Abstract
Due to enormous advances in quantitative proteomics and in immunohistochemistry (pathology), the two research areas have now reached the state to be successfully interwoven in order to tackle challenges in toponostics and to open tumor-targeted systems pathology approaches. In this study the differential expressions of candidate proteins nucleophosmin, nucleoside diphosphate kinase A/B (NDKA/B), osteoinducive factor (mimecan), and pyru-vate kinase M2 from a quantitative proteome signature for invasive ductal breast cancer were determined by immunohistochemistry on 53 tissue slices from formalin-fixed and paraffin-embedded tumor and control tissue samples from ten patients and fourteen controls. In addition, 87 images from the Human Protein Atlas representing seven tumor and nine normal breast tissue samples were investigated by computer-assisted semi-quantitative density measurements on nucleophosmin, nucleoside diphosphate kinase A/B (NDKA/B), osteoinducive factor (mimecan), pyruvate kinase M2, glyceraldehyde-3-phosphate dehydro-genase (GAP-DH), and mimecan (osteoinductive factor). Both IHC data sets match well to each other and support the quantitative proteome analysis data. Determining spatial distribution of signature protein expressions by protein imaging on morphologically intact tissue samples at the sub-cellular level and, hence, keeping all topological information, presents an added value to quantitative proteome data. Such comprehensive data sets are needed for both, pathway analyses and for “next generation clinical diagnostics” approaches.
Keywords: Breast carcinoma, proteome analysis, proteomics, protein expression signature, mass spectrometry, immunohistochemistry, tissue microarrays, image analysis, toponomics, toponostics
Introduction
Proteome research involves the detection of protein expression differences in a given entity, termed discovery phase, as well as the verification of the suggested protein expression differences, called validation. Having defined a quantitative proteome signature for a distinct disease status essentially asks to test its reliability and robustness, ideally using different methods and/or technology platforms in conjunction with increasing the number of investigated samples. While this request may sound trivial, accomplishing this duty with clinical samples presents in many cases a serious challenge, not only because the analysis of large sample numbers by proteomics generates a time-consuming and expensive research project.
It is widely accepted that antibody-based protein detection provides a powerful analytical means for validating proteome expression analyses e.g. by western blotting and immunohistochemistry (IHC), respectively [1]. These methods can be applied as soon as the target protein is known and functioning, i.e. specific antibodies which are directed against the protein of interest without cross-reactivity to unknown proteins are available. Especially the application of immunohistochemistry on formalin-fixed and paraffin-embedded (FFPE) tissue sections is an established, cost and time effective method in pathology bearing the great advantage of detecting the protein expression on morphologically intact tissue directly at the cellular level, hence, keeping all topological information [2]. After four decades of application and more recently, with the use of standardized protocols and automated staining systems, IHC has found its application in the diagnosis and classification of tumors including the high proportion of cancers affecting the female breast [3, 4]. The analysis of IHC images is mostly performed by visual inspection of images by pathologists that determine the staining intensity using a semiquantitative, frequently a three-point scoring system. This method inherently includes human subjectivity leadingto intra-observer and inter-observer variability [5].
Due to the enormous advances in both fields, quantitative proteomics on one side and immunohistochemistry on the other side, the two research areas are now ready to become interconnected on a routine basis. In order to establish comparability of the results from IHC with quantitative proteomics, several prerequisites have to be fulfilled. First, an important advancement for validation of proteome signatures as well as for diagnostic biomarkers and therapeutic targets is the development of tissue microarrays (TMAs). IHC staining on TMAs combined with computer-assisted image analysis provide a high-throughput method for the analysis of large collections of tissue samples enhancing both, precision and reproducibility. Keeping the importance of a high quality tissue as starting point in mind, these methods allow a standardized assessment of the huge reservoir of FFPE tissues of different origin and diseases stored in the pathology departments world-wide [1, 6]. Second, in order to standardize the image analysis process, IHC methods based on automated image analysis are recommended. Publicly available web applications and commercially available software systems handle several virtual slide formats and partly integrate both, scanning and image analysis tools [7, 8]. The analysis tools provide algorithms to examine a digital image on the pixel level resulting in a grouping of pixels into objects based on their color and/or shape. This allows a quantification of all different features in an image and therefore correlation with other quantitative data of the same sample [9]. Third, the availability of high quality immunohistochemically stained tissue images via the internet, particularly from the Human Protein Atlas (HPA; http://www.proteinatlas.org), provide an excellent source to be analyzed in the context of a proteome signature for a given disease status [10].
