Abstract
The Snf1/AMP-activated protein kinase family has broad roles in transcriptional, metabolic, and developmental regulation in response to stress. In Saccharomyces cerevisiae, Snf1 is required for the response to glucose limitation. Snf1 kinase complexes contain the α (catalytic) subunit Snf1, one of the three related β subunits Gal83, Sip1, or Sip2, and the γ subunit Snf4. We present evidence that the β subunits regulate the subcellular localization of the Snf1 kinase. Green fluorescent protein fusions to Gal83, Sip1, and Sip2 show different patterns of localization to the nucleus, vacuole, and/or cytoplasm. We show that Gal83 directs Snf1 to the nucleus in a glucose-regulated manner. We further identify a novel signaling pathway that controls this nuclear localization in response to glucose phosphorylation. This pathway is distinct from the glucose signaling pathway that inhibits Snf1 kinase activity and responds not only to glucose but also to galactose and sucrose. Such independent regulation of the localization and the activity of the Snf1 kinase, combined with the distinct localization of kinases containing different β subunits, affords versatility in regulating physiological responses.
Keywords: Snf1/AMPK kinases, yeast, nuclear localization, glucose signaling
The Snf1/AMP-activated protein kinase (AMPK) family is essential for metabolic regulation in response to stress in fungi, plants, and animals (for review, see Hardie et al. 1998; Kemp et al. 1998). In mammals, AMPK controls metabolic enzymes in response to stresses that affect the cellular energy supply, including nutrient limitation, hypoxia, heat shock, and exercise. Increases in the cellular AMP : ATP ratio activate AMPK, which then stimulates ATP-producing pathways and inhibits ATP-consuming pathways. The recent identification of an AMPK mutation in pigs that affects the glycogen content of muscle, and hence meat quality, provides genetic evidence for an important role of AMPK (Milan et al. 2000). AMPK has been implicated further in exercise-stimulated, insulin-independent glucose transport in muscle, which is relevant to the beneficial effects of exercise for patients with type II diabetes (Hayashi et al. 1998). AMPK also inhibits glucose-activated transcription in liver and pancreatic β cells (Silva Xavier et al. 2000; Woods et al. 2000).
Like its mammalian counterparts, the Snf1 kinase of the yeast Saccharomyces cerevisiae has an essential role in metabolic regulation in response to stress. Snf1 is required for the adaptation of cells to glucose limitation (Gancedo 1998; Carlson 1999) and also has been implicated in the response to salt stress, heat shock, and starvation for other nutrients (Thompson-Jaeger et al. 1991; Alepuz et al. 1997). Snf1 affects not only metabolic adaptation to different environmental conditions but also developmental processes. Mutations in the kinase complex cause defects in meiosis and sporulation (Honigberg and Lee 1998), filamentation and invasive growth (Cullen and Sprague 2000), survival in stationary phase (Ashrafi et al. 1998), and life span and aging (Ashrafi et al. 2000).
The role of Snf1 in the metabolic adaptation to glucose depletion and to growth on alternate carbon sources is understood in some detail. The Snf1 kinase is activated when glucose is low or absent by an unidentified signal (Woods et al. 1994; Wilson et al. 1996; Jiang and Carlson 1996); the AMP : ATP ratio may be important during carbon source transitions (Wilson et al. 1996) but does not appear to be the primary signal during steady-state growth (Gancedo 1998). Snf1 regulates the transcription of metabolic genes by controlling transcriptional repressors and activators (Carlson 1999) and RNA polymerase II holoenzyme function (Kuchin et al. 2000). Snf1 also regulates the activity of enzymes involved in fatty acid and glycogen metabolism (Hardy et al. 1994; Mitchelhill et al. 1994; Woods et al. 1994), which in some cases may have effects that extend beyond metabolic control; for example, acetyl-CoA carboxylase activity affects nuclear envelope morphology and mRNA transport (Schneiter and Kohlwein 1997).
The diverse roles of the Snf1/AMPK kinases raise the question: how is such versatility achieved? Genetic studies in yeast suggest that isoforms of the β subunit are important for functional specificity. Snf1/AMPK kinase complexes comprise three subunits, and in S. cerevisiae, each complex contains the α (catalytic) subunit Snf1, one of the three β subunits Sip1, Sip2, or Gal83, and the γ subunit Snf4. The α, β, and γ subunits all have roles in regulating the catalytic activity. In glucose-grown cells, the regulatory domain of Snf1 autoinhibits the catalytic domain, and in response to glucose limitation, Snf4 binds to the Snf1 regulatory domain and stabilizes the kinase in an open, active conformation (see Fig. 8; Jiang and Carlson 1996). The β subunit interacts with both Snf1 and Snf4 to maintain their association in a complex (Yang et al. 1992, 1994; Jiang and Carlson 1997), and a sip1Δ sip2Δ gal83Δ triple null mutant is defective for Snf1 function; this point was clarified recently by findings that the original gal83Δ allele is partially functional (Erickson and Johnston 1993; Schmidt and McCartney 2000). Genetic evidence indicates that the β subunit also confers functional specificity to the kinase (Yang et al. 1994; Schmidt and McCartney 2000). In particular, Gal83 has been shown to mediate the physical interaction of Snf1 with Sip4, a transcriptional activator of gluconeogenic genes (Vincent and Carlson 1999), and Sip2 has been implicated in control of life span and aging (Ashrafi et al. 2000).
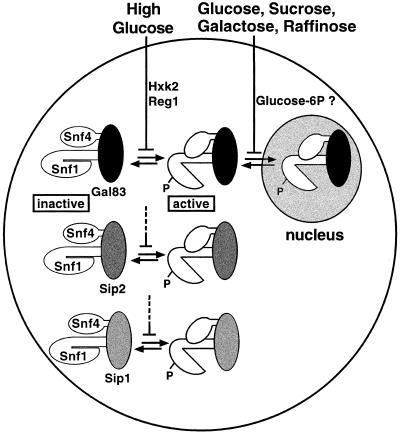
Figure 8.
Model for regulation of Snf1 kinase activity and localization by two distinct glucose signaling pathways. Snf1 complexes contain one of the three β subunits: Gal83 (black), Sip2 (dark gray), or Sip1 (light gray). Inactive Snf1 kinase complexes are depicted in a closed conformation in which the Snf1 catalytic domain is autoinhibited by the Snf1 regulatory domain. Active Snf1 complexes are shown in an open conformation and are phosphorylated (P). High glucose inhibits (indicated by a bar) the activation of the Snf1 kinase, and Hxk2 and Reg1 are required for this inhibition. Glucose and other fermentable carbon sources inhibit the nuclear localization of Snf1 complexes containing Gal83, and evidence cited in the text suggests that glucose-6-phosphate (Glucose-6P) is necessary and sufficient for this inhibition. This pathway also may regulate the vacuolar localization of Sip1 (not shown; see text).
In this study, we present evidence that the β subunits regulate the subcellular localization of the kinase. Sip1, Sip2, and Gal83 show distinct localization patterns, and the glucose-regulated nuclear localization of Snf1 depends on Gal83. These findings imply that the β subunits regulate the activity of Snf1 toward differently localized substrates. We further identify a novel signaling pathway that controls the nuclear localization of Gal83 and Snf1. This pathway is distinct from the glucose signaling pathway that controls Snf1 kinase activity and responds not only to glucose but also to other fermentable carbon sources such as galactose. We suggest that such independent regulation of the activity and localization of Snf1 provides the versatility needed for the Snf1 kinase to mediate diverse responses to stress.
