Abstract
To control organ shape, plant cells expand differentially. The organization of the cellulose microfibrils in the cell wall is a key determinant of differential expansion. Mutations in the COBRA (COB) gene of Arabidopsis, known to affect the orientation of cell expansion in the root, are reported here to reduce the amount of crystalline cellulose in cell walls in the root growth zone. The COB gene, identified by map-based cloning, contains a sequence motif found in proteins that are anchored to the extracellular surface of the plasma membrane through a glycosylphosphatidylinositol (GPI) linkage. In animal cells, this lipid linkage is known to confer polar localization to proteins. The COB protein was detected predominately on the longitudinal sides of root cells in the zone of rapid elongation. Moreover, COB RNA levels are dramatically upregulated in cells entering the zone of rapid elongation. Based on these results, models are proposed for the role of COB as a regulator of oriented cell expansion.
Keywords: Arabidopsis, cell expansion, COBRA, cellulose, GPI-anchor
Because there is no morphogenetic movement of plant cells, control of the three-dimensional structure of organs is only through regulation of cell division and cell expansion. Distinct from most other eukaryotes, after division plant cells dramatically increase their size achieving volumes that can be hundreds of times their original size (Cosgrove 1997). For plant organs to attain their final morphology and function properly, constituent cells must regulate tightly the way in which they expand. The orientation and extent of an individual cell's expansion are key parameters in determining its size and shape, yet little is known about the molecular mechanisms that regulate either aspect of cell expansion.
Although cell expansion is driven by cell turgor, all evidence indicates that neither water flow nor solute influx to maintain turgor is the primary determinant of the extent or direction of cell expansion (Pritchard 1994); rather, the plant cell wall is believed to be the regulator of both. The plant cell wall comprises an array of para-crystalline cellulose microfibrils, which are associated with cross-linking glycans (e.g., hemicellulose) and embedded in a matrix of pectin and small amounts of protein (McCann and Roberts 1991; Carpita and Gibeaut 1993). The polysaccharides of the growing plant cell wall are mostly long-chained polymers that form a cohesive network through noncovalent lateral associations and physical entanglements (Cosgrove 1999). The cell wall's ability to withstand enormous osmotic pressure while readjusting the arrangement of these constituent polymers appears to be critical to the expansion process.
Regulation of the direction in which a cell expands involves oriented control of cell wall extension as well as polarized deposition of new wall materials (Carpita and Gibeaut 1993). Biophysical considerations indicate that there must be a component in the expanding walls that resists the osmotic pressure, thereby channeling the direction of cell elongation. Most evidence points to cellulose microfibrils as the primary load-bearing component of the expanding cell wall performing this function (Pritchard 1994). In many cell types, cellulose microfibrils have been shown to be oriented perpendicular to the primary direction of expansion and to be analogous to hoops around a barrel (Green 1980; Giddings and Staehelin 1991). Therefore, to regulate the orientation of cell expansion, the cell must be able to control the deposition and spatial organization of cellulose microfibrils as well as to rearrange bonds to allow the wall to yield to or resist osmotic pressure.
Unlike pectins and cross-linking glycans, which are made in the cytoplasm and transported out to the wall via the Golgi apparatus (Gibeaut and Carpita 1994), cellulose microfibrils are synthesized at the cell membrane/cell wall interface (Delmer 1999). The cellulose microfibrils that are spooled around plant cells are generated by multimeric protein complexes in the plasma membrane commonly referred to as the “terminal complexes” or “particle rosettes.” About three dozen individual polymer chains of (1→4)-β-d-glucans are synthesized and subsequently crystallized into a microfibril (Delmer 1999). The process of microfibril crystallization may be facilitated by a subunit of the rosette complex (Delmer and Amor 1995). Recently, genes have been identified that are involved either in the synthesis of cellulose (Pear et al. 1996; Turner and Somerville 1997; Arioli et al. 1998; Taylor et al. 1999; Fagard et al. 2000) or of noncellulosic polysaccharide components at the Golgi apparatus or involved in their secretion to the wall (Lukowitz et al. 1996; Bonin et al. 1997; Nicol et al. 1998; Edwards et al. 1999; Perrin et al. 1999; Gibeaut 2000). Genes and their products have also been identified that function in rearranging bonds in the cell wall to allow for extensibility (Cosgrove 1999). In particular, expansins are a family of proteins involved in the disruption of the noncovalent bonds between cellulose microfibrils and cross-linking glycans, causing rapid induction of wall extension (McQueen-Mason and Cosgrove 1994). However, regulation of the orientation or extent of expansion is still poorly understood at the molecular level.
To further understand the molecular mechanisms involved in cell expansion, we have cloned the COBRA (COB) gene and determined its pattern of expression in roots. Initially, cobra, a member of the conditional root expansion (CORE) class of mutants, was isolated in a screen for Arabidopsis seedlings with abnormally expanded roots (Benfey et al. 1993). The phenotype of all CORE mutants is conditional on the root growing in the presence of high concentrations of sucrose or other conditions that stimulate rapid root growth (Hauser et al. 1995). Root cells in cob appear to be expanded more in a radial than in a longitudinal orientation while maintaining cell volume, indicating a role for COBRA in regulating the orientation of cell expansion. Here we report that COB encodes a putative GPI-anchored protein that is localized primarily in the plasma membrane of the longitudinal sides of root cells. We propose models based on the mutant phenotype, the COB RNA expression pattern, and COB's subcellular localization in which COBRA plays a key role in determining the orientation of cell expansion.
Results
Mutations in COBRA result in misoriented cell expansion
Generation of cell files in the Arabidopsis root requires initial cells and their immediate progeny to go through a continuous process of division and expansion in the growing region of the root. Cell divisions displace cells upward in the cell file. In wild type, cells undergo two distinct expansion phases (Fig. 1A). The first involves both longitudinal and radial expansion, whereby the ultimate root radius is established (Fig. 1C). The second phase is marked by a cessation of radial expansion and a dramatic increase in longitudinal expansion (Fig. 1C; Beemster and Baskin 1998). The region in which cells undergo primarily longitudinal expansion is known as the elongation zone. In striking contrast to wild type, cells in this region of the cob mutant do not undergo highly polarized, longitudinal expansion (Fig. 1B); rather, these cells expand radially to a far greater extent than those of the wild type (Fig. 1D). Although the cob phenotype is most obvious in the epidermal cell layer, where the cell volume is approximately the same as wild type, cells in the cortex and endodermis also expand more radially and less longitudinally than in wild type (Hauser et al. 1995). The three alleles of cob, which we have isolated, have a nearly identical phenotype. Although initially characterized as a semidominant mutant, the penetrance of the semidominant phenotype requires very specific growth conditions; therefore, cob generally behaves as a fully recessive mutation. There is no apparent phenotype in the aerial part of the cob mutant when grown in light. There appears to be a slight decrease in hypocotyl length when grown in the dark.
Figure 1.
