Abstract
Soil is predicted to contain thousands of unique bacterial species per gram. Soil DNA libraries represent large reservoirs of biosynthetic diversity from which diverse secondary metabolite gene clusters can be recovered and studied. The screening of an archived soil DNA library using primers designed to target oxy-tryptophan dimerization genes allowed us to identify and functionally characterize the first indolotryptoline biosynthetic gene cluster. The recovery and heterologous expression of an environmental DNA derived gene cluster encoding the biosynthesis of the antitumor substance BE-54017 is reported here. Transposon mutagenesis identified two monooxygenases, AbeX1 and AbeX2, as being responsible for the transformation of an indolocarbazole precursor into the indolotryptoline core of BE-54017.
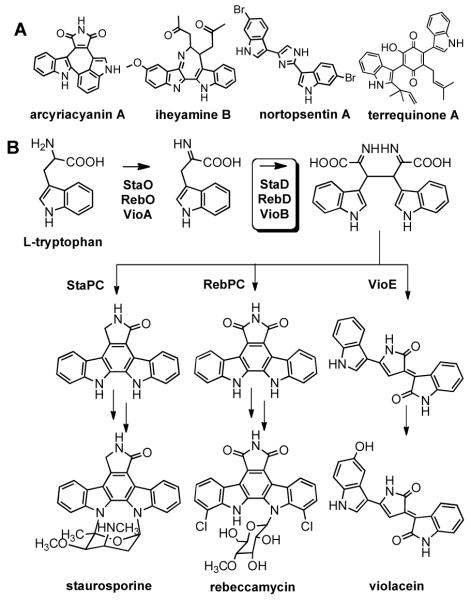
A number of structurally diverse and pharmacologically interesting natural products are thought to arise from the dimerization of tryptophan (Figure 1A and 1B).1,2 Biosynthetic gene clusters for violacein3 as well as representative bisindolyquinone (terrequinone A4) and indolocarbazole5-8 (Figure 1B) based metabolites have been cloned and extensively characterized. Most members of this diverse class of metabolites, however, have not been investigated biosynthetically. Soil contains an extraordinarily diverse collection of both cultured and as yet uncultured bacteria.9,10 DNA isolated from natural populations of soil bacteria (environmental DNA, eDNA) should therefore represent a reservoir from which additional tryptophan-dimer biosynthetic gene clusters can be cloned and characterized. Here we report the cloning and heterologous expression of an eDNA derived gene cluster that encodes the biosynthesis of the antitumor substance BE-54017 (1) (Figure 2).11 BE-54017 is part of a small family of indolotryptoline (indolotetrahydro-β-carboline) based natural products that show potent activity against tumor cell lines.1,11 Transposon mutagenesis of the abe (antitumor substance BE-54017) gene cluster revealed that the indolotryptoline core of BE-54017 arises from two successive oxidations of an indolocarbazole precursor.
Figure 1.
(A) Structurally diverse natural products appear to arise from the dimerization of two tryptophans. (B) Overview of indolocarbazole (staurosporine/rebeccamycin) and violacein biosynthesis. In these pathways, homologous genes (StaD/RebD/VioB) are responsible for the dimerization of oxy-tryptophan.
Figure 2.
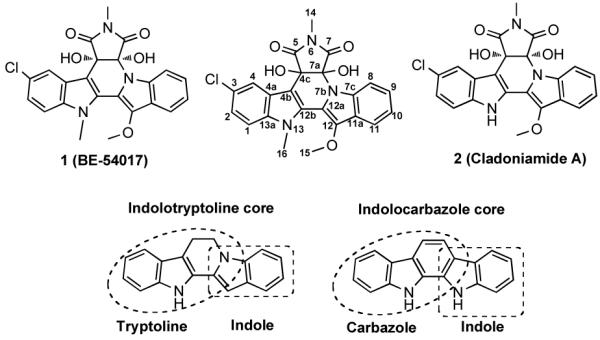
The eDNA-derived abe gene cluster encodes the biosynthesis of BE-54017 (1). BE-54017 and the cladoniamides share an indolotryptoline core. Here we show that indolotryptolines arise from indolocarbazole precursors.
