Abstract
Suppression of macroautophagy, due to mutations or through processes linked to aging, results in the accumulation of cytoplasmic substrates that are normally eliminated by the pathway. This is a significant problem in long-lived cells like neurons, where pathway defects can result in the accumulation of aggregates containing ubiquitinated proteins. The p62/Ref(2)P family of proteins is involved in the autophagic clearance of cytoplasmic protein bodies or sequestosomes. These unique structures are closely associated with protein inclusions containing ubiquitin as well as key components of the autophagy pathway. In this study we show that detergent fractionation followed by western blot analysis of insoluble ubiquitinated proteins (IUP), mammalian p62 and its Drosophila homologue, Ref(2)P can be used to quantitatively assess the activity level of aggregate clearance (aggrephagy) in complex tissues. Using this technique we show that genetic or age-dependent changes that modify the long-term enhancement or suppression of aggrephagy can be identified. Moreover, using the Drosophila model system this method can be used to establish autophagy-dependent protein clearance profiles that are occurring under a wide range of physiological conditions including developmental, fasting and altered metabolic pathways. This technique can also be used to examine proteopathies that are associated with human disorders such as frontotemporal dementia, Huntington and Alzheimer disease. Our findings indicate that measuring IUP profiles together with an assessment of p62/Ref(2)P proteins can be used as a screening or diagnostic tool to characterize genetic and age-dependent factors that alter the long-term function of autophagy and the clearance of protein aggregates occurring within complex tissues and cells.
Key words: p62, Ref(2)P, insoluble ubiquitinated proteins, aggregates, neural degeneration, Alzheimer disease, aging, macroautophagy
Introduction
Under normal physiological conditions there is a balance between the production of new cellular components or proteins and the elimination of redundant or damaged constituents. However, in the case of many neural degenerative disorders, there appears to be a progressive decline in the ability of cells to eliminate misfolded proteins or damaged organelles.1 These defects can eventually lead to the formation of intracellular inclusions. Often these pathological structures include ubiquitinated proteins along with the p62/Ref(2)P proteins (SQSTM1).2 Ubiquitin is a well-known targeting signal that directs extraneous or misfolded proteins to be degraded by the proteosome system and is involved with aspects of the ER stress response.3–5 There is a growing body of evidence showing that when several clearance pathways become less efficient or are overwhelmed by damaged cellular components, macroautophagy plays a vital role in sequestering and trafficking cytoplasmic material to the lysosome for elimination.1
Macroautophagy (hereafter autophagy) is a highly dynamic vesicle-based pathway that is involved with the turnover of a wide range of intracellular material.5,6 The pathway plays a significant role in many physiological processes that are involved with cellular homeostasis and is essential for the elimination of redundant or damaged proteins and organelles. This function is a particularly critical feature of the pathway in long-lived, nondividing cells such as neurons.1,7 Previously, we found an inverse correlation occurs between the decreased expression of key autophagy genes in the aging nervous system and the accumulation of ubiquitinated proteins.8 Over time these insoluble ubiquitinated proteins (IUP) form aggregates and become relatively insoluble cellular inclusions or proteopathies.8,9 Genetic manipulation of key autophagy components often alters IUP profiles in neuronal tissues.8–12
A second marker of aggregates, p62 also accumulates in cells when autophagy is inhibited.13–16 The p62 and Ref(2)P proteins contain multiple interaction motifs, including an UBA domain that binds poly-ubiquitinated proteins, as well as an MAP-LC3-interacting region (LIR).15,17 Recently a study has shown the build-up of ubiquitinated aggregates in autophagy mutants is not dependent on a specific Ub-Ub linkage type, but can involve all multi-ubiquitin chain topologies.18 This and the finding that the isolated UBA domain of p62 has minimal interaction with ubiquitin, suggests that full-length p62/Ref92)P protein has a low affinity but specific interaction with a wide range of aggregating proteins.18 Thus, p62 has the capacity to link ubiquitinated proteins to autophagic machinery and facilitate the targeting and degradation of many ubiquitinated protein aggregates. We have previously shown that the Drosophila p62 homologue, Ref(2) P, also shares these key structural elements and associates with ubiquitinated neural aggregates in exceptionally old control flies or autophagy mutants.17,19
Human p62 is an abundant constituent of protein inclusions that are often associated with many progressive neurological diseases.20–23 Immunohistological imaging of tissues prepared from patients suffering from Alzheimer disease (AD) and other neurological disorders indicates human p62 is a component of ubiquitin-containing aggregates.2,20,24 Studies of neural preparations taken from AD patients also show there is an accumulation of multiple autophagic vesicle subtypes within neurons.25,26 Given that p62 is a component of the autophagic pathway and protein aggregates, we examined the p62 and Ref(2)P proteins via western blot analysis together with IUP profiles from a diverse series of neuronal and cell culture preparation. Our hypothesis is that as autophagy is suppressed, and substrates that are normally cleared by the pathway begin to accumulate, then changes in the levels and/or solubility profiles of p62/Ref(2)P will serve as an accurate in vivo measure of long-term autophagic activity and substrate clearance (aggrephagy) in the nervous system.8,17,19,27
Here we report that using Drosophila genetic techniques, the level and solubility profiles of the Ref(2)P protein show a direct correlation with the build-up of IUP profiles and an inverse relationship with autophagic activity in different ages or mutant backgrounds. Immunoblot analysis of Ref(2)P from aged bchs mutants also show an accelerated accumulation profile consistent with transmission electron microscopy (TEM) images showing the formation of abnormal cytoplasmic structures and inclusions in mutant neurons. We also show that Ref(2)P and IUP profiles are altered in other autophagy and lysosomal trafficking mutants, which are associated with vesicle trafficking and protein clearance. Interestingly, we find that the normal age-dependent accumulation of neuronal Ref(2)P and IUP is significantly reduced in long-lived insulin signaling mutants. This suggests that autophagic function is enhanced in this mutant background at a time when the pathway shows an inability to clear protein aggregates or aggrephagy. Further, western blot analysis of human neural tissues prepared from control and Alzheimer disease patients finds that there is a close correlation between accumulated p62 and the number of senile plaques characterized for a particular individual. Consistent with previous findings, cell culture studies also show that direct suppression of autophagy (Atg5−/−KO) or altering ESCRT-III complex function (Vps24 siRNA) results in the accumulation of protein inclusions and insoluble p62. Further, expressing a mutant protein linked to progressive neurological disorders (CHMP2B) also results in the formation of insoluble inclusions containing p62. These studies indicate that changing solubility profiles of p62, Ref(2)P and IUP can be used as an effective measure of substrate turnover and the in vivo activity level of autophagy in complex neural tissues.
Results
Ref(2)P profiles reflect age-dependent and genetic changes to neuronal autophagy.
Antibodies directed against ubiquitin and p62 have been used as histological markers of protein inclusions in many neural degeneration studies.16,28 Previously, we have shown the Drosophila p62 homologue, Ref(2)P co-localizes with ubiquitin positive neural inclusions in Atg8a and bchs mutants, as well as in very old wild-type flies at a time when autophagic function is suppressed (8 weeks).17,19 To determine if Ref(2)P shows a shift in solubility pattern that is similar to IUP profiles we examined Ref(2)P levels from neural preparation taken at different ages and from flies were autophagic activity was genetically altered. For this study, heads from wild-type flies at 1-day, 1-, 2- and 4-weeks of age were collected, serial detergent extracted and the Triton-X and SDS soluble protein fractions used for western blot analysis of the Ref(2)P, ubiquitin and actin proteins (Fig. 1A and B). While Ref(2)P levels in the Triton-X fraction show a significant age-dependent increase in wild-type flies, both IUP and Ref(2)P have a more dramatic buildup in the SDS fraction from 4-week-old wild-type flies (>30 fold increase).