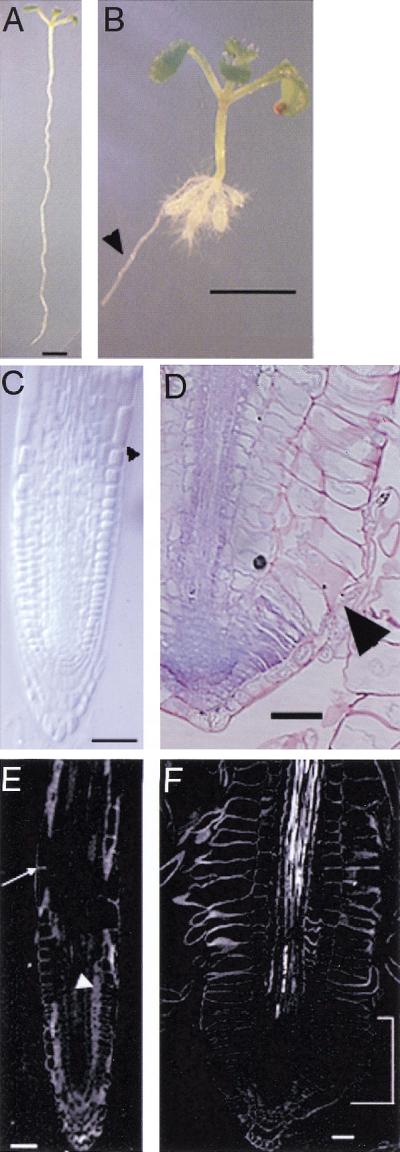
Phenotypic analysis of cobra. (A) Wild-type and (B) cob-1 seedlings oriented vertically grown for 10 d on nutrient agar medium containing 4.5% sucrose. The conditional nature of cob can be seen in the lateral root, which has grown into the agar, thereby slowing its growth (arrowhead). (C) Cleared whole mount of wild-type root viewed with Nomarski optics. Arrowhead indicates the region where rapid longitudinal expansion begins. (D) Longitudinal section of cob root stained with toluidine blue. Abnormal lateral expansion is most apparent in the epidermis (arrowhead). (E,F) Polarized-light micrographs. (E) In wild type, strong retardance is observed in cell walls throughout the root, both in cross-sectioned walls (arrow) and in longitudinal faces of cells in the plane of section (arrowhead). (F) In cob-1, there is little retardance detected in the growing region of the root, in either the crosswalls or the longitudinal faces (bracket), indicating a reduction in crystalline cellulose microfibrils in this region. Bars in A and B = 0.25 cm. Bars in C–F = 50μm.
There is a conditional defect in cellulose production in cobra
Because cellulose microfibrils are thought to orient the direction of cell expansion, we investigated whether the role of COBRA in polar longitudinal expansion is mediated through cellulose deposition. Cell walls were isolated from wild type and cob roots. Quantification of crystalline cellulose was determined as cell wall material resistant to acid hydrolysis. Measurements of five independent replicates showed that there was a highly significant difference (P < 0.003) between wild-type roots, which had a mean value of 133 ± 28 μg cellulose (mg cell wall)−1, and cob roots which had a mean value of 89 ± 2 μg cellulose (mg/cell wall) (Table 1). The decreased amount of cellulose in the root tissue suggests that COB directly or indirectly plays an important role in cellulose deposition.
Table 1.
Crystalline cellulose analysis. Cellulose was defined as cell wall material resistant to hydrolysis in 2 M trifluoroacetic acid. The cellulose was converted to mono- and oligosaccharides by Saeman hydrolysis, and determined by the phenol-sulfuric acid assay (Dubois et al. 1956)
| Sample
|
Average OD (490 nm)
|
Glucose quantity* (μg)
|
Cell wall starting material (mg)
|
Glucose quantity (μg)/cell wall starting material (mg)
|
|---|---|---|---|---|
| Wild type A | 0.757 | 224.58 | 1.8 | 124.77 |
| Wild type B | 0.656 | 194.1 | 1.7 | 114.17 |
| Wild type C | 0.682 | 202.01 | 1.4 | 144.29 |
| Wild type D | 0.276 | 79.79 | 0.5 | 159.58 |
| Wild type E | 0.429 | 125.84 | 1.0 | 125.84 |
| cobra A | 0.494 | 145.33 | 1.7 | 85.49 |
| cobra B | 0.496 | 145.63 | 1.7 | 85.66 |
| cobra C | 0.406 | 118.69 | 1.4 | 84.78 |
| cobra D | 0.170 | 47.58 | 0.5 | 95.16 |
| cobra E | 0.169 | 47.35 | 0.5 | 94.70 |
To determine whether there was any spatial specificity to the loss of cellulose in the root, we used a polarized light microscopy, which detects the presence of crystalline cellulose on a microscopic scale. Under polarized light, the amount of birefringent retardance is a measure of the amount of crystalline cellulose microfibrils in the light path as well as their average degree of alignment (Preston 1974). To assess the retardance of cell walls in cob, longitudinal sections of wild-type and mutant roots were observed through a microscope that uses circularly polarized light and digital imaging to produce images in which the intensity at each pixel is linearly proportional to the retardance, regardless of the orientation of the optical axis of the crystallites (Oldenbourg and Mei 1995). Thus, cell walls with aligned microfibrils will appear brighter than background for any direction of net alignment. In differentiated regions of the root, retardance in wild type and cob appeared similar; however, in the growing region, the genotypes differed, with retardance in cob being almost undetectable (cf. Fig. 1E,F). The decreased retardance indicates a deficit in the amount of crystalline cellulose or in its net alignment. However, a decrease in alignment is unlikely as the exclusive explanation because retardance decreased not only in walls lying in the plane of the section but also in cross-sectioned walls (cf. cross walls in Fig. 1E,F). A cell wall viewed edge on (as are the cross walls in Fig. 1) and containing random microfibrils gives rise to detectable retardance, because all microfibrils are parallel to the wall and, therefore, possess a net alignment from the point of view of light propagating through the wall from edge to edge.
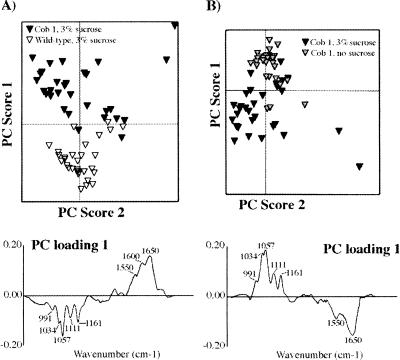
To confirm that the reduction in retardance was due to a loss in cellulose, we used Fourier transform infrared (FTIR) microspectroscopy to detect specific cell wall macromolecules and their orientation by their absorbance of infrared (IR) radiation (McCann et al. 1993). Because the identified alleles of cob are conditional, of interest was the state of cellulose under both restrictive and permissive conditions. Cell walls were prepared from 40 excised roots of wild-type and cob-1 plants, grown in the presence or absence of 3% sucrose. In each case, the pellet of cell walls was spread thinly onto a barium fluoride window, dried, and areas of 50 by 50 μm were selected for analysis by FTIR microspectroscopy (McCann et al. 1997). Thirty spectra were obtained from different areas of the barium fluoride window in each case and were compared by exploratory principal component analysis (PCA) (Kemsley 1998), a statistical method that reduces the dimensionality of the data from more than a hundred variates (one every 8 cm−1 from 1800 to 850 cm−1) to only a few principal components (PCs). The PCs are ordered in terms of decreasing variance. Each observation (spectrum) has a corresponding set of PC scores, which describes the variance of that spectrum relative to the mean of the population for each PC. The PC scores of the spectra can then be plotted against one another to reveal patterns or structure in the data (Kemsley 1998).
