Abstract
A yeast two-hybrid screen using the conserved carboxyl terminus of the nuclear receptor corepressor SMRT as a bait led to the isolation of a novel human gene termed SHARP (SMRT/HDAC1 Associated Repressor Protein). SHARP is a potent transcriptional repressor whose repression domain (RD) interacts directly with SMRT and at least five members of the NuRD complex including HDAC1 and HDAC2. In addition, SHARP binds to the steroid receptor RNA coactivator SRA via an intrinsic RNA binding domain and suppresses SRA-potentiated steroid receptor transcription activity. Accordingly, SHARP has the capacity to modulate both liganded and nonliganded nuclear receptors. Surprisingly, the expression of SHARP is itself steroid inducible, suggesting a simple feedback mechanism for attenuation of the hormonal response.
Keywords: SMRT, HDAC1, SRA, attenuation of hormonal response
The transcription action of steroids, retinoids, and thyroid hormone and their cognate receptors (NRs) (Mangelsdorf and Evans 1995; Mangelsdorf et al. 1995) are modulated by an extensive set of nuclear receptor cofactors (McKenna et al. 1999; Glass and Rosenfeld 2000; Westin et al. 2000). A great deal of effort has focused on the identification and characterization of the constituents of these complexes to understand the mechanistic basis of the regulated events. The recruitment of coactivator complexes is a critical step in hormone induction, whereas the recruitment of corepressor complexes mediates active repression of unliganded nuclear receptors. SMRT and N-CoR have been identified as nuclear receptor corepressors (Chen and Evans 1995; Horlein et al. 1995; Ordentlich et al. 1999). Various lines of evidence suggest that at least one mechanism underlying the repression activity of SMRT and N-CoR is through their recruitment of a histone deacetylase complex containing mSin3A and HDAC1 (Alland et al. 1997; Hassig et al. 1997; Heinzel et al. 1997; Laherty et al. 1997; Nagy et al. 1997; Zhang et al. 1997). Direct interaction of SMRT with the class II histone deacetylase (HDAC 4–7) independent of Sin3A provides yet another mechanism for SMRT-mediated transcriptional repression (Huang et al. 2000; Kao et al. 2000). Recruitment of histone deacetylase complexes by corepressors has been proposed to cause a local change in the chromatin structure, therefore resulting in transcriptional repression (Knoepfler and Eisenman 1999).
A search for cofactors that mediate ligand-dependent transactivation by nuclear receptors led to the identification of coactivators such as CBP/p300, PCAF, and the p160 family members including SRC-1, GRIP1/TIF2, and ACTR/RAC3/p/CIP (Onate et al. 1995; Hong et al. 1996; Kamei et al. 1996; Yao et al. 1996; Chen et al. 1997; Torchia et al. 1997; Blanco et al. 1998). Among these factors, CBP, PCAF, SRC-1, and ACTR have been shown to possess intrinsic histone acetyltransferase activity, consistent with a role for induced histone acetylation in transcriptional activation (Bannister and Kouzarides 1996; Ogryzko et al. 1996; Yang et al. 1996; Chen et al. 1997; Spencer et al. 1997). Targeted deletion of SRC-1 or p/CIP causes partial hormone insensitivity, suggesting a critical physiological role of coactivators (Xu et al. 1998, 2000). More recently, SRA has been identified as an RNA cofactor (Lanz et al. 1999), associating with activator complexes to potentiate steroid hormone action.
Kinetic studies have revealed that transcriptional responses to stimulation by steroid hormones are often attenuated after an initial activation (Brown et al. 1984; Sasaki et al. 1984; Cavailles et al. 1988; Dubik and Shiu 1988). In the case of the estrogen receptor, it has been proposed that attenuation of the hormonal response is caused by a rapid, yet transient, increase of histone acetylation, followed by a prolonged decrease of histone acetylation, despite the continuous presence of hormone stimuli (Chen et al. 1999). The molecular mechanism sustaining this repression remains largely unclear.
We identified a novel SMRT-interacting protein termed SHARP. SHARP is an unusual transcriptional repressor, which recruits histone deacetylase activity and also interacts with the RNA coactivator SRA through three conserved RNA recognition motifs. Overexpression of SHARP compromises SRA-potentiated transactivation by ER and GR, presumably by competition for a limiting amount of SRA transcripts. Moreover, SHARP is estrogen-inducible, providing a potential autoregulatory mechanism to attenuate the hormonal response.
Results
Isolation of SHARP as a SMRT-interacting protein
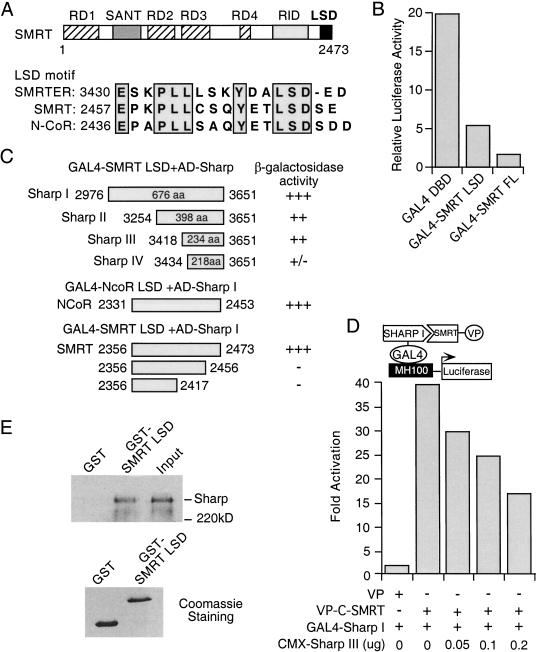
Previous studies have shown that the repression domains (RDs) of SMRT reside in the amino-terminal half of the protein, whereas the receptor interaction domain (RID) in the carboxy-terminal half. However, sequence comparison revealed that the extreme carboxyl terminus of SMRT contains an LSD (lysine-serine-aspartic acid) motif (Fig. 1A), which is conserved among mammalian SMRT and N-CoR, and Drosophila SMRTER (Tsai et al. 1999). To examine the potential function of this domain, we fused amino acid residues 2356 to 2473 of mSMRT containing the LSD motif to the GAL4 DNA binding domain (DBD). The GAL4–SMRT LSD construct was transfected into CV-1 cells along with MH100-tk-luc containing five copies of GAL4 DNA binding sites upstream of a luciferase reporter gene. Activity of the GAL4 fusions was measured relative to the activity of the GAL4 DBD alone (Fig. 1B). The SMRT LSD domain represses the GAL4 DBD-mediated transcription by 3- to 4-fold, as compared to a 12-fold repression by the full-length SMRT. Similar transcriptional repression was observed when the LSD domain of N-CoR (amino acid [aa] 2446 to 2511) was used (data not shown), thus revealing the presence of an additional repression domain at the extreme carboxyl terminus of SMRT and N-CoR.