In this study we show that the expression of candidate proteins from a predefined breast cancer proteome signature [11, 12] was successfully localized in the tissue to either tumor cells or respective differentiated cells, e.g. duct cells in the controls, by semi-quantitative analysis of immunohistochemically stained proteins on tissue slices from FFPE tumor tissue and control tissue samples. Furthermore, analysis of IHC images from the Human Protein Atlas showed that this source provides an excellent set of protein expression data as an independent sample source that may be regarded as a reference for proteome research projects in general.
Material and methods
Tissue samples
The HRO-UFK Mammakarzinom study was approved by the Institutional Review Board. Tumor samples of invasive breast carcinoma and control samples from the same breast were taken from ten postmenopausal women (A-J) with an age between 67 and 92 years (mean 77.3 years) and subjected to routine pathological examination. In four cases (patients K to N) only control tissue was available for further studies (Table 1). Samples were prepared after modified radical mastectomy at the women's hospital, Südstadt Clinical Center Rostock (Germany), after informed consent was given. No preoperative core biopsy was performed. Tumor samples had a histological grading of two or three and tumor size ranged between 14 mm and 55 mm. All tumors were classified as invasive ductal carcinoma. Axillary lymph node involvement was detected in four cases.
Table 1.
Clinical and pathological parameters of sample group
| Patient | Age [years] | Tumor diameter [mm] | Tissue specimen a | Histologic grading b | pN c | Lnd | ERe | PRf | HER2g |
|---|---|---|---|---|---|---|---|---|---|
| A | 75 | 38 | T/G | 3 | 0/12 | L1 | 2 | 2 | 1+ |
| B | 85 | 14 | T/G | 3 | 0/7 | L1 | 12 | 9 | 2+ |
| C | 85 | 28 | T/G | 2 | 1/4 | L1 | 0 | 0 | 1+ |
| D | 72 | 40 | T/G | 3 | 13/15 | L1 | 4 | 2 | 1+ |
| E | 92 | 46 | T/G | 2 | 0/20 | L1 | 12 | 0 | 1+ |
| F | 75 | 40 | T/G | 3 | 0/- | L1 | 0 | 0 | 3+ |
| G | 73 | 55 | T/G | 2 | 4/6 | L1 | 12 | 9 | 1+ |
| H | 67 | 52 | T/G | 3 | 0/- | L1 | 8 | 0 | 1+ |
| 1 | 75 | 15 | T/G | 2 | 5/21 | L1 | 9 | 6 | 3+ |
| J | 76 | 20 | T/G | 3 | 0/- | L1 | 6 | 6 | 2+ |
| K | 67 | - | -/G | - | - | - | - | - | - |
| L | 73 | - | -/G | - | - | - | - | - | - |
| M | 95 | - | -/G | - | - | - | - | - | - |
| N | 67 | - | -/G | - | - | - | - | - | - |
tissue images from invasive ductai carcinoma (T) and/or from healthy breast tissue (G)
tumor grades 1-3 [16]
pathologic axillary nodal status
present Iymphangiosis [16]
estrogen receptor (ER), immunoreactive score: 0-12 [17]
progesterone receptor (PR), immunoreactive score: 0-12 [17]
human epidermal growth factor receptor 2 (HER2), immunohistochemistry score: 0 - 3+.