It is possible to mathematically derive a “spectrum” (called a PC loading) from a PC to identify molecular factors responsible for the separation of groups of spectra (Chen et al. 1998; Kemsley 1998). The analysis showed that spectra from wild-type plants grown on sucrose can be separated from spectra of cob1 grown on sucrose by PC1, accounting for 78% of the total variance in the combined populations (Fig. 2A). The loading for PC1 depicted in Figure 2A showed characteristics of crystalline cellulose in the fingerprint region (peaks at 991, 1034, 1057, 1111, and 1161 cm−1 (Tsuboi 1957; Liang and Marchessault 1959), and of protein (peaks at 1650 and 1550 cm−1). The former peaks are negatively correlated with the latter. Because the PC scores of spectra of cob1 cell walls were negative relative to the mean, the data suggest that the cell walls of cob1 are relatively richer in protein and poorer in cellulose than wild-type cell walls when the plants have been grown in the presence of sucrose. However, the increase in protein may derive from some cytoplasmic contamination of the cell walls.
Figure 2.
FTIR analysis of cell walls prepared from wild-type and cob1 roots grown in the presence or absence of sucrose. Exploratory PCA (see Materials and Methods) was performed using 30 FTIR spectra from each population. (A) PC1 clearly discriminates wild-type spectra from cob1 spectra when the plants have been grown in the presence of 3% sucrose. Axes represent the mean values for the population. The corresponding loading (below) has features characteristic of cellulose and protein (peaks of interest are marked). Cellulose peaks are negatively correlated, indicating that cob1 is cellulose-deficient relative to wild type. (B) PC1 discriminates between spectra obtained from cob1 roots grown in the presence or absence of sucrose. The corresponding loading also has features of cellulose and protein (peaks of interest are marked). Cellulose peaks are positively correlated indicating that cob1 grown in the presence of sucrose is cellulose-deficient relative to cob1 grown in the absence of sucrose.
Similarly, a comparison between PC loadings obtained from cob1 cell walls grown in the presence or absence of sucrose revealed that plants grown in the presence of sucrose were deficient in crystalline cellulose but had increased protein relative to plants grown in the absence of sucrose (Fig. 2B). A slight but consistent difference in cellulose content was observed between wild type and cob1 grown in the absence of sucrose (data not shown) but of much less magnitude than when the plants were grown in the presence of sucrose. In summary, these data show that the conditional phenotype of cob1 grown in the presence of sucrose is cellulose deficient relative to wild-type plants or to cob1 plants grown in the absence of sucrose. These data suggest the possibility of functional redundancy under slower growth conditions, which could compensate at least partially for the loss of COB function.
Map-based cloning of COBRA
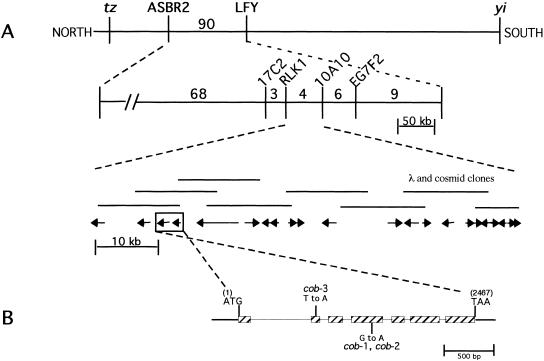
To better understand COB's role in cell expansion, we employed a map-based cloning approach to isolate the COB gene. Three point crosses were used to map the cobra locus to a region on chromosome 5 near the LEAFY locus. Molecular analysis of recombinant plant lines allowed us to map the COBRA locus between the molecular markers RLK1 and 10A10 (Fig. 3A). This region had been physically mapped and covered by a contig of two cosmids and six lambda clones (Bender and Fink 1998). Subsequently, the sequence of chromosome 5 was determined through the Arabidopsis Genome Initiative, allowing cobra to be localized within a 74 kb region. To identify the COBRA gene, we sequenced from the cob-1 mutant each of the open reading frames identified in this region. A missense mutation was found in one open reading frame, and we then found mutations in the other two cob alleles within the predicted gene (Fig. 3B). Both cob-1 and cob-2 have an identical mutation, suggesting that this may be a mutational hot spot. However, we cannot rule out the possibility that these are siblings of the same mutant stock. We note that because the molecular lesions in all cob alleles are missense mutations, these alleles may not represent the null phenotype, even though their phenotypes are almost identical.
Figure 3.
Cloning of COB and analysis of its deduced amino acid sequence. (A,B) Summary of the chromosome walk. (A) Initial mapping placed COB near the LFY locus. Three point crosses using yi tz and cob-1 plants identified crossover events between the CAPS markers ASBR2 and LFY (vertical lines). Analysis of internal CAPS markers placed COB in the 74 kb interval between the RLK1 and the 10A10 CAPS markers. (Numbers represent the number of recombinants isolated between the markers shown). The 22 putative genes or open reading frames in this region were sequenced from the cob-1 background until a mutation was revealed in a putative gene (shown in the boxed region; arrows indicate the direction of transcription). (B) Genomic organization of COB (hatched boxes represent exons). Positions of the translational start (ATG) and stop (TAA) of the predicted coding sequence are shown. The position and nucleotide changes in the mutant alleles are also shown. (C) The predicted amino acid sequence is shown directly below the cDNA sequence. Numbers to the right refer to the positions of nucleotides or amino acid residues. Triangles represent the positions of introns. Missense point mutations in cob-1, cob-2, and cob-3 alleles are shown above the nucleotide sequence. Underlined is the putative cleavable N-terminal signal sequence (as determined with pSORT [http://psort.nibb.ac.jp/]). Underlined and in bold is the motif, which in addition to the N-terminal signal sequence and hydrophilic central portion, appears to meet all of the sequence requirements for processing and GPI linkage and the GPI prediction site (http://mendel.imp. univie.ac.at/). In bold are the amino acids, which would be changed in the mutant alleles. (D) The GPI linkage motif in COB includes the predicted cleavage (ω) site, which is the residue to the left of the cleavage (arrow). ω +1 and ω +2 sites immediately following the cleavage site, which are part of a six amino acid spacer region (boxed), and a 19 amino acid hydrophobic tail (see text for consensus residues). (E) Hydropathy analysis of COB shows the hydrophobic N- and C-terminals and the hydrophilic central portion.
Rescue of the cob mutant phenotype (both in cob-1 and cob-3) was achieved by transformation of either the genomic region or the COB cDNA driven by the Cauliflower Mosaic Virus 35S promoter, which is constitutively active in most plant tissues (Odell et al. 1985). Genomic DNA blot analysis and database searching of the complete genome sequence indicated that there are no genes highly related to COB in Arabidopsis (data not shown). However, database comparisons have identified several genes with similar sequences in other plant species but not in other kingdoms (see Materials and Methods). This may indicate that COB is a member of a small plant-specific gene family that has been conserved in plant evolution.