To recover tryptophan dimer biosynthetic gene clusters from the environment, we screened a previously archived soil eDNA cosmid library by PCR using degenerate primers designed to recognize conserved regions in known oxy-tryptophan dimerization genes (StaD/RebD/VioB etc).12,13 Oxy-tryptophan dimerization enzymes were chosen as our probes because this enzyme family is used in the biosynthesis of structurally diverse tryptophan dimers. Both indolocarbazole biosynthetic gene clusters (e.g. staurosporine, rebeccamycin, K-252a, and AT2433) and violacein biosynthetic gene clusters contain homologous enzymes that carry out the oxidation (StaO/RebO/VioA) and subsequent dimerization (StaD/RebD/VioB) of tryptophan (Figure 1B). Cosmid clones associated with unique eDNA derived StaD-like homologs were recovered from the eDNA library using whole-cell PCR on serially diluted library aliquots. Each recovered cosmid was then de novo sequenced and annotated.
One eDNA derived cosmid, AB1650 was found to contain a complete set of conserved indolocarbazole biosynthetic genes (abeO, D, C, P) as well as two monooxygenases (abeX1, X2), three methyltransferases (abeM1, M2, M3) and a halogenase (abeH ) (Figure 3). The presence of the two predicted monooxygenases was unprecedented in known indolocarbazole biosynthetic pathways, suggesting that this gene cluster likely encoded the biosynthesis of an oxidized or rearranged indolocarbazole based metabolite. To investigate this possibility, cosmid AB1650 was retrofitted with the genetic elements required for conjugation, selection and stable integration into Streptomyces, and then conjugated into Streptomyces albus for heterologous expression studies.
Figure 3.
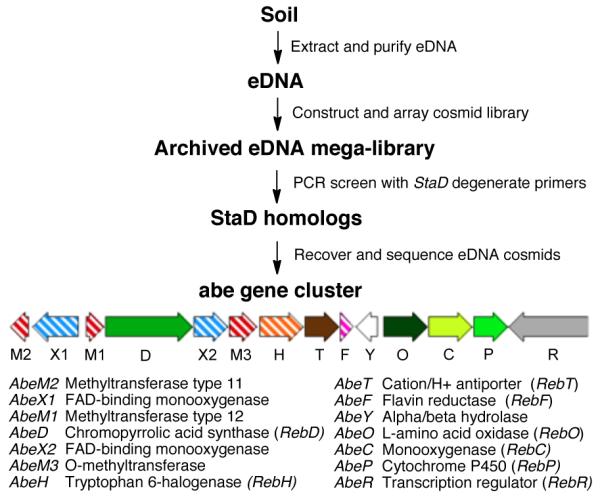
The BE-54017 biosynthetic gene cluster (abe) (GenBank accession JF439215) was recovered from an eDNA library using primers that recognize StaD-like oxy-tryptophan dimerization genes.
Cultures of S. albus transformed with AB1650 produce one major clone specific metabolite as well as four minor clone specific metabolites. 1H NMR, 13C NMR, UV and HR-ESI-MS14 data for the major metabolite (1) are identical to data reported for the antitumor substance BE-54017 (Figure 2).11 Minor metabolites 3 and 4 each differ from 1 by the loss of 28 mass units or “CO”.14 Recently reported derivatives of the structurally related metabolite cladoniamide A (2), in which the N-methylsuccinimide has been hydrolyzed, show a similar loss of “CO”.15 As would be expected for correspondingly hydrolyzed BE-54017 derivatives, compounds 3 and 4 both contain a new COSY spin system between an amide nitrogen and a methyl singlet that now shows an HMBC correlation to only one of the two carbonyl carbons in the structure. HMBC correlations from the C-4c hydroxyl proton to C-7a, -5, -4c and -4b in compound 3 and from the C-7a hydroxyl proton to C-7, -7a and -4c in compound 4 allowed us to define the position of the N-methylamide in both structures. Based on comparisons of HRMS14 and 2D NMR data, compounds 5 and 6 were determined to be the deschloro analogues of compounds 1 and 4, respectively. Compounds 3 - 6 are novel secondary metabolites (Figure 4).
Figure 4.
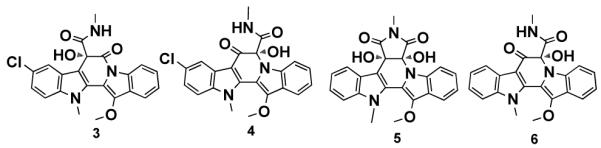
Minor compounds isolated from the culture broth of S. albus transformed with the abe gene cluster.
It has been suggested that the indolotryptoline core seen in BE-54017 and the cladoniamides might arise either from the oxidation of an indolocarbazole precursor or from an indolocarbazole independent pathway.1,15 To elucidate the origin of the indolotryptoline substructure and to assign specific functions to the individual genes found in the abe gene cluster, we carried out a transposon mutagenesis study on cosmid AB1650. Individual transposon mutants were sequenced to identify clones with insertions in key biosynthetic genes (Figure 5). This collection of transposon mutants was then conjugated back into S. albus and the major clone specific metabolites found in the culture broth extracts of each mutant were structurally characterized.
