Abstract
Yeast defective in the checkpoint kinase Rad53 fail to recover from transient DNA replication blocks and synthesize intact chromosomes. The effectors of Rad53 relevant to this recovery process are unknown. Here we report that overproduction of the chromatin assembly factor Asf1 can suppress the Ts phenotype of mrc1rad53 double mutants and the HU sensitivity of rad53 mutants. Eliminating silencing also suppresses this lethality, further implicating chromatin structure in checkpoint function. We find that Asf1 and Rad53 exist in a dynamic complex that dissociates in response to replication blocks and DNA damage. Thus, checkpoint pathways directly regulate chromatin assembly to promote survival in response to DNA damage and replication blocks.
Keywords: Asf1, Rad53, checkpoint kinases, chromatin assembly
DNA damage and DNA replication interference represent life threatening situations to which cells have evolved sophisticated responses (Lindahl and Wood 1999). The repair of DNA damage requires not only recognition of the altered DNA structure, but also the recruitment of repair factors to the site of damage. Furthermore, it may be necessary to reorganize the chromatin to optimally initiate repair. After repair, the chromatin structure of the repaired DNA must be reassembled. It is not understood how chromatin reorganization takes place during repair, however, a number of chromatin remodeling complexes such as the Swi/Snf and Rsc complexes exist that can facilitate chromatin reorganization (Cairns 1998). Likewise, chromatin assembly factors such as the CAF1 complex and RCAF (Asf1) can act to deposit chromatin onto newly synthesized DNA during DNA replication and possibly during repair and replication fork reorganization. Subunits of CAF1 have been shown to bind to PCNA and localize to DNA in response to DNA damage, presumably during resynthesis following excision repair (Shibahara and Stillman 1999).
This response to replication interference and DNA damage is orchestrated through checkpoint pathways that carry out a number of important functions to promote survival (Zhou and Elledge 2000). These include cell cycle arrest, inhibition of late origins of replication, transcriptional induction of DNA repair genes, post-translational modification of repair proteins, and alteration of chromatin structure. In budding yeast, central among the proteins involved in this response are the checkpoint kinases Mec1, Rad53, Chk1, and Dun1.
Mec1 is the central transducer of these stress-response signals and it acts to turn on the other three kinases (Zhou and Elledge 2000). Both Rad53 and Mec1 are key proteins involved in the response to replication blocks and they act together with a novel regulator of Rad53, Mrc1, that is also required for a subset of Rad53 responses (A. Alcasabas et al., in prep.). Cells lacking Rad53 or Mec1 die rapidly when treated with hydroxyurea, an inhibitor of dNTP synthesis, due primarily to an inability to recover and properly synthesize complete chromosomes after the replication block is removed (Desany et al. 1998). How Rad53 promotes the synthesis of complete chromosomes after replication insults is unclear, but the DNA damage response pathway has been linked to the control of chromatin organization. In response to DNA damage, certain proteins that are normally localized to silent telomeric chromatin, such as the Sir3 silencing factor, relocalize to sites of DNA damage (Martin et al. 1999; Mills et al. 1999). This relocalization is dependent on Mec1. In rad53 mutants, telomeric silencing is enhanced, whereas in mec1 mutants it is disrupted (Craven and Petes 2000; Longhese et al. 2000).
Another connection to silencing comes from checkpoint studies in meiotic cells. Mutants in dmc1 or zip1 result in pachytene arrest that is dependent on the DNA damage checkpoint (Grushcow et al. 1999). In addition, mutations in three genes involved in silencing, PCH1/DOT1, PCH2, or SIR2 bypass the pachytene arrest, thereby linking chromatin structure to checkpoint control (San-Segundo and Roeder 1999; Roeder and Bailis 2000). How chromatin structure controls checkpoints and how checkpoints control chromatin structure is not known. In this work we discover that the chromatin assembly factor ASF1 resides in a complex with Rad53 that dissociates in the presence of replication blocks or DNA damage. We propose that in response to DNA replication blocks and DNA damage, Rad53 directs Asf1 to regulate nucleosome deposition and thereby links Rad53 to chromatin assembly.