Figure 1.
Isolation of SHARP as a SMRT-interacting protein. (A) Schematic representation of SMRT domain structure and the LSD motif sequences. (B) The SMRT LSD domain represses basal transcription. CV-1 cells were transfected with GAL4 DBD or GAL4-fusion constructs, together with MH100-tk-luc reporter and CMX-β-gal. The reporter luciferase activity was normalized with the internal control β-galactosidase activity. The results represent the average of triplicate assays. (C) SHARP interacts with the LSD domain of SMRT and N-CoR. GAL4 fusion of SMRT LSD (residues 2356–2473), N-CoR LSD (residues 2331–2453), or the SMRT LSD deletion mutants were transformed into yeast cells along with an activation domain (AD) fusion of various SHARP fragments (I to IV), followed by a liquid β-galactosidase assay. Two viable transformants were picked for each assay. (D) SMRT–SHARP interaction in mammalian two-hybrid assay. CV-1 cells were cotransfected with GAL4–SHARP I and VP alone or VP-C-SMRT with the indicated amounts of CMX-SHARP III, together with MH100-tk-luc and CMX-β-gal. Fold activation is determined by the relative reporter luciferase activity normalized with the β-galactosidase activity and represents an average of three independent experiments. (E) Full-length SHARP interacts with SMRT. 35S-labeled in vitro translated full-length SHARP was incubated with purified GST or GST–SMRT LSD fusion on glutathione beads. The bound proteins were analyzed by SDS-PAGE and visualized by fluorography. Lower panel is the Coomassie staining of GST and GST–SMRT LSD fusion protein.
To investigate the properties of the LSD domain, a yeast two hybrid screen was employed to identify putative LSD interacting proteins. SHARP was pulled out repeatedly with the SMRT LSD domain as a bait. One cDNA clone isolated encodes a 676 aa protein (I), which interacts strongly with the SMRT LSD domain as shown in a liquid β-galactosidase assay in yeast (Fig. 1C). A similar interaction of SHARP with the N-CoR LSD domain (aa 2331–2453) was also observed in the same assay (Fig. 1C). Fragments II and III encoded by two shorter cDNA clones retain strong interaction with the SMRT LSD domain, whereas a smaller fragment (IV) has much weaker interaction, suggesting SHARP III spans the minimal SMRT interaction domain (SID). To examine whether the conserved LSD motif of SMRT is important for its interaction with SHARP, we generated a SMRT LSD mutant in which the last 17 amino acids (aa 2457–2473) were deleted. This deletion mutant loses the ability to interact with SHARP (Fig. 1C). Further deletion to residue 2417 results in a similar loss of the interaction, demonstrating that the LSD motif is necessary for its interaction with SHARP.
The interaction between SMRT and SHARP was further examined in a mammalian two hybrid assay (Fig. 1D). GAL4–SHARP I (aa 2976–3651) was transfected into CV-1 cells together with a VP16 activation domain construct (VP) or a carboxy-terminal SMRT fused to the VP16 activation domain (VP-C-SMRT), along with the MH100-luc reporter. As shown in Figure 1D, transfection of VP-C-SMRT but not VP alone led to a significant increase of the reporter activity due to the interaction of SHARP with C-SMRT. Overexpression of increasing amount of SHARP III (aa 3418–3651) resulted in a dose-dependent decrease in reporter activity, presumably by sequestering VP-C-SMRT from binding to GAL4–SHARP I, further supporting the association between SHARP and SMRT. The interaction of full-length SHARP with SMRT was also demonstrated in a GST-pull down assay (Fig. 1E). Full-length SHARP was specifically bound to GST–SMRT LSD, but not to GST alone. These data together demonstrate that SHARP interacts directly with SMRT both in vivo and in vitro.
Expression pattern of SHARP
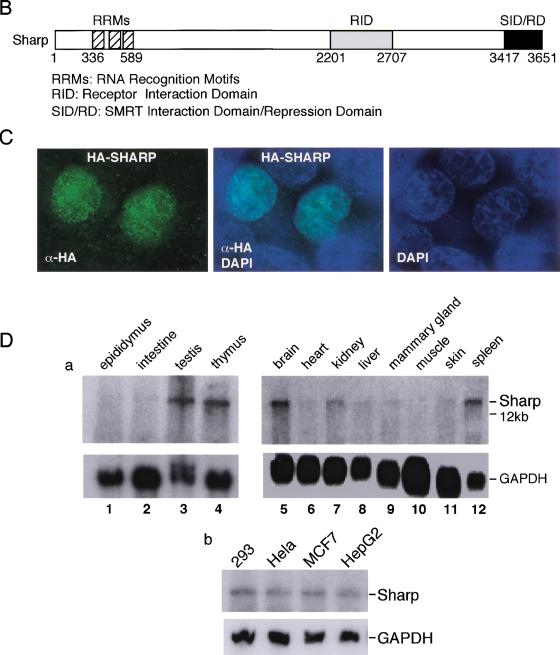
The cDNAs obtained from the yeast two hybrid screen were used as probes to screen for the full-length SHARP from a human pituitary cDNA library and a human liver cDNA library. The surprisingly large clone contains an in-frame stop codon upstream of its first methionine proceeded by a consensus Kozak sequence. The deduced amino acid sequence of SHARP is shown in Figure 2A. This 3651 amino acid polypeptide is predicted as a 400-kD protein rich in proline and serine residues. The SMRT interaction domain (SID), which is also the RD, (see below), is located at the most carboxy-terminal end of the protein (Fig. 2B). An RNA-binding domain with three RNA recognition motifs (RRMs) was predicted at its amino terminus. In addition, there are four consensus nuclear localization signals (NLSs), suggesting that SHARP is a nuclear protein. Indeed, the nuclear localization of SHARP is confirmed by an immunofluorescence assay, revealing a broad, granular staining pattern (Fig. 2C). Blast search revealed that Mint (Newberry et al. 1999), a homeodomain repressor Msx2 interacting protein, is likely a mouse homolog of SHARP. The region homologous to the Mint Msx2-interaction domain (MID) is located towards the center of the protein, corresponding to SHARP amino acid residues 2117–2451.
