395D, GFP–p53 remained largely nuclear (Fig. 7A, panel i).
Figure 7.
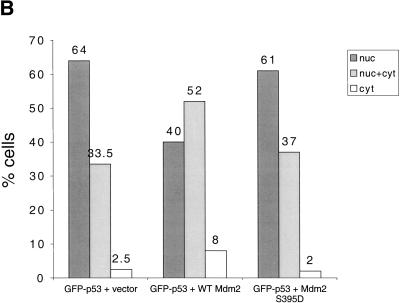
Effect of wild-type Mdm2 and Mdm2 S395D on p53 localization. (A) U2-OS cells were transiently transfected with 1 μg of GFP–p53 expression plasmid together with 2 μg of expression plasmid encoding either wild-type or S395D Mdm2, or with empty vector, as indicated. Cells were processed exactly as in Fig. 6B, except that harvesting was done after 29 h only. (B) Graphic representation of the effect of the various forms of Mdm2 on the intracellular distribution of p53. Transfected cultures such as shown in A were scored under the fluorescent microscope for green fluorescence, as in Fig. 7A (panels c,f,i). Cells were empirically divided into three categories: (1) those where GFP–p53 was exclusively or very predominantly nuclear (nuc), (2) those where GFP–p53 was easily detectable in both nucleus and cytoplasm (nuc + cyt), and (3) those where GFP–p53 was very predominantly cytoplasmic (cyt). Cells were scored independently, in a double-blind fashion, by three individuals. In each case, 100–200 positive cells were scored for each combination of transfected plasmids. The number above each column represents the average value obtained for the percentage of cells falling within the indicated category.
For a more quantitative assessment of the data, cells positive for transfected Mdm2 were scored in three categories: strictly nuclear GFP–p53, GFP–p53 easily detectable in both nucleus and cytoplasm, and predominantly cytoplasmic GFP–p53 (Boyd et al. 2000; Geyer et al. 2000). Figure 7B represents the average of three independent double-blind counts. In the absence of cotransfected Mdm2 (vector), only 36% of the transfected U2-OS cells had significant amounts of cytoplasmic GFP–p53. This may already be an overestimate, reflecting some contribution of the endogenous Mdm2 whose transcription is activated by the transfected GFP–p53. Cotransfection with wild-type Mdm2 resulted in 60% of the cells showing significant cytoplasmic p53 staining, attesting to augmented cytoplasmic export. In contrast, the picture with S395D was much closer to that seen without transfected Mdm2: Only 39% of the cells exhibited conspicuous cytoplasmic GFP–p53. Thus, S395D is markedly less able to promote p53 cytoplasmic export; this may account for its reduced ability to promote p53 degradation. By analogy, phosphorylation of Mdm2 on S395 is predicted to also have a similar effect.
Discussion
The Mdm2 protein plays a dual role in the inactivation of p53. On the one hand, it interferes with the ability of p53 to function as a transcription factor. On the other hand, as a p53-specific E3 ubiquitin ligase, Mdm2 drives the continuous proteasomal degradation of p53, thereby enabling the cell to maintain very low steady state levels of p53.
Signals leading to the rapid activation of p53 must disrupt the Mdm2-dependent autoinhibitory feedback loop that normally keeps p53 latent. In the case of DNA damage, this is achieved partly through posttranslational modification of p53, most notably phosphorylation (for review, see Fuchs et al. 1998; Giaccia and Kastan 1998; Jayaraman and Prives 1999; Meek 1999; Oren 1999; Prives and Hall 1999; Kapoor et al. 2000). However, p53 phosphorylation is unlikely to be the sole mechanism through which DNA damage leads to p53 accumulation. In fact, in some situations p53 phosphorylation does not appear to contribute at all to its stabilization (Ashcroft et al. 1999; Blattner et al. 1999; Dumaz and Meek 1999). Hence, modifications on other proteins are also likely to contribute to p53 stabilization. The most natural candidate is Mdm2.