Immunohistochemistry
After mastectomy, tissue blocks were fixed in 4 % neutral buffered formalin at room temperature prior to dehydration with ascending alcohol concentrations from 70 % to 100 % (6 steps in 5 h). Excess of solvent was removed and xylene was added for fixation (3.5 h) before soaking the samples with paraffin at 60° C (2.75 h) for embedding. For generation of tissue slices the paraffin blocks were pre-cooled at −10°C. Tissue slices of 3-5 µm thickness were generated using a rotation microtome (Leica Microsystems, Wetzlar, Germany) and dried for 10 min at 80° C. Slices were deparaffinized in xylene and rehydrated before immunohistochemical staining. The /6000™ Automated Staining System (BioGenex, San Ramon, CA, USA) was used for simultaneous processing of slides according to manufacturers protocols. Slides were immunostained with the following monoclonal antibodies in a dilution of 1:100 for 60 min: mouse anti human nm23 (anti nm23) (AbD Serotec, Oxford, UK), mouse anti nucleophosmin/B23 (anti NCP) (Invitrogen, Carlsbad, CA, USA), and mouse anti human and rat pyruvate kinase type M2 (anti PKM2) (ScheBo Biotech AG, Gießen, Germany). The Link Label IHC Detection System for human tissue (BioGenex) containing a bioti-nylated secondary antibody and avidin-conjugated horseradish peroxidase, diamino-benzidine as substrate (DAB), was used to identify antibody binding. Slides were counter-stained with hematoxylin (0.1 % hematoxylin, 0.02 % NalO3, 5 % potash alum, 5 % hydrochloric acid, and 0.1 % citric acid, all dissolved in distilled water). Stained slices were evaluated by pathologists and immunohistochemical staining was considered positive when at least 10 % of breast cells were positively stained as compared to negative controls.
Digitalization of proprietary micrographs
All micrographs were digitized using standardized conditions, i.e. the same magnification (twentyfold), exposure time (7.76 ms), and homogeneous illumination (standard setting). Stained tissue sections were digitized applying a mosaic approach to obtain a representative tissue area at a resolution necessary for successive pixel classification (0.37 µm/pixel edge length). A three axis motorized digital microscope (Olympus BX51, Hamburg, Germany) with a CX9000 camera (mbf Bioscience, Vermont, USA) was controlled by the virtual slide module Stereolnvestigator v8 (mbf Bioscience, Vermont, USA). The upper left corner of a rectangular region containing relevant structures for pixel classification was determined as a starting point for grabbing four images during meandric shift of the object stage. Each image tile was corrected for inhomogeneous background (shading correction). The four image tiles were merged to a single high resolution image representing a section area of 2.832 mm2. The resulting image was stored as a TIF-file in 24 bit RGB color mode containing 1580 × 2360 pixels. High resolution printouts were re-evaluated by pathologists prior to digital image analysis. Pixels from regions that contained no tissue, e.g. lumen of milk ducts, were manually replaced by white pixels.
Images from the Human Protein Atlas
Images from tissues stained with one of 9 different antibodies detecting 5 different proteins: nucleoside diphosphate kinase A/B (gene name: NME1-NME2), nucleophosmin (gene name: NPM1), pyruvate kinase M1/M2 (gene name: PKM2), glyceraldehyde-3-phosphate dehydrogenase (gene name: GAPDH), and mime-can/osteoinductive factor (gene name: OGN) were downloaded as JPEG-files from the Human Protein Atlas (HPA) Version 6.0 (http://www.proteinatlas.org). Images were scanned at twentyfold magnification and contained about 9 million pixels per image in 24 bit RGB color mode. Patient information, such as age and tissue classification was available for each image [13]. In total, we analyzed 66 images from ductal breast carcinoma tissue from 7 different patients (1-7) with an age between 61 and 93 (mean 77.4 years) and 20 images from normal breast tissue originating from 9 different individuals (8-16) with an age between 23 and 75 years (mean 43.4 years) (Table 2). It should be noted that CAB002169, CAB012983, and CAB016392 are monoclonal antibodies from mice for which the antigen sequence is not known. CAB005197 is a so-called monospecific antibody with unknown antigen sequence. CAB019421 is a polyclonal antibody from rabbit with unknown antigen sequence. The four HPA antibodies are polyclonal antibodies from rabbit for which PrEST (protein epitope signature tag used for immunization) information are given by the HPA.
Table 2.