Results
Distinct subcellular localization of the β subunits Sip1, Sip2, and Gal83
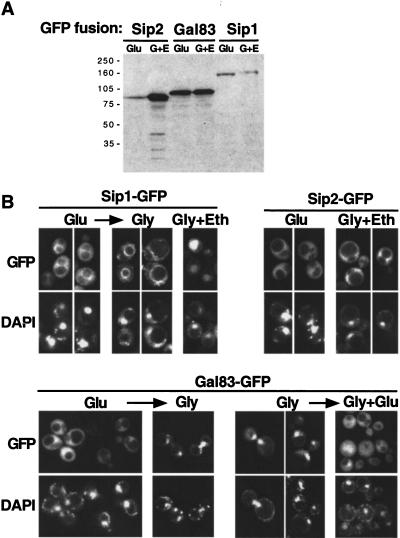
To determine the localization of Sip1, Sip2, and Gal83, we fused green fluorescent protein (GFP) to the C terminus of each protein, which was expressed from its own promoter on a centromere-containing plasmid. Immunoblot analysis confirmed the presence of full-length fusion proteins and showed that Gal83–GFP is the most abundant of the three β subunits in glucose-grown cells (Fig. 1A). In addition, expression of Sip2–GFP was found to be glucose regulated. All three fusion proteins are functional in several assays (see Materials and Methods).
Figure 1.
Subcellular localization of Sip1, Sip2, and Gal83. Strains MCY2649, MCY2700 (sip2Δ), and MCY4458 (gal83Δ) were transformed with centromeric plasmids expressing Sip1–GFP, Sip2–GFP, or Gal83–GFP, respectively; plasmids were pOV90, pRT9, and pRT12. (A) Cells were grown in synthetic medium with 2% glucose (Glu) or 2% glycerol plus 3% ethanol (G + E). Protein extracts were prepared, separated by SDS-PAGE, and immunoblotted with anti-GFP antibody. (B) The subcellular localization of each GFP fusion protein was examined by fluorescence microscopy in cells grown on synthetic medium with 2% glucose (Glu), 2% glycerol plus 3% ethanol (Gly + Eth), or 5% glycerol (Gly). The arrows indicate that cells were shifted from the growth medium to a different carbon source and incubated for 20 min before observation. Cells grown in glucose were shifted to 2% or 5% glycerol, and cells grown in glycerol were shifted to 5% glycerol plus 2% glucose. Nuclear export of Gal83 also was observed when cells were shifted from 5% glycerol to 2% glucose (data not shown). A low level of autofluorescence was observed with untransformed strains (not shown). DNA was stained with DAPI. (GFP) green fluorescent protein.
We examined the localization of the fusion proteins in both glucose-repressed and -derepressed cells (Fig. 1B). In glucose-grown cells, all three proteins were in the cytoplasm and were largely excluded from the nucleus. When cells were grown on glycerol and/or ethanol, however, each of the three fusion proteins showed different patterns. For Sip1–GFP, fluorescence was detected mainly in the vacuole; moreover, when glucose-grown cells were shifted to glycerol, fluorescence intensified at the periphery of the vacuole within 20 min. Sip2–GFP remained cytoplasmic and excluded from the nucleus. Gal83–GFP was located predominantly in the nucleus of glycerol-grown cells, with only low-level fluorescence detectable in the cytoplasm. When glucose-grown cells were shifted to glycerol (or shifted to water; data not shown), Gal83–GFP was imported into the nucleus within 20 min (within 5 min in other experiments; data not shown); conversely, when glucose was added to glycerol-grown cells, Gal83–GFP was exported rapidly from the nucleus.
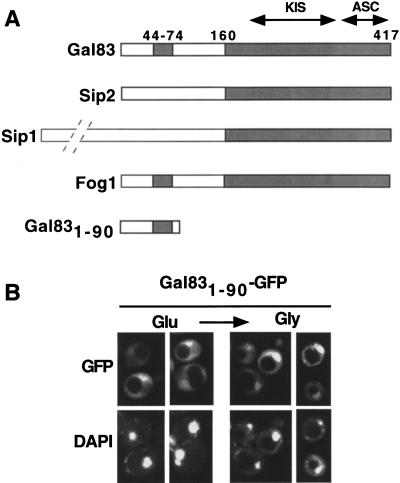
Sip1, Sip2, and Gal83 have similar C termini (Fig. 2A), suggesting that the sequences responsible for differential localization lie in the divergent N-terminal regions. Sip2 is myristoylated (Ashrafi et al. 1998), which most likely affects its localization. Because residues 44–74 of Gal83 are conserved (67% identity) in the Kluyveromyces lactis homolog Fog1 (Goffrini 1996), we expressed the N-terminal 90 residues of Gal83 fused to GFP. Gal831–90–GFP was partially excluded from the nucleus in glucose-grown cells and to some extent was enriched in the nucleus after a shift to glycerol (Fig. 2B). Thus, the N terminus of Gal83 confers regulated localization, but not as effectively as the full-length protein.
Figure 2.
N terminus of Gal83 confers regulated localization. (A) Sip1, Sip2, Gal83, and Fog1 are represented schematically. Shading indicates conserved sequences, which include two domains that interact with Snf1 and Snf4, designated KIS and ASC, respectively (Yang et al. 1994; Jiang and Carlson 1997). Numbers indicate amino acid residues in Gal83. Sip1 is substantially larger than the other subunits, with 863 residues, as indicated by the broken lines. (B) Fluorescence microscopy was performed with strain MCY4458 (gal83Δ) expressing Gal831–90–GFP from the GAL83 promoter on the centromeric plasmid pOV92. Cells were grown to mid-log phase on synthetic medium with 2% glucose (Glu) and shifted for 20 min to 5% glycerol (Gly). Immunoblot analysis confirmed that Gal831–90–GFP is intact. (GFP) green fluorescent protein.
The distinct localizations of Sip1, Sip2, and Gal83 in glucose-depleted cells imply that Snf1 kinase complexes containing these β subunits also show different localizations and, presumably, serve different functions. The nuclear localization of Gal83 and the increased expression of Sip2 during growth in glycerol and/or ethanol are consistent with genetic evidence that Sip2 and Gal83 function under these conditions; the sip2Δ gal83Δ double mutant shows a growth defect (Schmidt and McCartney 2000), and the gal83Δ mutant grows slowly after a shift (Vincent and Carlson 1999). Gal83 is the only subunit that translocates to the nucleus, suggesting a specific role in mediating the effects of Snf1 in the nucleus. We therefore have focused on Gal83.
