Abstract
Intracellular pH affects protein structure and function, and proton gradients underlie the function of organelles such as lysosomes and mitochondria. We engineered a genetically-encoded pH sensor by mutagenesis of the red fluorescent protein mKeima, providing a new tool to image intracellular pH in live cells. This sensor, named pHRed, is the first ratiometric, single-protein red fluorescent sensor of pH. Fluorescence emission of pHRed peaks at 610 nm while exhibiting dual excitation peaks at 440 nm and 585 nm that can be used for ratiometric imaging. The intensity ratio responds with an apparent pKa of 6.6 and a greater than 10-fold dynamic range. Furthermore, pHRed has a pH-responsive fluorescence lifetime that changes by ~0.4 ns over physiological pH values and can be monitored with single wavelength two-photon excitation. After characterizing the sensor, we tested pHRed’s ability to monitor intracellular pH by imaging energy-dependent changes in cytosolic and mitochondrial pH.
Intracellular pH plays a vital role in cell biology. pH modulates protein structure and function, and organelles such as lysosomes and mitochondria require proton gradients to function properly. Intracellular pH is also coupled to processes such as the cell cycle and apoptosis.1,2 Currently, there are several small-molecule sensors of pH, but these indicators can suffer from leakage from cells and poor control over subcellular targeting.3 There are also several genetically-encoded pH sensors based on green fluorescent protein (GFP) variants or fluorescence resonance energy transfer between two proteins.4 However, for multicolor imaging these GFP-based pH sensors have limited compatibility with GFP-based sensors of other analytes because of overlapping excitation or emission bands.5 For example, we are interested in imaging neuronal energy metabolism with GFP-based genetically-encoded ATP sensors.6,7 Changes in energy metabolism often correlate with pH changes, and it would be useful to monitor both ATP and pH simultaneously. Critically, the ATP sensors, like many GFP-based probes,8–10 are also pH sensitive. Thus, monitoring pH dynamics is also necessary to correct for pH artifacts. Unfortunately, there are no GFP-based pH sensors with wavelength requirements that are compatible with those of the ATP sensors. Although the development of red fluorescent proteins (RFPs) has greatly improved prospects for multicolor imaging,5,11,12 until now there were no ratiometric RFP-based pH sensors.13
To this end, we developed pHRed, a genetically-encoded sensor engineered from the long Stokes shift RFP mKeima. Excited-state proton transfer (ESPT)14,15 was recently described in mKeima,16 suggesting it could serve as a scaffold for engineering a sensor that is excitation ratiometric.9,17 In the development of mKeima, an early-stage precursor exhibited two pH-dependent fluorescence excitation peaks at 452 and 580 nm, and the 452 nm peak was attenuated with four mutations: S61F, I92T, F158Y and S213A.16 Of these mutations, the side chains of residues 92 and 158 are exterior facing, and residues 61 and 92 were further mutagenized in subsequent stages of development. Residue 158 is adjacent to D157, a residue important for ESPT;14,15 however, the F158Y mutation is sterically conservative. In contrast, the side chain of residue 213 projects towards the chromophore,14,15 and therefore we rationalized that reversing the S213A mutation would be a good starting point for recovering the 452 nm peak. Indeed, the mKeima-A213S mutant exhibits dual excitation peaks that respond ratiometrically to pH (Figure 1a). Because of proximity to the chromophore, we also conducted random mutagenesis of residues 60 and 61, but no improved variants were identified. The mKeimaA213S mutant, which we have named “pHRed”, has proven useful for monitoring pH, and in this report we characterize the pH-dependent photophysical properties of pHRed. The fluorescence response of this pH sensor purified in solution correlated well with its response when expressed in live cells, and we also imaged energy dependent changes in mitochondrial and cytosolic pH of live cells.