Results
mrc1-1 rad53-21 mutants have a temperature-sensitive lethal phenotype
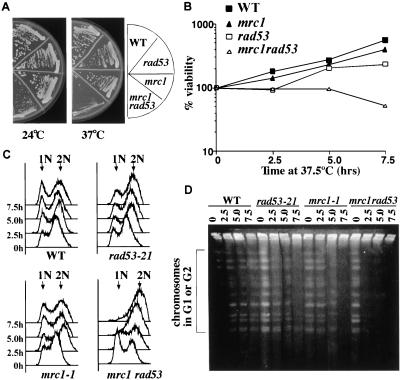
Mrc1 is a new component of the S-phase checkpoint pathway that is required to prevent spindle elongation in response to HU treatment. Although epistasis studies suggest that Mrc1 and Rad53 function in the same pathway in the S-phase checkpoint pathway with respect to spindle elongation in HU-arrested cells, the relationship between MRC1 and RAD53 is complex (Alcasabas et al., in prep.). For example, mrc1Δrad53-21 double mutants are lethal, and the lethality cannot be suppressed by RNR1 overexpression, suggesting Mrc1 has at least one function distinct from Rad53. mrc1-1 rad53-21 double mutants grow very slowly at room temperature and they are temperature sensitive for growth at 37°C, whereas the single mutants are not (Fig. 1A). The double mutants lose viability after growth at 37.5°C (Fig. 1B).
Figure 1.
mrc1-1 rad53-21 double mutants arrest at the nonpermissive temperature with incompletely replicated chromosomes. (A) Temperature sensitivity of mrc1-1 rad53-21 double mutants. Yeast strains of the indicated genotypes were streaked out on YPD plates and were either grown at room temperature for 4 d or grown at 37°C for 2 d. (B–D) After growth at 37.5°C, mrc1-1 rad53-21 double mutants lose viability and arrest with incompletely replicated chromosomes. Asynchronous log-phase culture at 24°C were shifted to 37.5°C and samples were taken every 2.5 h to examine viability (B), to analyze DNA content (C), or to prepare DNA for PFGE analysis (D).
To investigate the lethality of mrc1 rad53 double mutants, we analyzed the cell cycle progression of mrc1-1 rad53-21 mutants at the nonpermissive temperature. Asynchronous log phase cultures at 24°C were shifted to 37.5°C and samples were taken every 2.5 h. From FACS analysis, we observed that the majority of mrc1-1 rad53-21 mutants delay in S phase. After 5 h at 37.5°C, they became arrested with a G2 DNA content (Fig. 1C) and short spindles (data not shown). To determine whether this arrest was in S phase or in G2, we examined whether the mrc1-1 rad53-21 mutant had finished DNA replication using pulsed-field gel electrophoresis (PFGE). Replicating chromosomes cannot enter the gel because of secondary structures caused by replication intermediates and, therefore, remain in the well. However, fully replicated chromosomes will migrate into the gel. We can detect fully replicated chromosome signals in the gel from wild-type cells at all time points tested. However, mrc1-1 rad53-21 double mutants fail to enter the gel within 2.5 h after the shift (Fig. 1D) and show an accumulation of S phase cells (Fig. 1C). Thus, mrc1-1 rad53-21 double mutants have a severe defect in DNA replication and arrest with incompletely replicated chromosomes at the nonpermissive temperature. The failure of chromosomes to migrate could also be caused by unresolved recombination intermediates. However, given the accumulation of cells with an S-phase DNA content at 2.5 h after the shift, we feel incomplete replication is the simplest explanation.