Snf1 shows regulated, Gal83-dependent nuclear localization
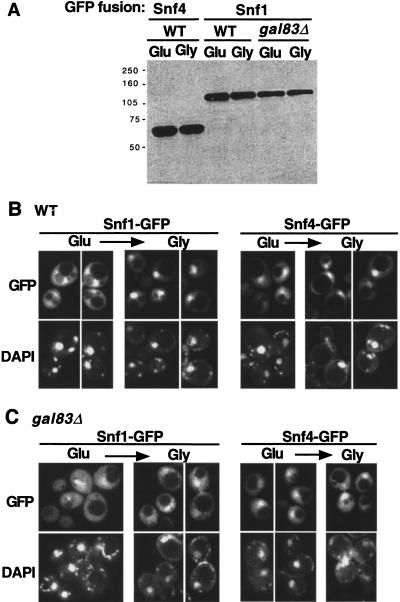
It is not known whether the Snf1 kinase complex is a relatively stable entity or rather undergoes a dynamic process of disassembly and reassembly. The observed translocation of Gal83 into and out of the nucleus may represent the translocation of an entire kinase complex, or Gal83 may move as a separate subunit and reassemble with Snf1 and Snf4 to form a new kinase complex. To assess the localization of Snf1 and Snf4, we fused GFP to the C terminus of each protein, which was expressed from its own promoter on a centromeric plasmid. Both fusion proteins are stable during growth in glucose and after a shift to glycerol (Fig. 3A), and both restore wild-type function in the cognate mutant strain.
Figure 3.
Glucose-regulated localization of Snf1–GFP and Snf4–GFP. Strains MCY4455 (wild type, WT) and MCY4458 (gal83Δ) were transformed with centromeric plasmids pOV84 and pOV76, expressing Snf1–GFP and Snf4–GFP, respectively. Cells were grown to mid-log phase on synthetic medium with 2% glucose (Glu) and shifted to 2% glycerol (Gly) for 20 min. (A) Protein extracts were prepared from the indicated cultures, separated by SDS-PAGE, and immunoblotted with anti-GFP antibody. (B,C) Wild-type (B) and gal83Δ mutant (C) cells expressing the indicated proteins were examined by fluorescence microscopy. (GFP) green fluorescent protein.
In glucose-grown wild-type cells, Snf1–GFP was in the cytoplasm and was largely excluded from the nucleus (Fig. 3B). When cells were shifted to glycerol, most of the Snf1–GFP was translocated rapidly to the nucleus. Some fusion protein remained in the cytoplasm, presumably because of its association with Sip1 or Sip2, which are present at much lower levels than Gal83 during growth in glucose (Fig. 1A). We note that these results differ from those of a previous study, in which this differential localization probably was lost during fixation with formaldehyde (Celenza and Carlson 1986).
To determine whether this regulated localization requires Gal83, we examined a gal83Δ strain (Fig. 3C). No nuclear exclusion was detected in glucose-grown cells, suggesting that Gal83 serves to retain Snf1 in the cytoplasm under these conditions. When the mutant strain was shifted to glycerol for 20 min, the extent of nuclear enrichment of Snf1–GFP was strongly reduced relative to the wild type.
We also examined cells during steady state growth in glycerol plus ethanol. In the wild type, Snf1–GFP was enriched in the nucleus, but more fluorescence was detected in the cytoplasm than after a shift; in a gal83Δ mutant, Snf1–GFP was largely excluded from the nucleus (data not shown). These differences between steady state growth and a shift most likely reflect the increased expression of Sip2 during growth in glycerol plus ethanol (Fig. 1A). Together, these findings indicate that the nuclear localization of Snf1 is regulated and strongly dependent on Gal83.
In contrast, Snf4–GFP showed partial nuclear enrichment in glucose-grown wild-type cells, with a slight increase after a shift to glycerol. In the gal83Δ strain, Snf4–GFP was still partially enriched in the nucleus in glucose-grown cells; we are uncertain whether enrichment increased after the shift. These results indicate that Gal83 does not significantly affect the balance of nucleocytoplasmic localization for Snf4 and suggest that in glucose-grown cells the nucleus contains Snf4 proteins that are not associated with other subunits of the kinase complex.
Snf1 and Gal83 function together in the nucleus
The translocation of Snf1 and Gal83 to the nucleus in response to glucose depletion suggests that Snf1 kinase complexes containing Gal83 have specific nuclear functions under these conditions. Two lines of evidence support this idea.
First, we examined the localization of Sip4, a transcriptional activator of gluconeogenic genes (Lesage et al. 1996; Vincent and Carlson 1998). Gal83 mediates the physical interaction of Snf1 with Sip4, and Gal83 is required for the Snf1-dependent phosphorylation of Sip4 and up-regulation of its activator function in response to glucose limitation (Vincent and Carlson 1999). We expressed GFP–Sip4 from the SIP4 promoter on a 2-μm plasmid and found that GFP–Sip4 is a nuclear protein when cells are grown in glucose, shifted to glycerol, or grown in glycerol plus ethanol (data not shown). These findings imply that the Gal83-dependent interaction of Snf1 with Sip4 occurs in the nucleus.
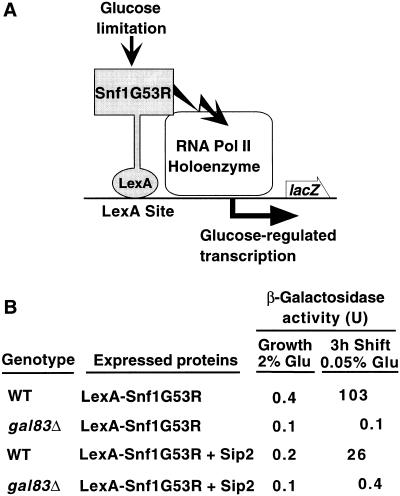
Second, previous studies provided evidence for a regulatory shortcut between Snf1 and the RNA polymerase II holoenzyme. One line of evidence was that LexA–Snf1G53R, a catalytically hyperactive Snf1 kinase, stimulates transcription of a lacZ reporter with LexA sites in glucose-limited cells, dependent on the kinase activity (Kuchin et al. 2000; Fig. 4A; see Materials and Methods). We tested whether this stimulation depends on Gal83. In filter assays for β-galactosidase activity, LexA–Snf1G53R produced blue color in wild-type and sip2Δ cells but not in gal83Δ cells. In quantitative assays, β-galactosidase activity was high in wild-type cells after a 3-h shift to 0.05% glucose (which is rapidly consumed) but was undetectable in gal83Δ mutant cells (Fig. 4B). Overexpression of Sip2 decreased activation in the wild type, presumably because Sip2 and Gal83 compete for binding to Snf1, and did not compensate for the absence of Gal83 in the mutant. Thus, Gal83 is specifically required for the nuclear function of LexA–Snf1G53R in stimulating transcription. We note that this requirement is unlikely to reflect a simple dependence on Gal83 for nuclear import as overexpression of LexA proteins appears to partially bypass regulation of nuclear localization (O. Vincent, unpubl.).
Figure 4.