Figure 2.
The amino acid sequence and expression pattern of SHARP. (A) The amino acid sequence of SHARP predicted from the cDNA sequence. The amino-terminal three RRMs, the receptor interaction domain (RID, aa 2201–2707), and the carboxy-terminal SID/RD are boxed and shaded individually. The IXXI/V motifs (or its variants) in the RID are underlined. (B) Schematic representation of the SHARP functional domains. (C) Nuclear-localization of SHARP. CMX-HA-SHARP was transfected into 293 cells. The transfected cells were immuno-stained with HA-specific antibody and FITC-conjugated secondary antibody. Also shown are nuclear staining by DAPI and merged FITC and DAPI staining. No FITC staining was seen in HA antibody-incubated nontransfected cells. (D) Expression pattern of SHARP. (a) Mouse tissue Northern blots were probed with a SHARP cDNA probe. (b) Expression of SHARP in human tumor cells.
Northern blot analysis revealed that SHARP is expressed as a >12-kb transcript at relatively high levels in brain, testis, spleen, and thymus and lower levels in kidney, liver, mammary gland, and skin (Fig. 2D). SHARP was also detected in a variety of human tumor cells examined, including 293 cells (kidney), HeLa cells (cervix), MCF-7 cells (breast), and HepG2 cells (liver).
SHARP recruits histone deacetylase activity
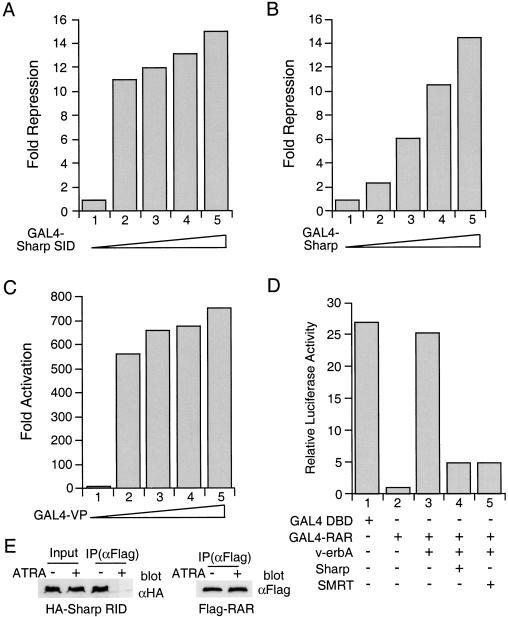
The interaction of SHARP with SMRT prompted us to examine whether SHARP is involved in transcriptional repression. We first fused the SHARP SID to the GAL4 DBD and analyzed its effect on the basal activity of the GAL4-dependent reporter in a transient transfection assay in CV-1 cells. Increasing amounts of the SHARP SID result in robust repression of basal activity (Fig. 3A). The full-length SHARP exhibits similar repression activity (Fig. 3B). In contrast, transfection of increasing amounts of GAL4-VP leads to transcriptional activation (Fig. 3C). Together these data suggest that SHARP is a potent transcriptional repressor and an RD resides in its carboxy-terminal SID.
Figure 3.
SHARP represses transcription. (A) SHARP SID represses basal transcription. Increasing amounts of GAL4–SHARP SID were transfected into CV-1 cells along with MH100-tk-luc and CMX-β-gal. Fold repression was determined relative to the activity of GAL4 DBD and represents an average of triplicate assays. (B) The full-length SHARP represses basal transcription. The experimental procedure was the same as in A except that the GAL4 fusion of full-length SHARP was used. (C) GAL4-VP does not repress basal transcription. The experimental procedure was the same as in A except that the GAL4 fusion of VP16 activation domain was used. (D) SHARP restores the repression activity of RAR. GAL4–RAR was transfected into CV-1 cells with v-erbA, a combination of v-erbA and SHARP, or v-erbA and SMRT, together with MH100-luc and CMX-β-gal. The transcriptional activity of GAL4–DBD and GAL4–RAR was determined as relative luciferase reporter activity and represents an average of triplicate assays. (E) SHARP interacts with RAR. 293 cells transfected with CMX-HA-SHARP RID and CMX-Flag-RAR were treated with or without ATRA. The cell lysates were immunoprecipitated with an anti-Flag antibody. The immunoprecipitates (IP) were immunoblotted with an anti-HA or an anti-Flag antibody. Cell lysate inputs were included in the anti-HA blot.
Having established that SHARP represses basal transcription in a GAL4 system, we next examined whether SHARP regulates nuclear receptor-mediated transcription. GAL4–RAR was transfected into CV-1 cells along with v-erbA and SHARP (Fig. 3D). GAL4–RAR represses transcription (Fig. 3D, lane 2) as expected (Chen and Evans 1995). Expression of an excess amount of v-erbA fully relieved RAR repression (Fig. 3D, lane 3), presumably by titrating out a limiting amount of free corepressors (Chen and Evans 1995). In contrast, overexpression of the full-length SHARP substantially restores RAR repression activity (Fig. 3D, lane 4), suggesting that SHARP may be present in limiting concentration. Because the SMRT interaction domain of SHARP is a potent transcriptional repressor itself, SMRT binding presumably contributes to SHARP repression.