Dosage suppressors of the mrc1-1 rad53-21 Ts phenotype
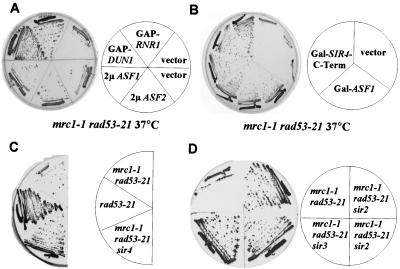
To understand the synthetic lethality of the mrc1 rad53 double mutant, we isolated high-copy suppressors of their Ts phenotype. Four suppressors, SLT2, PCL1, YER033C, and ASF1 were found in this screen. SLT2 encodes a MAP kinase. PCL1 encodes a G1 cyclin. Slt2 and Pcl1 are involved in activating the SWI transcription complex (Igual et al. 1996; Madden et al. 1997), which controls RNR1 and RNR3 expression. Because the phenotype of mrc1-1 rad53-21 is similar to a rad53 null mutation and the lethality of rad53Δ can be suppressed by RNR1 overexpression (Desany et al. 1998), we asked whether RNR1 could suppress mrc1-1 rad53-21 mutants. Both RNR1 and its positive regulator DUN1 suppress mrc1-1 rad53-21 mutants when overproduced (Fig. 2A), suggesting that SLT2 and PCL1 may work through RNR1 regulation. Unlike RNR1, overexpression of ASF1 does not suppress the lethality of rad53Δ. Therefore, in this respect, mrc1-1 rad53-21 does not fully mimic the rad53Δ mutation.
Figure 2.
Suppression of the mrc1-1 rad53-21 Ts phenotype by RNR1 and DUN1 overexpression and anti-silencing. (A,B) mrc1-1 rad53-21 mutants containing the indicated plasmids were streaked out on YPD plates (A) or YP-GAL plates (B) and grown at 37°C for 2 d. (C,D) Yeast strains of indicated genotypes were streaked out on YPD plates and grown at 37°C for 2 d.
Disrupting silencing can suppress the lethality of mrc1-1 rad53-21 mutants
ASF1 has been implicated in histone deposition during DNA replication (Tyler et al. 1999). asf1 mutants are sensitive to HU and DNA-damaging agents (Le et al. 1997; Tyler et al. 1999). Both overproduction and deletion of ASF1 lead to silencing defects at telomere and mating-type loci (Le et al. 1997; Singer et al. 1998). In addition, Rad53 has been implicated in regulation of chromatin adjacent to telomeres (Craven and Petes 2000; Longhese et al. 2000). Therefore, we asked whether the suppression of the mrc1 rad53 double mutants might reflect the anti-silencing property of ASF1 by expressing other anti-silencing factors in mrc1-1 rad53-21 mutants, such as ASF2 and the carboxyl terminus of SIR4. Overexpression of ASF2 causes silencing defects at the HML and HMR loci (Le et al. 1997) and overexpression of carboxyl termini of SIR4 disrupts Sir complex assembly, thereby interfering with silencing (Marshall et al. 1987; Cockell et al. 1995). Surprisingly, overexpression of either ASF2 or of the carboxyl terminus of SIR4 suppresses mrc1-1 rad53-21, but the vector alone cannot (Fig. 2A,B). Furthermore, sir2, sir3, or sir4 null mutants, which cause loss of silencing, can also suppress mrc1-1 rad53-21 (Fig. 2C,D). Thus, anti-silencing can suppress mrc1-1 rad53-21. We have also checked the suppression of mrc1-1 rad53-21 Ts phenotype by overexpressing other DOT genes identified as dosage disruptors of telomeric silencing (Singer et al. 1998). Unlike SIR4 carboxyl terminus or ASF1, overexpression of all other DOT genes fail to suppress (data not shown). This suggests that either the degree of anti-silencing must be beyond a threshold level to suppress, or that ASF1 may possess a special property relevant to Rad53 function. The suppression of the Ts phenotype of the mrc1 rad53 double mutant by ASF1 overproduction is not due to the psuedo-diploid nature of these mutants resulting from ASF1's anti-silencing properties because the homozygous mrc1 rad53 diploid is still Ts, although less so than the haploid. Overproduction of ASF1 still suppresses the Ts phenotype.