Gal83 is required for transcriptional activation of a reporter by LexA–Snf1G53R. (A) The hyperactive kinase LexA–Snf1G53R is bound to LexA sites 5′ to the promoter of a lacZ reporter. Previous studies showed that when cells are limited for glucose, transcription of the reporter is stimulated, dependent on Snf1 catalytic activity (Kuchin et al. 2000). (B) CTY10–5d (WT) and its isogenic derivative MCY4024 (gal83Δ) were transformed with pRJ216 expressing LexA–Snf1G53R (Kuchin et al. 2000) and either pOV65, which overexpresses Sip2 from the ADH1 promoter or its parent vector pSK134, which does not express any protein (Vincent and Carlson 1999). Transformants were grown selectively to mid-log phase in 2% glucose and shifted to 0.05% glucose for 3 hr. β-galactosidase activity was assayed in permeabilized cells and expressed in Miller units (Miller 1972). Values are the average β-galactosidase activity of at least three transformants. Standard errors were <8% for values >0.5. Immunoblot analysis confirmed the stable expression of LexA–Snf1G53R in a gal83Δ mutant.
These findings together suggest that the nuclear import of Snf1 and Gal83 in response to glucose depletion correlates with a requirement for Snf1 and Gal83 for specific nuclear functions.
Regulated localization of Gal83 is independent of Snf1, Hxk2, and Reg1
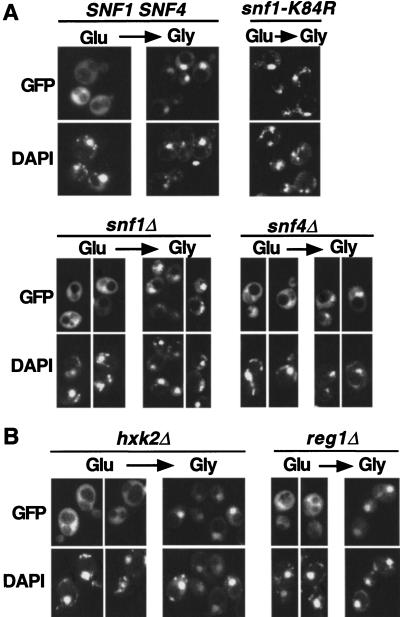
We next addressed the mechanisms that regulate the localization of Gal83. To test whether the Snf1 catalytic activity or the Snf1 or Snf4 proteins are required, we examined a snf1-K84R mutant, which encodes a kinase-dead protein, and snf1Δ and snf4Δ strains. When Gal83–GFP was expressed from a centromeric plasmid, the fluorescence was barely detectable, and immunoblot analysis showed that levels of Gal83 were a fewfold reduced relative to the wild type. We therefore transferred the gene fusion to a 2-μm plasmid and confirmed that the localization pattern in wild-type cells was as expected (Fig. 5A).
Figure 5.
Glucose-regulated localization of Gal83–GFP in mutant strains. (A) The subcellular localization of Gal83–GFP expressed from the 2-μm plasmid pRT14 was monitored in strains with the indicated genotypes (MCY4458, MCY2634, MCY2916, and MCY2693). Cells were grown to mid-log phase on synthetic medium with 2% glucose (Glu) and shifted for 20 min to 5% glycerol (Gly). (B) Gal83–GFP was expressed from pRT12 in strain MCY3541 (hxk2Δ) and from pRT14 in MCY4408 (reg1Δ). Cells were grown in 2% glucose and shifted for 20 min to 2% glycerol. (GFP) green fluorescent protein.
Examination of the snf1-K84R strain showed that the Snf1 catalytic activity is not required for nuclear import of Gal83–GFP on a shift to glycerol (Fig. 5A). In both snf1Δ and snf4Δ strains, Gal83–GFP was at least partially excluded from the nucleus in glucose-grown cells and accumulated in the nucleus after a shift to glycerol, indicating that the Snf1 and Snf4 proteins are not required for regulated localization of Gal83. However, after the shift, the fluorescence patterns of the mutants appeared more punctate than those of the wild type.
We then tested whether regulatory proteins that control Snf1 kinase activity also control the localization of Gal83 (Fig. 5B). Snf1 kinase activity is regulated by the hexokinase Hxk2 and by protein phosphatase 1 (PP1), which is targeted to Snf1 by the Reg1 subunit; hxk2Δ and reg1Δ mutations abolish glucose regulation of the conformation of the Snf1 kinase complex (Ludin et al. 1998; Sanz et al. 2000) and abolish glucose repression of many genes (for review, see Gancedo 1998). Gal83–GFP was still excluded from the nucleus in glucose-grown hxk2Δ cells and partially excluded in reg1Δ cells. In both mutants, Gal83–GFP was translocated to the nucleus after a shift to glycerol. Thus, Hxk2 and Reg1 regulate Snf1 kinase activity but not the nuclear localization of Gal83. These findings are consistent with evidence above that localization of Gal83 is independent of Snf1 activity and suggest the existence of a distinct regulatory mechanism.
Because components of the Snf1 pathway are not responsible for regulated localization of Gal83, we examined the roles of two other signaling cascades that control nuclear localization of transcription factors in response to nutrients, the TOR and cAMP-dependent protein kinase (PKA) pathways (Gorner et al. 1998; Beck and Hall 1999). Treatment with rapamycin, which inhibits the TOR signaling cascade and mimics nutrient starvation, did not induce nuclear import of Gal83–GFP in glucose-grown cells (data not shown). Regulated nuclear translocation of Gal83–GFP occurred in a strain (ASY62) lacking all three catalytic subunits of PKA (data not shown).
Nuclear exclusion of Gal83 in glucose-grown cells depends on glucose phosphorylation
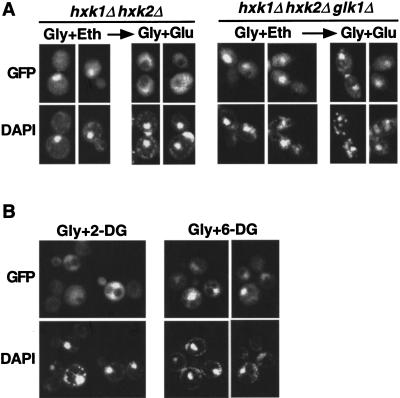
Although Hxk2 is required for glucose repression of many genes, phosphorylation of glucose by any of the three glucose kinases Hxk1, Hxk2, or Glk1 is sufficient to trigger some glucose repression mechanisms (De Winde et al. 1996; Sanz et al. 1996; Yin et al. 1996). To test whether glucose phosphorylation is required for the glucose-dependent nuclear exclusion of Gal83, we examined hxk1Δ hxk2Δ and hxk1Δ hxk2Δ glk1Δ mutants. Cells were grown on glycerol plus ethanol, and then glucose was added to the medium. Gal83–GFP was exported to the cytoplasm in the double mutant within 20 min but was not exported in the triple mutant lacking glucose kinases (Fig. 6A), even after 90 min (data not shown).
Figure 6.
Glucose phosphorylation is required for nuclear export of Gal83–GFP. (A) Strains MCY4043 (hxk1Δ hxk2Δ) and WAY.78-1 (hxk1Δ hxk2Δ glk1Δ) expressed Gal83–GFP from pRT12. Cells were grown to mid-log phase on synthetic medium with 2% glycerol plus 3% ethanol (Gly + Eth) and shifted to 2% glycerol + 2% glucose (Gly + Glu) for 20 min. (B) Strain MCY4458 (gal83Δ) expressing Gal83–GFP from pRT12 was grown on 5% glycerol and shifted to 5% glycerol + 0.02% 2-deoxyglucose (Gly + 2-DG) or 5% glycerol + 0.02% 6-deoxyglucose (Gly + 6-DG) for 20 min. Nuclear export of Gal83–GFP occurred normally in wild-type cells isogenic to WAY.78-1 (ENY-WA-1A; not shown). (GFP) green fluorescent protein.