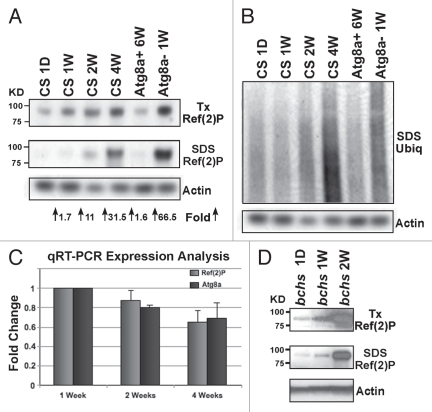
Figure 1.
Accumulation of IUP, Ref(2)P and cytoplasmic inclusions associated with neuronal aging and autophagy defects. (A) Wild-type flies were aged between 1 day and 4 weeks (1 D, 1 W, 2 W, 4 W) before heads were collected and serially detergent extracts prepared for western blot analysis. Neural extracts were also prepared from 6-week-old flies with enhanced Atg8a expression (Atg8a+, APPL-GAL4/UAS-Atg8a) or from 1-week-old Atg8a−/− mutant flies (Atg8a1/Atg8a2). Immunoblots were probed with the Ref(2)P and actin antibodies and the fold increase in SDS soluble Ref(2)P levels compared to 1-day controls. (B) Matching IUP profiles. (C) qRT-PCR analysis was used to examine the expression profiles of the Ref(2)P and Atg8a gene from multiple head RNA extractions were taken from wild-type heads at 1 (n = 5), 2 (n = 2) and 4 weeks (n = 3) of age. Atg8a and ref(2)P mRNA values were normalized to individual actin•5c profiles. 1-week values were set at 1- and 2- and 4-week readings averaged and standard deviations calculated. Both Ref(2)P and Atg8a mRNA levels show a similar age-dependent decline. (D) Western blot analysis of neural extracts prepared from 1 day-, 1 week- and 2-week-old bchs−/− mutant flies, shows Ref(2)P accumulation profiles in both the Triton-X and SDS protein fractions.
The timing of protein accumulation (between 3 and 4 weeks) is consistent with our previous findings that show neuronal expression of key autophagy genes is suppressed, which results in a reduction in the turnover of substrates by the pathway in the Drosophila CNS.8 Generally a close correlation exists between global levels of Atg8a in the adult brain and autophagic activity.8 In line with this observation, we find that both IUP and Ref(2) P show a similar pattern, with lower levels of both proteins accumulating in old animals having enhanced expression of Atg8a (Fig. 1A and B and Atg8a+ 6-weeks).8 Conversely, young 1-week-old animals that have genetic inhibition of the pathway show an accelerated build-up of both markers in neuronal tissues (Fig. 1A and Atg8a1/2 1-week).8 The age-dependent accumulation of the Ref(2)P protein in wild-type flies could be due to its increased expression in older flies. To clarify this point we prepared neural RNA from male flies at 1-, 2- and 4-weeks of age and examined the expression profiles of actin 5C (loading control), Atg8a and ref(2)P genes. Corrected 1-week values were set at 1.0 and the relative message levels of Atg8a and ref(2)P at different ages are illustrated in Figure 1C. As seen previously, Atg8a message levels decrease in older flies as does ref(2)P expression. This indicates that the age-dependent build-up and solubility changes of the Ref(2)P protein are likely due to an overall decline in basal rates of neuronal autophagy and not from increased gene expression.
Previously we found that mutations in the blue cheese (bchs) gene reduces adult longevity and accelerates neural degeneration.9,29 These phenotypes are accompanied by IUP accumulation and the formation of amyloid protein precursor-like protein (APPL) and ubiquitin neural inclusions in the Drosophila brain.9 The human homologue Alfy is also required for the degradation of substrates or selective aggrephagy.27,30 For bchs mutant animals the suppression of autophagic capacity results in the inability of relatively young flies to clear neuronal IUP (also see Sup. Fig. 3).9,17 The accumulation of autophagic substrates is further confirmed by western blot analysis of the Ref(2)P protein. Adult bchs mutant flies (bchs6/bchs3CyO) were aged for 1 day, or 1 or 2 weeks, before their heads were collected and processed via detergent extraction.9 Immunoblots of the Triton-X and SDS fractions were probed for Ref(2)P and actin (Fig. 1D). We find a dramatic accumulation of Ref(2)P in both protein fractions, which is consistent with our previous findings on IUP profiles.9,17 These results indicate that reduced autophagic flux due to aging or genetic mutations to the pathway (Atg8a and bchs mutants) can be quantitatively assessed for in vivo studies by using western blot analysis of IUP and Ref(2)P profiles.
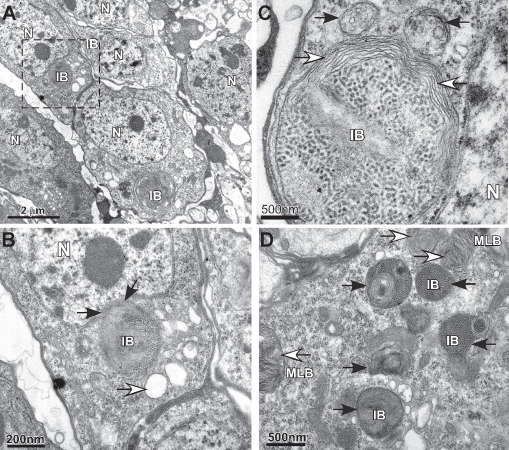
A second method used to identify autophagy or substrate clearance defects is electron microscopy, which is used to detect the formation of abnormal subcelluar structures and protein aggregates. Using transmission electron microscopy (TEM) we find the formation of many abnormal cytoplasmic structures in neurons prepared from 2-week-old bchs mutants (Fig. 2). In a single image several neuronal cell bodies (i.e., soma) can be found that contain large perinuclear features (Fig. 2A–C) that are similar to abnormal cytoplasmic structures termed aggresomes or inclusion bodies (IB).25,31 Typically these perinuclear structures are a strong indication of misfolded proteins and progressive clearance defects and as with protein aggregates are usually positive for ubiquitin and p62.3,15,17,32,33 Similar structures are rarely found in neurons from age-matched wild-type flies (Sup. Fig. 1A–C). In addition, smaller inclusion bodies (IB) and multilamellar bodies (MLB) can be found in other bchs mutant neurons (Fig. 1D). The marked accumulation of intracellular defects (qualitative) as well as altered IUP and insoluble Ref(2)P profiles (quantitative) in bchs mutants strengthens the correlation between western blot analysis and the long-term in vivo capacity of autophagy to clear targets from complex organs, tissues and cells.
Figure 2.
TEM analysis of bchs mutant neurons. (A) TEM image of 2-week-old adult bchs−/− mutant flies (bchs3/bchs6) shows three nerve cells within a single image that contain inclusion bodies (IB) located near the nucleus (N). These perinuclear structures show hallmark features of aggresomes and morphology of other subcellular organelles are abnormal. (B) Enlargement shows the indentation of the nucleus by the inclusion body (black arrows) and the formation of vacuoles (white arrows). (C) In a different neuron the inclusion body is surrounded by multiple membrane layers (white arrows) and is adjacent to two large double membrane organelles (black arrows). (D) Multiple abnormal structures are detected in a different neuronal cell body that include multilamellar bodies (MLB) and electron dense structures that may represent a population of autophagic vesicles or inclusion body-like structures.
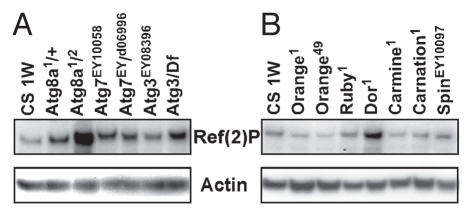
Mutations in other autophagy and lysosomal genes are known to reduce longevity and result in the enhanced accumulation of ubiquitinated proteins.8,9,11,12,29 To determine if changing Ref(2) P protein levels is an effective tool for the detection of autophagic trafficking defects we examined adult neural profiles that were prepared without detergent fractionation. Total Ref(2)P profiles were examined from 1-week-old controls (CS) and adult flies with different allelic combinations of the Atg8a, Atg7 and Atg3 genes (8 heads per genotype). As seen previously a strong allelic combination of Atg8a (Atg8a1/2) produces adult flies with a significant build-up of Ref(2)P at 1 week. Different genetic combinations of Atg7 and Atg3 (Atg7EY10058/d06996 and Atg3EY08396/Df) also show a significant increase in Ref(2)P levels (Fig. 3A and quantified values see Sup. Fig. 2A).