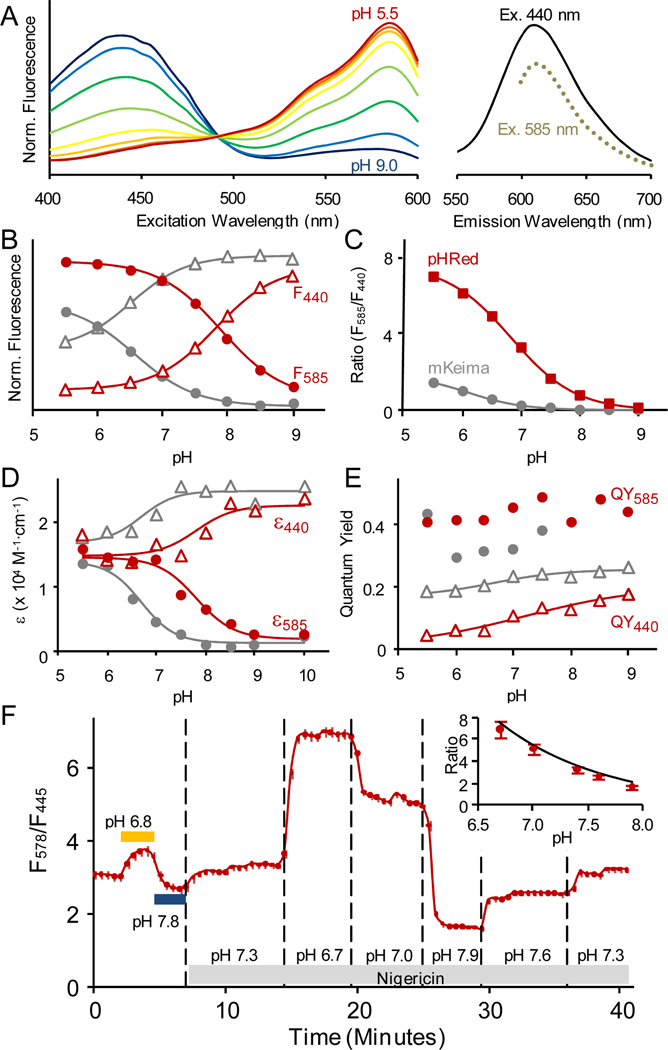
Figure 1.
(a) Fluorescence excitation and emission spectra of purified pHRed in solution. pH response of the (b) 440 nm and 585 nm excitation peak intensities with 620 nm emission, (c) the F585/F440 ratio, (d) extinction coefficients, and (e) quantum yields. (a–b) Fluorescence intensity is normalized to total integrated intensity. (b–e) mKeima data is shown in grey for comparison. (f) pHRed reports intracellular pH in live Neuro2A cells. Changes in extracellular pH without permeabilization caused minor changes to intracellular pH (horizontal bars). The protonophore nigericin was used to manipulate intracellular pH. n=43 cells, bars indicate standard error. Inset: pH calibration in cells agrees well with purified protein (line).
We first characterized pHRed purified in solution. As for mKeima, the “reverse” pH dependence14 of pHRed’s fluorescence response is reminiscent of the behavior of the GFP-derived pHluorins,9 and it is opposite the trend observed for avGFP.4 In mKeima the reverse pH dependence is a result of chromophore interactions with S142 and D157.14 The carboxylate of D157 stabilizes the protonated chromophore in a trans conformation, but under acidic conditions in which D157 is protonated, hydrogen bonding with S142 stabilizes the anionic chromophore in a cis conformation.14 However, at alkaline pH D157 may also stabilize the cis-protonated chromophore, and the role of the isomerization is not clear.15 Furthermore, residues S142 and D157 also provide a proton transfer network for ESPT following excitation of the protonated chromophore, and excitation of either ionization state results in red fluorescence emission.
pHRed’s peak fluorescence emission occurs at 610 nm, and there are excitation peaks at 440 nm (protonated neutral chromophore) and 585 nm (anionic chromophore) (Figure 1a).4,14 Acidification from pH 9 to 6 causes a 7-fold increase in the 585 nm peak intensity (F585) and a 4-fold decrease in the 440 nm peak intensity (F440), and both peaks respond with a pKa of 7.8 (Figure 1b). The intensity ratio (F585/F440) increases >10-fold with acidification and responds with a lower pKa of 6.6 because the 585 nm peak has a greater absolute intensity change (Figure 1c, Supplemental Figure S1). The ratio response is also insensitive to differences in buffer ion composition (K+, Na+, Cl−, Mg2+, Ca2+, HCO3−), oxidative stress (H2O2, dithiothreitol), and temperature (21°C to 37°C) (Supplemental Figure S1). Hence, pHRed is a specific sensor for monitoring pH.
To compare its brightness to mKeima with excitation at 440 and 585 nm, we measured the extinction coefficients (ε440 and ε585) and quantum yields (QY440 and QY585) of pHRed (Supplemental Figures S2 and S3, Supplemental Table S1). At pH 7.5, pHRed’s ε440 is 1.7-fold lower and ε585 is 3.4-fold larger compared to mKeima. Likewise, pHRed’s QY440 is 1.8-fold lower and the QY585 is 1.3-fold larger. As a result, at pH 7.5 the brightness (ε · QY) of pHRed is 3-fold lower than mKeima at 440 nm but 4-fold higher at 585 nm.