We then tested the effect of the glucose analog 2-deoxyglucose, which is phosphorylated but not further metabolized, and 6-deoxyglucose, which cannot be phosphorylated. Addition of 0.02% 2-deoxyglucose to glycerol-grown cells resulted in rapid nuclear export of Gal83–GFP; in contrast, 0.02% 6-deoxyglucose had no effect (Fig. 6B). Together with the genetic evidence, these results suggest that phosphorylation of glucose is necessary and sufficient for the glucose-regulated localization of Gal83.
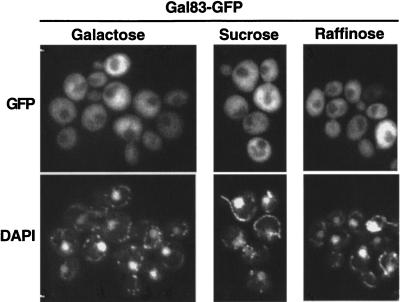
Gal83 is excluded from the nucleus in cells grown on fermentable carbon sources other than glucose
The Snf1 kinase is inhibited by glucose but is active and required for growth on alternate fermentable carbon sources such as sucrose and galactose. However, repression of some Snf1-dependent promoters is triggered not only by glucose but also by sucrose and galactose (Schöler and Schüller 1994; Gancedo 1998; Rodriguez and Gancedo 1999), suggesting the existence of an additional repression mechanism besides that regulating Snf1 kinase activity. We therefore tested the effect of these carbon sources on the localization of Gal83. We examined cells growing on sucrose, raffinose, or galactose, and Gal83–GFP in all cases was largely excluded from the nucleus (Fig. 7). These results suggest that nuclear localization of Gal83 is inhibited by any fermentable carbon source and confirm the evidence from hxk2 and reg1 mutants that Gal83 is nuclear excluded in some conditions when the Snf1 kinase is active. Thus, regulation of the kinase activity and localization of Gal83 are not coupled but rather are controlled by two distinct signaling pathways, one that responds to glucose and another that responds to glucose and also alternate fermentable sugars.
Figure 7.
Localization of Gal83–GFP in cells grown in different fermentable carbon sources. Fluorescence microscopy was performed with strain MCY4416 (gal83Δ) expressing Gal83–GFP from plasmid pRT12. Cells were grown to mid-log phase on synthetic medium with either 2% sucrose, 2% raffinose, or 2% galactose. (GFP) green fluorescent protein.
With respect to the functional roles of Snf1 and Gal83 during growth on sucrose, raffinose, or galactose, these data do not rule out the possibility that a small fraction of Gal83–GFP, and hence Snf1, is in the nucleus. We also note that Gal83 is not essential for growth on any of these carbon sources (Schmidt and McCartney 2000; O. Vincent, unpubl.).
Discussion
Previous studies showed that the β subunit plays a structural role in the Snf1 kinase complex and confers specificity to the kinase in its interactions with target proteins. Here, we present evidence that the β subunits also regulate localization of the kinase in response to the carbon source. In cells grown on nonfermentable carbon sources, the β subunits Sip1, Sip2, and Gal83 are each differently localized, and Gal83 specifically directs the Snf1 kinase to the nucleus. We further present evidence that localization of Snf1 complexes is regulated by a novel signaling pathway distinct from the glucose signaling pathway that regulates Snf1 kinase activity, thereby revealing another layer of complexity in the regulatory response of yeast cells to carbon source.
First, we show that GFP fusions to all three β subunits reside in the cytoplasm in glucose-grown cells. When cells are shifted to a nonfermentable carbon source, these subunits assume distinct subcellular localizations: Gal83 translocates to the nucleus, Sip1 is directed to the vacuole, and Sip2 remains cytoplasmic. The N terminus of Gal83, which is not conserved in the other two β subunits, has a role in localization. We then show that Snf1 is in the cytoplasm during growth on glucose and becomes enriched in the nucleus on a shift to a nonfermentable carbon source. This nuclear translocation is strongly dependent on Gal83, indicating that the β subunit directs Snf1 to the nucleus and suggesting that Snf1 and Gal83 are associated during the import process. Evidence that nuclear exclusion of Snf1–GFP is defective in glucose-grown gal83Δ mutant cells further suggests that Gal83 serves to anchor Snf1 in the cytoplasm. This proposed anchoring function may be shared by other β subunits as a sip1Δ sip2Δ mutant has a partial defect in nuclear exclusion of Snf1 during growth in glucose (O. Vincent and R. Townley, unpubl.). Finally, we observed that Gal83 does not significantly affect the nucleocytoplasmic distribution of Snf4, suggesting that in glucose-grown cells the nucleus contains Snf4 that is not associated with Snf1 or Gal83.
The differential localization of the three β subunits supports a role for these subunits in mediating the interaction of Snf1 with specific substrates located in different cellular compartments. Gal83 is the only subunit that translocates from the cytoplasm to the nucleus, suggesting that Gal83 mediates the effects of Snf1 in the nucleus. We present two lines of evidence supporting such a role. First, the transcriptional activator Sip4 is an exclusively nuclear protein, indicating that the Gal83- and Snf1-dependent phosphorylation and up-regulation of Sip4 (Vincent and Carlson 1999) reflect nuclear functions of the kinase. Second, stimulation of reporter transcription by a hyperactive Snf1 that is bound to the promoter requires Gal83. These findings suggest that Snf1 complexes containing Gal83 have specific nuclear functions. We expect that further study of Sip1 and Sip2 will reveal roles in Snf1 kinase function that are consistent with the localization of these β subunits. These findings also raise the possibility that the β subunits confer specificity primarily by providing proximity to specific subsets of differently localized substrates, rather than by mediating sequence-specific interactions with substrates, although the two mechanisms are not mutually exclusive.
We also identify a previously unrecognized signaling pathway that controls the nuclear localization of Gal83 and Snf1. Several lines of evidence indicate that this pathway is distinct from the glucose signaling pathway responsible for glucose inhibition of the Snf1 kinase activity (Fig. 8). First, glucose-regulated localization of Gal83 does not require Reg1 or the hexokinase Hxk2, whereas both regulate the conformation and activity of the Snf1 kinase. Second, phosphorylation of glucose is necessary and sufficient for its inhibitory effect on nuclear localization of Gal83; the effect of glucose requires glucose kinase activity, and 2-deoxyglucose, which is phosphorylated but not further metabolized, is sufficient for nuclear exclusion of Gal83. In contrast, glucose phosphorylation is not sufficient for inhibition of Snf1 kinase activity, as inhibition is lost in an hxk2Δ mutant despite the presence of two other glucose kinases. Third, the Snf1 kinase is active during growth in sucrose, raffinose, or galactose, but Gal83 is still excluded from the nucleus. Thus, the mechanism responsible for regulating localization responds not only to glucose but also to other fermentable carbon sources that do not inhibit Snf1 kinase activity. The fact that galactose has the same effect as glucose on the localization of Gal83 further excludes a specific function of the glucose kinases in this signaling pathway because metabolism of galactose is independent of glucose kinases. As expected, galactose causes nuclear exclusion of Gal83 in a hxk1Δ hxk2Δ glk1Δ triple mutant (O. Vincent and R. Townley, unpubl.). We note that metabolism of galactose also yields glucose-6-phosphate, suggesting this metabolite as a candidate for a signaling role.