Mrc1 is required for silencing
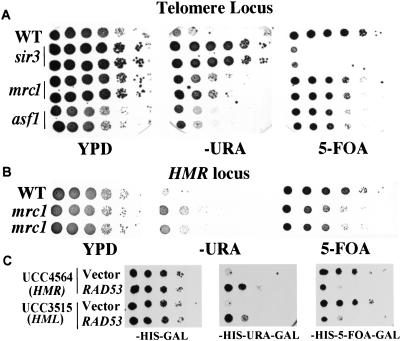
The involvement of silencing in the temperature sensitivity of mrc1 rad53 mutants led us to examine whether Mrc1 or Rad53 has a role in silencing. It has been reported that rad53 mutants have a modest increase of telomeric silencing (Craven and Petes 2000; Longhese et al. 2000), implicating Rad53 in control of telomeric chromatin. To test whether Mrc1 also has a role, the mrc1 mutation was introduced into test strains that have a URA3 reporter at either telomeric or HMR loci. mrc1 mutants have partial loss of silencing at both loci, which is reflected by increased growth on SC-ura plates and reduced growth on 5-FOA plates (Fig. 3A,B). This silencing defect is not as dramatic as in sir mutants. Overexpression of RAD53 also causes partial loss of silencing (Fig. 3C), whereas overexpression of either DUN1 or RNR1 does not (data not shown). Thus, both Rad53 and Mrc1 can affect silencing and may be involved in chromatin regulation.
Figure 3.
Role of Mrc1 and Rad53 in silencing. (A) mrc1 mutants have a telomere silencing defect. Wild-type (UCC3505), isogenic sir3Δ, or mrc1Δ mutants were grown to saturation. Serial 10X dilutions of these cultures were made and spotted onto YPD, SC-ura, or SC 5-FOA plates and were grown for 4–5 d at 30°C. (B) mrc1 mutants have a HMR-silencing defect. The test strain used is UCC4564. (C) Overproduction of Rad53 causes a silencing defect at both HML and HMR loci. Saturated cultures of URA3 reporter strains UCC4564 or UCC3515 containing either vector alone or HIS3–GAL–RAD53 plasmids were diluted and spotted onto SC-his and SC-his,ura plates or SC-his plates containing 5-FOA as indicated, and grown as in A.
Connections between silencing and checkpoint control
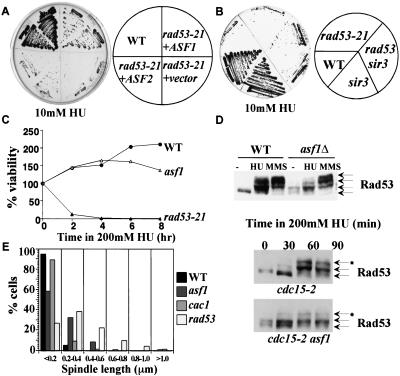
Due to the connection between anti-silencing and the suppression of the mrc1-1 rad53-21 Ts phenotype, we next asked whether anti-silencing could also suppress the sensitivity of rad53 mutants to HU. Interestingly, overexpression of ASF1 can strongly suppress the HU sensitivity of both rad53-21 and rad53Δ mutants on 10 mM HU, whereas the vector control cannot (Fig. 4A). Overexpression of ASF2 (Fig. 4A) or mutation of SIR3 (Fig. 4B) can also weakly suppress the HU sensitivity of rad53 mutants. The weak suppression by sir3 mutants indicates that the suppression by ASF1 cannot be attributed solely to its anti-silencing property.
Figure 4.
Connection between silencing and checkpoint control. (A) Suppression of the HU sensitivity of rad53 mutants by ASF1 or ASF2 overexpression. Yeast strains of indicated genotypes were streaked out on YPD plates containing 10 mM HU and grown for 2 d at 30°C. (B) sir3Δ can weakly suppress the HU sensitivity of rad53 mutants on 10 mM HU. (C) asf1 mutants lose viability after prolonged exposure to HU. A total of 200 mM HU was added to asynchronous cultures at 30°C and samples were taken every 2 h to examine viability. (D) Activation of Rad53 in asf1 mutants. (Top) Wild-type (WT) or asf1 mutant cells were grown to log phase at 30°C. Protein extracts were made from cells untreated (−), treated with 200 mM HU for 1.5 h (HU), or treated with 0.05% MMS for 1 h (MMS). (Bottom) Log phase culture of cdc15-2 or cdc15-2 asf1 mutant cells at room temperature were shifted to 37°C for 2 h. Those telophase-arrested cells were then shifted back to room temperature in YPD medium containing 200 mM HU. Samples were taken every half-hour and protein extracts were made. Proteins were resolved on 8% SDS-PAGE. Anti-Rad53 antibody (1:250 dilution) was used to detect Rad53p. (E) Spindle phenotypes of asf1 mutants in HU. Asynchronous culture of wild-type, asf1, cac1, or rad53-21 mutant cells were treated with 200 mM HU for 3 h at 30°C and spindle lengths were measured.