The pH dependence of ε440 and ε585 correlates well with the differing pKa’s of the fluorescence intensity responses of pHRed (pKa ~ 7.8) and mKeima (pKa ~ 6.6) (Figure 1d). Interestingly, pHRed’s QY585 is not strongly pH dependent, but the QY440 increases 3-fold between pH 6 and 9 (Figure 1e). The QY440 of mKeima is also pH dependent, increasing 1.4-fold from pH 6 to 9. pHRed generally has a lower QY440 than mKeima, but the difference decreases as pH increases. This might suggest that mKeima, with a lower pKa, is closer to saturation in its QY440 pH response, but it may also be that the A213S mutation simply increases the rate of non-radiative decay of the 440 nm absorbing neutral chromophore in pHRed. The pH dependence of the QY440 itself is not likely due to a pH-dependent change in ESPT efficiency because we do not observe any green emission, as would be expected if pH-dependent ESPT were a mechanism (Supplemental Figure S4).18
The spectroscopic data suggest that the A213S mutation also increases the apparent pKa of pHRed relative to mKeima. It is possible that a serine hydroxyl at position 213 could interact with the adjacent carboxylate of E211. The crystal structures of mKeima14,15 show that E211 directly interacts with the chromophore’s imidazolinone moiety, and E211 may indirectly interact with S142 and D157 via waters and R193. This might provide a network by which a hydrogen bonding interaction between S213 and E211 could cause a pKa shift.
The purified protein characterizations demonstrate that pHRed is better suited than mKeima for intracellular pH sensing. pHRed has two well-matched peak intensities that facilitate ratio imaging, and it has a larger ratio response dynamic range that is also better matched to the physiological pH range. Therefore, we next characterized the pH response of the intensity ratio when pHRed is expressed in live cells. Heterologously expressed in Neuro2A cells, pHRed reports intracellular pH. To manipulate the intracellular pH, live cells were permeabilized using the K+/H+-ionophore nigericin and incubated in buffer with high extracellular KCl.19 The pH dependence of pHRed’s intensity ratio in cells correlated well with the pH response of purified protein in solution (Figure 1f).
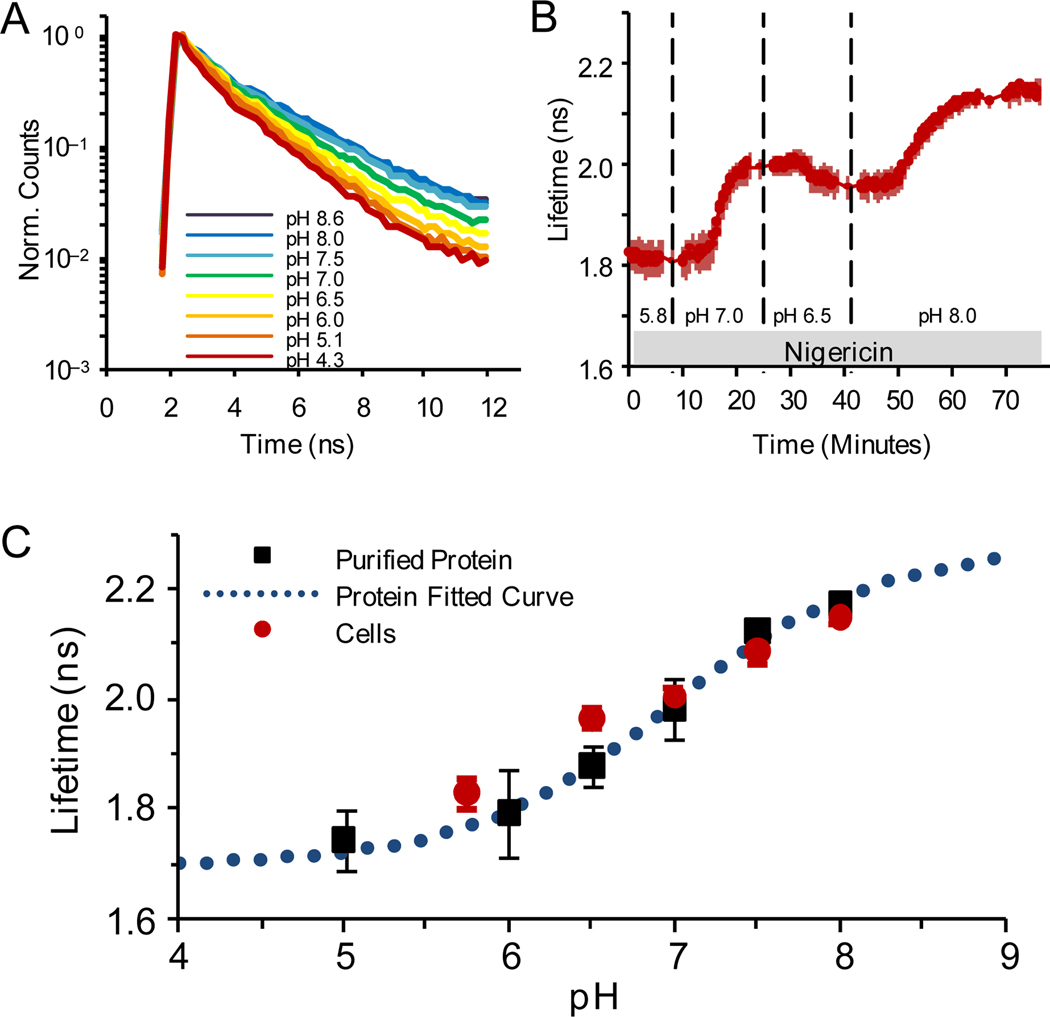