Sequence analysis of SHARP revealed that there are several conserved IXXI/V motifs that have been shown to be receptor-interaction domains for SMRT and N-CoR. We therefore examined whether the IXXI/V motifs-containing region in SHARP (aa 2201–2707, referred to as RID) interact with the nuclear receptor RAR. Lysates from cells cotransfected with CMX-HA-SHARP RID and CMX-Flag-RAR were immunoprecipitated with an anti-Flag antibody, and analyzed for the presence of HA-SHARP in the RAR immunocomplex by Western blot analysis with an anti-HA antibody. SHARP associates with RAR in the absence of ligand, whereas addition of the RAR ligand ATRA (all-trans retinoic acid) disrupts the interaction (Fig. 3E). This result provides a molecular explanation for the rescue of RAR repression by SHARP.
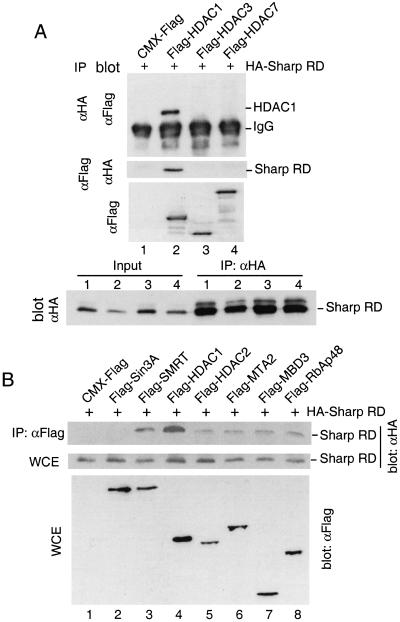
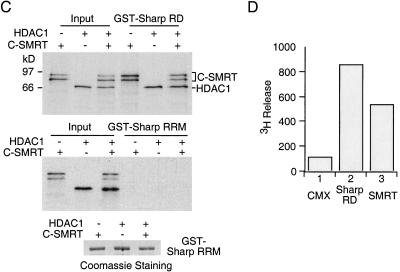
Recruitment of HDACs has been proposed to be responsible for the repression activity of SMRT (Alland et al. 1997; Hassig et al. 1997; Heinzel et al. 1997; Laherty et al. 1997; Nagy et al. 1997; Zhang et al. 1997; Huang et al. 2000; Kao et al. 2000). By analogy, we wish to determine if SHARP also associates with HDACs. Flag-tagged HDAC1, HDAC3, and HDAC7, members of two distinct HDAC families, were cotransfected with HA-tagged SHARP RD into human 293 cells. Cell lysates of the transfected cells were immunoprecipitated with HA-specific antibody and subjected to Western blot analysis for HDACs. As shown in Figure 4A, HDAC1 was readily coimmunoprecipitated with SHARP although no association with HDAC3 and HDAC7 was found. Furthermore, the same result was obtained in a reciprocal coimmunoprecipitation assay (Fig. 4A), indicating that SHARP interacts with HDAC1 specifically.
Figure 4.
SHARP is associated with SMRT and HDAC1. (A) SHARP interacts with HDAC1. Human 293 cells were transfected with CMX-Flag, or Flag tagged-HDAC1, HDAC3, and HDAC7, each along with CMX-HA-SHARP RD. Half of the cell lysates were immunoprecipitated with anti-HA agarose and the immunoprecipitates were subjected to Western blot analysis with Flag-specific antibody. Another half of the cell lysates were immunoprecipitated with anti-Flag agarose and immunoblotted with HA-specific antibody. One aliquot of the cell lysates was assayed directly by anti-Flag Western blot. An aliquot of the cell lysates and anti-HA immunoprecipitates were assayed by anti-HA Western blot. (B) SHARP is associated with components of the NuRD complex. Flag-tagged Sin3A, SMRT (short version of human SMRT, corresponding to mSMRT amino acids 1060–2473; Chen and Evans 1995), HDAC1, HDAC2, MTA2, MBD3, RbAp48, or CMX-Flag were transfected into 293 cells along with CMX-HA-SHARP RD. The cell lysates were immunoprecipitated with anti-Flag agarose and the immunoprecipitates were analyzed by Western blot with HA-specific antibody. Aliquots of the cell lysates were also assayed directly by Western blot with HA-specific antibody or Flag-specific antibody. (C) SHARP interacts with SMRT and HDAC1 in vitro. 35S-labeled in vitro translated HDAC1 and SMRT (residues 1851–2473) were incubated with purified recombinant GST–SHARP RD or GST–SHARP RRMs on glutathione beads individually, or together. The bound proteins were analyzed by SDS-PAGE and fluorography. Lower panel is Coomassie staining of the GST–SHARP RRMs protein. (D) SHARP is associated with histone deacetylase activity. Lysates prepared from cells expressing vector alone, HA-SHARP RD, or Flag-SMRT were immunoprecipitated with anti-HA or anti-Flag agarose. The immunoprecipitates were resuspended in deacetylase assay buffer for histone deacetylase assays.
To examine if SHARP interacts with other proteins, Flag-tagged HDAC1, HDAC2, MTA2, MBD3, and RbAp48, members of the NuRD complex (Zhang et al. 1999), and Flag-Sin3A were individually transfected into 293 cells along with HA-tagged SHARP RD. Flag-SMRT is included as a positive control. The cell lysates were immunoprecipitated with an anti-Flag antibody and immunoblotted with anti-HA antisera to test the presence of SHARP RD in the immuno-complex. SHARP appears to coimmunoprecipitate with at least five members of the NuRD complex (HDAC1, HDAC2, MTA2, MBD3, and RbAp48) (Fig. 4B). SMRT also interacts with SHARP as expected, although no association was detected between SHARP and Sin3A.