The strong suppression of rad53 HU sensitivity by Asf1 overproduction suggests that Asf1 may have a role in the RAD53-dependent DNA replication stress-response pathway. We examined the viability of asf1Δ mutants in response to HU treatment. Compared with wild-type cells, asf1 mutants lose viability after prolonged exposure to HU (Fig. 4C).
We also examined the activation of Rad53 in response to HU and MMS in asf1 mutants. Asynchronous cultures of wild-type and asf1 mutant cells were treated with HU and MMS and Rad53 modification status was examined by Western blotting. Regulation of Rad53 by MMS was unaffected, but there was a significant reduction in the accumulation of the hyperphosphorylated form of Rad53 in response to HU in asf1 mutants (Fig. 4D). To evaluate the kinetics of Rad53 phosphorylation, we examined synchronized cultures. Due to their silencing defects, asf1 mutants cannot be synchronized with α-factor. Instead, we used the cdc15 mutation to synchronize cells in telophase and released the cells into 200 mM HU. Rad53p shows a partial mobility shift during an unperturbed cell cycle in asf1 mutants, suggesting that asf1 mutants may be defective in DNA replication and accumulate DNA damage that activates Rad53. In response to HU, Rad53 gets further activated in asf1 mutants, but the kinetics and extent of hyperphosphorylation are delayed compared with wild- type cells (Fig. 4D). This could be a result of the slow cell cycle progression of asf1 mutants or may mean that Asf1 plays a more direct role in activating Rad53. Consistent with a delay in Rad53 activation, we find that asf1 mutants treated with HU have a partial checkpoint defect with more cells displaying partially elongated spindles (Fig. 4E). This defect is not as severe as seen in rad53 mutants with respect to either percentage of elongated spindles or the length of spindle elongation (Fig. 4E).
Physical interaction between Asf1 and Rad53 modulated by DNA damage
Although ASF1 overexpression suppresses rad53's HU sensitivity, it cannot suppress mec1-21, dun1Δ, or mrc1Δ mutants (data not shown). This suggests that Asf1 overproduction is supplying a particular function normally carried out specifically by Rad53 as opposed to the pathway in general. The suppression of HU sensitivity of rad53 mutants by increased Asf1 levels cannot be attributed solely to the anti-silencing effect of Asf1 overproduction because direct abrogation of silencing by sir mutations provide only a marginal suppression. Together, these observations suggest that this suppression may reflect specific interactions between Asf1 and Rad53.
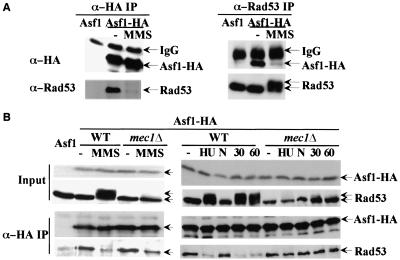
To explore this hypothesis, we tested for a physical interaction. We tagged the genomic ASF1 gene with a triple HA tag and immunoprecipitated Asf1 with anti-HA antibodies. We detected Rad53 in the IP product, but not in the control IP in which Asf1 is not tagged (Fig. 5A). Moreover, the amount of Rad53 pulled down by HA antibodies is much less in the presence of the DNA damage agent, MMS (Fig. 5A) or in the presence of HU (Fig. 5B). We observed only the unphosphorylated form of Rad53 in the IP, although the majority of Rad53 is phosphorylated in the presence of MMS. We can also detect Asf1 in the reciprocal immunoprecipitations using anti-Rad53 antibodies (Fig. 5A). We hypothesize that in the presence of DNA damage, activation of the damage checkpoint leads to phosphorylation of Rad53 and dissociation of Asf1 and Rad53.
Figure 5.