Data of sample group from the Human Protein Atlas
| No. | Patient ID | Age | Tissue classification |
|---|---|---|---|
| 1 | 1432 | 84 | ductal breast cancer |
| 2 | 1785 | 93 | ductal breast cancer |
| 3 | 1874 | 80 | ductal breast cancer |
| 4 | 1910 | 61 | ductal breast cancer |
| 5 | 1916 | 62 | ductal breast cancer |
| 6 | 1939 | 87 | ductal breast cancer |
| 7 | 2428 | 75 | ductal breast cancer |
| 8 | 1447 | 45 | normal breast tissue |
| 9 | 1801 | 39 | normal breast tissue |
| 10 | 1916 | 62 | normal breast tissue |
| 11 | 2042 | 75 | normal breast tissue |
| 12 | 2259 | 23 | normal breast tissue |
| 13 | 2773 | 23 | normal breast tissue |
| 14 | 3158 | 52 | normal breast tissue |
| 15 | 3286 | 27 | normal breast tissue |
| 16 | 3544 | 45 | normal breast tissue |
Image analysis
The image analysis algorithms, based on CNT (Cognition Network Technology), were provided by the Definiens Developer XD 1.1 (http://www.definiens.com). Because DAB and hematoxylin staining was used for our images as well as for the images of the HPA, we developed one master-ruleset for all images. Because the tissue on the images of the HPA is present in a circular area (tissue cores) surrounded by background it was necessary to add an option for finding these tissue cores in the ruleset for the HPA images. First, images were scaled down to 50 % of size by summarizing RGB values of neighboring pixels resulting in 790 × 1180 pixel for proprietary images and in 1500 × 1500 pixel for images from the HPA. The key steps of the master-ruleset were 1) segmentation into image objects, 2) classification of image objects, and 3) measuring output parameters on the object classes. First, a color channel for the brownish DAB staining of the protein of interest as a proportion between the components of the RGB color model was created representing the equation: R > G > B. A second color channel, that fulfills the inverse equation, represents the bluish hematoxylin (Hx) staining of the cell nuclei. Second, the major classes of tissue and background were classified using automatically generated thresholds. Classification into protein staining and staining of the nuclei was done by using logical operators (AND, OR, <, =, >) for R, G, and B and manual setting of thresholds for the unmixed color channels of DAB and Hx. Finally, the output parameters were defined as (i) number of pixels for all classes (tissue, background, brown staining, and blue staining) and (ii) mean of brown intensity. The pixel classification (brown: protein staining, blue: cell staining, gray: unstained tissue, and green: tissue-free background) resulted in a false-color image that represented the classification image. The applied ruleset was validated manually with a subset of representative images by comparison of the original image with the classification image. Afterwards, the ruleset was stored and applied in a batch mode for analyzing all images that were included in the study in an automated fashion. The batch process is a part of the Definiens Server XD software package and allows an average performance time of 12 s (range 9 s to 19 s) per image for the self-made image set and of 32 s (range 18 s to 80 s) for the HPA image set. Pixel numbers and intensity values of each classified image were stored in tables. For quantification of the protein expression from each image, the number of brown pixels, representing the staining of the protein of interest, was summed up and divided through the sum of all pixels representing tissue (sum of all pixels not being background pixels). Color intensities in pixels were not examined at this time.
Bioinformatics analysis
Box and whisker plots [14] were generated with the experimentally determined data using the Origin software (Version 6.1G, OriginLab Corporation, Northampton, MA, USA). Statistical significance of IHC stainings was tested with a two-sample two-tailed t test assuming unequal variances (Welch's ttest) with the significance level at alpha =0.05 [15].
Results
Clinical samples and starting materials
In the first set of experiments, tissue samples from healthy breasts (gland) and tumors were collected upon full removal of the gland from fourteen patients (patients A-N) with breast carcinoma (Table 1). From them ten were diagnosed with invasive ductal carcinoma (patients A-J). Both control and tumor tissues were obtained from the same breast in each case. Additional tissue samples from just healthy breast were included from four more patients (K-N) as here other tumor types were diagnosed. Histological tumor grade was determined using an established scoring system [16]. Estrogen receptor and progesterone receptor score was estimated according to internationally accepted guidelines [17]. Lymphangiosis was present in all patients. Four patients (patients C, D, G, and I) showed involved axillary lymph nodes. Mean age of patients was 77.3 years for patients A-J and 76.9 for patients A-N, respectively.