Figure 3.
Defects in autophgy-lysosomal trafficking promotes Ref(2)P accumulation in the adult nervous system. Ref(2)P immunoblot were prepared from total protein extracts taken from 1-week-old control, autophagy and lysosomal trafficking mutant flies (eight heads per genotype). (A) Autophagy mutations that produced viable adults include Atg8a (Atg8a1/Atg8a2), Atg7 (Atg7EY10058, Atg7d06990) and Atg3 (Atg3EY08386). (B) Viable adult mutations in lysosomal trafficking genes were also examined. At 1 week Deep orange mutants (Dor1) show elevated neuronal levels of Ref(2)P, while other lysosomal trafficking mutants (orange1, orange49, ruby1, carmine1, carnation1 and spinsterEY10097) are not significantly different from that of age-matched controls (Canton-S, CS). Quantification of Ref(2)P levels are illustrated in Supplemental Figure 2A and B.
Some lysosomal trafficking mutants show altered Ref(2)P profiles.
Previously we had identified several lysosomal mutants with genetic interactions with bchs that also have shortened adult life spans and early protein trafficking defects.8,29 To further assess the role of lysosomal genes in autophagic clearance, trafficking mutants were collected, aged for 1 week and adult head extracts used for western blot analysis. When compared with age-matched controls, mutations in the orange (Orange1, Orange4), ruby (Ruby1), carmine (Carmine1), carnation (Carnation1) and spinster (Spinster1) genes show normal Ref(2)P levels. This suggests autophagic flux is normal in these mutants, while deep orange (dor) immunoblots show Ref(2)P accumulation (Fig. 3B and quantified values see Sup. Fig. 2B).29,34 We also examined the IUP profiles from several lysosomal trafficking mutants (Sup. Fig. 3) and found that like bchs and Atg8a mutants there is an early accumulation of ubiquitinated proteins in several mutant backgrounds. The disparity between IUP and Ref(2)P profiles in these mutants may result from these proteins having a primary role in endosomal trafficking and a minimal involvement in autophagy or aggrephagy.
To further clarify these findings we examined the formation of LysoTracker® Red positive puncta in larval fat body tissues. This tissue contains large cells that have both fasting- and hormonal-induced forms of autophagy. Multiple studies have used LysoTracker to characterize and quantify the induction of autophagy in this tissue.11,34,35 Following a 3-hour fast, wild-type (Canton-S) 2nd instar larvae show a marked accumulation of LysoTracker Red positive puncta (Sup. Fig. 4A). Larvae expressing an autophagy specific RNAi construct (Cg-Gal4/UAS-dsAtg4, Sup. Fig. 4B) show few if any LysoTracker positive puncta following a fast, while larvae expressing a deep orange RNAi construct (Cg-Gal4/UAS-dsDor, Sup. Fig. 4C) show the formation of positive vesicles with significantly altered morphology. The dor results are consistent with previous findings.34 Fat body cells from wandering 3rd instar larvae also demonstrate a hormone induced form of autophagy (i.e., ecdysone) that is independent of fasting. When 3rd instar wild-type fat body tissues were stained with LysoTracker Red, abundant vesicles could be detected throughout the cell (Sup. Fig. 5A). As seen previously dor4 mutant males (Vps18p, E3 ubiquitin ligase) at this stage show defects in vesicle and lipid droplet morphology (Sup. Fig. 5B).34 In carnation1 mutant animals (Vps 33, sec1 protein) the number and morphology of puncta and lipid droplets are very similar to that of wild type controls (Sup. Fig. 5C).29 This indicates the function of the Carnation protein is primarily involved with endosomal trafficking and likely has minimal effect on autophagy. Imaging results, Ref(2)P and IUP profiles (Sup. Fig. 3) are consistent with our previous findings showing the dual role of Dor in the autophagic and endosomal pathways.34 These results also indicate that western blot analysis of total Ref(2)P levels is an effective screening tool to characterize progressive defects due to altered autophagic-lysosomal function in the adult nervous system.
Ubiquitin and Ref(2)P profiles following caloric restriction.
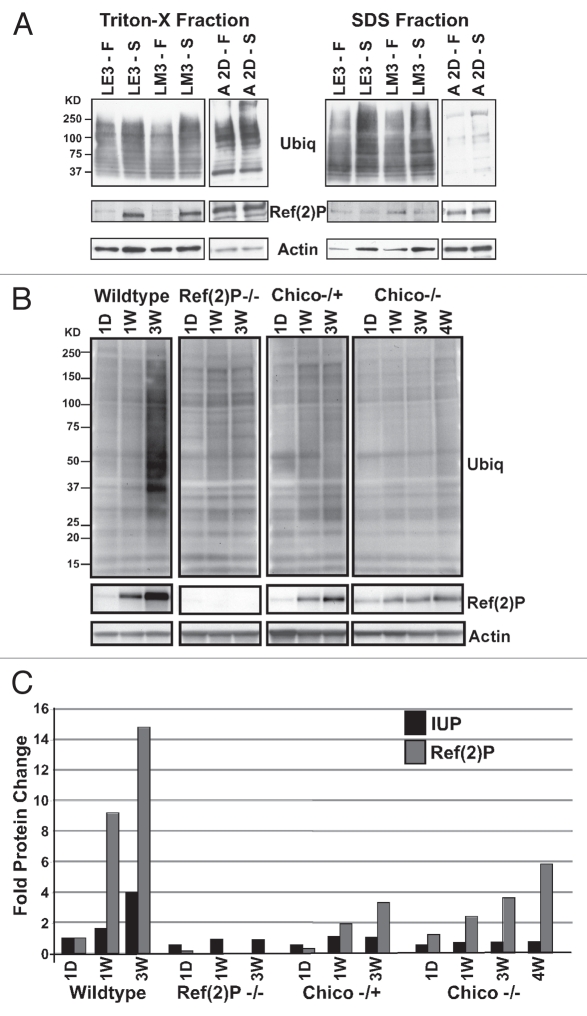
Outside of the nervous system many Drosophila tissues show a dynamic autophagic response that is triggered by developmental cues (Sup. Figs. 6 and 7) or environmental factors such as caloric restriction or fasting. Studies using cultured primary neurons indicate the pathway can be induced following nutrient deprivation,36 while whole animal in vivo studies suggest the intact nervous system shows little change in basal autophagic levels following a fast.7,37 To examine whether ubiquitin or Ref(2)P profiles change in neuronal and non-neuronal tissues following a fast, early 3rd and middle 3rd instar larvae (LE3 and LM3) and 2-day-old adult wild-type flies were collected and kept under normal culturing conditions (Fed, F) or were fasted (Starved, S). Serial detergent extractions were prepared from larval fat body tissues or adult heads, immunoblotted and probed for ubiquitin, Ref(2)P and actin. IUP profiles in both fractions are slightly elevated in fasted larval tissues, following the activation of autophagy.34,35 Triton-X soluble Ref(2)P levels are elevated in fasted larvae, while SDS profiles show a reduction (Fig. 4A and see Sup. Fig. 5 for quantification). The build-up of soluble Ref(2)P is consistent with other results showing p62 role in specifying substrates for autophagic clearance (for quantification see Sup. Fig. 8A and B).13,17,38,39 However, the minor change in soluble and insoluble ubiquitinated protein profiles suggests cytoplasmic inclusions or aggregates are not formed during a fast. This indicates that following acute induction of the pathway, trafficking or flux of substrates remains relatively effective. Adult neural tissues show different protein profiles following a fast. Soluble and IUP profiles show only a modest increase, while Ref(2)P has little or no change in either fraction (Fig. 4A and for quantification see Sup. Fig. 8A and B). This indicates that autophagic flux is not dramatically altered in neural tissues from animals placed under fed and fasted conditions and is consistent with in vivo mouse studies showing neurons have a limited ability to activate autophagy above basal levels following fasting.7,37
Figure 4.