The interaction of SHARP with HDAC1 was further demonstrated in a GST-pull down assay (Fig. 4C). Both SMRT and HDAC1 could bind directly to the SHARP RD, either individually or together, whereas none of them bind to the SHARP RRMs (Fig. 4C). Based on its associated proteins in Figure 4A–C, the SHARP immunocomplex should possess histone deacetylase activity. Accordingly, HA-tagged SHARP RD was expressed in 293 cells, immunoprecipitated, and assayed for histone deacetylase enzymatic activity with purified 3H-labeled acetate histones. The immunocomplex displays histone deacetylase activity as measured by the release of 3H-labeled acetate (Fig. 4D), further strengthening the association with HDACs and SHARP repression.
SHARP binds the RNA coactivator SRA
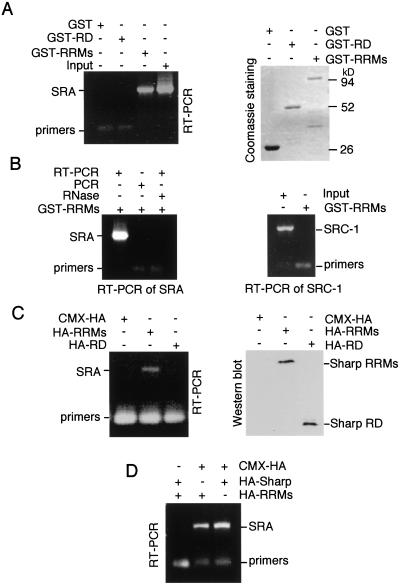
In addition to the repression domain, SHARP also contains three RNA recognition motifs (RRMs), prompting us to examine whether SHARP could interact with the steroid receptor RNA cofactor SRA (Lanz et al. 1999). SRA has been shown to potentiate steroid hormone receptor transcriptional activity as an RNA transcript. The interaction of SHARP and SRA was first examined in a GST-pull down assay. In vitro-transcribed SRA mRNA was incubated with GST–SHARP RRMs (aa 1–608) on glutathione beads. After extensive washes, the beads were added to an RT–PCR reaction using SRA-specific primers. The presence of SRA transcript on the GST–SHARP RRM beads was revealed by the production of an SRA DNA fragment on RT–PCR (Fig. 5A). No SRA transcripts were bound to GST alone or to the GST–SHARP RD fusion, despite a similar amount of proteins used in the binding assay. As controls, a no-RT–PCR reaction lacks to produce any complementary SRA DNA (Fig. 5B, left panel). Similarly, prior RNase treatment obliterates the SRA RT–PCR signal, again indicating that SHARP directly binds SRA RNA. To demonstrate the specificity of the SHARP–SRA interaction, in vitro-transcribed SRC-1 mRNA was included in a parallel GST-pull down assay followed by an RT–PCR reaction. No SRC-1 mRNA was associated with GST–SHARP RRMs even though the input RNA produces a strong RT–PCR signal (Fig. 5B, right panel). No interaction was detected between the SHARP RRMs and synthetic RNAs polyU and poly(CUG) either (data not shown). These data together suggest that the interaction of SHARP with SRA appears to be specific. The interaction of SHARP with SRA was further substantiated by an immunoprecipitation assay (Fig. 5C,D). CMX-HA-SHARP RRMs, SHARP RD, or full-length SHARP was cotransfected into 293 cells with CMV-SRA. The cell lysates were immunoprecipitated with HA-specific antibody. The immuno-complexes of HA-tagged SHARP were examined for the presence of SRA mRNA by RT–PCR. Both RRMs and full-length SHARP bound to the SRA mRNA, whereas no binding was detected between the SHARP RD and SRA. These results provide the first example in which a cofactor binds to both a coactivator and a corepressor.
Figure 5.
SHARP binds to SRA. (A) SHARP interacts with SRA in vitro. In vitro-transcribed SRA mRNA was incubated with GST, GST–SHARP RD, or GST–SHARP RRMs on glutathione beads. After extensive washes, the beads were added to a RT–PCR reaction with SRA specific primers. The RT–PCR products (700 bp) were analyzed with 1% TAE agarose gel. Right panel is Coomassie staining of an aliquot of the GST proteins. (B) Specificity of the SHARP–SRA interaction. In vitro-transcribed SRA mRNA was incubated with GST–SHARP RRMs on glutathione beads. After extensive washes, the beads were either added to a RT–PCR reaction, or to a PCR reaction without reverse transcriptase, or incubated with RNase prior to RT–PCR. In a parallel experiment, in vitro-transcribed SRC-1 mRNA was incubated with GST–RRMs and subjected to RT–PCR using SRC-1 specific primers. An aliquot of the SRC-1 RNA input was included in the RT–PCR reaction. (C) The SHARP RRMs interact with SRA in vivo. 293 cells were transfected with CMX, CMX-HA SHARP RRMs, or CMX-HA SHARP RD, along with SCT1–SRA2 (Lanz et al. 1999). The cell lysates were incubated with anti-HA agarose and the immunoprecipitates were analyzed by RT–PCR with SRA specific primers. Right panel is anti-HA Western blot of the cell lysates. (D) The full-length SHARP interacts with SRA. The experimental procedure is the same as in C except that CMX-HA full-length SHARP was transfected.
SHARP represses SRA-potentiated steroid receptor activity
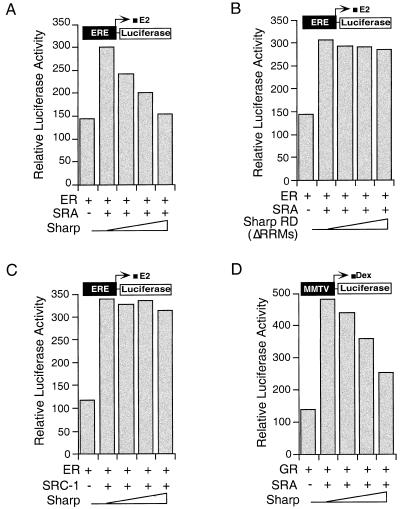
The dynamic interaction of SHARP with corepressor and coactivator prompted us to examine whether SHARP expression could inhibit SRA-stimulated steroid receptor transcriptional activity. Indeed, the full-length SHARP but not the RD repressed the SRA-potentiated ER transcriptional activity as demonstrated with an ER-responsive reporter ERE-luc (Fig. 6A,B). In contrast, no effects were seen on SRC-1-enhanced ER activity (Fig. 6C), indicating that SHARP specifically reversed SRA activation. Similar repression by SHARP was observed on SRA-stimulated GR transcriptional activity (Fig. 6D). Taken together, these results provide an unusual way in which a repressor that does not directly bind a liganded receptor may attenuate hormone action.