Interaction between Asf1p and Rad53p. (A) Coimmunoprecipitation of Asf1–HA3 and Rad53p. Protein extracts from ASF1, ASF1–HA3, or ASF1–HA3 cells treated with 0.05% MMS for 1 h were immunoprecipitated with either anti-HA or anti-Rad53 antibodies. The immunoprecipitates were resolved by SDS-PAGE and probed with either anti-HA or anti-Rad53 antibodies as indicated. (B) Coimmunoprecipitation of Asf1–HA3 and Rad53p in mec1Δ mutants in response to DNA damage and replication blocks. Asynchronous cultures of wild-type or mec1Δ mutants were untreated (−), treated with 0.05% MMS for 1 h, or 200 mM HU for 1.5 h (HU). Separate cultures were arrested with nocodazole and either untreated (N) or γ-irradiated (6 Krad), while maintaining the nocodazole arrest. Samples were taken either 30 min (30) or 60 min (60) after irradiation and immunoprecipitation studies were performed as in A.
Dissociation of Rad53 and Asf1 is regulated by Mec1
It is not clear whether the dissociation of Asf1 and Rad53 occurs because of Rad53 activation or merely correlates with it. Therefore, we examined the association of Rad53 and Asf1 in mec1Δ mutants, in which the phosphorylation of Rad53 is significantly reduced. mec1Δ cells show reduced dissociation of the complex (Fig. 5B), indicating that this response is partially Mec1-dependent in asynchronous cells. However, there was still a significant amount of dissociation of Asf1 and Rad53 in mec1 mutants upon MMS treatment (Fig. 5B). We also examined the dissociation of Asf1 and Rad53 in mec1 mutants in response to HU. Whereas in wild-type cells treated with HU there is very little association of Asf1 and Rad53, significant association is maintained in mec1 mutants treated with HU (Fig. 5B).
The differences in wild-type and mec1 mutants could be due to the fact that MEC1 function is required for dissociation or could result from the fact that wild-type cells treated with HU or DNA damage are arrested in the cell cycle, whereas mec1 mutants progress to different stages of the cycle. To control for cell cycle differences, we first arrested cells at G2/M using nocodazole and then introduced DNA damage using γ-irradiation while maintaining the G2/M block. In this experiment, the dissociation of Asf1 and Rad53 is totally abolished in mec1 mutants. We conclude that in response to checkpoint signals during replication blocks and DNA damage, Mec1 activation leads to activation of Rad53 and dissociation of Asf1.
Discussion
In this study we provide evidence for several connections between checkpoint pathway and chromatin assembly. First, Mrc1 is required for efficient silencing. We have also found that, unlike the silencing defect of mec1 mutants (Craven and Petes 2000; Longhese et al. 2000), the defect in mrc1 mutants is not suppressed by overproducing RNR1 (data not shown) and is likely to be more direct than that in mec1 mutants. Secondly, we identified ASF1 as a dosage suppressor of rad53 mrc1 mutants. This lethality can also be suppressed by interfering with silencing by mutations in Sir2, Sir3, and Sir4. This suggests that heterochromatic structures may be incompatible with these mutants. We can only speculate as to the nature of this incompatibility. One possibility is that heterochromatic regions are difficult to replicate through and provide a significant challenge to checkpoint defective mutants. By eliminating silencing, these regions are converted to euchromatic regions that present less of a problem. A second possibility is that certain repair factors are sequestered at telomeric chromatin through the silencing machinery (Martin et al. 1999; Mills et al. 1999) and these are released in response to damage and may be important for overcoming the DNA replication problems experienced by the mrc1 rad53 double mutants. By interfering with silencing, these factors may be released to facilitate repair of the replication defect. Whereas mrc1 mutants themselves interfere with silencing, they do so to a lesser degree than the mutants capable of suppressing the lethality of the double mutants. A third possibility is that without silencing, simultaneous expression of MAT a/MAT α genes could change the efficiency of repair pathways (Astrom et al. 1999) and thus affect the viability of mrc1-1 rad53-21 double mutants. The mrc1-1 rad53-21 homozygous mutant diploids are still Ts and the Ts can be suppressed by overexpression of ASF1. It should be noted that the diploid is not as Ts as the haploid, and interfering with Sir function does not enhance growth (data not shown), therefore, diploid status might have a partial effect on the temperature sensitivity but cannot explain the suppression by ASF1.