We also found that pHRed exhibited a pH-dependent fluorescence lifetime that could be used to image intracellular pH using fluorescence lifetime imaging microscopy (FLIM) with near-infrared two-photon excitation. Imaging thick samples such as brain slices is greatly hindered by light penetration and light scatter, but this problem can be overcome by the use of two-photon excitation at infrared wavelengths.17,20 However, two-photon excitation profiles are generally broad, and collecting an intensity ratio for pHRed would be challenging given the long wavelength of pHRed’s second excitation peak. FLIM offers an alternative solution. Purified pHRed in solution can be excited in two-photon mode at a single wavelength of 860 nm,17 and its fluorescence lifetime decreased upon acidification (Figure 2a). To test its pH response in cells, we again used the KCl-nigericin method to manipulate the intracellular pH of live Neuro2A cells expressing pHRed (Figure 2b). We found that the pH response of the fluorescence lifetime of pHRed expressed in cells correlated well with that of purified protein in solution (Figure 2c). The fluorescence lifetime of pHRed increased upon changing from pH 5 to 8 with a half-maximal response at pH 6.9 ± 0.2 (n=6), making pHRed a well-tuned fluorescence lifetime sensor for intracellular pH. Furthermore, pHRed’s lifetime changed by ~0.4 ns, a substantial lifetime difference that can be well detected in FLIM.21,22 As with intensity ratios, the fluorescence lifetime is independent of expression levels. Thus, pHRed is a versatile probe that could be used to monitor pH in both thin and thick samples.
Figure 2.
pH response of pHRed fluorescence lifetime (630 nm emission) with 860 nm two-photon excitation. (A) pH response of peak normalized fluorescence lifetime decays of purified pHRed in solution. (B) Intracellular pH in live Neuro2A cells imaged with FLIM. The nigericin method was used to manipulate pH. (C) pH response of pHRed fluorescence lifetime in cells (n=6) and protein in solution (n=3) in solution agreed well with an apparent pKa of 6.9 ± 0.2, similar to the F575/F440 intensity ratio response.
To test the utility of pHRed for live-cell microscopy, we used pHRed in two experimental regimes related to cellular energy metabolism: first, pHRed was targeted to the mitochondrial matrix and used to probe mitochondrial function; second, pHRed was used in conjunction with the sensor Perceval6 to simultaneously monitor intracellular ATP and pH, demonstrating energy-dependent changes in cytosolic pH.