Figure 6.
SHARP represses SRA-potentiated ER and GR activity. (A–C) SHARP represses SRA- but not SRC-1-potentiated ER activity. CMX-ER, ERE-luc, and CMX-β-gal were transfected into CV-1 cells along with SCT1–SRA2 or CMX-SRC-1, and increasing amount of CMX–SHARP or CMX–SHARP RD (ΔRRMs). The transfected cells were treated with 10 nM 17β-estradiol (E2) for 24 h before luciferase assay. (D) SHARP represses SRA-stimulated GR activity. CMX-GR, MMTV-luc, and CMX-β-gal were transfected into CV-1 cells along with SCT1–SRA2 and increasing amount of CMX–SHARP. The transfected cells were treated with 50 nM dexamethasone (Dex) for 24 h before luciferase assay.
SHARP expression is hormone inducible
It has been shown that hormone-induced gene expression is attenuated rapidly, coinciding with a transient histone hyperacetylation at target genes (Cavailles et al. 1988; Dubik and Shiu 1988; Chen et al. 1999). Chen et al. have proposed recently that the dissociation of a coactivator ACTR from liganded-ER might account for the attenuation of histone hyperacetylation. The ability of SHARP to interact with the SRA coactivator and repress SRA-potentiated ER transcriptional activity shown above raises the possibility that hormone-induced SHARP expression could contribute to the attenuated response. In this view, it is possible that E2 activation induces the expression of a corepressor such as SHARP as part of a self-limiting response. To test this hypothesis, MCF-7 cells were treated with estrogen E2 for different time periods up to 24 h. The mRNA level of SHARP at each time point was analyzed by a Northern blot analysis. As shown in Figure 7, SHARP mRNA was induced 3-fold within 1 h following E2 treatment, and 4.2-fold after 6 h, indicating that activated ER induces SHARP expression. Interestingly, the message level of SHARP declined after 24 h treatment, suggesting that SHARP expression also attenuates with decreased E2 action. The ability of E2 to induce SHARP suggests a potential mechanism to attenuate the hormonal response.
Figure 7.
SHARP expression is estrogen-inducible. (A) MCF-7 cells were treated with E2 for indicated periods of time. Northern blot on total RNA was probed for SHARP and GAPDH. (B) Fold induction of SHARP was determined by PhosphorImager quantitation of the SHARP message level at each time point and normalized by the mRNA level of GAPDH.
Discussion
We described the isolation and functional characterization of SHARP, a 3651 aa protein, which interacts with both the nuclear receptor corepressor SMRT, HDAC1, and the coactivator SRA. SHARP exerts repression activity at least in part by recruitment of HDACs via its carboxy-terminal RD. In addition, SHARP interacts with the RNA coactivator SRA through its RRMs to repress SRA-potentiated transcriptional activity. By sequestering an activator the repressor acquires a novel means to repress agonist activation.
SHARP was originally identified as a SMRT-interacting protein in a yeast two hybrid assay. The minimal SMRT-interaction domain corresponds to the SHARP repression domain, which interacts with HDACs and components of the NuRD complex. However, no association of SHARP with Sin3A was detected in our coimmunoprecipitation assay, suggesting that the repression by SHARP is either Sin3A-independent or the interaction is unstable under our conditions. A Sin3A-independent repression has also been reported for SMRT (Huang et al. 2000; Kao et al. 2000).
Sequence analysis of SHARP revealed that there are several putative IXXI/VI motifs, the core NR-interaction sequences found in corepressors SMRT and N-CoR (Nagy at al. 1999; Perissi et al. 1999). Indeed, these motifs mediate the interaction of SHARP with unliganded RAR. The presence of NR-interacting motifs in both SHARP and SMRT and their ability to interact with each other is similar to interactions by NR-coactivators such as CBP and p160. Both CBP and p160 contain the NR-interacting LXXLL motifs and interact with each other (Chen et al. 1997; McInerney et al. 1998). Accordingly, like coactivators, corepressors could achieve a concerted regulatory effect by mutual recruitment.
Both SMRT and Sin3A are large proteins that can function as platforms for the recruitment of multi-protein complexes. By analogy, the large size (400 kD) of SHARP suggests that it might also act as a scaffold protein to recruit different protein complexes. The ability of SHARP to interact with HDACs and components of the NuRD complex supports such a notion. In addition to the hormonal response (Heinzel et al. 1997; Nagy et al. 1997), Sin3A has been shown to modulate other signaling pathways such as the Mad/Max transcriptional regulation by associating with HDACs (Ayer et al. 1995; Hassig et al. 1997). Whether SHARP is involved in additional regulatory pathways remains to be examined, but its large size could encompass many interactions. Indeed, a mouse homolog (Mint) of SHARP has been shown to interact with a homeodomain repressor Msx2 (Newberry et al. 1999). The Msx2 interaction domain (MID) of Mint is well conserved in SHARP (65% homology in amino acid sequence). It will be interesting to know whether SHARP can interact with homeodomain transcription factors and modulate their activity. Further dissection of the functional domains of SHARP will facilitate understanding of its roles in other regulatory pathways.
Other conserved sequences in SHARP include three RRMs located at its amino terminus. The RRM motif is one of the best-studied RNA binding domains, initially characterized by a 75 aa RNA binding module in proteins involved in RNA processing (Burd and Dreyfuss 1994; Siomi and Dreyfuss 1997). SHARP binds to the RNA cofactor SRA through its RRMs. Deletion mutants of SHARP lacking the RRMs (Fig. 5; data not shown) lose the interaction with SRA, suggesting that the RRMs are necessary for the SHARP-SRA association. Moreover, the interaction of SHARP and SRA appears to be specific, as SHARP does not bind to the SRC-1 mRNA.