Metabolic Changes and Altered IUP and Ref(2)P Neural Profiles. (A) Early and mid 3rd instar larvae (LE3, LM3) were maintained under fed (F) or starvation (S) conditions, as were 2-day-old adult Drosophila. Larval fat tissues and adult heads were collected after 3 hours or 12 hours of fasting, respectively. Immunoblots were prepared from the Triton-X and SDS fractions and probed with ubiquitin, Ref(2)P and actin antibodies. (B) SDS head extract immunoblots were prepared from 1-day-old, 1-, 3- or 4-week-old wild-type, Ref(2)P−/− mutants (Ref(2)Pc/e) and heterozygous−/+ and homozygous chico−/− mutant flies (chico1/+, chico1/2). Both IUP and Ref(2)P profiles show similar age and genotype-dependent accumulation patterns with the exception of Ref(2)P null flies. (C) IUP, Ref(2)P and actin values for each age and genotype were quantified using ImageJ. The corrected IUP and Ref(2)P values for 1-day-old wild-type flies were set at one, other values adjusted and graphed using these values.
IUP and Ref(2)P profiles indicate enhanced neural autophagy and longevity.
The reduced levels of IUP and Ref(2)P detected in older Atg8a+ flies (Fig. 1A and APPL-GAL4/UAS-Atg8a) indicates that detergent extraction coupled with western blot analysis could be used to detect conditions were neuronal autophagy is enhanced. To explore this possibility we examined adult flies containing mutations in the chico gene (chico1/2, insulin receptor substrate homologue, IRS).40,41 Highly conserved signaling pathways, defects in both the insulin and IGF (insulin-like growth factor) signals are known to enhance longevity and link caloric restriction with life-span extension.42,43 The cellular mechanisms mediating these effects are not entirely clear, but both pathways are known to enhance TOR signaling (target of rapamycin), which in turn directly suppresses the activation of autophagy.44–46 For this study, wild-type, ref(2)Pc/e, chico1/+ and chico1/2 adults were collected (20 per vial) and aged for 1 day or for 1, 3 or 4 weeks. Flash frozen heads were collected, detergent extracted and used for analysis of IUP, Ref(2)P and Actin profiles. Wild-type flies show a normal age-dependent accumulation of IUP and Ref(2)P, while ref(2)Pc/e null mutants have reduced neuronal IUP levels at 3 weeks of age (Fig. 4B). This is consistent with previous findings that examined ubiquitin profiles Ref(2)P mutants and p62 knockout mice.13,18,19 Heterozygous and homozygous chico mutants show a similar decrease in neural IUP levels. Further, long-lived chico mutants also have reduced Ref(2)P levels at the critical 3–4 week time point.8,41 This suggests that basal autophagy rates are enhanced in neurons in which insulin signaling is suppressed and clearance of autophagic substrates is enhanced.40,41 Ref(2)P and Actin densitometry readings were used to generate corrected values to illustrate protein levels of fly extracts prepared from different genotypes and ages (Fig. 4C). An examination of Atg8a and ref(2)P expression profiles in young homozygous chico mutants shows that both genes start at significantly lower levels than those found in age-matched controls (1 week, Sup. Fig. 9) and over time both mRNA profiles are further reduced (4 week, Sup. Fig. 9). The initial low expression levels of Atg8a and ref(2)P may reflect the early developmental and metabolic defects that are caused by insulin signaling defects.41,42 The further decline in expression is also reflected in Atg8a profiles found in 1-week-old and 4-week-old ref(2)P mutants, suggesting the decline in autophagy gene expression is a consistent feature of neuronal aging (Sup. Fig. 9).
p62 accumulation associates with progressive human neurological disorders.
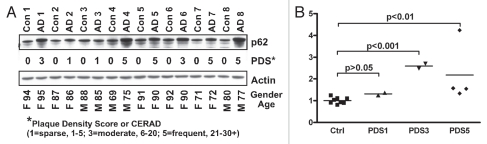
Immunohistochemical studies of human tissues show ubiquitin and p62 are common components of pathological inclusions that are associated with neurological disorders such as Alzheimer disease (AD).20,21,47,48 As a quantitative assessment of aggregate formation, western blot analysis of human neural samples was preformed using p62 as potential marker of neural degeneration. Flash frozen post-mortem tissues, from AD patients or from age- and gender-matched controls, were obtained from the USC brain bank.49 Previously, adjacent tissues had undergone a histological examination and were assigned plaque density (PDS) and CERAD scores.47,49,50 Adjacent flash frozen tissue samples were processed directly in 2% SDS extraction buffer and western blots were probed with p62 and Actin antibodies (Fig. 5A).49,51 When compared with the different PDS we find greater p62 accumulation in tissues with higher PDS (Fig. 5B and PDS3, p < 0.001 and PDS5, p < 0.01). Autophagic defects and the ability of the pathway to clear intracellular neural aggregates are implicated as compounding factors in progressive neural degeneration.1,25,52,53 Results from complex human tissue preparations suggest that western blot analysis of p62 levels closely correlates with progressive loss of autophagic function and AD plaque accumulation.20,21
Figure 5.
p62 profiles from human Alzheimer disease neural tissues. (A) Mid-frontal cortical tissue samples were taken from AD patients or control subjects (age and gender matched), flash frozen in liquid nitrogen and stored at −80°C. Protein extracts (2% SDS) were examined for p62 and actin. Previous histological examinations were performed on adjacent tissue samples and plaque density score (PDS) for a particular individual were assigned. PDS, age (years) and gender (M = male or F = female) are indicated for each sample. (B) Densitometry values for the two major p62 bands (arrows) were corrected using actin and normalized to the average p62 levels detected in control samples (PDS = 0, N = 8). Individuals in the PDS1 group had p62 levels that were not substantially different than controls (n = 2, p > 0.05), while samples from individuals with higher PDS showed a significant accumulation of p62 (one way ANOVA with Bonferroni post-test, p < 0.01, PSD3 N = 2; PDS N = 4).
Autophagic defects alter p62 profiles in mammalian tissue culture cells.
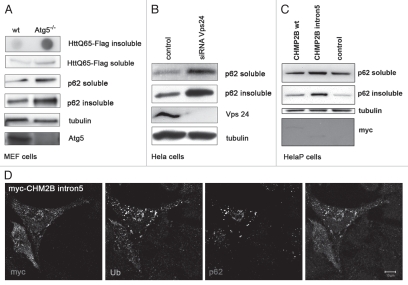
The results from AD samples are also corroborated by work involving mammalian tissue culture cells. Previously, several studies have shown a strong correlation between the inhibition of autophagy and the accumulation of IUP, p62 and other proteins associated with neurodegenerative disease. For this study, Atg5+/+ (wild-type) and Atg5−/− mouse embryonic fibroblast (MEF) cells were transfected with an aggregate-prone PolyQ-containing protein that is associated with Huntington disease (Flag-Htt-Q65).10,27,54 Cells were cultured for 5 days and detergent extracts used for immunoblots that were probed using p62, Flag and α-tubulin antibodies (Fig. 6A). There is marked Htt-Q65 and p62 accumulation in the insoluble protein fraction.27 In contrast there was little or no change in the level of soluble HttQ65 between wild-type and Atg5−/− KO cells.27 Atg5 western blots confirm loss of the gene in the Atg5−/− KO MEF cells, and LC3 immunoblots show inhibition of the pathway (data not shown).10,54
Figure 6.