COB encodes a putative GPI-anchored protein
Alignment of the full-length cDNA sequence with genomic sequence indicates that COB is composed of six introns and seven exons (Fig. 3C). The deduced COB gene product is 454 amino acids in length and contains several motifs indicative of subcellular localization (Fig. 3C). These include an N-terminal signal sequence for secretion, a highly hydrophobic C-terminal, a hydrophilic region in between both termini, and a stretch of residues found in proteins in which the C-terminal is processed and replaced with a glycosylphosphatidylinositol (GPI) anchor (Udenfriend and Kodukula 1995a).
The attachment of a GPI moiety to a protein is predicted by conserved amino acid residues at the C-terminal. The site of attachment has been termed ω and only Ser, Asn, Ala, Gly, Asp, and Cys are found at this site, while only Ala, Gly, Thr, and Ser are found at the ω + 2 site (Udenfriend and Kodukula 1995b). The ω + 2 residue of known GPI-anchored proteins is usually followed by a spacer of five to seven amino acids rich in charged and/or Pro residues followed by a stretch of 10–30 hydrophobic residues. COBRA meets all of these requirements (Fig. 3D). Although there is only one highly charged residue following the ω + 2 residue, this was also found to be true of a bona fide GPI-linked arabinogalactan protein recently isolated from Nicotiana alata (Youl et al. 1998).
Proteins destined to be GPI anchored contain hydrophobic sequences at both their N- and C-terminals, with the remainder of the protein sequence being essentially hydrophilic (Udenfriend and Kodukula 1995a). A hydropathy plot of the deduced amino acid sequence of COBRA confirms these three motifs (Fig. 3E). Apart from their C-terminal domains specifying GPI attachment, the protein moieties attached by GPI anchors have little in common structurally or functionally (Thompson and Okuyama 2000).
Expression of COBRA RNA and subcellular localization of COBRA protein in the root
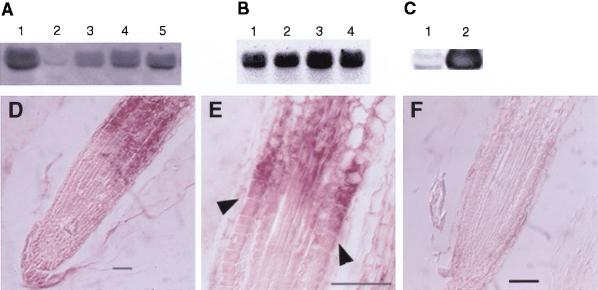
To determine if there is organ-specific expression of COB, RNA blot analysis was performed. COB RNA was found in all organs tested, with significantly lower levels in siliques (Fig. 4A). Expression in the mutant alleles appeared similar to wild type (Fig. 4B). As would be expected, plants overexpressing COB contained greatly elevated levels of COB mRNA (Fig. 4C).
Figure 4.
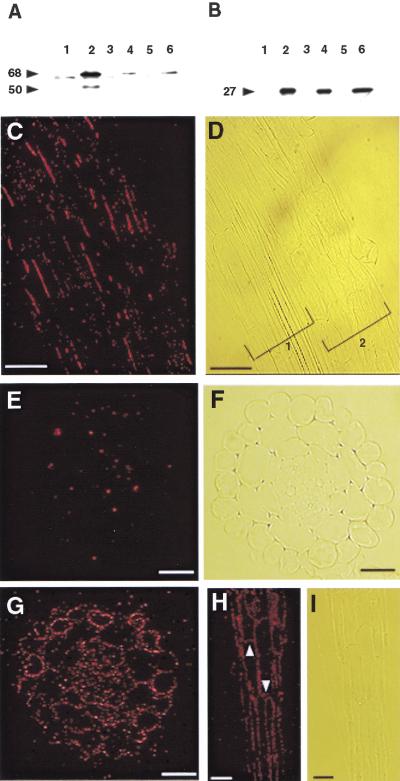
COB expression analysis. (A–C) RNA blot analysis. Total RNA from different organs or plants was isolated and RNA blot analysis was performed with a digoxigenin-labeled COB probe. Equal loading was assessed using ethidium bromide stained 18S and 25S rRNA (data not shown). (A) flowers (lane 1), siliques (lane 2), stems (lane 3), leaves (lane 4), and roots (lane 5). (B) wild-type Columbia (lane 1), cob-1 (lane 2), cob-2 (lane 3), and cob-3 (lane 4). (C) Wild-type WS (lane 1), and 35S::COB transgenic in cob-3 (lane 2). The exposure time for the blot in C was significantly shorter than in A and B to highlight the different expression levels. (D–F) In situ hybridization analysis of COB gene expression in wild-type Columbia roots. Root longitudinal sections were probed with either anti-sense (D,E) or sense (F) digoxigenin-labeled, in vitro-transcribed RNA from a portion of COB. Upregulation of COB occurs in cells that are undergoing rapid longitudinal expansion (arrowheads in E). Bar, 50 μm.
To analyze the cell specificity of COB expression, RNA in situ hybridization was performed on longitudinal root sections. Strikingly, expression levels increased dramatically in cells in the elongation zone (Fig. 4D). In fact, expression appeared to increase abruptly as cells entered the zone of rapid longitudinal expansion (Fig. 4E).
To determine the localization of COB protein both at the cellular and subcellular level, we raised polyclonal antisera to a portion of COB fused to the maltose-binding protein. The antisera, after affinity purification, were tested against soluble and microsomal protein fractions, the identity of the latter verified by the presence of a tonoplast marker, α-TIP (Fig. 5B). The antiCOB serum recognized a polypeptide with an apparent molecular mass of 68 kD in the microsomal membranes (Fig. 5A). Although the predicted size of COB based on its amino acid sequence is 49 kD, modifications, such as the potential GPI anchor, may alter its gel mobility. In plants that overexpressed COB, we detected a minor species of polypeptide at ∼50 kD, not detected in wild type, which may represent the unmodified protein (Fig. 5A).
Figure 5.
COB protein localization in roots. (A,B) Immunoblot analysis. Even numbered lanes are total microsomal membrane fractions, odd numbered lanes are soluble protein fractions. Plants analyzed: 35S::COB transgenic (lanes 1,2), wild-type Columbia (lanes 3,4), and cob-1 (lanes 5 and 6). (A) Affinity purified COB antibodies. (B) Antibodies to the tonoplast specific α-TIP. Arrowheads indicate size in kD. (C–I) COB immunolocalization in root tissue sections by epifluoresence microscopy. Roots were incubated with affinity purified anti-COB antibodies and fluorescently labeled secondary antibodies (red signal). In wild type, binding was detected predominately on portions of the longitudinal sides of cells in all cell layers (C), corresponding light image is in (D) (brackets 1 and 2 denote two separate roots). The signal was not continuous around the circumference of the cells, but rather it was found in patches, shown in transverse section (E), corresponding light microscope image is in (F). Analysis of plants overexpressing COB (35S::COB genomic) detected the protein in a more uniform pattern in transverse sections (G), and localized it to the apical and basal sides of cells (white arrowheads) (H) as well as to the longitudinal sides, corresponding light image in (I). G–I are rescued cob-3 mutants). Scale bars in E, F, and G = 25 μm;. Scale bars in C, D, H, and I = 50 μm.