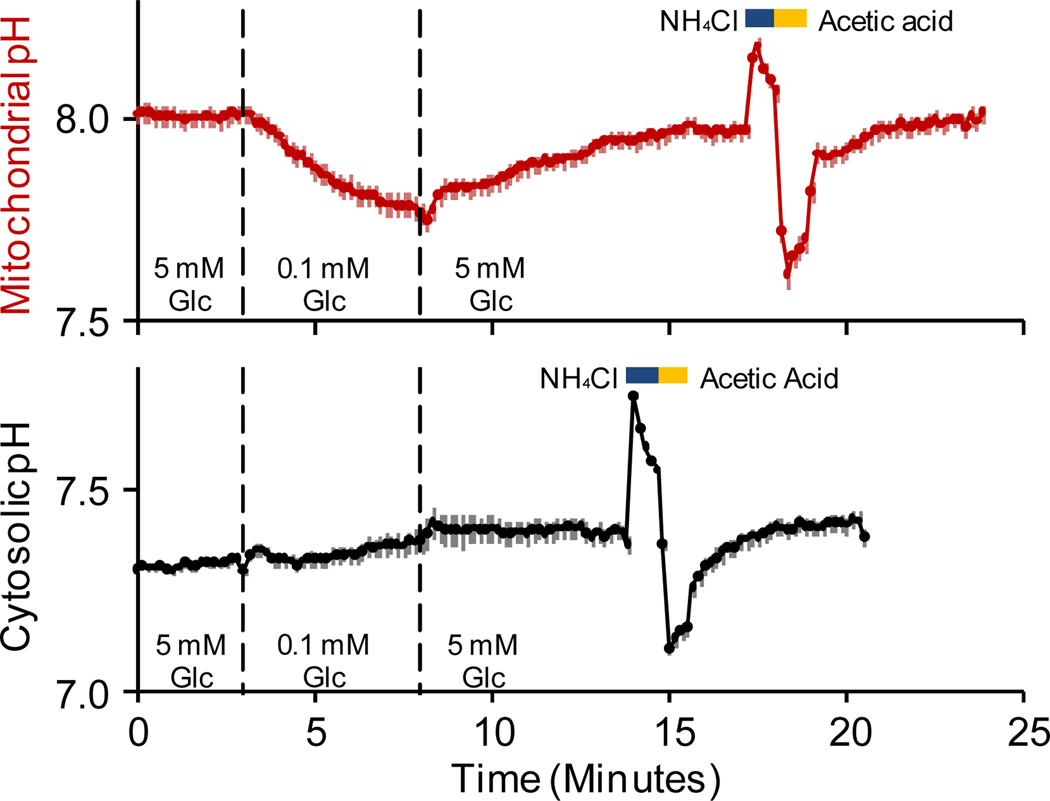
The cytochrome c oxidase subunit VIII (COX8) signal sequence was used to target pHRed to the mitochondrial matrix,23,24 and glucose-dependent mitochondrial function was reported by COX8-pHRed. COX8-pHRed reported that the matrix was alkaline, resting at pH 8.0 in the presence of 5 mM extracellular glucose. When the extracellular glucose concentration was decreased from 5 to 0.1 mM, the matrix acidified, and this acidification could be reversed by increasing the extracellular glucose concentration. The function of pHRed was verified at the end of each experiment by applying consecutive pulses of 10 mM NH4Cl and 10 mM acetic acid. Ammonium chloride alkalinizes the cell while acetic acid causes an acidification (Figure 3).25–27 Without an appended signal sequence, pHRed expressed throughout the cell and reported that the cytosol did not acidify when extracellular glucose concentration was lowered, resting near pH 7.3 (Figure 3). The mitochondrial uncoupler p-trifluoromethoxy carbonyl cyanide phenyl hydrazone similarly caused an acidification of the mitochondrial matrix but not the cytosol (Supplemental Figure S5). Acidification of the mitochondrial matrix is consistent with a decrease in mitochondrial metabolism when glucose levels are decreased. Lowered glucose causes a decrease in substrates available to enter the tricarboxylic acid cycle and fuel the electron transport chain, causing a collapse of the inner membrane potential and mitochondrial pH gradient.23,25 These data illustrate that targeted expression of pHRed can be used to monitor the pH of different organelles and their pH-dependent function.
Figure 3.
Decreased glucose concentration depletes mitochondrial substrates causing a loss of inner membrane potential, and COX8-pHRed reported a decrease in matrix pH (red, n=17). Cytosolic pH reported by pHRed did not acidify when glucose is lowered (black, n=3). At the end of each experiment, consecutive 10 mM NH4Cl and 10 mM acetic acid pulses were used to verify that pHRed correctly reported an induced intracellular alkalinization and acidification, respectively. Note that the mitochondrial matrix (pH 8.0) rested alkaline relative to the cytosol (pH 7.3).