Figure 5.
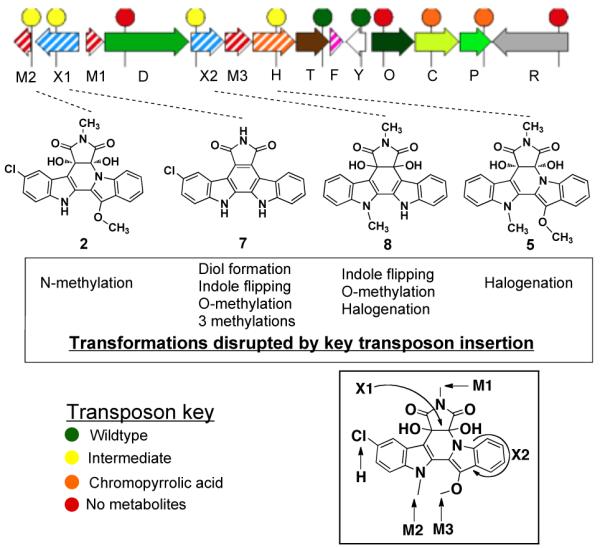
The major metabolites produced by select transposon mutants. Sites on BE-54017 where key biosynthetic enzymes are predicted to act are indicated in the inset.
As would be predicted from what is known about the biosynthesis of indolocarbazoles (Figure 1B), transposon insertions in predicted indolocarbazole biosynthetic genes result in either the absence of organic extractable clone specific metabolites (abeO, D, R) or in the production of the known indolocarbazole intermediate 3-chloro-chromopyrrolic acid (abeC, P) (Figure 5). Homologs of the two predicted oxi-doreductases, abeX1 and abeX2, do not appear in any known bisindole biosynthetic gene clusters. Disruption of abeX2 results in the accumulation of compound 816 and disruption of abeX1 leads to the accumulation of the simple indolocarbazole, 3-chloro-arcyriaflavin (7).17 The isolation of compound 7 confirmed the indolocarbazole origin of BE-54017.
Compound 8 contains the C-4c/C-7a diol seen in BE-54017, but has not undergone a rearrangement of the indolocarbazole core, while compound 7 contains neither the diol nor flipped indole. AbeX1 and AbeX2 show homology to class A flavoprotein monooxygenases and therefore the oxidation reactions they carry out are predicted to proceed through epoxides (Figure 6).18 The mutagenesis results coupled with homology arguments allow us to construct a biosynthetic scheme for the formation of the indolotryptoline core in BE-54017 from the indolocarbazole 7 (Figure 6). Our scheme is consistent with the proposed biogenesis of the cladoniamides outlined by Williams, et. al.15 In this scheme the diol is introduced by AbeX1 through the epoxidation of the C-4c/C-7a double bond. AbeX2 is then responsible for promoting the rearrangement of the indole via the introduction of a second epoxide. The opening of this epoxide is accompanied by the fragmentation of the C-7a/C-7b bond, followed by the rotation of the indole around C-12a/C-12b and finally the formation of the C-7a/N-7b bond.
Figure 6.
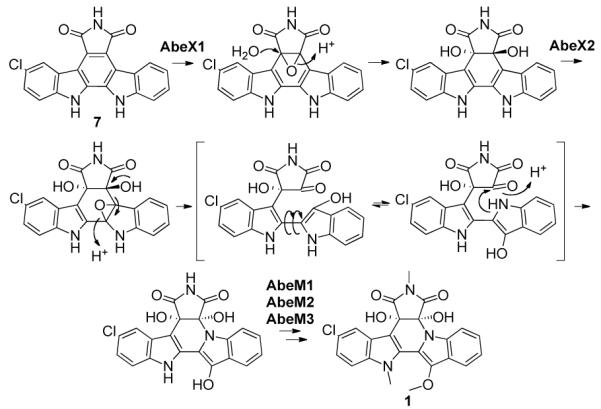
Two monooxygenases, AbeX1 and AbeX2, are responsible for the conversion of an indolocarbazole precursor into the indolotryptoline core of BE-54017. BE-54017 requires three methylations; the exact timing of these transformations is not known.