A distinction of SHARP from other corepressors is its interaction with the coactivator SRA, independent of its association with the corepressor SMRT and HDACs. Although SRA does not bind SRC-1, it has been shown to exist in a SRC-1-containing ribonucleoprotein complex, which is recruited by steroid receptors following ligand activation (Lanz et al. 1999). Expression of SRA potentiates steroid receptor-mediated transactivation. The interaction of SHARP with SRA could therefore provide several possible means to modulate steroid receptor activity, in particular, to attenuate the hormonal response. As shown previously, estrogen-induced gene expression rapidly attenuates after the initial induction (Brown et al. 1984; Sasaki et al. 1984; Cavailles et al. 1988; Dubik and Shiu 1988), which coincides well with a transient hyperacetylation pattern of nuclear receptor target DNA (Chen et al. 1999). It has been suggested that acetylation of the coactivator ACTR leads to disruption of the receptor-coactivator interaction, which might account for the attenuation. Our results suggest that the interaction of SHARP and SRA might contribute to the repression of steroid receptor transcriptional activity by sequestering the coactivator SRA and its associated cofactor complex from the receptors, or via the ability of SHARP to recruit histone deacetylase activity. This provides an alternative but not mutually exclusive explanation for attenuation of the hormonal response.
Surprisingly, the expression level of SHARP is up-regulated by E2 treatment (Fig. 7). As shown in Figure 6, the increased level of SHARP coincided with repression of the ER promoter. In this view, the inducible expression of a corepressor might serve as a self-limiting mechanism to attenuate the hormonal response. Another example of inducible expression of a transcriptional repressor is the orphan nuclear receptor SHP. The increased expression of SHP on bile acid induction effectively inhibits bile acid synthesis by repressing CYP7A1 gene expression (Goodwin et al. 2000; Lu et al. 2000). In parallel to SHP, which keeps bile acid levels in check, SHARP might help maintain estrogen homeostasis.
A profound connection has been established between transcriptional repression and fundamental aspects of cell biology including proliferation, differentiation, and cancer. Isolation of SHARP as a novel estrogen-inducible nuclear receptor corepressor and its ability to modulate transcriptional activity of liganded ER would provide more insights into estrogen physiology and potentially its role in human diseases.
Materials and methods
Yeast two-hybrid and cDNA library screen
Yeast two-hybrid assays were carried out by the lithium acetate method (Kao et al. 2000). Mouse embryonic 17-day yeast two-hybrid library (Stratagene) and pGBT9-mSMRT (aa 2356–2473) were cotransformed into yeast strain Y190. Approximately 5 × 106 yeast transformants were screened and selected on yeast minimal medium Leu-Trp-His plates containing 40 mM 3-AT (Sigma). Colonies were picked seven days after transformation and confirmed by β-galactosidase assays. Plasmids were recovered from yeast and retransformed into yeast along with the bait construct. Positive clones were subjected to sequencing. Liquid β-galactosidase assays were carried out as described (Kao et al. 2000). A human pituitary cDNA ZAP II library and a human liver cDNA ZAP II library (Stratagene) were used to screen for the full-length SHARP cDNA sequence.
Plasmids
The plasmids CMX, CMX-GAL4 DBD, MH100-tk-luc, CMX-mSin3A, CMX-Flag-SMRTα, CMX-Flag-HDAC1, CMX-Flag-HDAC2, CMX-Flag-HDAC3, CMX-Flag-HDAC7, CMX-Flag-MBD3, CMX-Flag-MTA2, CMX-Flag-RbAp48, SCT1, SCT1-SRA2 have been described previously (Lanz et al. 1999; Downes et al. 2000). GAL4–SHARP I was constructed by subcloning a EcoRI and SalI fragment of SHARP from the pGAD4–SHARP obtained from the yeast two-hybrid screen into the same enzyme-digested CMX-GAL4 DBD vector. CMX-HA-SHARP RD was constructed by PCR of SHARP residues 3417–3651 with a carboxy-terminal HA-epitope tag and cloned into CMX vector. The full-length SHARP cDNA was assembled in the pBluescript vector and subsequently cloned into CMX-GAL4 DBD and CMX vector to get GAL4–SHARP and CMX–SHARP, respectively. To generate the SMRT LSD deletion mutants, PCR products of SMRT aa residues 2356–2456 and SMRT residues 2356–2417 were cloned into pGBT9 vector. All constructs were verified by DNA sequencing to confirm identity and reading frame.
Northern blot analysis
Total RNA from mouse tissues or cultured cells was isolated using Trizol reagent (Life Technology). Northern blot and hybridization were carried out as described (Xie et al. 2000). The probe for SHARP mRNA detection encompasses SHARP cDNA from nucleotide 8968 to 10995.
GST pull-down assay
Glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli BL21 strain and affinity purified by glutathione-sepharose 4B beads (Pharmacia). Protein–protein interaction assays were carried out by incubating GST fusion proteins with 35S-labeled in vitro-translated protein products in TEN100 buffer (Shi et al. 1998) at 4°C for 1 h. SHARP–SRA or SRC-1 RNA interaction assays were performed by incubating GST fusion proteins with in vitro-transcribed RNA in RNA binding buffer (50 mM KCl, 1.5 mM MgCl2) at 4°C for 1 h. After extensive washes, protein samples were analyzed by SDS-PAGE and visualized by fluorography. RNA samples were used as templates for RT–PCR assay.
Coimmunoprecipitation assay
Human 293 cells in a 10-cm plate were transfected with 10 μg of appropriate plasmids with Targefect F1 (Targeting Systems). Cells were harvested 48 h posttransfection and lysed in RIPA buffer (Shi et al. 1998). Cell lysates were incubated with either HA-agarose (Santa Cruz) or M2 (Flag)-agarose (Sigma) at 4°C for 2 h. After extensive washes, the agarose-bound proteins were analyzed by Western blot analysis using ECL detection system (Amersham).
Immunofluorescence assay
Human 293 cells in 6-well plates were transfected with 0.5 μg of CMX-HA-SHARP with Targefect F1. Cells were fixed 24 h posttransfection and incubated with HA-specific antibody, followed by a subsequent staining with FITC-conjugated secondary antibody.