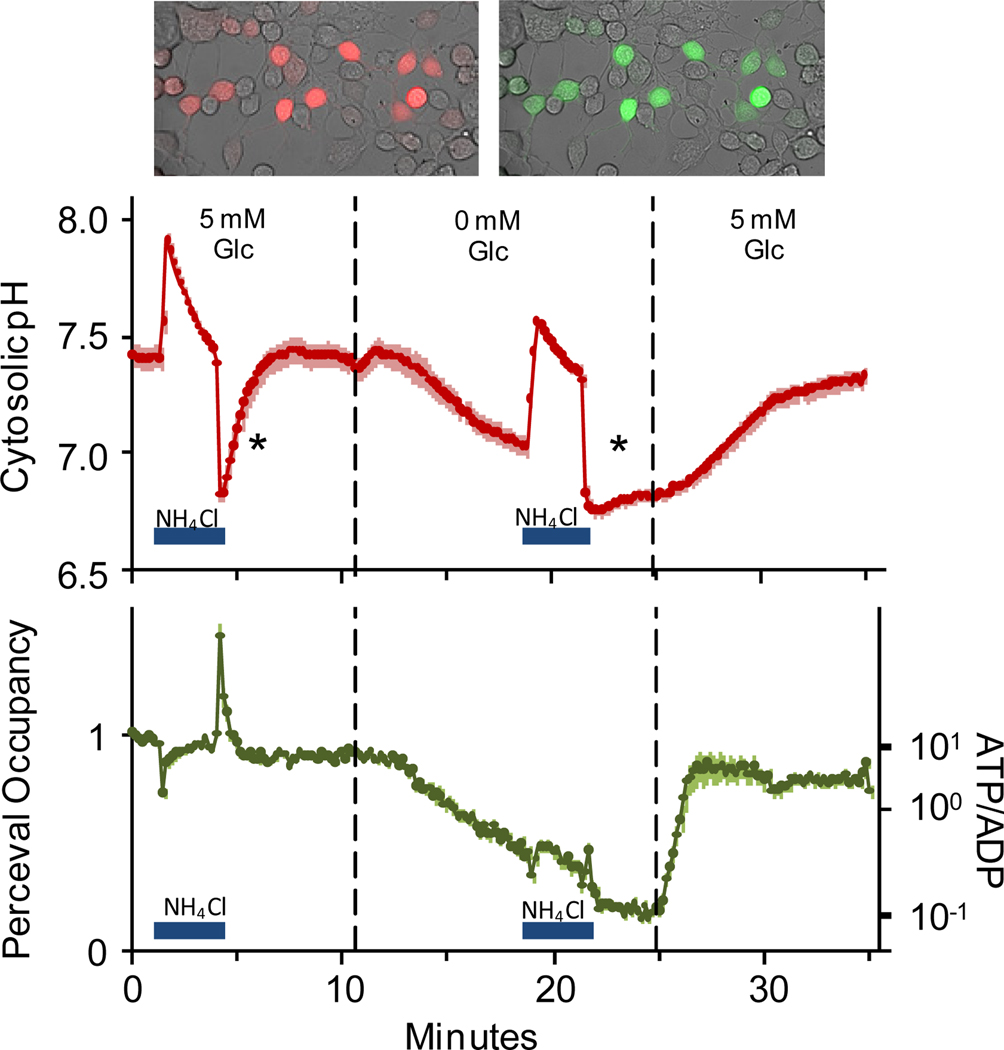
We also demonstrated that pHRed can be used in multicolor experiments to simultaneously image intracellular ATP and pH and to facilitate correction for pH sensitivity of the ATP sensor. pHRed was co-expressed with Perceval, a sensor of the intracellular ATP/ADP ratio,6 and the effect of acute glucose starvation on intracellular pH and ATP was monitored. As described above, lowering the extracellular glucose concentration to 0.1 mM causes mitochondrial acidification but not cytosolic acidification. These observations suggest that 0.1 mM extracellular glucose provides sufficient energy to maintain cytosolic pH despite attenuated mitochondrial function, at least for short periods of time in Neuro2A cells. In contrast, complete glucose starvation causes a cellular energy crisis that leads to oxidative stress, apoptosis, and loss of pH regulation.2,28 Whereas increasing extracellular glucose can alkalinize the cytosol, complete glucose withdrawal can acidify the cytosol because ATP depletion attenuates the function of Na+/H+-exchangers, the Na+/K+-ATPase, H+-ATPases, and other transporters that act together in H+ homeostasis.26,27,29–31 By co-expressing pHRed and Perceval, ATP depletion and its effect on intracellular pH could be simultaneously imaged. With 5 mM extracellular glucose ATP was high in Neuro2A cells, but ATP was quickly depleted with complete glucose starvation (Figure 4). The onset of ATP depletion correlated with the onset of intracellular acidification as the cells entered energy crisis (Figure 4). Additionally, recovery from acidification induced by a 10 mM NH4Cl prepulse differed in high glucose/high ATP versus no glucose/low ATP conditions. In the NH4Cl prepulse method, extracellular NH4Cl causes intracellular alkalinization followed by a transient acidification when it is removed. This method is commonly used to assay the function of membrane transporters involved in H+ homeostasis.27 High ATP supported a rapid recovery from acidification, but low ATP prevented a rapid recovery phase (Figure 4). Furthermore, glucose re-feeding promptly raised intracellular ATP, but recovery from acidification was delayed until ATP reached near original levels reported by Perceval (Figure 4). These data are consistent with other studies showing that acid/base transporters and pH homeostasis require ATP for energy or as an allosteric modulator for transporter function.27,30,31
Figure 4.