Phosphorylation of Mdm2 may affect its ability to promote p53 ubiquitination. The p53-binding domain and the RING finger, residing near the amino and carboxyl termini of Mdm2, respectively, are attractive targets for such modifications. However, other domains, including the nuclear localization and nuclear export signals, also participate in the in vivo regulation of Mdm2 function (Roth et al. 1998; Lain et al. 1999; Tao and Levine 1999a,b). The ability of Mdm2 to ubiquinate p53 is negatively regulated by ARF (Pomerantz et al. 1998; Honda and Yasuda 1999; Zhang and Xiong 1999; Fang et al. 2000), suggesting that modifications affecting ARF binding might also modulate Mdm2 function.
Mdm2 can be phosphorylated in vitro by DNA-PK on serine 17 (S17), within the p53-binding domain, resulting in reduced binding of Mdm2 to p53 in vitro (Mayo et al. 1997). However, there is no indication so far that S17 is phosphorylated in vivo, nor is there evidence for a role of DNA-PK in p53 stabilization (Jimenez et al. 1999). Moreover, ATM-dependent, DNA damage-induced phosphorylation of Mdm2 in vivo is DNA-PK-independent (Khosravi et al. 1999). Mdm2 is also an in vitro target of CKII (Gotz et al. 1999), but the relevance remains unknown.
We now propose that S395 of Mdm2 is a site of rapid in vivo phosphorylation in response to IR and radiomimetic drugs. ATM phosphorylates Mdm2 on S395 in vitro. Moreover, S395 appears to be phosphorylated in an ATM-dependent manner in vivo. ATM activity is crucial for the rapid stabilization of p53 following ionizing radiation (Kastan et al. 1992). Our data suggests strongly that the p53-stabilizing effect of ATM is exerted in part through phosphorylation of the other arm of the autoregulatory negative feedback loop, namely Mdm2. Substitution of S395 to aspartic acid (S395D), presumably mimicking phosphorylation, diminishes the capacity of Mdm2 to drive p53 degradation. Furthermore, S395D is inefficient in promoting the cytoplasmic export of p53, crucial for effective p53 degradation. This lends strong support to the notion that ATM-dependent phosphorylation of Mdm2 on S395 plays a significant role in enabling the rapid stabilization and activation of p53 in response to particular types of DNA damage.
S395D still retains some ability to mediate p53 degradation. A trivial explanation is that the substitution mimics phosphorylated S395 only partially. However, it is perhaps more likely that S395 phosphorylation is but one of several modifications and/or conformational changes that together contribute to abrogation of Mdm2 activity by stress signals. Further work should resolve this issue.
The precise mechanism through which S395 phosphorylation attenuates Mdm2 function is unclear. S395 and its adjacent amino acids display a high degree of evolutionary conservation, supporting the functional importance of this part of the molecule. Yet, S395 does not reside within any recognizable structural motif of Mdm2 (Momand et al. 2000). The proximity of S395 to the RING finger of Mdm2, crucial for E3 activity (Kubbutat et al. 1997; Fang et al. 2000), might suggest that its phosphorylation affects directly this enzymatic activity of Mdm2. However, in a cell-free ubiquitination assay, the E3 activity of a GST–Mdm2 S395D fusion protein was not significantly different from that of its wild-type counterpart (data not shown). It therefore appears plausible that S395 phosphorylation impinges on a more complex interaction, involved in regulation of Mdm2 activity in vivo. For instance, it may affect the association of Mdm2 with proteins that regulate its ability to ubiquinate p53 and promote its cytoplasmic export.
Our data does not exclude the possibility that the other 2A10 epitope, spanning positions 258–260, may also be targeted by a kinase, particularly if this requires prior phosphorylation of S395 by ATM. It is also noteworthy that the residues around S260 and S395 possess unusual similarity (Fig. 2B), highly conserved in evolution. This raises the intriguing possibility that both sites attract not only the same antibody, but also a much more relevant cellular factor involved in the physiological regulation of Mdm2 function.
Overall, our findings imply that the simultaneous ATM-dependent phosphorylation of both p53 and Mdm2 ensures a well-coordinated p53 response.