Mammalian cell culture proteopathies models and p62 profiles. (A) Atg5+/+ (wild type) and Atg5−/− mouse embryonic fibroblasts (MEFs) were transfected with Flag-HttQ65 (48 hrs) or were untreated before extraction in 1.0% Triton-X (soluble) and 2% SDS buffers (insoluble). Samples were immunoblotted with anti-Flag, anti-Atg5 and anti-p62 antibodies. Equal loading was verified using anti-α-Tubulin and Atg5 protein levels confirmed using anti-Atg5 antibodies. Clearance of SDS-soluble Flag-HttQ65 inclusions was further analyzed by filter-trap assays (upper part). (B) HeLa cells were transfected with control or Vps24 siRNAs for 5 days and then processed for p62 western blot analysis. Knockdown efficiencies and loading were verified using Vps24 and α-Tubulin levels. (C) HeLa cells were transfected for 48 hrs with myc-tagged constructs containing wild-type myc-CH MP2B or a mutant myc-CHMP2Bintron5 construct and mock-transfected cells were used as control. Cells were extracted, immunoblotted and probed with p62, myc and α-Tubulin antibodies. Cells expressing mutant CHMP2B show p62 accumulation, whereas control and cells expressing wild-type myc-CHMP2B have similar low levels of p62. Anti-myc and anti-α-Tubulin immunoblots were used as expression and loading controls. (D) HeLa cells were transfected for 48 hrs with myc-CHMP2Bintron5, fixed and processed for immunofluorescence analysis. Cells were labeled with antibodies against myc, ubiquitin and p62/SQSTM1 and imaged using confocal microscopy. Co-localization is indicated in white (right panel, scale bar = 10 µm).
Inhibition of autophagosome maturation also leads to accumulation of IUP and p62.17 Interestingly, recent studies have identified a link between mutations found in patients suffering from a familial form of frontotemporal dementia (FTD3) and abnormal autophagosome maturation.55 FTD3 patients have extensive and early onset neurological disorders that include the formation of neural aggregates containing ubiquitin.28,56 To examine this further we ectopically expressed in HeLa cells a mutant form of CHMP2B (ESCRT-III subunit, CHMP2B-intron5), which corresponds to a genetic defect found in FTD3 patients.22 Expression of mutant CHMP2B in HeLa cells causes a significant accumulation of p62 and ubiquitin, as analyzed by immunoblots and immunoflourescence microscopy. Cells expressing the native protein are similar to controls.22 Equal expression and loading were verified using anti-Myc and anti-Tubulin antibodies (Fig. 6C). Moreover, as seen with other intracellular aggregate models, p62 closely co-localized with large inclusions that contain both the mutant CHMP2B protein and ubiquitin (Fig. 6D).17,22 To determine whether siRNA-mediated suppression of autophagy could also change p62 solubility profiles, cells were transfected with siRNA against Vps24, a second subunit of the ESCRT-III complex (endosomal sorting complex required for transport).22,57,58 Western blots show that Vps-24-deficient cells have marked p62 accumulation in both detergent fractions, indicating the protein becomes less soluble as Vps24 becomes depleted and autophagosome-lysosmal fusion and substrate flux is suppressed (Fig. 6B).17 In conclusion, our studies show a clear association between the autophagy levels, accumulation of p62/Ref(2)P and IUP and progression of neurological disorders both in vivo and in vitro.
Discussion
Recent work has shown that the p62/SQSTM1/Ref(2)P protein family plays a central and expanding role in the autophagy pathway, in part by serving as a selective receptor for substrates marked with ubiquitin. The unique combination of LIR, UBA motifs and other interacting domains in p62 and Ref(2)P allows these proteins to interface with autophagy components (Atg8/LC3) as well as ubiquitinated intracellular substrates that are destined for elimination.17,38,59 p62 has been shown to promote the clearance of other ubiquitinated substrates including bacteria and vacuolar remnants.59–61 The ability of p62 to shuttle between the nucleus and cytoplasm as well as its role in the formation of cytoplasmic p62 bodies suggests the protein may be a key component of a complex signaling process that occurs during periods of cellular stress and aggregate formation.17,38,62 Along these lines, a recent study has shown p62 modulates the activity of the stress responsive Nrf2 transcription factor and may also be involved with the regulation of ARE-containing genes.18,63 In neural degenerative disorders, which are associated with the formation of protein aggregates or inclusions, p62 serves as an effective histological marker of pathological structures.
In this report we show that measuring insoluble ubiquitinated proteins as well as p62 and Ref(2)P profiles can be used to detect the formation of protein inclusions and changes in autophagic activity under a wide range of cellular and physiological conditions. Previously, we had shown that detergent fractionation of proteins from complex neural tissues can be used to generate a reproducible series of samples that contain ubquitinated proteins with different solubility profiles.8,9,64 Coupling this technique with western blot analysis of Ref(2)P and p62, we found that progressive defects in autophagy, due to age or loss-of-function mutations, could be readily detected. Based on these results we expanded this technique to include immunoblot analysis of p62 or Drosophila Ref(2)P profiles to examine aggregate formation in neural tissues and cell culture models. Using this method we demonstrated that both IUP and Ref(2)P protein profiles show a marked accumulation and solubility shift in neural samples prepared from older flies (Figs. 1 and 4B). This time-dependent build-up is consistent with the natural reduction of autophagy gene expression profiles that occurs in the aging Drosophila nervous system.8 The age-dependent decrease in the ref(2)P message indicates insoluble Ref(2)P accumulation in older flies is likely the result of a progressive global decline in autophagy and not due to enhanced production of the protein (Fig. 1C). Further, genetic and transgenic suppression or enhancement of neuronal autophagy also has a profound effect on the in vivo levels of insoluble Ref(2)P and IUP (SDS fraction). The extent of Ref(2)P accumulation indicates that western blot analysis could be used as part of a genetic screen for mutations that accelerate neuronal aging or effect aggrephagy rates. Consistent with this hypothesis we find a marked change in neuronal IUP levels in a diverse series of lysosomal trafficking mutants. However, only dor mutations (Vps18, class C complex) show a marked increase in neuronal Ref(2)P levels (Fig. 3B), IUP profiles (Sup. Fig. 3) as well as defects in autophagosome formation (Sup. Figs. 4 and 5).29,34
Western blot analysis of Drosophila samples from different fasting and developmental conditions, shows Ref(2)P and ubiquitin profiles change dramatically at times consistent with activation of autophagy.35,65–67 In terms of inclusion body or aggregate formation, we identified two exceptions. The first is that the soluble and insoluble profiles of both markers increase at a time when hormonally triggered autophagy is occurring (Sup. Figs. 7 and 8C and D). The second is that fasting does not increase the insoluble Ref(2)P or IUP levels in adipose tissues and neither marker is significantly altered in adult neural extracts following caloric restriction (Fig. 4A and Sup. Fig. 8A and B). This suggests that flux or clearance of substrates in adipose tissues is not impeded following an acute induction of the pathway and in turn may prevent the accumulation of cytoplasmic insoluble inclusions. Further, the minor change in neural IUP and Ref(2)P profiles in adults following a fast is consistent with previous work showing that neurons have a limited ability to increase in autophagosome formation following caloric restriction.7,37 However, we did find that IUP and Ref(2)P neural profiles are reduced when insulin signaling is suppressed. Mutations in chico (IRS, insulin receptor substrate) are similar to other mutations in the insulin/IGF signaling pathways in that small but long-lived and stress resistant individuals are produced.68,69 The small size of chico results in part from changes in downstream TOR signaling, which can also regulate basal rates of autophagy via TORC1 signaling.35,44,45,70 Our western blot results are consistent with multiple studies showing that defects in insulin/IGF signaling as well as caloric restriction can slow cellular aging, potentially by enhancing autophagy rates in critical tissues like the nervous system.35,44,45,70
In cultured cells we find that p62 solubility profiles are altered under conditions where autophagy is either directly suppressed (Atg5−/− KO, Fig. 6A), when ESCRT-III complex function is inhibited (Vps24 siRNA, Fig. 6B), or when disease associated proteins such as the CHMP2B protein is expressed (mutant CHMP2Bintron5, Fig. 6C).10,22,23 As seen in Atg8a and bchs Drosophila mutants, functional loss of Atg5 in the mouse CNS results in the accumulation of ubiquitinated proteins10 and inability of cells to effectively sequester aggregate prone proteins (i.e., HttQ65) and accumulation of p62.17,27 Another disorder, frontotemporal dementia (FTD), is the most common form of neural degeneration occurring in people before the age of 60.22,71 The ESCRT-III complex, which functions primarily in late endosomal trafficking events, is associated with familial cases of FTD as well as the accumulation of ubiquitin-positive protein deposits in affected neurons.22 Mutations in individual members like CHMP2B and suppression of Vps24 are known to inhibit autophagy and exacerbate the accumulation of autophagosomes and p62 in cultured cell models of the disorder.22,23,71
Finally our work with human tissues indicates that western blot analysis of p62 is potentially a useful measure of proteopathies associated with Alzheimer disease and plaque formation (Fig. 5). There appears to be an activation of neuronal autophagy during the early phases of the disease but as the disorder progresses the pathway becomes increasingly abnormal resulting in the accumulation of cytoplasmic vesicles and inclusions.25,26,53,72 Western blot analysis of p62 shows a strong correlation with the long-term accumulation of protein aggregates in human neural tissues. Recent findings demonstrate that ubiquitinated inclusions contain a diverse complement of Ub-Ub isoforms suggest autophagy has the capacity to recognize and respond to a diverse array of aggregating proteins.18,63 Cellular assays designed to measure autophagosome formation (i.e., GFP-LC3) and LC3-I/LC3-II ratios have been instrumental in assessing the acute activation of autophagy and shaping our understanding of the complex role the pathway plays in cellular homeostasis. The work presented in this report suggests that analysis of ubiquitinated proteins as well as p62/Ref(2)P profiles can also be used to establish the long-term level of autophagic activity that is occurring in complex tissues like the nervous system.