To immunolocalize COB, we incubated the affinity-purified antibodies with longitudinal (Fig. 5C) and transverse (Fig. 5E) sections of wild-type roots. We consistently observed far greater binding of the antibodies to the longitudinal sides of the cells than to the apical or basal sides. Moreover, the signal was detected primarily in discrete regions along the longitudinal cell surfaces, which is consistent with the animal data indicating that GPI linkage can direct proteins to microsomal rafts located on specific sides of polarized cells (Rodriguez-Boulan and Powell 1992).
Consistent with the RNA expression pattern, protein was undetectable in the distal region of the root tip (data not shown). Antibody binding was first detected somewhat proximal to the region in which RNA is first detected, which probably reflects the time required for detectable amounts of protein to accumulate at the cell surface.
Ectopic expression of COB
In transgenic plants containing the 35S promoter driving COB cDNA, we detected binding of antiCOB antibodies to the apical and basal cell surfaces as well as to the longitudinal sides of root cells (Fig. 5G,H). Ectopic expression of COB did not appear to alter the expansion profile of these cells. Moreover, the ectopic expression of COB in cells at the root tip, which do not normally contain COB, did not alter the phenotype in a detectable fashion. The rescue of the cob mutant phenotype in the context of this overexpression suggests that COB is necessary, but not sufficient for highly oriented cell elongation.
Discussion
To achieve correct organ morphology, plants must regulate the orientation and extent of cell expansion. Here we report that cobra, a member of the CORE class of expansion mutants, synthesizes lower amounts of cellulose in the root, with a concomitant altered orientation of cell expansion. The marked reduction in cellulose in the cob root tip, coupled with the lateral expansion of root cells in the mutant, suggest that COB's regulation of oriented cell expansion is associated with the deposition of cellulose.
COB is expressed in cells undergoing highly oriented longitudinal expansion
COB RNA is upregulated markedly in cells in the root elongation zone. In fact, there appears to be a good correlation between increased COB expression and cells entering the zone of rapid longitudinal expansion. This suggests that COB function is not required until the transition to rapid longitudinal expansion. This would argue against a role for COB as a general factor required for cell expansion. Rather, the expression pattern is consistent with a role for COB in regulating oriented cell expansion. COB appears to be expressed in all tissue layers, which correlates with abnormal expansion throughout the root (Benfey et al. 1993).
Although COB RNA levels are greatly upregulated in the expansion zone, the mutant phenotype extends to the tip of the root. One possible explanation is that the root is capable of altering its overall architecture in response to individual cell shape changes. Support for this hypothesis comes from condition shift experiments. The first cells that expand aberrantly are those entering the zone of rapid longitudinal expansion. At later time points, cells closer to the root tip progressively begin to expand (Hauser et al. 1995).
COBRA encodes a putative GPI-anchored protein which is polarly localized
The importance of covalent lipid modification as a means of regulating protein activity or cellular localization is becoming increasingly apparent (Thompson and Okuyama 2000). One such lipid modification is the addition of a GPI anchor to a protein. GPI lipid substitutions have been shown to anchor a diverse group of secreted proteins in the plasma membrane of protozoa, yeast, and animals, possibly increasing their lateral mobility (Rodriguez-Boulan and Powell 1992). While some GPI-anchored proteins have been documented in plants (Schultz et al. 1998; Sherrier et al. 1999), none have so far been associated with mutant phenotypes. The COBRA gene product contains a putative GPI anchor as suggested by its amino acid sequence and its hydropathy plot.
The presence of a GPI anchor is frequently associated with polar protein sorting in animal cells (Matter and Mellman 1994). There is evidence that GPI anchors act as sorting signals in the Golgi to target proteins specifically into apical portions of the cell membrane in both neuronal and epithelial cells of mammals (Rodriguez-Boulan and Powell 1992). Recently, it was also shown in animal cells that not only are GPI proteins specifically targeted within the membrane, but also that they are organized into microdomains at the cell surface (Friedrichson and Kurzchalia 1998; Varma and Mayor 1998).
Immunolocalization of COB protein indicates that it is primarily localized in discrete regions along the longitudinal cell surfaces in the root. If it is demonstrated that this is dependent on the GPI-anchor motif, this would provide evidence for a shared protein sorting signal between plants and animals.
Evidence from animals and plants indicates that some GPI-anchored proteins can have the lipid moiety cleaved and thus release the polypeptide, which could then act as a diffusible signal. From the protein blot analysis, COB was found primarily in the microsomal fraction, which was consistent with the immunolocalization results. Nevertheless, we cannot rule out the possibility that some COB protein is cleaved from its putative GPI anchor.
Isolation of a partial COB cDNA was reported to complement a Saccharomyces pombe mutant defective in phytochelatin synthesis (Leuchter et al. 1998). Phytochelatins are small oligopeptides produced enzymatically from glutathione, which are necessary to protect cells from divalent heavy metals, such as cadmium, by binding and rendering them insoluble and nontoxic in the cytoplasm (Ortiz et al. 1992). The partial COB cDNA, which was able to confer resistance to cadmium in S. pombe, did not contain the first 94 amino acids of COB. These missing amino acids contain the putative N-terminal signal sequence as well as the domain in which the cob-3 mutation is found. Although this reported phytochelatin synthesis activity may not be directly related to COB's in planta function, it is possible that COB binds divalent metals, and this feature could have functional relevance.
COBRA's role in aerial organs
COB RNA is present in stems, leaves, flowers, and siliques, yet the cob alleles we have isolated show no apparent light-grown shoot phenotype. One explanation for the lack of a phenotype in the aerial portions of cob mutants is that we do not have a null allele even though the three alleles (representing two different mutations) are phenotypically almost identical. Alternatively, there may be genes with functions redundant to COB in the aerial organs.
The absence of an aerial phenotype may also be due to cell expansion being controlled by different genetic programs in different organs. The angustifolia and rotundifolia3 mutations result in abnormal expansion only in leaves and floral organs (Kim et al. 1998). Furthermore, these mutations provide evidence that polar expansion may be regulated by factors that control expansion only in specific directions. The angustifolia mutant has defective cell expansion in the leaf-width direction, resulting in thicker cells, while the rotundifolia3 mutant affects cell elongation in the leaf-length direction. The ROTUNDIFOLIA gene was shown to encode a member of the P450 cytochrome gene family (Kim et al. 1998), although the precise mechanism for its function remains to be determined. Another possibility is that the rate of cell expansion in the shoot is generally less than that of the root and that COB function is only required in cells elongating at very high rates.
The conditional nature of the cobra mutation
Under permissive conditions (slower growth), the roots of cob appear to be similar to wild type. We have determined that under these conditions there are nearly wild-type levels of cellulose as well as COB RNA (data not shown). Moreover, the conditional phenotype is lost in double-mutant combinations with other CORE loci (quill, lion's tail, pompom-1, and pompom-2) (Hauser et al. 1995). The recent identification of LION'S TAIL as encoding a member of the endo-1,4–β-D-glucanase (EGase) family, allelic to KORRIGAN (Nicol et al. 1998) and QUILL as a member of the cellulose synthase gene family allelic to PROCUSTE (Fagard et al. 2000) suggests a possible explanation for the conditional phenotype. These proteins may act as part of complex and/or in the process of cellulose deposition and confer a degree of functional redundancy.