Materials and methods
Cell culture, transfections, infections, and plasmids
Primary MEFs were grown for four passages at 37°C in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with nonessential amino acids, 60 μM β-mercapto-ethanol, and 10% heat inactivated fetal bovine serum (FBS, Sigma). 174-2 cells (p53−/−, Mdm2−/− MEFs) were grown at 37°C in DMEM supplemented with 10% heat inactivated FBS. AP29 (p53−/− MEFs) were grown at 37°C in DMEM supplemented with 15% heat inactivated FBS. U2-OS cells (human osteosarcoma, wild-type p53) were maintained at 37°C in DMEM supplemented with 10% heat inactivated FBS. H1299 cells (human lung adenocarcinoma, p53 null) were maintained at 37°C in RPMI medium supplemented with 10% heat inactivated FBS. U2-OS and H1299 cells were transfected using the Fugene protocol (Boehringer Mannheim), according to the manufacturer's instructions. 174-2 cells were transfected using PEI (Sigma). AP29 cells were infected with pBabe puro recombinant retroviruses encoding either wild-type or S395A Mdm2.
Human wild-type Mdm2 cDNA and p53 cDNA, cloned in pCMV-neo-Bam, were kindly provided by Dr. B. Vogelstein. Derivatives of this expression plasmid, carrying substitutions at amino acid position 395, were prepared with the aid of the QuikChange Site-Directed Mutagenesis kit (Stratagene). The GFP–p53 expression plasmid (Stommel et al. 1999) was kindly provided by Drs. Jane Stommel and Geoff Wahl. The expression plasmid for FLAG-tagged wild-type ATM was as described (Canman et al. 1998).
Isolation of epitope-presenting phage from a hexapeptide epitope library
The anti-Mdm-2 monoclonal antibody 2A10 (mAb 2A10) was purified on a column containing 1 ml immobilized protein A and protein G (Schleicher & Schuel). 2A10 hybridoma supernatant (100 ml) was applied to the column, and after washing with 50 ml phosphate buffered saline (PBS), the IgG fraction was eluted with 10 ml of 0.1 M glycine at pH 2.8. Aliquots (0.7 ml) were collected and instantly neutralized with 70 μl of 1 M Tris-HCl at pH 8.0. IgG concentration was calculated from absorbance at 280 nm (1.4 OD = 1 mg/ml pure IgG). Biotinylation of the purified mAb 2A10 was carried out by incubating 100 mg of the antibody (in 0.1 M NaHCO3 at pH 8.6) with 5 μg biotin amidocaproate N-hydroxysuccinimide ester (Sigma B2643) from a stock solution of 1 mg/ml in dimethylformamide. The mixture was incubated for 2 h at room temperature and then dialyzed overnight against PBS at 40°C, using a cellulose membrane with a molecular weight cutoff of 12,000–14,000.
The hexapeptide epitope library was kindly provided by George P. Smith (University of Missouri, Columbia, MO) and was constructed by using the phage fd-derived vector fUSE5 as described (Scott and Smith 1990). The library consists of ∼70% of the total theoretical number of phage particles (6.4 × 107) representing the entire combinatorial diversity of hexapeptides. Each phage clone contains a hexapeptide fused to the amino terminus of the minor coat protein P3 (ADGAX6GAAGA … ).
Phage clones binding to mAb 2A10 were selected from the hexapeptide phage epitope library according to Scott and Smith 1990. A sample containing 1010 infectious phage particles was subjected to the first round of selection and amplification (biopanning) using biotinylated mAb 2A10 (4 μg/ml) as a selector. For the subsequent second and third rounds of biopanning, a sample of 1010 phage particles, obtained from the former round, was incubated with a reduced concentration of mAb 2A10 (0.4 μg/ml). In each round the reaction mixture was incubated for 15 min on a streptavidin-coated 60 mm polystyrene Petri dish. Unbound phage particles were removed by extensive washing (10 washes, 10 min each) with PBS containing 0.5% Tween-20. Bound phage particles were eluted by incubation with glycine-HCl at pH 2.2 for 5 min, followed by neutralization with Tris buffer. Amplification of the eluted phage particles was performed by infecting E. coli K91 cells. After the second and third rounds of biopanning, individual amplified phage clones were tested by ELISA (Balass et al. 1993) for specific binding to mAb 2A10. Positive clones were subjected to DNA sequencing.