The final connection between checkpoints and chromatin is the physical association between Rad53 and the chromatin assembly factor Asf1. Whereas eliminating silencing was found to provide weak suppression to rad53 mutants, overproduction of ASF1 provided a very strong suppression of the lethality of rad53 mutants experiencing replication blocks. This suppression is likely to be direct because Asf1 and Rad53 exist together in a dynamic complex that dissociates in the presence of DNA damage or replication blocks. The dissociation is dependent on the activation of the checkpoint pathway, because the interaction of Asf1 and Rad53 is maintained in mec1 mutants in response to replication blocks or DNA damage. The phosphorylation of Rad53 by Mec1 is correlated with the dissociation, but it is unclear whether Rad53 phosphorylation is the critical factor.
ASF1 overproduction suppresses rad53-21 mutants and, although there is a physical interaction between Rad53 and Asf1, we did not see restoration of Rad53-21 kinase activity by ASF1 overproduction, nor increase in its abundance (data not shown). We have also not detected phosphorylation of Asf1 by Rad53 in vivo or in vitro. However, as ASF1 overproduction is such a strong suppressor of the rad53 mutants's HU sensitivity, it suggests that ASF1 is an effector of Rad53, carrying out part of Rad53's function in response to DNA replication blocks. This hypothesis is supported by the fact that asf1 mutants are sensitive to HU and DNA damage as are rad53 mutants, suggesting a similar function for these proteins. This leads to the question of the role of Rad53 in Asf1 regulation. The most plausible explanation for the relationship between Asf1 and Rad53 is that they work together to repair damage and alleviate DNA replication problems. In response to DNA damage, Rad53 may activate Asf1 or confer special properties on it, causing it to leave its complex and contribute to repair. Rad53 is a chromatin-associated protein (Frei and Gasser 2000), and it is possible that Rad53 localizes Asf1 to sites of damage, where it is released to facilitate chromatin assembly. This might explain why Asf1 overproduction suppresses rad53 mutants, because when overexpressed, Asf1 now can locate these sites without the aid of Rad53. Alternatively, Asf1 release in response to replication blocks and damage may allow it to sequester free nucleosomes whose excess might interfere with repair processes by binding to DNA. Although this is a plausible explanation, if this were true, one might expect ASF1 overproduction to suppress mec1 mutants, which it does not.
The above model represents the simplest explanation for the relationships between Asf1 and Rad53, but more complex models exist. For example, Asf1 by virtue of its ability to interact with nucleosomes could help localize Rad53 to replication forks and help it find substrates or facilitate its activation. This would be consistent with the delayed kinetics of Rad53 hyperphosphorylation and the partial checkpoint defect in asf1 mutants in response to HU treatment. In this model, the suppression of the HU sensitivity of rad53 mutants by ASF1 overproduction would be indirect. It is also possible that both models could be partially correct and these proteins could mutually regulate one another. Whereas the precise biochemical relationship between these proteins remains to be fully elucidated, their genetic and physical interactions strongly suggest that they function together to facilitate DNA repair and replication. Future investigation of their common functions should provide significant insights into the connection between chromosome structure and checkpoint function.
Materials and methods
Yeast strains and cytological techniques
All strains used in this study are isogenic with the W303-derived Y300 strain (Zhou and Elledge 1993) except for the silencing test strains UCC3505, UCC3515, and UCC4564 (Singer et al. 1998). SIR3 and carboxy-terminal 2561 bp of MRC1 were deleted in UCC3505 or UCC4564 by use of a PCR-based gene disruption method (Longtine et al. 1998) and confirmed by PCR analysis. mrc1-1 rad53-21 sir mutants were constructed by deleting SIR genes in mrc1-1 rad53-21 mutants by use of the same method (Longtine et al. 1998). Genomic HA3-tagged ASF1 and asf1Δ mutants were constructed according to the same method (Longtine et al. 1998). All other stains are constructed by standard genetic cross.