Overexpression of COB
Expression of COB under the control of the constitutive 35S promoter was able to rescue the cob mutant phenotype. However, expression from this construct in wild-type plants did not result in any detectable phenotype. This result suggests the existence of other proteins that are necessary for COB function, and these cofactors are limiting either spatially or quantitatively.
When driven by the 35S promoter, COB levels are elevated and COB protein is now found more uniformly distributed on the cell surface. The generation and maintenance of distinct cell surface polarity involves extensive molecular sorting (Matter and Mellman 1994) and perhaps the mechanisms that localize COB cannot keep up with the overproduction in these transgenic lines. The mislocalization without any aberrant phenotype is further evidence that COB is necessary, but not sufficient to cause a shift in expansion.
Possible models for COB's role in oriented cell expansion
The specification of cell shape requires regulation of the production and arrangement of cell wall constituents. The deficit in cellulose in cobra indicates that the COBRA gene product is either directly or indirectly involved in cellulose synthesis.
However, the role of COB is likely to be more complicated than simply providing a component of the cellulose deposition machinery. The cob phenotype is distinct from the phenotypes of the mutants rsw1 (Arioli et al. 1998) and procuste (Fagard et al. 2000), in which the affected genes encode catalytic subunits of cellulose synthase, or of korrigan, in which the affected gene encodes an endo-glucanase that is involved in cellulose synthesis (Nicol et al. 1998). The cob phenotype suggests that COB functions specifically in the transition between the arrest of radial expansion and the initiation of the rapid longitudinal expansion phase. We suggest two possible models for COB function. The localization of COB to the longitudinal surfaces of expanding cells in a nonuniform fashion raises the possibility that COB acts to recruit cellulose synthesizing complexes to discrete positions on the cell surface. This could allow for highly polarized longitudinal expansion in the root. Alternatively, COB may act through an as yet undefined mechanism to alter the orientation of cell expansion by resisting radial wall expansion and/or promoting longitudinal wall expansion. In this model, the effect on cellulose deposition in the mutant would be indirect, resulting from insufficient numbers of cellulose synthesizing complexes being recruited to keep pace with the outward expansion of the cell.
Materials and methods
Growth of plants
Arabidopsis plants were grown as described previously (Benfey et al. 1993). Except as noted, plant growth medium was supplemented with 4.5% sucrose, and the pH (5.7) was adjusted after the addition of agar. cob-1 and cob-2 are in the Columbia ecotype and cob-3 is in the Wassilewskija (W.S.) ecotype.
Cellulose analysis
Five-day-old seedling root tissue weighing 2–5 g were frozen at −80°C, ground in a glass-glass grinder and incubated in 95% ethanol for 30 min at 65°C. Samples were cooled to room temperature and pelleted (5000 rpm). Pellets were washed twice in 95% ethanol and then extracted overnight in methanol/chloroform (2/3 v/v). Extracted material was pelleted and washed five times in 95% ethanol. The cell wall pellet was dried overnight at 65°C and then freeze dried. Wall material (0.5–1.8 mg) was hydrolyzed in 1 mL of 2 M trifluoroacetic acid at 120°C for 90 min. The undigested material, mostly crystalline cellulose, was pelleted (4000 rpm). The cellulosic material was digested to glucose monomers by Saeman hydrolysis as described in Shatalov et al. (1999), and total sugar was determined by the phenol-sulfuric method (Dubois et al. 1956).
Polarized light microscopy
To assess birefringent retardance of the cell walls, roots were fixed and embedded in plastic as described below and sectioned at 3 μm thickness. Retardance was assessed on a polarized-light microscope (Jenapol, Zeiss) equipped for circularly polarized light quantitative digital imaging (LC pol Scope, Cambridge Research Instruments).
For plastic embedding, 1-2-wk-old seedlings were fixed in 4% paraformaldehyde in PBS overnight at 4°C and rinsed two times in PBS. The roots were subsequently pre-embedded in 1% agarose, dehydrated in ethanol, and infiltrated with Historesin (Technovit 7100, Kulzer). Plastic sections were mounted on Superfrost slides (Fisher). In Figure 1D, the root was stained by submersion in 0.05% toluidine blue for 3 min.
FTIR spectroscopy
Cell wall material was prepared by excising roots from 40 plants of each of Columbia and cob1 grown on 3% or no sucrose. The plants were ground in an Eppendorf tube using a miniature homogenizer. The homogenate was centrifuged at 13,000 rpm for 10 min and the supernatant discarded. The pellet was resuspended in distilled water and washed three times. The final pellet was resuspended in 100 μL of distilled water and pipetted onto a barium fluoride window.
The wet pellets of cell walls were dried at 37%°C for 1 h on barium fluoride windows (Crystran Ltd.). The windows were supported on the stage of a UMA500 microscope accessory of a Bio-Rad FTS175c FTIR spectrometer equipped with a liquid nitrogen-cooled mercury cadmium telluride detector. An area of wall (50×50 μm) was selected for spectral collection in transmission mode. Collected were 128 interferograms with 8 cm−1 resolution and co-added to improve the signal-to-noise ratio for each sample. Thirty spectra were collected from different areas of each window. All data sets were baseline corrected and area normalized before statistical methods were applied. Exploratory PCA was carried out using Win-Discrim software (Kemsley 1998). Reference IR absorption spectra of cellulose were used for peak assignments (Tsuboi 1957; Liang and Marchessault 1959).
Cloning of COB
Initial analysis of 120 F2 plants from a cob-1 (Col) x wild type (Ler) cross-positioned the COB locus on the distal portion of chromosome 5, 0.63 cM north of the LEAFY (LFY) cleaved amplified polymorphic sequence (CAPS) molecular marker (Konieczny and Ausubel 1993). Initial genetic data indicated that COB was contained in the approximate 15 cM region between the YI (yellow inflorescence) and TZ (thiazole requiring) loci. Three point crosses between the yi tz double mutant (Ler) and cob-1 (Col) were preformed to generate recombination events on both sides of the COB gene. Thirteen F1 plant lines were propagated to the F2 generation and ∼16,000 plants were scored for the yi phenotype in the absence of exogenous thiamine (needed by tz plants). A total of 383 plants were scored in this category, which is indicative of a recombination event between yi and tz.
Because the mutations in cob-1(cob-2) and cob-3 created polymorphisms with NlaIV and AluI, respectively, we were able to confirm the mutations by PCR of genomic DNA and restriction analysis. PCR amplification and digestion were performed at two independent times.
Southern blotting was performed as described (Sambrook et al. 1989). Arabidopsis genomic DNA was isolated according to the procedure described by (Ausubel et al. 1987). For detection of DNA, nucleic acids were immobilized onto nylon membranes by UV crosslinking and were hybridized with a single-stranded digoxigenin (DIG)-labeled probe made using PCR (Finckh et al. 1991).