Microinjection, antibodies, and immunostaining
Primary MEFs were grown for four passages and plated on 18 mm glass cover slips 24 h before microinjection. Microinjection into cell nuclei was done with the aid of the Eppendorf system (Microinjector 5242, Micromanipulator 5171). Cells were injected with either 2A10 or 4B2 ascites fluid. For transient transfection, U2-OS cells were plated on 18 mm glass cover slips at ∼50% confluence, 24 h before transfection. Microinjected and transfected cells were fixed for 30 min in methanol at −20°C. Following staining as indicated in the corresponding figure legends, cover slips were mounted on microscope slides and fluorescence was visualized with the aid of a Zeiss Axioskop fluorescent microscope, using a 40× objective.
The Mdm2-specific monoclonal antibodies 2A10, 2A9, and 4B2 (Chen et al. 1993) were a generous gift of Dr. Arnold Levine. The p53-specific monoclonal antibodies DO-1 and PAb1801 were generous gifts of Drs. David Lane and Lionel Crawford, respectively.
Production of GST-fusion proteins
GST fusion peptide expression vectors were constructed as described earlier (Kim et al. 1999). Briefly, complementary oligonucleotides encoding Mdm2 SQ sites (aa 11–24, aa 380–393, aa 390–402, and aa 401–414) were cloned into the BamHI–SmaI sites of pGEX-2T (Amersham Pharmacia Biotech). The constructs were confirmed by DNA sequencing. To construct GST-conjugated full-length Mdm2 protein, Mdm2 was amplified by PCR with Pfu polymerase using the following primers: 5′ sense, 5′-AGGAGCGGATCCATGTGCAATACCAACATG-3′, 3′ antisense, 5′-CTAGGGGAAATAAGTTAGCACAAT-3′. The amplified PCR product was cloned into the BamHI–SmaI sites of pET41 (Novagen). The GST-conjugated Mdm2 S395A mutant was generated using the QuikChange Site-Directed Mutagenesis kit (Stratagene). The GST fusion proteins were expressed in BL21(DE3). After isopropyl-β-d-thiogalactoside induction for 3 h (0.4 mM final concentration), GST-conjugated proteins were purified by affinity chromatography using glutathione-Sepharose beads (Sigma) as described previously (Kim et al. 1999).
In vitro kinase assays
In vitro kinase assays for ATM were performed as described previously (Canman et al. 1998). Briefly, 293T cells were transfected with 10 μg of FLAG–ATM. Cell lysates were prepared by resuspension in modified TGN buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 1% Tween 20, 0.3% Nonidet P-40, 1 mM sodium fluoride, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor mixture from Roche Molecular Biochemicals). Cell lysates were incubated with anti-FLAG M2 antibody (Sigma) and protein A/G-agarose for 2 h at 4°C. The precipitated beads were washed with TGN buffer followed by TGN buffer plus 0.5 M LiCl, and two washes with kinase buffer (20 mM HEPES at pH 7.5, 50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, and 10 mM MnCl2). Finally, the pellet was resuspended in 50 μl of kinase buffer containing 10 μCi of [γ-32P]ATP and 1 μg of GST-fusion substrate. The kinase reaction was conducted at 30°C for 20 min and stopped by the addition of SDS-PAGE protein sample buffer. Proteins were separated by SDS-PAGE, transferred to nitrocellulose and visualized by Fast Green staining. Immunoprecipitated FLAG–ATM was confirmed by Western blotting with α-FLAG M2 monoclonal antibody. 32P incorporation into GST–MDM2 substrates was visualized and quantified with the aid of a PhosphorImager (Molecular Dynamics).