Plasmids
The plasmid used for GAP–RNR1 was pBAD70 (Desany et al. 1998). The plasmid used for GAP–DUN1 was pZZ74-2 (Zhou and Elledge 1993). GAL-carboxy-terminal SIR4 and GAL–ASF1 were kindly given as pTRP10 and pTRP53 (Singer et al. 1998) by Dr. Gottschling (Division of Basic Sciences, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA). pARF24 containing 2μ ASF2 (Le et al. 1997) was kindly given by Dr. Sternglanz (Department of Biochemistry and Cell Biology, State University of New York, Stony Brook, New York). pFH10 was used as 2μ ASF1 in this study. PCR reactions using primers 5′-CAGGATAT TGTCTTGGTGGCGTC and 5′-ACGTAGCTGTTGCCCTAACCCG were performed to amplify the ASF1 gene. This genomic fragment was then cloned into PCR2.1-TOPO (Invitrogen) to give pFH8. The EcoRI fragment of pFH8 containing ASF1 was cloned into pRS426 to give pFH10.
Protein Techniques
Protein extracts for Western blots were made by use of the TCA precipitation method as described (Paciotti et al. 1998). For immunoprecipitations, log phase cultures of yeast cells were pelleted, resuspended in buffer containing 50 mM Tris7.5, 150 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, and proteinase inhibitors and broken open by bead beating. A total of 5 mg of protein extract was diluted to 1 mL with the same buffer and precleared by incubating with Pro-G beads for 1 h at 4°C. For α-Rad53 immunoprecipitations, 30 μL of anti-Rad53 antibodies were added to precleared supernatant and rotated at 4°C overnight. After spinning for 10 min at 14 K, 40 μL of Protein-A beads were added to the supernatant and rotated for another hour. For α-HA immunoprecipitation, 40 μL of anti-HA matrix was added to precleared supernatant and rotated at 4°C overnight. Beads were then washed with 1 mL buffer four times. SDS-loading dye was added and samples were boiled and resolved on SDS-PAGE.
Acknowledgments
We thank Dr. Gottschling and Dr. Sternglanz for yeast strains and plasmids, Jeff Bachant, Lee Zou, Tom Petes, Lorraine Pillus, and Jim Kadonaga for helpful discussions. This work was supported by National Institutes of Health Grant GM44664 to S.J.E. S.J.E. is an Investigator with the Howard Hughes Medical Institute and Senior Scholar of the Ellison Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL selledge@bcm.tmc.edu; FAX (713) 798-8717.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.873201.
References
- Astrom SU, Okamura SM, Rine J. Yeast cell-type regulation of DNA repair. Nature. 1999;397:310. doi: 10.1038/16833. [DOI] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling machines: Similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig AJ, Gasser SM. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: Evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RJ, Petes TD. Involvement of the checkpoint protein Mec1p in silencing of gene expression at telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:2378–2384. doi: 10.1128/mcb.20.7.2378-2384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes & Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C, Gasser SM. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes & Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- Grushcow JM, Holzen TM, Park KJ, Weinert T, Lichten M, Bishop DK. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics. 1999;153:607–620. doi: 10.1093/genetics/153.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igual JC, Johnson AL, Johnston LH. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 1996;15:5001–5013. [PMC free article] [PubMed] [Google Scholar]
- Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- Longhese MP, Paciotti V, Neecke H, Lucchini G. Checkpoint proteins influence telomeric silencing and length maintenance in budding yeast. Genetics. 2000;155:1577–1591. doi: 10.1093/genetics/155.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Madden K, Sheu YJ, Baetz K, Andrews B, Snyder M. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science. 1997;275:1781–1784. doi: 10.1126/science.275.5307.1781. [DOI] [PubMed] [Google Scholar]
- Marshall M, Mahoney D, Rose A, Hicks JB, Broach JR. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4441–4452. doi: 10.1128/mcb.7.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- Paciotti V, Lucchini G, Plevani P, Longhese MP. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder GS, Bailis JM. The pachytene checkpoint. Trends Genet. 2000;16:395–403. doi: 10.1016/s0168-9525(00)02080-1. [DOI] [PubMed] [Google Scholar]
- San-Segundo PA, Roeder GS. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999;97:313–324. doi: 10.1016/s0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Elledge SJ. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell. 1993;75:1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]