Co-expression of pHRed and Perceval for simultaneous imaging of the intracellular pH and the ATP/ADP ratio in live Neuro2A cells. Top: pHRed 629 nm (left) and Perceval 525 nm (right) emission in the same cells. Middle: In high glucose, cells rested at pH 7.4 and showed rapid recovery from an acid load induced by a 10 mM NH4Cl prepulse (first asterisk). Complete glucose withdrawal caused acidification, and recovery from an acid load was attenuated (second asterisk). Bottom: Glucose withdrawal caused a decrease in ATP that was promptly reversed with re-feeding. The Perceval signal was pH corrected using pHRed. Small errors in the correction remain at the start and end of the NH4Cl pulses. n=14.
This multicolor imaging experiment also illustrates the utility of pHRed in providing a simultaneous pH signal that can be used to correct the pH sensitivity of a green sensor. Sensitivity to pH is a critical problem for GFP-based sensors, particular circularly-permuted GFP sensors, that can cause severe artifacts if not taken into account.6,8,10 Perceval is pH-sensitive, and changes in intracellular metabolism and ATP/ADP ratio often occur concurrently with pH changes. Rather than requiring parallel experiments, simultaneous imaging of pHRed with Perceval allowed pH artifacts to be corrected on a cell-by-cell basis (Supplemental Figure S6). In this experiment, an empirical correlation between the green Perceval signal and the red pHRed signal was measured when cells were exposed to a mild pH manipulation not associated with a change in intracellular ATP. The empirical linear correlation was used to normalize Perceval for the pH changes reported by pHRed. This empirical method works well to remove the large pH artifacts observed with the uncorrected Perceval signal (Supplemental Figure S6). The ATP dynamics that were on the time scale of the metabolic changes monitored here were well corrected, despite small transient errors that remain in the corrected Perceval signal at the start and end of the ammonium pulse (Figure 4). Hence, pHRed facilitates imaging of multiple cellular parameters and can be valuable for providing a pH signal to correct for the pH-sensitivity of other sensors.
In conclusion, we engineered a new sensor called pHRed, a mutant of mKeima, that we used to image intracellular pH in live-cell microscopy. A pH sensor also using an RFP has been previously published, but this sensor provides intensity-based measurements that require corrections for expected photobleaching.13 To our knowledge, this is the first sensor based on a single RFP that is excitation ratiometric. We demonstrated that pHRed uniquely allows us to image intracellular ATP and pH simultaneously because of its spectral compatibility with the GFP-based ATP sensor Perceval. We also demonstrated that in principle pHRed can be used in two-photon FLIM, potentially providing a new tool for imaging pH in thick samples or possibly even in vivo. With the great progress made in engineering RFPs and ESPT in RFPs,17,32 this ratiometric sensing strategy could potentially be applied to create a variety of new sensor color variants and greatly enhance multicolor live-cell microscopy.
Supplementary Material
Acknowledgments
We thank Tanya Abramson for technical assistance. We also thank Bernardo Sabatini, his group, and Rebecca Mongeon for providing instrumentation and assistance with two-photon FLIM and Ryohei Yasuda for providing FLIM software. We thank the Harvard NeuroDiscovery Center Optical Imaging Program for providing instrumentation for FLIM. M.T. is supported by a Postdoctoral Fellowship from the NIH (F32NS066613), and this work was supported by a research grant from the NIH (R01NS055031) to G.Y.