The hydropathy plot was generated using the Weizmann Institute of Sciences Genome and Bioinformatics Web site (http://bioinformatics.weizmann.ac.il/hydroph/plotfft_hydroph.html) with Kyte and Doolittle parameters and a window size of 17 amino acids.
RNA analysis
RNA was isolated using a phenol extraction protocol (Jackson and Larkins 1976) or the RNeasy kit (QIAGEN). For RNA blot analysis, prehybridization, hybridization, and detection were performed as described in Di Laurenzio et al. (1996). Blots were hybridized with a single-stranded digoxigenin (DIG)-labeled probe (Finckh et al. 1991). To make the probe, a single-stranded 975 bp antisense COB DNA fragment was created by PCR (bp 1688–2663, including 64 bases of the 3′ untranslated region).
In situ hybridization analysis was performed as described in Di Laurenzio et al. (1996). To generate COB-specific antisense and sense RNA probes, a PCR-amplified DNA fragment of the COB cDNA (base pairs 220–766) was inserted into pCR2.1 (Invitrogen). After digestion with BamHI, a fragment (bp 361–766) was subcloned into the BamHI site of pBluescript II SK (+) (Stratagene) in both orientations. After linearizing each construct with NotI, in vitro transcription and DIG-labeling using the DIG RNA Labeling Kit (Boehringer Mannheim) was performed.
35S::COB transgene and Agrobacterium-mediated transformation
The 35S::COB cDNA and genomic constructs were made by placing sequences between the 35S promoter and the nopaline synthase polyadenylation sequence. To make the 35S::COB cDNA construct, the full-length cDNA was excised from λPRL2 (EST 163D2T7, GenBank accession no. R29979) as a HindIII, EcoRI fragment, subcloned into pBluescript II KS (+), excised by XbaI, XhoI and inserted into the corresponding sites of the plasmid V7 (from Dr. T. Brears). The 35S::COB genomic construct was made using a 4.1 kb HindIII fragment (containing the entire COB genomic sequence + 2 kb downstream) of a λ clone from the chromosome walk, subcloned into the HindIII site of pBluescript II KS (+). The fragment was excised by digestion with HindIII, XbaI and inserted into the corresponding sites of the plasmid W104 (from Dr. T. Brears). Expression plasmids were transformed into cob-1 and cob-3 plants by the floral dipping method (Clough and Bent 1998).
Production of COB-specific polyclonal antiserum
To generate COB-specific polyclonal antibodies, we ligated, after PCR amplification and addition of a 5′ EcoRI site and a 3′ HindIII site, a fragment of the COB cDNA encoding amino acids 67 through 193 into the bacterial expression vector pMAL-c2 (New England BioLabs). After expression in Escherichia coli strain BL21, the recombinant protein was affinity purified on an amylose resin column (New England BioLabs), according to manufacturer's instructions, and injected into two New Zealand White rabbits (Covance). Polyclonal antisera were affinity purified against the recombinant COB protein coupled to NHS-Activated Sepharose 4 fast flow (Amersham Pharmacia Biotech), according to manufacturer's instructions.
Immunolocalization
Microsome fractions were prepared as in Jinn et al. (2000). Isolation of microsomes was assessed using the microsome-specific anti α-TIP antibody (Johnson et al. 1990). Protein samples were resolved by SDS-PAGE as in Laemmli (1970). Under restrictive conditions, 10- to 14-d-old plants were grown (Benfey et al. 1993), fixed, embedded, and sectioned at 8 μm. Antibody incubation and immunohistochemistry were performed as described (Di Laurenzio et al. 1996) with the following modifications: Root sections were incubated with affinity-purified antiCOB at room temperature for 1 h. Incubation with secondary antibodies conjugated to Alexa Fluor 568 (Molecular Probes, 1:250) was done at ambient temperature for 45 min to 1 h. Goat serum was added to all incubations (1:50, v/v).
Acknowledgments
We thank J. Bender for clones from the contig and unpublished CAPS markers, which expedited COB cloning, J. Rogers for his generous gift of the α-TIP antisera, J. Malamy for guidance, support and the wild-type image in Figure 1C. We also thank S. Pagant, H. Höfte, and members of his lab, D. Shoue and M. Madson from the Carpita lab for helpful discussions, and members of the Benfey lab for critically reading the manuscript. This project was funded in part by a grant to T.I.B. from the U.S. Department of Energy (award no. 94ER20146), which does not constitute endorsement by that department of views expressed herein, funding in part to N.C. from the USDA-NRICGP. M.C.M. is funded by a Royal Society University Research Fellowship and P.D. acknowledges the support of a BBSRC special studentship. This project was funded primarily from a grant to P.N.B. from the NSF. G.S. was supported by a Dean's Dissertation Fellowship from the Graduate School of Arts and Science at New York University.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
GenBank accession nos. of clones most homologous to COB are: cotton, AI730765; rice, C74834; soybean, AI416924; tomato, AI773156; maize, AI861761; aspen, AI166150; Brassica napus, H74542; Medicago truncatula, AW559872; Lotus japonicus, AW720520; ice plant, BE130595; Loblolly pine, AA556615.
The COBRA GenBank accession no. is AF319663. The truncated COB cDNA clone, which was isolated based on its ability to complement a S. pombe mutant deficient in phytochelatin synthesis, has the accession no. AJ006787 (Leuchter et al. 1998).
Footnotes
E-MAIL philip.benfey@nyu.edu; FAX (212) 995–4204.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.879101.