The pathway that controls localization of Gal83 also may regulate the localization of Sip1. Sip1 is located in the cytoplasm in glucose-grown cells, remains cytoplasmic when cells are grown in sucrose (O. Vincent and R. Townley, unpubl.), and is directed to the vacuole when cells are grown in glycerol plus ethanol. This similar pattern of carbon source responsiveness is consistent with regulation by the same pathway.
The distinct regulation of Snf1 activity and localization may affect transcriptional control of particular genes differently, depending on whether the Snf1-regulated activators and repressors of the gene are restricted to the nucleus or shuttle between the nucleus and cytoplasm. For example, the gluconeogenic genes are still subject to glucose repression in an hxk2 mutant (Gancedo 1998), suggesting that nuclear exclusion of Snf1 is sufficient for repression even when the kinase is active. It is noteworthy that one of the Snf1-dependent activators of gluconeogenic genes, Sip4, is restricted to the nucleus (the activator Cat8 has not been localized but is a related zinc-cluster protein). Thus, nuclear localization of Snf1 during growth in nonfermentable carbon sources may facilitate activation of gluconeogenic genes; the reduced growth rate of a gal83Δ mutant after a shift to glycerol plus ethanol (Vincent and Carlson 1999) supports this view and is also consistent with the presence of some fraction of Snf1 in the nucleus. In contrast with the gluconeogenic genes, SUC2 is not glucose repressible in an hxk2 mutant, indicating that repression of SUC2 requires inhibition of Snf1 kinase activity. Correspondingly, the repressor, Mig1, shuttles between the nucleus and cytoplasm and could be phosphorylated by Snf1 in either compartment (DeVit et al. 1997; Ostling and Ronne 1998; Treitel et al. 1998). The predominantly cytoplasmic localization of Snf1 during growth in sucrose suggests that the function(s) of Snf1 that are required for SUC2 transcription primarily occur in the cytoplasm, although a small fraction of Snf1 may be nuclear and have important functions.
The Snf1 kinase has very broad roles in cellular responses to carbon source availability; Snf1 is required not only for metabolic adaptation to glucose limitation and growth on alternate carbon sources but also for various developmental processes, including invasive growth, meiosis, and control of life span (Gancedo 1998; Carlson 1999; Ashrafi et al. 2000; Cullen and Sprague 2000). The findings reported here can account, at least in part, for the versatility of the Snf1 kinase in regulating diverse responses to stress. We show that the Snf1 kinase is regulated by two distinct pathways that independently regulate the localization and activity of Snf1 complexes. These two pathways respond to different carbon source signals. In addition, the three β subunits show different localization patterns. The combinations of two signaling pathways and three β subunits provide mechanisms by which different inputs to the kinase result in phosphorylation of distinct sets of substrates, depending on their subcellular localization.
The mammalian AMPK also has broad roles in cellular regulation, and evidence suggests that localization of AMPK may be similarly important in regulating AMPK activity toward specific substrates. AMPK containing the β2 subunit appears to colocalize with its substrate creatine kinase to the M line of myofibrils (Ponticos et al. 1998), and the β1 subunit of AMPK is myristoylated, as is Sip2 (Mitchelhill et al. 1997; Ashrafi et al. 1998). In addition, AMPK containing the α2 subunit, but not α1, shows nuclear enrichment in some cell types (Salt et al. 1998). Our findings in yeast further raise the possibility that specific signaling pathways control localization of AMPK in mammalian cells.
Materials and methods
Strains and genetic methods
The S. cerevisiae strains used are listed in Table 1. sip1Δ::KanMX6 is a complete deletion of the SIP1 coding sequence created by homologous recombination with the KanMX6 marker by using the method of PCR synthesis of marker cassettes (Wach 1996). sip2Δ3::LEU2 was described previously (Yang et al. 1994), and gal83Δ::TRP1 (Vincent and Carlson 1999) is a null allele, distinct from the gal83Δ::URA3 allele (Erickson and Johnston 1993) shown to confer partial function (Schmidt and McCartney 2000). Inactivation of SIP1, SIP2, and GAL83 by the alleles used here was confirmed by the growth defect of sip2Δ gal83Δ mutants on glycerol plus ethanol and the growth defect of sip1Δ sip2Δ gal83Δ mutants on raffinose, sucrose, or galactose (Schmidt and McCartney 2000). Standard genetic methods were followed, and yeast cultures were grown in synthetic complete (SC) medium lacking appropriate amino acids to maintain selection for plasmids (Rose et al. 1990).
Table 1.
Strains used in this study
| Strain
|
Genotype
|
Source
|
|---|---|---|
| MCY2634 | MATa snf4Δ2 his3-Δ200 leu2-3,112 ura3-52 | This lab |
| MCY2649 | MATahis3-Δ200 leu2-3,112 ura3-52 | This lab |
| MCY2693 | MATα snfl-K84R his3-Δ200 leu2-3,112 ura3-52 | This lab |
| MCY2700 | MATα sip2Δ3::LEU2 his3-Δ200 leu2-3,112 ura3-52 | This lab |
| MCY2916 | MATasnf1Δ10 his3-Δ200 leu2-3,112 ura3-52 | This lab |
| MCY3541 | MATα hxk2Δ::URA3 his3-Δ200 lys2-801 | This lab |
| MCY4040 | MATα sip1Δ::KanMX6 sip2Δ3::LEU2 gal83Δ::TRP1 his3-Δ200 leu2-3,112 trp1Δ1 ura3-52 lys2-801 | This study |
| MCY4043 | MATα hxk1Δ::HIS3 hxk2Δ::TRP1 his3Δ200 leu2Δ1 trp1Δ63 ura3-52 | This study |
| MCY4408 | MATα reg1Δ::HIS3 his3Δ200 leu2Δ1 trp1Δ63 ura3-52 | This lab |
| MCY4416 | MATα gal83Δ::TRP1 his3Δ200 leu2Δ1 trp1Δ63 ura3-52 | This study |
| MCY4455 | MATα his3-Δ200 leu2-3,112 trp1Δ1 ura3-52 lys2-801 | This study |
| MCY4456 | MATα sip2Δ3::LEU2 gal83Δ::TRP1 his3-Δ200 leu2-3,112 trp1Δ1 ura3-52 lys2-801 | This study |
| MCY4458 | MATα gal83Δ::TRP1 his3-Δ200 leu2-3,112 trp1Δ1 ura3-52 lys2-801 | This study |
| WAY.78-1 | MATα hxkl::HIS3 hxk2::LEU2 glk1::LEU2 leu2-3,112 ura3-52 trpl-289 MAL2-8c MAL3 SUC3 | P. Sanz |
| ENY-WA-1A | MATα leu2-3,112 ura3-52 trp1-289 his3-Δ1 MAL2-8c MAL3 SUC3 | P. Sanz |
| ASY62 | MATatpk1Δ::ADE8 tpk2::HIS3 tpk3::TPR1 msn2Δ::HIS3 msn4Δ::LEU2 ura3-52 his3 leu23,112 trpl ade8 | S. Garrett |
| CTY10-5d | MATa ade2-101 his3-Δ200 leu2-Δ1 trp1-Δ901 gal4 gal80 URA3::lexAop-lacZ | R. Sternglanz |
| MCY4024 | CTY10-5d gal83Δ::TRP1 | This lab |
Plasmids
GFP fusion plasmids pRT9 (Sip2), pRT12 (Gal83), pOV76 (Snf4), pOV84 (Snf1), and pOV90 (Sip1) were constructed in a centromere-containing vector by using the following strategy. Two DNA fragments, one containing the ORF and at least 600 bp of 5′ sequence, and the other containing at least 380 bp 3′ to the ORF, were generated by PCR with Vent DNA polymerase (New England Biolabs), genomic clones as templates, and different sets of primers (sequences are available on request). The primers 5′ to the promoter and 3′ to the terminator create restriction sites that were used for cloning in the pRS313 polylinker (Sikorski and Hieter 1989). The other two primers create a NotI site immediately 5′ to the stop codon. PCR fragments and pRS313 were digested with the appropriate enzymes, and a triple ligation was performed, resulting in a plasmid containing the promoter, ORF, and terminator with a NotI site 5′ to the stop codon. Finally, a NotI fragment from pSFGP1 (Kim and Hirsch 1998) containing GFP sequence was inserted in the NotI site of the construct, creating an in-frame fusion between the ORF and GFP. In pRT9 and pRT12, the first 843 bp of SIP2 and 1114 bp of GAL83 were finally replaced by genomic DNA. pOV72 and pRT14 are derivatives of pRS316 and pRS426, respectively, containing the entire gene fusion on a PvuI fragment from pRT12.