Footnotes
Supporting Information Available. Materials and methods, supplemental figures, tables, and discussion. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Casey JR, Grinstein S, Orlowski J. Nat. Rev. Mol. Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 2.Lagadic-Gossmann D, Huc L, Lecureur V. Cell Death Differ. 2004;11:953–961. doi: 10.1038/sj.cdd.4401466. [DOI] [PubMed] [Google Scholar]
- 3.Han J, Burgess K. Chem. Rev. 2010;110:2709–2928. doi: 10.1021/cr900249z. [DOI] [PubMed] [Google Scholar]
- 4.Bizzarri R, Serresi M, Luin S, Beltram F. Anal. Bioanal. Chem. 2009;393:1107–1122. doi: 10.1007/s00216-008-2515-9. [DOI] [PubMed] [Google Scholar]
- 5.Palmer AE, Qin Y, Park JG, McCombs JE. Trends Biotechnol. 2011;29:144–152. doi: 10.1016/j.tibtech.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg J, Hung YP, Yellen G. Nat. Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H. Proc. Natl. Acad. Sci. U.S.A. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Nat. Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 9.Miesenbock G, De Angelis DA, Rothman JE. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 10.Nagai T, Sawano A, Park ES, Miyawaki A. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaner NC, Steinbach PA, Tsien RY. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 12.Verkhusha VV, Lukyanov KA. Nat. Biotechnol. 2004;22:289–296. doi: 10.1038/nbt943. [DOI] [PubMed] [Google Scholar]
- 13.Johnson DE, Ai HW, Wong P, Young JD, Campbell RE, Casey JR. J. Biol. Chem. 2009;284:20499–20511. doi: 10.1074/jbc.M109.019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Violot S, Carpentier P, Blanchoin L, Bourgeois D. J. Am. Chem. Soc. 2009;131:10356–10357. doi: 10.1021/ja903695n. [DOI] [PubMed] [Google Scholar]
- 15.Henderson JN, Osborn MF, Koon N, Gepshtein R, Huppert D, Remington SJ. J. Am. Chem. Soc. 2009;131:13212–13213. doi: 10.1021/ja904665x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kogure T, Karasawa S, Araki T, Saito K, Kinjo M, Miyawaki A. Nat. Biotechnol. 2006;24:577–581. doi: 10.1038/nbt1207. [DOI] [PubMed] [Google Scholar]
- 17.Piatkevich KD, Hulit J, Subach OM, Wu B, Abdulla A, Segall JE, Verkhusha VV. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5369–5374. doi: 10.1073/pnas.0914365107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson GT, McAnaney TB, Park ES, Rendell ME, Yarbrough DK, Chu S, Xi L, Boxer SG, Montrose MH, Remington SJ. Biochemistry. 2002;41:15477–15488. doi: 10.1021/bi026609p. [DOI] [PubMed] [Google Scholar]
- 19.Thomas JA, Buchsbaum RN, Zimniak A, Racker E. Biochemistry. 1979;18:2210–2218. doi: 10.1021/bi00578a012. [DOI] [PubMed] [Google Scholar]
- 20.Kogure T, Kawano H, Abe Y, Miyawaki A. Methods. 2008;45:223–226. doi: 10.1016/j.ymeth.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Esposito A, Gralle M, Dani MA, Lange D, Wouters FS. Biochemistry. 2008;47:13115–13126. doi: 10.1021/bi8009482. [DOI] [PubMed] [Google Scholar]
- 22.Harvey CD, Ehrhardt AG, Cellurale C, Zhong H, Yasuda R, Davis RJ, Svoboda K. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19264–19269. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filippin L, Abad MC, Gastaldello S, Magalhaes PJ, Sandona D, Pozzan T. Cell Calcium. 2005;37:129–136. doi: 10.1016/j.ceca.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Abad MF, Di Benedetto G, Magalhaes PJ, Filippin L, Pozzan T. J. Biol. Chem. 2004;279:11521–11529. doi: 10.1074/jbc.M306766200. [DOI] [PubMed] [Google Scholar]
- 26.Demaurex N, Romanek RR, Orlowski J, Grinstein S. J. Gen. Physiol. 1997;109:117–128. doi: 10.1085/jgp.109.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demaurex N, Grinstein S. J. Exp. Biol. 1994;196:389–404. doi: 10.1242/jeb.196.1.389. [DOI] [PubMed] [Google Scholar]
- 28.Jelluma N, Yang X, Stokoe D, Evan GI, Dansen TB, Haas-Kogan DA. Mol. Cancer Res. 2006;4:319–330. doi: 10.1158/1541-7786.MCR-05-0061. [DOI] [PubMed] [Google Scholar]
- 29.Shepherd RM, Henquin JC. J. Biol. Chem. 1995;270:7915–7921. doi: 10.1074/jbc.270.14.7915. [DOI] [PubMed] [Google Scholar]
- 30.Dechant R, Binda M, Lee SS, Pelet S, Winderickx J, Peter M. EMBO J. 2010;29:2515–2526. doi: 10.1038/emboj.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura S. Am. J. Physiol. Cell. Physiol. 2004;287:C97–C105. doi: 10.1152/ajpcell.00469.2003. [DOI] [PubMed] [Google Scholar]
- 32.Piatkevich KD, Malashkevich VN, Almo SC, Verkhusha VV. J. Am. Chem. Soc. 2010;132:10762–10770. doi: 10.1021/ja101974k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.