Materials and Methods
Drosophila stocks and culturing conditions.
The Canton-S, Df(2L)clot7/CyO, Atg8a1, Atg82/FM7C, light1, orange1, orange4, ruby1, deep orange1, carmine1, carnation1, chico1/CyO, chico2/CyO and spinster1, stock lines were identified using Flybase and obtained from the Bloomington Stock Center (flybase.bio.indiana.edu). The Ref(2)Pc03993 and Ref(2)Pe00482 lines were obtained from the Harvard Medical School Exelixis Stock Collection, the bchs3 and bchs6 alleles have been previously described in reference 9, the hook1 line was a gift from Dr. H. Kramer (University of Texas, Southwestern Medical Center, Texas) and the APPL-Gal4 pan-neural driver line was a gift from Dr. K. White (Brandeis University, MA).8 The Atg8a1 and Atg8a2 alleles (Atg8a−) and the Atg8a+ genotype (APPL-Gal4/UAS-Atg8a) have been previously reported in reference 8. To age adults, female or male flies were collected and maintained separately at 25°C (25 flies per vial) on standard Drosophila media for the indicated time. Flies were transferred to vials containing fresh media every 3 to 4 days until collected and used for protein extracts.8,9
Antibodies.
Primary antibodies were from the following sources: rabbit polyclonal anti-Ref(2)p (1:3,000 dilution, Dr. Contamine, Versailles, France); mouse monoclonal anti-ubiquitin (1:1,000 dilution, Cell Signaling Technologies, 3933); poly-ubiquitin antibodies (clone FK1 and FK2, Affinity); β-actin (1:10,000 dilution Cell Signaling Technologies, 4967); mouse monoclonal anti-tubulin (Sigma, T8203); mouse monoclonal anti-myc (ATCC, 9E10); polyclonal goat anti-Atg5 (Santa Cruz Biotechnology, sc-133158); rabbit polyclonal anti-APG5L (Abgent, AP1812a); monoclonal SQSTM1/p62 (Abgent, AT3836a); guinea pig SQSTM1/p62 (US Biological, P1001-33Y); mouse monoclonal anti-Htt (Chemicon, MAB5492); rabbit polyclonal anti-LC3B (Abcam, AP1802b). Secondary antibodies Cy2-, Cy3- and Cy5-labeled antibodies were from Jackson ImmunoResearch Laboratories.
SDS-PAGE and western blot analysis.
Briefly, to detect the accumulation of insoluble p62/Ref(2)P and ubiquitinated proteins (IUP) in cultured cells and fly heads (∼20 per genotype or age) were collected and flash frozen or processed immediately. Samples were first homogenized in 1.0% Triton-X, PBS containing protease inhibitors (4°C) and centrifuged (14,000 rpm) for 10 minutes (4°C). The supernatants were collected for each data set and saved as the Triton-X soluble fraction. The remaining protein pellets were washed with 1.0% Triton-X, PBS, followed by a second 2% SDS buffer extraction.8,9 Between 10 to 20 µg of protein were loaded for each sample and resolved on 4–12% gradient gels (BioRad, 345-0124), followed by electroblotting on to Immobilon-P membranes (Millipore, IPVH00010). For analysis of total proteins, heads from lysosomal mutants (20 per age and genotype) were homogenized directly in a 2% SDS buffer (protease inhibitors at 4°C, Roche, 11 836 170 001). 20 µg of protein were loaded for each sample and resolved on 4–12% gradient gels. As loading controls we used anti-Histone2B, anti-Actin5c or anti-a-Tubulin antibodies.
A similar extraction technique was used for cultured cells to establish the cellular levels of different proteins and their solubility profiles. Cells were extracted in ice-cold lysis buffer (50 mM NaCl, 10 mM Tris, 5 mM EDTA, 0.1% SDS, 1% Triton X-100 + protease and phosphatase inhibitor cocktails), centrifuged (14,000 rpm) for 10 minutes and the supernatants (soluble fraction) collected. The remaining protein pellets were washed with phosphate buffered saline (PBS) and further extracted with 2% SDS-containing sample buffer (insoluble fraction). Protein concentrations in the soluble fractions were determined and approximately 20 µg of protein per sample was loaded and resolved on 15% or 4–20% gradient gels, and electroblotted on to Immobilon-P membranes. Preparation of human samples involved the homogenization of ∼100 mg of tissues in extraction buffer (50 mM Tris pH 7.5, 2% SDS, PBS and protease inhibitors at 4°C). 10 µg of protein was resolved for each sample on a 10% SDS-Page gel (BioRad 345-0112) and immunoblots were sequentially probed with p62 and actin antibodies. Protein concentrations were determined using the detergent compatible Lowry assay (BioRad, 500-0111). The blots were probed with specific antibodies and detected using standard ECL reagents (Pierce® ECL, 32106). Band intensities were quantified using the software provided by the ChemiGenius imaging system (Syngene) or digitally scanned using a Calibrated Desitometer and Quantity One imaging and analysis software (BioRad, GS-800). Corrected protein values were graphed using Microsoft Excel or Graphpad software. Statistical analysis was done by ANOVA followed by Bonferroni's test and p-values <0.05 were considered significant.
Quantitative RT-PCR analysis.