Sip1–GFP, Sip2–GFP, and Gal83–GFP each restored the growth of a sip1Δ sip2Δ gal83Δ triple mutant on raffinose, sucrose, or galactose, and Sip2–GFP and Gal83–GFP each restored the growth of a sip2Δ gal83Δ strain on glycerol plus ethanol. Gal83–GFP restored activation by LexA–Snf1G53R in a gal83Δ mutant (see Fig. 4). Snf1-GFP and Snf4-GFP both function to restore growth of the corresponding mutants on sucrose, raffinose, galactose, and glycerol plus ethanol.
To construct pOV92 (Gal831–90–GFP), we amplified a DNA fragment containing 600 bp of 5′ sequence and 90 codons of the GAL83 ORF by PCR. The 3′ primer created an in-frame fusion with the first 50 bp of the GFP gene. Yeast cells were cotransformed with the PCR product and gapped pRT12 lacking the GAL83 sequence from position 243 to 1114. Gap repair of the plasmid resulted in an in-frame fusion between codon 90 of Gal83 and GFP.
Immunoblot analysis
Protein extracts were prepared as described previously for immunoprecipitation assays (Vincent and Carlson 1999). Extracts (30–50 μg) were analyzed by SDS-PAGE in 7.5% polyacrylamide and immunoblotting with monoclonal anti-GFP (Boehringer Mannheim). Antibody was detected by enhanced chemiluminescence with ECLPlus reagents (Amersham).
Fluorescence microscopy
Yeast strains expressing GFP fusion proteins were grown to mid-log phase in synthetic media containing the appropriate carbon source. Cells from 1-mL cultures were harvested by centrifugation and resuspended in 1 mL nonfluorescent medium (0.9 g/L KH2PO4, 0.23 g/L K2HPO4, 0.5 g/L MgSO4, 3.5 g/L [NH4]2SO4) containing the same carbon source, except that cells grown in glycerol plus ethanol were resuspended in glycerol to minimize background fluorescence. Nuclei were stained by addition of 0.8 μg/mL of DAPI to the cell suspension for 5 min. Cells were collected by centrifugation and resuspended in ∼20 μL of the residual medium, and 1.4 μL of the suspension was placed on a microscope slide. GFP localization in live cultures was monitored by direct fluorescence within 5 min. For shift experiments, cells were grown and harvested as above but resuspended in 1 mL of nonfluorescent medium containing a different carbon source, as indicated. Cells were incubated for 10 min, DAPI was added, and cells were incubated for an additional 5 min. Cells were then collected for examination as above. Cells were viewed using a Nikon Eclipse E800 fluorescent microscope. Images were taken with an Orca100 (Hamamatsu) camera using Open Lab (Improvision) software, and processed in Adobe Photoshop 2.5.1.
Assay for transcriptional activation by LexA–Snf1G53R
This assay has been described previously (Kuchin et al 2000). Briefly, strain CTY10–5d and its gal83Δ derivative contain a chromosomal lacZ reporter with LexA-binding sites 5′ to the promoter. Cells were transformed with plasmid pRJ216 expressing LexA–Snf1G53R, a functional LexA fusion to a catalytically hyperactive mutant of Snf1, which contains a Gly-to-Arg substitution that is found in natural plant homologs of Snf1. When cells were grown in high glucose, LexA–Snf1G53R did not stimulate reporter transcription. Cells that were limited for glucose produced high-level β-galactosidase activity, dependent on the binding of the fusion protein to the promoter and its catalytic activity. Expression of Snf1G53R without a LexA moiety did not activate transcription, and the kinase-dead derivative LexA–Snf1G53R,K84R also did not activate.
Acknowledgments
We are grateful to Fred Chang and Phong Tran for help with microscopy and to Saul Silverstein for providing a microscope. We thank Pascual Sanz and Steve Garrett for strains. This work was supported by NIH grant GM34095 to M.C. R.T. also received support from NIH training grant T32 GM08224.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
EMAIL mbc1@columbia.edu; FAX (212) 305-1741.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.879301.