References
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seiden JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: John Wiley; 1987. [Google Scholar]
- Beemster GT, Baskin TI. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 1998;116:1515–1526. doi: 10.1104/pp.116.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J, Fink GR. A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc Natl Acad Sci. 1998;95:5655–5660. doi: 10.1073/pnas.95.10.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis. Development. 1993;119:57–70. doi: 10.1242/dev.119.Supplement.57. [DOI] [PubMed] [Google Scholar]
- Bonin CP, Potter I, Vanzin GF, Reiter WD. The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D- mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc Natl Acad Sci. 1997;94:2085–2090. doi: 10.1073/pnas.94.5.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Chen L, Carpita NC, Reiter WD, Wilson RH, Jeffries C, McCann MC. A rapid method to screen for cell-wall mutants using discriminant analysis of Fourier transform infrared spectra. Plant J. 1998;16:385–392. doi: 10.1046/j.1365-313x.1998.00301.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol. 1997;13:171–201. doi: 10.1146/annurev.cellbio.13.1.171. [DOI] [PubMed] [Google Scholar]
- ————— Enzymes and other agents that enhance cell wall extensibility. Annu Rev Plant Phys. 1999;50:391–417. doi: 10.1146/annurev.arplant.50.1.391. [DOI] [PubMed] [Google Scholar]
- Delmer DP. Cellulose biosynthesis: Exciting times for a difficult field of study. Annu Rev Plant Phys. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- Delmer DP, Amor Y. Cellulose biosynthesis. Plant Cell. 1995;7:987–1000. doi: 10.1105/tpc.7.7.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles K, Hamilton J, Rebers P, Smith F. A colorimeteric method for the determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- Edwards ME, Dickson CA, Chengappa S, Sidebottom C, Gidley MJ, Reid JSG. Molecular characterisation of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J. 1999;19:691–697. doi: 10.1046/j.1365-313x.1999.00566.x. [DOI] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 2000;12:2409–2424. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finckh U, Lingenfelter PA, Myerson D. Producing single-stranded DNA probes with the Taq DNA polymerase: A high yield protocol. Biotechniques. 1991;10:35–36. [PubMed] [Google Scholar]
- Friedrichson T, Kurzchalia TV. Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature. 1998;394:802–805. doi: 10.1038/29570. [DOI] [PubMed] [Google Scholar]
- Gibeaut D M. Nucleotide sugars and glycosyltransferases for synthesis of cell wall matrix polysaccharides. Plant Physiol Biochem. 2000;38:69–80. [Google Scholar]
- Gibeaut DM, Carpita NC. Biosynthesis of plant cell wall polysaccharides. FASEB J. 1994;8:904–915. doi: 10.1096/fasebj.8.12.8088456. [DOI] [PubMed] [Google Scholar]
- Giddings THJ, Staehelin LA. Microtubule-mediated control of microfibril deposition: A re-examination of the hypothesis. In: Lloyd CW, editor. The Cytoskeletal basis of plant growth and form. London: Academic; 1991. pp. 85–99. [Google Scholar]
- Green PB. Organogenesis—A biophysical view. Ann Rev Plant Physiol. 1980;31:51–82. [Google Scholar]
- Hauser MT, Morikami A, Benfey PN. Conditional root expansion mutants of Arabidopsis. Development. 1995;121:1237–1252. doi: 10.1242/dev.121.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AO, Larkins BA. Influence of ionic strength, pH, and chelation of divalent metals on isolation of polyribosomes from tobacco leaves. Plant Physiol. 1976;57:5–10. doi: 10.1104/pp.57.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes & Dev. 2000;14:108–117. [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Hofte H, Chrispeels MJ. An intrinsic tonoplast protein of protein storage vacuoles in seeds is structurally related to a bacterial solute transporter (GIpF) Plant Cell. 1990;2:525–532. doi: 10.1105/tpc.2.6.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemsley EK. Discriminant analysis and modelling of spectroscopic data. Chichester, UK: John Wiley; 1998. Chemometric methods for classification problems; pp. 1–47. [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes & Dev. 1998;12:2381–2391. doi: 10.1101/gad.12.15.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leuchter R, Wolf K, Zimmermann M. Isolation of an Arabidopsis cDNA complementing a Schizosaccharomyces pombe mutant deficient in phytochelatin synthesis. Plant Physiol. 1998;117:1526. [Google Scholar]
- Liang CY, Marchessault RH. Infrared spectra of crystalline polysaccharides II. Native celluloses in the region from 640 to 1700 cm−1. J Polym Sci. 1959;39:269–278. [Google Scholar]
- Lukowitz W, Mayer U, Jurgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Matter K, Mellman I. Mechanisms of cell polarity: Sorting and transport in epithelial cells. Curr Opin Cell Biol. 1994;6:545–554. doi: 10.1016/0955-0674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- McCann MC, Roberts K. Architecture of the primary cell wall. In: Lloyd CW, editor. The cytoskeletal basis of plant growth and form. London: Academic Press; 1991. pp. 109–129. [Google Scholar]
- McCann MC, Stacey NJ, Wilson R, Roberts K. Orientation of macromolecules in the walls of elongating carrot cells. J Cell Sci. 1993;106:1347–1356. doi: 10.1242/jcs.106.4.1347. [DOI] [PubMed] [Google Scholar]
- McCann M, Chen L, Roberts K, Kemsley EK, Séné C, Carpita NC, Stacey NJ, Wilson RH. Infrared microspectroscopy: Sampling heterogeneity in plant cell wall composition and architecture. Physiol Plant. 1997;100:729–738. [Google Scholar]
- McQueen-Mason S, Cosgrove DJ. Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci. 1994;91:6574–6578. doi: 10.1073/pnas.91.14.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Hofte H. A plasma membrane-bound putative endo-1,4-b-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 1998;17:5563–5576. doi: 10.1093/emboj/17.19.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua NH. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature. 1985;313:810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- Oldenbourg R, Mei G. New polarized light microscope with precision universal compensator. J Microsc. 1995;180:140–147. doi: 10.1111/j.1365-2818.1995.tb03669.x. [DOI] [PubMed] [Google Scholar]
- Ortiz DF, Kreppel L, Speiser DM, Scheel G, McDonald G, Ow DW. Heavy metal tolerance in the fission yeast requires an ATP-binding cassette-type vacuolar membrane transporter. EMBO J. 1992;11:3491–3499. doi: 10.1002/j.1460-2075.1992.tb05431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K. Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science. 1999;284:1976–1979. doi: 10.1126/science.284.5422.1976. [DOI] [PubMed] [Google Scholar]
- Preston RD. The physical biology of plant cell walls. London: Chapman & Hall; 1974. pp. 75–108. [Google Scholar]
- Pritchard J. The control of cell expansion in roots. New Phytol. 1994;127:3–26. doi: 10.1111/j.1469-8137.1994.tb04255.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Powell SK. Polarity of epithelial and neuronal cells. Annu Rev Cell Biol. 1992;8:395–427. doi: 10.1146/annurev.cb.08.110192.002143. [DOI] [PubMed] [Google Scholar]
- Sambrook F, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schultz C, Gilson P, Oxley D, Youl JJ, Bacic A. GPI-anchors on arabinogalactan-proteins: Implications for signalling in plants. Trend Plant Sci. 1998;3:426–431. [Google Scholar]
- Shatalov AA, Evtuguin DV, Neto CP. (2-O-a-D-galactopyranosyl-4-O-methyl-a-D-glucurono)-D-xylan from Eucalyptus globulus. Labill. Carbohydr Res. 1999;320:93–99. doi: 10.1016/s0008-6215(99)00136-6. [DOI] [PubMed] [Google Scholar]
- Sherrier DJ, Prime TA, Dupree P. Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis. 1999;20:2027–2035. doi: 10.1002/(SICI)1522-2683(19990701)20:10<2027::AID-ELPS2027>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell. 1999;11:769–780. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GA, Jr, Okuyama H. Lipid-linked proteins of plants. Prog Lipid Res. 2000;39:19–39. doi: 10.1016/s0163-7827(99)00014-4. [DOI] [PubMed] [Google Scholar]
- Tsuboi M. Infrared spectrum and crystal structure of cellulose. J Polym Sci. 1957;25:159–171. [Google Scholar]
- Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udenfriend S, Kodukula K. How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu Rev Biochem. 1995a;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- ————— Prediction of omega site in nascent precursor of glycosylphosphatidylinositol protein. Methods Enzymol. 1995b;250:571–581. doi: 10.1016/0076-6879(95)50098-7. [DOI] [PubMed] [Google Scholar]
- Varma R, Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- Youl JJ, Bacic A, Oxley D. Arabinogalactan-proteins from Nicotiana alata and Pyrus communis contain glycosylphosphatidylinositol membrane anchors. Proc Natl Acad Sci. 1998;95:7921–7926. doi: 10.1073/pnas.95.14.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]