Histone deacetylase assay
Histone deacetylase assays were performed according to Heinzel et al. (1997). Briefly, 60,000 cpm of 3H-labeled histones were incubated with immunoprecipitates for 2 h at 37°C. Reactions were stopped by addition of acetic acid/HCl to a final concentration of 0.12/0.72 N and extracted with 2 volume of ethyl acetate. Samples were centrifuged for 1 min and the supernatant was counted in a scintillation counter. Each reaction represents ∼½ of the cells in a 10-cm plate.
In vitro transcription and RT–PCR reaction
In vitro RNA synthesis was performed using T7 RNA polymerase and 2.5 mM rNTPs with linearized CMV-SRA or CMX-SRC-1 plasmid DNA as template. Following RNA synthesis, DNase treatment was performed to destroy the template DNA. RNA was recovered by ethanol precipitation after phenol:chloroform extraction. RT–PCR reaction was performed using SuperScript One-Step RT–PCR System (Life Technologies). RNA was incubated at 45°C for 30 min prior to cDNA amplification. PCR was performed as following: 2 min at 95°C, 36 cycles of 1 min at 95°C, 2 min at 56°C, and 3 min at 72°C, and a final step of 10 min at 72°C. PCR products were visualized on 1% TAE agarose gel. The SRA specific primers for RT–PCR are SRA RT1: 5′-CGC GGC TGG AAC GAC CCG CCG C-3′ and SRA RT2: 5′-AGG AGA TGG TGT CCG GTG AGT CTG-3′. The SRC-1 specific primers for RT–PCR are SRC RT1: 5′-AGA TGA CAC TAA TTC TGG AAT G-3′ and SRC RT2: 5′-CTT TGT TAT CTT TGG ACT CAG C-3′.
Acknowledgments
We thank Drs. O'Malley and Lanz for the SRA cDNA, the Sanger Centre Clone Resources for genomic clone RPCI1, and Dr. Richard Lin for reading the manuscript and helpful discussion. Lita Ong, Shardi Ghandchi, and Henry Juguilon are acknowledged for technical help, and Elaine Stevens for administrative assistance. Y.S. and W.X. are fellows of the Susan G. Komen Breast Cancer Foundation, H.Y.K. is a fellow of the Leukemia and Lymphoma Society. Ronald M. Evans is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology. This work was supported by grants from 9R01DK57978 and HD27183.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL evans@salk.edu; FAX (858) 455-1349.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.871201.
References
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Ayer DE, Lawrence QA, Eisenman RN. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Blanco JC, Minucci S, Lu J, Yang XJ, Walker KK, Chen H, Evans RM, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes & Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AM, Jeltsch JM, Roberts M, Chambon P. Activation of pS2 gene transcription is a primary response to estrogen in the human breast cancer cell line MCF-7. Proc Natl Acad Sci. 1984;81:6344–6348. doi: 10.1073/pnas.81.20.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Cavailles V, Augereau P, Garcia M, Rochefort H. Estrogens and growth factors induce the mRNA of the 52K-pro-cathepsin-D secreted by breast cancer cells. Nucleic Acids Res. 1988;16:1903–1919. doi: 10.1093/nar/16.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors [see comments] Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Downes M, Ordentlich P, Kao HY, Alvarez JG, Evans RM. Identification of a nuclear domain with deacetylase activity. Proc Natl Acad Sci. 2000;97:10330–10335. doi: 10.1073/pnas.97.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubik D, Shiu RP. Transcriptional regulation of c-myc oncogene expression by estrogen in hormone-responsive human breast cancer cells. J Biol Chem. 1988;263:12705–12708. [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes & Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression [see comments] Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor [see comments] Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Huang EY, Zhang J, Miska EA, Guenther MG, Kouzarides T, Lazar MA. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes & Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, et al. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- Kao HY, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes & Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Eisenman RN. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: The second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney EM, Rose DW, Flynn SE, Westin S, Mullen TM, Krones A, Inostroza J, Torchia J, Nolte RT, Assa-Munt N, et al. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes & Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: Cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Love JD, Li C, Banayo E, Gooch JT, Krishna V, Chatterjee K, Evans RM, Schwabe JW. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes & Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry EP, Latifi T, Towler DA. The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry. 1999;38:10678–10690. doi: 10.1021/bi990967j. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Ordentlich P, Downes M, Xie W, Genin A, Spinner NB, Evans RM. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc Natl Acad Sci. 1999;96:2639–2644. doi: 10.1073/pnas.96.6.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Staszewski LM, McInerney EM, Kurokawa R, Krones A, Rose DW, Lambert MH, Milburn MV, Glass CK, Rosenfeld MG. Molecular determinants of nuclear receptor–corepressor interaction. Genes & Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Cripe TP, Koch SR, Andreone TL, Petersen DD, Beale EG, Granner DK. Multihormonal regulation of phosphoenolpyruvate carboxykinase gene transcription. The dominant role of insulin. J Biol Chem. 1984;259:15242–15251. [PubMed] [Google Scholar]
- Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as HSF1-specific transcriptional repressors. Genes & Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. RNA-binding proteins as regulators of gene expression. Curr Opin Genet Dev. 1997;7:345–353. doi: 10.1016/s0959-437x(97)80148-7. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, et al. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Kao HY, Yao TP, McKeown M, Evans RM. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol Cell. 1999;4:175–186. doi: 10.1016/s1097-2765(00)80365-2. [DOI] [PubMed] [Google Scholar]
- Westin S, Rosenfeld MG, Glass CK. Nuclear receptor coactivators. Adv Pharmacol. 2000;47:89–112. doi: 10.1016/s1054-3589(08)60110-6. [DOI] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Downes M, Blumberg B, Simon CM, Nelson MC, Neuschwander-Tetri BA, Brunt EM, Guzelian PS, Evans RM. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406:435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai MJ, O'Malley BW. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O'Malley BW. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc Natl Acad Sci. 2000;97:6379–6384. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- Yao TP, Ku G, Zhou N, Scully R, Livingston DM. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes & Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]