References
- Alepuz PM, Cunningham KW, Estruch F. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol Microbiol. 1997;26:91–98. doi: 10.1046/j.1365-2958.1997.5531917.x. [DOI] [PubMed] [Google Scholar]
- Ashrafi K, Farazi TA, Gordon JI. A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J Biol Chem. 1998;273:25864–25874. doi: 10.1074/jbc.273.40.25864. [DOI] [PubMed] [Google Scholar]
- Ashrafi K, Lin SS, Manchester JK, Gordon JI. Sip2p and its partner Snf1p kinase affect aging in S. cerevisiae. Genes & Dev. 2000;14:1872–1885. [PMC free article] [PubMed] [Google Scholar]
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- Celenza JL, Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF., Jr Glucose depletion causes haploid invasive growth in yeast. Proc Natl Acad Sci. 2000;97:13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVit MJ, Waddle JA, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Winde JH, Crauwels M, Hohmann S, Thevelein JM, Winderickx J. Differential requirement of the yeast sugar kinases for sugar sensing in establishing the catabolite-repressed state. Eur J Biochem. 1996;241:633–643. doi: 10.1111/j.1432-1033.1996.00633.x. [DOI] [PubMed] [Google Scholar]
- Erickson JR, Johnston M. Genetic and molecular characterization of GAL83: Its interaction and similarities with other genes involved in glucose repression in Saccharomyces cerevisiae. Genetics. 1993;135:655–664. doi: 10.1093/genetics/135.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffrini P, Ficarelli A, Donnini C, Lodi T, Puglisi PP, Ferrero I. FOG1 and FOG2 genes, required for the transcriptional activation of glucose-repressible genes of Kluyveromyces lactis, are homologous to GAL83 and SNF1 of Saccharomyces cerevisiae. Curr Genet. 1996;29:316–326. [PubMed] [Google Scholar]
- Gorner W, Durschschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes & Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- Hardy TA, Huang D, Roach PJ. Interactions between cAMP-dependent and SNF1 protein kinases in the control of glycogen accumulation in Saccharomyces cerevisiae. J Biol Chem. 1994;269:27907–27913. [PubMed] [Google Scholar]
- Hayashi T, Hirshman M, Kurth E, Winder W, Goodyear L. Evidence for 5′ AMP-activated protein kinase mediation of the effect of muscle contraction on glucose transport. Diabetes. 1998;47:1369–1373. doi: 10.2337/diab.47.8.1369. [DOI] [PubMed] [Google Scholar]
- Honigberg SM, Lee RH. Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:4548–4555. doi: 10.1128/mcb.18.8.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Carlson M. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes & Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- Jiang R, Carlson M. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol Cell Biol. 1997;17:2099–2106. doi: 10.1128/mcb.17.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp BE, Mitchelhill KI, Stapleton D, Michel BJ, Chen Z-P, Witters LA. Dealing with energy demand: AMP-activated protein kinase. Trends Biochem Sci. 1998;24:22–25. doi: 10.1016/s0968-0004(98)01340-1. [DOI] [PubMed] [Google Scholar]
- Kim J, Hirsch JP. A nucleolar protein that affects mating efficiency in Saccharomyces cerevisiae by altering the morphological response to pheromone. Genetics. 1998;149:795–805. doi: 10.1093/genetics/149.2.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin S, Treich I, Carlson M. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc Natl Acad Sci. 2000;97:7916–7920. doi: 10.1073/pnas.140109897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage P, Yang X, Carlson M. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: A new role for SNF1 in the glucose response. Mol Cell Biol. 1996;16:1921–1928. doi: 10.1128/mcb.16.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludin K, Jiang R, Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan D, Jeon J-T, Looft C, Amarger V, Robic A, Thelander M, Rogel-Gaillard C, Paul S, Iannuccelli N, Rask L, et al. A mutation in PRKAG3 associated with excess glycogen content in pig skeletal muscle. Science. 2000;288:1248–1251. doi: 10.1126/science.288.5469.1248. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Mitchelhill KI, Stapleton D, Gao G, House C, Michell B, Katsis F, Witters LA, Kemp BE. Mammalian AMP-activated protein kinase shares structural and functional homology with the catalytic domain of yeast Snf1 protein kinase. J Biol Chem. 1994;269:2361–2364. [PubMed] [Google Scholar]
- Mitchelhill KI, Michell B, House CM, Stapleton D, Dyck J, Gamble J, Ullrich C, Witters LA, Kemp BE. Posttranslational modifications of the 5′-AMP-activated protein kinase beta1 subunit. J Biol Chem. 1997;272:24475–24479. doi: 10.1074/jbc.272.39.24475. [DOI] [PubMed] [Google Scholar]
- Ostling J, Ronne H. Negative control of the Mig1p repressor by Snf1p-dependent phosphorylation in the absence of glucose. Eur J Biochem. 1998;252:162–168. doi: 10.1046/j.1432-1327.1998.2520162.x. [DOI] [PubMed] [Google Scholar]
- Ponticos M, Lu QL, Morgan JE, Hardie DG, Partridge TA, Carling D. Dual regulation of the AMP-activated protein kinase provides a novel mechanism for the control of creatine kinase in skeletal muscle. EMBO J. 1998;17:1688–1699. doi: 10.1093/emboj/17.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez C, Gancedo JM. Glucose signaling in yeast is partially mimicked by galactose and does not require the Tps1 protein. Mol Cell Biol Res Commun. 1999;1:52–58. doi: 10.1006/mcbr.1999.0112. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Salt I, Celler JW, Hawley SA, Prescott A, Woods A, Carling D, Hardie DG. AMP-activated protein kinase: Greater AMP dependence, and preferential nuclear localization, of complexes containing the α2 isoform. Biochem J. 1998;334:177–187. doi: 10.1042/bj3340177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P, Nieto A, Prieto JA. Glucose repression may involve processes with different sugar kinase requirements. J Bacteriol. 1996;178:4721–4723. doi: 10.1128/jb.178.15.4721-4723.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P, Alms GR, Haystead TA, Carlson M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol Cell Biol. 2000;20:1321–1328. doi: 10.1128/mcb.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MC, McCartney RR. β-Subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 2000;19:4936–4943. doi: 10.1093/emboj/19.18.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Kohlwein SD. Organelle structure, function, and inheritance in yeast: A role for fatty acid synthesis? Cell. 1997;88:431–434. doi: 10.1016/s0092-8674(00)81882-6. [DOI] [PubMed] [Google Scholar]
- Schöler A, Schüller H-J. A carbon source-responsive promoter element necessary for activation of the isocitrate lyase gene ICL1 is common to genes of the gluconeogenic pathway in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3613–3622. doi: 10.1128/mcb.14.6.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva Xavier Gd, Leclerc I, Salt IP, Doiron B, Hardie DG, Kahn A, Rutter GA. Role of AMP-activated protein kinase in the regulation by glucose of islet β cell gene expression. Proc Natl Acad Sci. 2000;97:4023–4028. doi: 10.1073/pnas.97.8.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Jaeger S, Francois J, Gaughran JP, Tatchell K. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treitel MA, Kuchin S, Carlson M. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6273–6280. doi: 10.1128/mcb.18.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O, Carlson M. Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 1998;17:7002–7008. doi: 10.1093/emboj/17.23.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O, Carlson M. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J. 1999;18:6672–6681. doi: 10.1093/emboj/18.23.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wilson WA, Hawley SA, Hardie DG. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP : ATP ratio. Curr Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- Woods A, Munday MR, Scott J, Yang X, Carlson M, Carling D. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J Biol Chem. 1994;269:19509–19516. [PubMed] [Google Scholar]
- Woods A, Assout-Marniche D, Foretz M, Stein SC, Lemarchand P, Ferre P, Foufelle F, Carling D. Characterization of the role of AMP-activated protein kinase in the regulation of glucose-activated gene expression using constitutively active and dominant negative forms of the kinase. Mol Cell Biol. 2000;20:6704–6711. doi: 10.1128/mcb.20.18.6704-6711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Hubbard EJA, Carlson M. A protein kinase substrate identified by the two-hybrid system. Science. 1992;257:680–682. doi: 10.1126/science.1496382. [DOI] [PubMed] [Google Scholar]
- Yang X, Jiang R, Carlson M. A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase complex. EMBO J. 1994;13:5878–5886. doi: 10.1002/j.1460-2075.1994.tb06933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Smith RJ, Brown AJP. Multiple signalling pathways trigger the exquisite sensitivity of yeast gluconeogenic mRNAs to glucose. Mol Microbiol. 1996;20:751–764. doi: 10.1111/j.1365-2958.1996.tb02514.x. [DOI] [PubMed] [Google Scholar]