Whether the proposed epoxide hydrolysis and indole rearangement reactions occur spontaneously, are catalyzed by the monooxygenases themselves or are catalyzed by other enzymes is not clear. AbeY shares the same α/β-hydrolase fold superfamily as most epoxide hydrolases and could therefore play a role in these reactions.19 Our studies suggest however that AbeY is dispensable for the biosynthesis of compounds 1 and 3 - 6, as all five compounds are produced by the abeY knockout mutant. The appearance of small quantities of low-molecular weight clone specific metabolites in extracts from cultures of the abeY mutant suggests that, while AbeY is not required, it may be involved in enhancing the efficiency of one or more biosynthetic transformations, similar to the reported function of StaC in staurosporine biosynthesis.20 Alternatively, the role of AbeY may be complemented by the host’s endogenous biosynthetic machinery.
A detailed accounting of the functionality that appears on the compounds produced in our transposon mutagenesis experiments allowed us to assign functions to the three methyltransferases and the predicted halogenase found in the abe gene cluster (Figure 5). Disruption of abeH, a homolog of the tryptophan halogenase found in the rebeccamycin pathway, led to the accumulation of the deschloro derivative 5, thereby confirming its role as a halogenase. The absence of the chloride substituent on compound 8 suggested that the transposon insertion in abeX2 disrupts the expression of downstream genes in the same operon. AbeM3 is positioned between abeX2 and abeH and is therefore predicted to be transcriptionally silenced in this transposon mutant. Since both the N-6 and N-13 methylations appear on 8, abeM3 is predicted to be responsible for the methylation of the C-12 hydroxyl in BE-54017. The regiospecificity of the two remaining N-methyltransferases, abeM1 and abeM2, was inferred from the accumulation of the O-12/N-6 dimethylated derivative 2 (cladoniamide A) in the abeM2 transposon mutant. AbeM1, AbeM2 and AbeM3 are therefore N-6, N-13 and O-12 specific methyltransferases, respectively (Figure 5 inset).
No gene in the abe cluster could be linked to the hydrolysis of the N-methylsuccinimide suggesting that this reaction is either carried out by the host or occurs spontaneously during the fermentation process.
Compounds 1 - 8 were assayed for antiproliferative activity against human colon cancer HCT116 cells. While 3 - 8 were not active below 8 μg/ml, 1 and 2 exhibited potent antiproliferative activities (IC50 μg/ml: 0.079 for 1 and 0.0088 for 2). Crystallographic studies have shown that the planar structure of indolocarbazoles is important for topoisomerase/DNA (rebeccamycin) and kinase (staurosporine) binding.21,22 The C-4c/C-7a diol seen in indolotryptolines causes the N-methylsuccinimide to bend out of the bisindole plane, which may afford these compounds the ability to bind different cellular targets for their antitumor activity. Considerable progress has been made in generating novel indolocarbazole analogues by combinatorial biosynthesis, and the identification of AbeX1 and AbeX2 provides new tools for producing structurally and functionally diverse bisindole metabolites.23
The cloning of DNA extracted directly from environmental samples provides a means of exploring the biosynthetic capacity of thousands of bacterial genomes simultaneously. Archived eDNA libraries represent permanent resources from which diverse gene clusters can be recovered and studied. In this study the screening of an archived eDNA library using primers designed to recognize oxy-tryptophan dimerization genes allowed us to clone and characterize the first indolotryptoline biosynthetic gene cluster.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIH GM077516. SFB is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
Supporting Information Available: Experimental details, annotation table for the abe gene cluster, tables of NMR assignments, and 1D and 2D NMR spectra for compound 1-8 are available free of charge at http://pubs.acs.org.