Protein analysis
Analysis of p53 degradation in transiently transfected H1299 cells was done as described previously (Haupt et al. 1997). Proteins were visualized with the aid of the ECL kit (Amersham), as directed by the manufacturer.
Acknowledgments
We thank Sylvie Wilder for expert technical assistance, Xinjang Wang and Jan Taplick for spending many hours by the fluorescent microscope, Jane Stommel and Geoff Wahl for the gift of the GFP–p53 expression plasmid, Gigi Lozano for the 174-2 cells, and Arnold Levine for many Mdm2-specific MoAbs. This work was supported in part by United States Public Health Service Grant RO1 CA40099, the Center for Excellence Program of the Israel Science Foundation, the German-Israel Project Cooperation (DIP), the Cooperation Program in Cancer Research of the DKFZ and Israel's Ministry of Science (MOS), the Bosch Foundation (Germany), and Yad Abraham Center for Cancer Diagnosis and Therapy (to M.O), and NIH grants CA71387 and CA21765 and the American Lebanese Syrian Associated Charities (ALSAC) of the St. Jude Children's Research Hospital (to M.B.K).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL moshe.oren@weizmann.ac.il; FAX 972-8-9465223.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.886901.
References
- Ashcroft M, Kubbutat MH, Vousden KH. Regulation of p53 function and stability by phosphorylation. Mol Cell Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft M, Taya Y, Vousden KH. Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol. 2000;20:3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balass M, Heldman Y, Cabilly S, Givol D, Katchalski-Katzir E, Fuchs S. Identification of a hexapeptide that mimics a conformation-dependent binding site of acetylcholine receptor by use of a phage-epitope library. Proc Natl Acad Sci. 1993;90:10638–10642. doi: 10.1073/pnas.90.22.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C, Tobiasch E, Litfen M, Rahmsdorf HJ, Herrlich P. DNA damage induced p53 stabilization: No indication for an involvement of p53 phosphorylation. Oncogene. 1999;18:1723–1732. doi: 10.1038/sj.onc.1202480. [DOI] [PubMed] [Google Scholar]
- Bottger A, Bottger V, Sparks A, Liu WL, Howard SF, Lane DP. Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr Biol. 1997;7:860–869. doi: 10.1016/s0960-9822(06)00374-5. [DOI] [PubMed] [Google Scholar]
- Boyd SD, Tsai KY, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- Buschmann T, Fuchs SY, Lee CG, Pan ZQ, Ronai Z. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumaz N, Meek DW. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 1999;18:7002–7010. doi: 10.1093/emboj/18.24.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- Freedman DA, Levine AJ. Regulation of the p53 protein by the MDM2 oncoprotein–-thirty-eighth G.H.A. Clowes memorial award lecture. Cancer Res. 1999;59:1–7. [PubMed] [Google Scholar]
- Fuchs SY, Fried VA, Ronai Z. Stress-activated kinases regulate protein stability. Oncogene. 1998;17:1483–1490. doi: 10.1038/sj.onc.1202184. [DOI] [PubMed] [Google Scholar]
- Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- Giaccia AJ, Kastan MB. The complexity of p53 modulation: Emerging patterns from divergent signals. Genes & Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- Gotz C, Kartarius S, Scholtes P, Nastainczyk W, Montenarh M. Identification of a CK2 phosphorylation site in mdm2. Eur J Biochem. 1999;266:493–501. doi: 10.1046/j.1432-1327.1999.00882.x. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Henning W, Rohaly G, Kolzau T, Knippschild U, Maacke H, Deppert W. MDM2 is a target of simian virus 40 in cellular transformation and during lytic infection. J Virol. 1997;71:7609–7618. doi: 10.1128/jvi.71.10.7609-7618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Jayaraman L, Prives C. Covalent and noncovalent modifiers of the p53 protein. Cell Mol Life Sci. 1999;55:76–87. doi: 10.1007/s000180050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez GS, Khan SH, Stommel JM, Wahl GM. p53 regulation by post-translational modification and nuclear retention in response to diverse stresses. Oncogene. 1999;18:7656–7665. doi: 10.1038/sj.onc.1203013. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Oren M. Mdm2: The ups and downs. Mol Med. 1999;5:71–83. [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Hamm R, Yan W, Taya Y, Lozano G. Cooperative phosphorylation at multiple sites is required to activate p53 in response to UV radiation. Oncogene. 