Heads were collected from male Canton-S flies at 1, 2 and 4 weeks of age. The Trizol RNA isolation reagents and techniques (Invitrogen, 15596-026) were used to prepare total RNA from each sample. Purity and quantity were measured by optical density.8 For each condition 1.0 µg of total RNA was used for cDNA synthesis using the Fermentas primers and cDNA synthesis kit (Fermentas, K1622). Real-time PCR was performed in 25 µl reactions containing 12.5 µl of 2x SensiMix™ SYBR and Fluorescein Kit (Bioline Inc., QT615-05), 25 ng cDNA and pretested gene specific primer sets into 96-well optical plates. The cycling conditions for the BioRad CFX96™ Real-Time PCR Detection System were 95°C for 5 min, 40 cycles of 94°C 5 sec, 55°C 20 sec and 72°C 20 sec.8 Real-time efficiencies were calculated from the slopes of standard dilution curves. RNA transcription levels were determined by the method of direct comparison of CT values (CT >35 rejected) TT and relative quantities were calculated by the ΔΔCT equation using Excel software.8 Transcripts were normalized to actin-5C values for each condition. All qRT-PCR reactions were done in using two to four independent experiments.
Cell culture, transfections and siRNA techniques.
Control Atg5+/+ and Atg5−/− KO mouse embryonic fibroblasts were a gift from Noboru Mizushima. Both cell types were left untreated or transfected with Flag-HttQ65 construct for 48 hrs.73 Cells were lysed in mild detergent (soluble), centrifuged and the remaining pellet extracted in 2% SDS (insoluble).64 Proteins were separated by SDS-PAGE and immunoblotted and probed with anti-Flag (1:100 dilution) and anti-p62 (1:1,000 dilution) antibodies. Equal loading was verified by anti-α-tubulin immunoblotting. The absence of the Atg5 protein and the inhibition of autophagy were determined using anti-Atg5 (1:100) and anti-LC3 (1:100) antibodies, respectively. Clearance of SDS-insoluble Flag-HttQ65 inclusions was analyzed by filter-trap assays. 48 h after siRNA-Vps24 transfection, HeLa htt25Q, 65Q- and 103Q-mCFP cells were exposed to 100 ng/ml dox for another 3 days to suppress htt-protein production and permit more than 50% of clearance. The effect of siRNA treatment on polyQ aggregate clearance was analyzed either by confocal quantification as described in reference 73, or by the membrane filter trap assay for detection of amyloid-like polyglutamine-containing protein aggregates as previously published in reference 27. For studies of the Vps24 gene, HeLa cells were transfected with control or Vps24 siRNA for 5 days and processed for western blot analysis as described. Immunoblotting using anti-p62 antibodies was performed for both in the soluble and insoluble fractions of Vps24-deficient and control cells. Knockdown efficiencies were analyzed using an anti-Vps24 antibody and equal loading verified using anti-a-tubulin antibody. HeLa cells were transfected for 48 hrs with myc-tagged wild-type CHMP2B or a mutant version that corresponds to the mutation found to cause disease in patients having FTD3 (myc-CHMP2Bintron5). Mock-transfected cells were used as controls. Cells were detergent fractionated and analyzed by immunoblotting using anti-p62 antibodies. Equal expression and loading levels were verified by anti-Myc and anti-α-Tubulin immunoblots, respectively.
Transmission electron microscopy (TEM) and confocal imaging techniques.
Wild-type (Canton-S) and bchs mutant flies were collected and aged between 1 and 15 days before heads were removed, dissected and fixed in 2% glutaraldehyde in 0.1 M Cacodylate buffer, pH 7.2. The tissue was post-fixed with 2% OsO4 and 1.5% KFeCN, dehydrated in a graded ethanol series and washed in a propylene oxide solution. Samples were oriented and embedded in Epon/Araldite resin and polymerize overnight at 60°C. Thin sections (40–60 nm) were cut on a Leica Ultracut, mounted on Parlodian coated copper slot grids before staining with Pb citrate.8,74 Sections were observed at 60 kV using a JEOL1230 electron microscope. Images were recorded with a Morada digital camera and prepared further using Adobe Photoshop. Fat body tissues were isolated from staged larvae (6–10 per genotype and condition) and stained in PBS containing LysoTracker® Red (1:1,000 dilution, Invitrogen, L7528) for 10 min.35 Tissues were rinsed with PBS, fixed and imaged immediately using a Lieca confocal microscope and lasers. The number of puncta contained within 50 µm2 area from several tissues were counted. Averages and standard deviations were calculated and graphed using Excel software. HeLa cells were transfected for 48 hrs with myc-CHMP2B-intron5 were fixed and labeled with antibodies against Myc (red), Ubiquitin (green) and p62 (blue) for immunofluorescence. Co-localization of the different markers is indicated in white (Scale bar = 10 mm). Adobe Photoshop CS3 and Canvas X imaging software were used to assemble figures.
Acknowledgements
We would like to thank Malcom Wood for his outstanding TEM studies of Drosophila neural tissues. We would also like to thank Didier Contamine and Sébastien Gaumer for their generous gift of antibodies and Helmut Kramer and Kalpana White for Drosophila lines and Noboru Mizushima for providing Atg5−/− KO MEF cells. We would also like to thank Dr. Natalie Gude and Dr. Mark Sussman (SDSU) for their imaging expertise and Lena van der Stap for her review of this manuscript. This work was supported by grants from the NIH/NIA R43AG033427 and R21AG030187 (K.D.F., B.J.B., H.S.), R01AG033283 (K.D.F., R.A.G.), and the FUGE program of the Norwegian Research Council (P.I., I.N., A.S.).
Abbreviations
- IUP
insoluble ubiquitinated proteins
- UBA domain
ubiquitin-associated domain
- ALFY
autophagy-linked FYVE protein
- bchs
blue cheese
- SQSTM1
sequestosome-1
- ESCRT
endosomal sorting complex required for transport
- IRS
insulin receptor substrate homologue
- CHMP2B
charged multivesicular body protein 2b
- FTD
frontotemporal dementia
- GFP
green fluorescent protein
Supplementary Material
References
- 1.Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- 2.Zerovnik E. Protein Conformational Pathology in Alzheimer's and other Neurodegenerative Diseases; New Targets for Therapy. Curr Alzheimer Res. 2009 doi: 10.2174/156720510790274437. [DOI] [PubMed] [Google Scholar]
- 3.Nagaoka U, Kim K, Jana NR, Doi H, Maruyama M, Mitsui K, et al. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J Neurochem. 2004;91:57–68. doi: 10.1111/j.1471-4159.2004.02692.x. [DOI] [PubMed] [Google Scholar]
- 4.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 5.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melendez A, Neufeld TP. The cell biology of autophagy in metazoans: a developing story. Development. 2008;135:2347–2360. doi: 10.1242/dev.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klionsky DJ, Cuervo AM, Dunn WA, Jr, Levine B, van der Klei I, Seglen PO. How shall I eat thee? Autophagy. 2007;3:413–416. doi: 10.4161/auto.4377. [DOI] [PubMed] [Google Scholar]
- 8.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 9.Finley KD, Edeen PT, Cumming RC, Mardahl-Dumesnil MD, Taylor BJ, Rodriguez MH, et al. blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration. J Neurosci. 2003;23:1254–1264. doi: 10.1523/JNEUROSCI.23-04-01254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 11.Juhasz G, Neufeld TP. Drosophila Atg7: required for stress resistance, longevity and neuronal homeostasis, but not for metamorphosis. Autophagy. 2008;4:357–358. doi: 10.4161/auto.5572. [DOI] [PubMed] [Google Scholar]
- 12.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 14.Lindmo K, Brech A, Finley KD, Gaumer S, Contamine D, Rusten TE, et al. The PI 3-kinase regulator Vps15 is required for autophagic clearance of protein aggregates. Autophagy. 2008;4:500–506. doi: 10.4161/auto.5829. [DOI] [PubMed] [Google Scholar]
- 15.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 16.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, et al. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010:6. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 18.Riley BE, Kaiser SE, Shaler TA, Ng AC, Hara T, Hipp MS. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J Cell Biol. 2010;191:537–552. doi: 10.1083/jcb.201005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoozemans JJ, van Haastert ES, Nijholt DA, Rozemuller AJ, Eikelenboom P, Scheper W. The unfolded protein response is activated in pretangle neurons in Alzheimer's disease hippocampus. Am J Pathol. 2009;174:1241–1251. doi: 10.2353/ajpath.2009.080814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waguri S, Komatsu M. Biochemical and morphological detection of inclusion bodies in autophagy-deficient mice. Methods Enzymol. 2009;453:181–196. doi: 10.1016/S0076-6879(08)04009-3. [DOI] [PubMed] [Google Scholar]