REFERENCES
- (1).Ryan KS, Drennan CL. Chem. Biol. 2009;16:351–64. doi: 10.1016/j.chembiol.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Sanchez C, Mendez C, Salas JA. Nat. Prod. Rep. 2006;23:1007–45. doi: 10.1039/b601930g. [DOI] [PubMed] [Google Scholar]
- (3).August PR, Grossman TH, Minor C, Draper MP, Mac-Neil IA, Pemberton JM, Call KM, Holt D, Osburne MS. J Mol. Microbiol. Biotechnol. 2000;2:513–9. [PubMed] [Google Scholar]
- (4).Balibar CJ, Howard-Jones AR, Walsh CT. Nat. Chem. Biol. 2007;3:584–92. doi: 10.1038/nchembio.2007.20. [DOI] [PubMed] [Google Scholar]
- (5).Onaka H, Taniguchi S, Igarashi Y, Furumai T. J. Antibiot. (Tokyo) 2002;55:1063–71. doi: 10.7164/antibiotics.55.1063. [DOI] [PubMed] [Google Scholar]
- (6).Sanchez C, Butovich IA, Brana AF, Rohr J, Mendez C, Salas JA. Chem. Biol. 2002;9:519–31. doi: 10.1016/s1074-5521(02)00126-6. [DOI] [PubMed] [Google Scholar]
- (7).Gao Q, Zhang C, Blanchard S, Thorson JS. Chem. Biol. 2006;13:733–43. doi: 10.1016/j.chembiol.2006.05.009. [DOI] [PubMed] [Google Scholar]
- (8).Kim SY, Park JS, Chae CS, Hyun CG, Choi BW, Shin J, Oh KB. Appl. Microbiol. Biotechnol. 2007;75:1119–26. doi: 10.1007/s00253-007-0924-x. [DOI] [PubMed] [Google Scholar]
- (9).Rappe MS, Giovannoni SJ. Annu. Rev. Microbiol. 2003;57:369–94. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- (10).Torsvik V, Goksoyr J, Daae FL. Appl. Environ. Microbiol. 1990;56:782–7. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Nakase K, Nakajima S, Hirayama M, Kondo H, Kojiri K, Suda H. 2000. JP 2000178274.
- (12).Feng Z, Kim JH, Brady SF. J. Am. Chem. Soc. 2010;132:11902–3. doi: 10.1021/ja104550p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Primers StaDVF: GTSATGMTSCAGTACCTSTACGC; StaDVR: YTCVAGCTGRTAGYCSGGRTG.
- (14).HR-ESI-MS: 1 m/z: C2 3H1 9ClN3O5 [M+H]+ calcd 452.1013; found 452.10193, 2 m/z: C2 2H1 7ClN3O5 [M+H]+ calcd 438.0778; found 438.0844, 3 m/z: C2 2H1 9ClN3O4 [M+H]+ calcd 424.1064; found 424.1048, 4 m/z: C2 2H1 9ClN3O4 [M+H]+ calcd 424.1064; found 424.1068, 5 m/z: C2 3H2 0N3O5 [M+H]+ calcd 418.1403; found 418.1383, 6 m/z: C2 2H2 0N3O4 [M+H]+ calcd 390.1454; found 390.1457, 7 m/z: C2 0H9ClN3O2 [M-H]− calcd 358.0462; found 358.0385.
- (15).Williams DE, Davies J, Patrick BO, Bottriell H, Tarling T, Roberge M, Andersen RJ. Org. Lett. 2008;10:3501–4. doi: 10.1021/ol801274c. [DOI] [PubMed] [Google Scholar]
- (16).The predicted molecular formula for 8 (HR-ESI-MS (m/z): C2 2H1 7N3O4Na [M+Na]+ calcd 410.1117; found 410.1119) differs from 5 by “CH2O”. A comparison of the 1H spectra from 5 and 8 showed that 8 does not contain the C-12 methoxy protons and that it has a new NH hydrogen. Most HMBC correlations seen in 8 also appeared in 5, however new HMBC correlations from the C-7a OH to C-7b and from the NH to C-7b, C-12a and C-11a indicate that the indole in 8 has not been flipped.
- (17).Sanchez C, Zhu L, Brana AF, Salas AP, Rohr J, Mendez C, Salas JA. Proc. Natl. Acad. Sci. U. S. A. 2005;102:461–6. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).van Berkel WJ, Kamerbeek NM, Fraaije MW. J Biotechnol. 2006;124:670–89. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- (19).Widersten M, Gurell A, Lindberg D. Biochim. Biophys. Acta. 2010;1800:316–26. doi: 10.1016/j.bbagen.2009.11.014. [DOI] [PubMed] [Google Scholar]
- (20).Howard-Jones AR, Walsh CT. J. Am. Chem. Soc. 2006;128:12289–98. doi: 10.1021/ja063898m. [DOI] [PubMed] [Google Scholar]
- (21).Staker BL, Feese MD, Cushman M, Pommier Y, Zembower D, Stewart L, Burgin AB. J. Med. Chem. 2005;48:2336–45. doi: 10.1021/jm049146p. [DOI] [PubMed] [Google Scholar]
- (22).Prade L, Engh RA, Girod A, Kinzel V, Huber R, Bossemeyer D. Structure. 1997;5:1627–37. doi: 10.1016/s0969-2126(97)00310-9. [DOI] [PubMed] [Google Scholar]
- (23).Salas JA, Mendez C. Curr. Opin. Chem. Biol. 2009;13:152–60. doi: 10.1016/j.cbpa.2009.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.