2000;19:358–364. doi: 10.1038/sj.onc.1203300. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Lim D. The many substrates and functions of ATM. Nat Reviews. 2000;1:179–186. doi: 10.1038/35043058. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, El-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Khanna KK, Keating KE, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, Lees-Miller SP, et al. ATM associates with and phosphorylates p53: Mapping the region of interaction. Nat Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci. 1999;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Lain S, Midgley C, Sparks A, Lane EB, Lane DP. An inhibitor of nuclear export activates the p53 response and induces the localization of HDM2 and p53 to U1A-positive nuclear bodies associated with the PODs. Exp Cell Res. 1999;248:457–472. doi: 10.1006/excr.1999.4433. [DOI] [PubMed] [Google Scholar]
- Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN. Phosphorylation of p53 serine 15 increases interaction with CBP. J Biol Chem. 1998;273:33048–33053. doi: 10.1074/jbc.273.49.33048. [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH. Identification of a cryptic nucleolar-localization signal in MDM2. Nat Cell Biol. 2000;2:179–181. doi: 10.1038/35004057. [DOI] [PubMed] [Google Scholar]
- Maki CG, Huibregtse JM, Howley PM. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- Maya R, Oren M. Unmasking of phosphorylation-sensitive epitopes on p53 and Mdm2 by a simple Western-phosphatase procedure. Oncogene. 2000;19:3213–3215. doi: 10.1038/sj.onc.1203658. [DOI] [PubMed] [Google Scholar]
- Mayo LD, Turchi JJ, Berberich SJ. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- Meek DW. Mechanisms of switching on p53: A role for covalent modification? Oncogene. 1999;18:7666–7675. doi: 10.1038/sj.onc.1202951. [DOI] [PubMed] [Google Scholar]
- Middeler G, Zerf K, Jenovai S, Thulig A, Tschodrichrotter M, Kubitscheck U, Peters R. The tumor suppressor p53 is subject to both nuclear import and export, and both are fast, energy-dependent and lectin-inhibited. Oncogene. 1997;14:1407–1417. doi: 10.1038/sj.onc.1200949. [DOI] [PubMed] [Google Scholar]
- Midgley CA, Desterro JM, Saville MK, Howard S, Sparks A, Hay RT, Lane DP. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312–2323. doi: 10.1038/sj.onc.1203593. [DOI] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- Momand J, Wu HH, Dasgupta G. MDM2 master regulator of the p53 tumor suppressor protein. Gene. 2000;242:15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- O'Neill T, Dwyer AJ, Ziv Y, Chan DW, Lees-Miller SP, Abraham RH, Lai JH, Hill D, Shiloh Y, Cantley LC, et al. Utilization of oriented peptide libraries to identify substrate motifs selected by ATM. J Biol Chem. 2000;275:22719–22727. doi: 10.1074/jbc.M001002200. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- Oren M. Regulation of the p53 tumor suppressor protein. J Biol Chem. 1999;274:36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin J, Potes L, Chen K, Orlow I, Lee HW, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Roth J, Dobbelstein M, Freedman DA, Levine AJ. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Weber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and ATR: Networking cellular responses to DNA damage. Curr Opin Genet Dev. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. DNA damage induces phosphorylation of the amino terminus of p53. Genes & Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: Regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Levine AJ. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci. 1999a;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci. 1999b;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH. p53. Death star. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Weber JD, Kuo ML, Bothner B, DiGiammarino EL, Kriwacki RW, Roussel MF, Sherr CJ. Cooperative signals governing ARF-mdm2 interaction and nucleolar localization of the complex. Mol Cell Biol. 2000;20:2517–2528. doi: 10.1128/mcb.20.7.2517-2528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm2 autoregulatory feedback loop. Genes & Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]