- 22.Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusten TE, Simonsen A. ESCRT functions in autophagy and associated disease. Cell Cycle. 2008;7:1166–1172. doi: 10.4161/cc.7.9.5784. [DOI] [PubMed] [Google Scholar]
- 24.Kuusisto E, Salminen A, Alafuzoff I. Early accumulation of p62 in neurofibrillary tangles in Alzheimer's disease: possible role in tangle formation. Neuropathol Appl Neurobiol. 2002;28:228–237. doi: 10.1046/j.1365-2990.2002.00394.x. [DOI] [PubMed] [Google Scholar]
- 25.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nixon RA. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007;120:4081–4091. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 27.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seelaar H, Klijnsma KY, de Koning I, van der Lugt A, Chiu WZ, Azmani A, et al. Frequency of ubiquitin and FUS-positive, TDP-43-negative frontotemporal lobar degeneration. J Neurol. 2009 doi: 10.1007/s00415-009-5404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonsen A, Cumming RC, Lindmo K, Galaviz V, Cheng S, Rusten TE, et al. Genetic modifiers of the Drosophila blue cheese gene link defects in lysosomal transport with decreased life span and altered ubiquitinated-protein profiles. Genetics. 2007;176:1283–1297. doi: 10.1534/genetics.106.065011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, et al. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 31.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 32.Garcia-Mata R, Gao YS, Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002;3:388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- 33.Johnston JA, Illing ME, Kopito RR. Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil Cytoskeleton. 2002;53:26–38. doi: 10.1002/cm.10057. [DOI] [PubMed] [Google Scholar]
- 34.Lindmo K, Simonsen A, Brech A, Finley K, Rusten TE, Stenmark H. A dual function for Deep orange in programmed autophagy in the Drosophila melanogaster fat body. Exp Cell Res. 2006;312:2018–2027. doi: 10.1016/j.yexcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Young JE, Martinez RA, La Spada AR. Nutrient deprivation induces neuronal autophagy and implicates reduced insulin signaling in neuroprotective autophagy activation. J Biol Chem. 2009;284:2363–2373. doi: 10.1074/jbc.M806088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A, Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 39.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 40.Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 41.Tu MP, Epstein D, Tatar M. The demography of slow aging in male and female Drosophila mutant for the insulin-receptor substrate homologue chico. Aging Cell. 2002;1:75–80. doi: 10.1046/j.1474-9728.2002.00010.x. [DOI] [PubMed] [Google Scholar]
- 42.Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 43.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arsham AM, Neufeld TP. Thinking globally and acting locally with TOR. Curr Opin Cell Biol. 2006;18:589–597. doi: 10.1016/j.ceb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 45.Diaz-Troya S, Perez-Perez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4:851–865. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- 46.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 47.Cole GM, Timiras PS. Ubiquitin-protein conjugates in Alzheimer's lesions. Neurosci Lett. 1987;79:207–212. doi: 10.1016/0304-3940(87)90698-7. [DOI] [PubMed] [Google Scholar]
- 48.Kadokura A, Yamazaki T, Lemere CA, Takatama M, Okamoto K. Regional distribution of TDP-43 inclusions in Alzheimer disease (AD) brains: their relation to AD common pathology. Neuropathology. 2009;29:566–573. doi: 10.1111/j.1440-1789.2009.01017.x. [DOI] [PubMed] [Google Scholar]
- 49.Cumming RC, Schubert D. Amyloid-beta induces disulfide bonding and aggregation of GAPDH in Alzheimer's disease. FASEB J. 2005;19:2060–2062. doi: 10.1096/fj.05-4195fje. [DOI] [PubMed] [Google Scholar]
- 50.Metsaars WP, Hauw JJ, van Welsem ME, Duyckaerts C. A grading system of Alzheimer disease lesions in neocortical areas. Neurobiol Aging. 2003;24:563–572. doi: 10.1016/s0197-4580(02)00134-3. [DOI] [PubMed] [Google Scholar]
- 51.Long J, Garner TP, Pandya MJ, Craven CJ, Chen P, Shaw B, et al. Dimerisation of the UBA domain of p62 inhibits ubiquitin binding and regulates NFkappaB signalling. J Mol Biol. 2010;396:178–194. doi: 10.1016/j.jmb.2009.11.032. [DOI] [PubMed] [Google Scholar]
- 52.Ling D, Song HJ, Garza D, Neufeld TP, Salvaterra PM. Abeta42-induced neurodegeneration via an age-dependent autophagic-lysosomal injury in Drosophila. PLoS ONE. 2009;4:4201. doi: 10.1371/journal.pone.0004201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang DS, Lee JH, Nixon RA. Monitoring autophagy in Alzheimer's disease and related neurodegenerative diseases. Methods Enzymol. 2009;453:111–144. doi: 10.1016/S0076-6879(08)04006-8. [DOI] [PubMed] [Google Scholar]
- 54.Lee JA, Gao FB. Inhibition of autophagy induction delays neuronal cell loss caused by dysfunctional ESCRT-III in frontotemporal dementia. J Neurosci. 2009;29:8506–8511. doi: 10.1523/JNEUROSCI.0924-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rollinson S, Rizzu P, Sikkink S, Baker M, Halliwell N, Snowden J, et al. Ubiquitin associated protein1 is a risk factor for frontotemporal lobar degeneration. Neurobiol Aging. 2009;30:656–665. doi: 10.1016/j.neurobiolaging.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rusten TE, Filimonenko M, Rodahl LM, Stenmark H, Simonsen A. ESCRTing autophagic clearance of aggregating proteins. Autophagy. 2007:4. [Google Scholar]
- 58.Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 59.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 60.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 61.Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6:137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 62.Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, et al. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- 63.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 64.Cumming RC, Simonsen A, Finley KD. Quantitative analysis of autophagic activity in Drosophila neural tissues by measuring the turnover rates of pathway substrates. Methods Enzymol. 2008;451:639–651. doi: 10.1016/S0076-6879(08)03235-7. [DOI] [PubMed] [Google Scholar]
- 65.Neufeld TP, Baehrecke EH. Eating on the fly: function and regulation of autophagy during cell growth, survival and death in Drosophila. Autophagy. 2008;4:557–562. doi: 10.4161/auto.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Juhasz G, Csikos G, Sinka R, Erdelyi M, Sass M. The Drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett. 2003;543:154–158. doi: 10.1016/s0014-5793(03)00431-9. [DOI] [PubMed] [Google Scholar]
- 67.Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 68.Papaconstantinou J, Deford JH, Gerstner A, Hsieh CC, Boylston WH, Guigneaux MM, et al. Hepatic gene and protein expression of primary components of the IGF-I axis in long lived Snell dwarf mice. Mech Ageing Dev. 2005;126:692–704. doi: 10.1016/j.mad.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 69.Tu MP, Yin CM, Tatar M. Mutations in insulin signaling pathway alter juvenile hormone synthesis in Drosophila melanogaster. Gen Comp Endocrinol. 2005;142:347–356. doi: 10.1016/j.ygcen.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 70.Bergamini E, Cavallini G, Donati A, Gori Z. The anti-ageing effects of caloric restriction may involve stimulation of macroautophagy and lysosomal degradation, and can be intensified pharmacologically. Biomed Pharmacother. 2003;57:203–208. doi: 10.1016/s0753-3322(03)00048-9. [DOI] [PubMed] [Google Scholar]
- 71.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 72.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 73.Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan S, Wood M, Maher P. Oxidative stress induces a form of programmed cell death with characteristics of both apoptosis and necrosis in neuronal cells. J Neurochem. 1998;71:95–105. doi: 10.1046/j.1471-4159.1998.71010095.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.