After pathological evaluation, FFPE tissue blocks were made available for immunohistochemical analysis of signature proteins. Tissue slices were decorated using commercially available monoclonal antibodies against three target proteins from our proteome signature, i.e. antibodies against nucleoside diphosphate kinase A (P15531), pyruvate kinase M2 (P14618), and nucleophosmin (P06748). Normal breast tissue typically contained intact milk ducts, fat cells, and connective tissue (exemplified in Figures 1B and 1D), whereas tumor tissue was characterized through the presence of a high density of equally shaped, undifferentiated tumor cells (cf. Figures 1A and 1C). In all cases, independent of the origin of the samples, immunohistochemical staining of the protein of interest yielded in brown/gray coloring. In addition, cells and extracellular material were stained unspecifically (hematoxylin staining, Hx) producing blue/gray color on top of the immunostain. In total 13 images (6 tumor (T) and 7 gland (G) images) were obtained with nucleoside diphosphate kinase A (P15531) decoration, and 22 images (9T/13G) were obtained with pyruvate kinase M2 (P14618) staining. Comparably, 18 images (9T/9G) with nucleophosmin (P06748) staining were obtained (cf. Figures 1A and 1B).
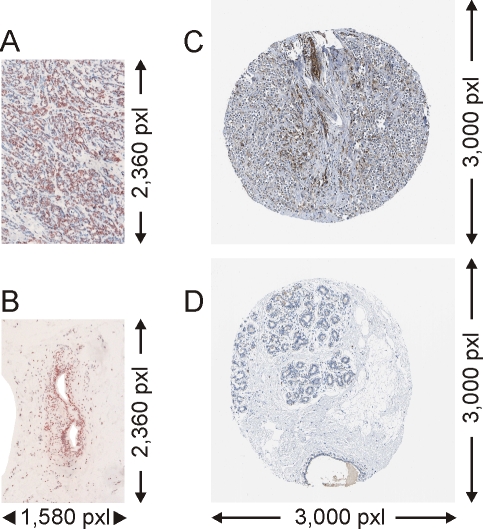
Figure 1.
Images of immunohistochemical protein staining of paraffin embedded tissues of invasive ductai breast carcinoma and of healthy breast tissue (gland). A. TIF image (24 bit) of immunohistochemical staining (anti NCP) of the protein nucleophosmin (P06748) in a proprietary tumor sample. B. Proprietary control sample. C. JPEG image (24 bit) of immunohistochemical staining (CAB019421) of the protein pyruvate kinase M2 (P14618) from the Human Protein Atlas. D. control sample. Cells are stained with hema-toxylin. 20-fold magnification.
Nucleophosmin is a nucleolar phosphoprotein resulting in brownish staining (protein staining) of the nucleus. In the images of our tumor tissues most of the cell nuclei showed positive protein staining in agreement with known protein localization. The minority of cell nuclei (blue staining) of the tumor cells as well as the lesser amount of connective tissue within the tumor (gray to white color) showed no specific nucleophosmin staining. By contrast, in the images of control tissues only endothelial cells of the milk ducts showed intense protein staining that again was localized in the nuclei. The surrounding connective and fat tissues were negative for both, protein and background staining.
All micrographs of our own stains were digitized using standardized conditions, i.e. the same magnification (twentyfold), exposure time (7.76 ms), and homogeneous illumination (standard setting) producing convenient images. The images of our self-made stains (1580 × 2360 pixels) were stored in lossless TIF format.
As an independent source for immunohistochemically stained images we mined the Human Protein Atlas (HPA, Table 2). For this second set of experiments, we downloaded images from tissue cores of ductal breast carcinoma from seven patients (patients 1-7) and normal breast tissue images from nine individuals (patients 8-16). We only included tumor images from patients that were age matched to our patient cohort resulting in a mean age of 77.4 years for the HPA-derived patients. However, the mean age in the control group was 43.4 years for the HPA patients because only images from younger individuals were provided.
The HPA tissue cores were stained with one of nine different antibodies directed against five proteins from our proteome signature: CAB002169 (3T/3G images) and HPA008467 (6T/2G) detecting nucleoside diphosphate kinase A/B, CAB012983 (8T/3G) and HPA011385 (8T/2G) targeting nucleophosmin, CAB019421 (8T/3G) and HPA029501 (12T/2G) binding to pyruvate kinase M2, CAB005197 (5T/2G) and CAB016392 (8T/2G) directed against glyceraldehyde-3-phosphate dehydrogenase (P04406), and HPA013132 (8T/2G) recognizing mimecan/osteoinductive factor (P20774). With 3000 x 3000 pixels the HPA-images covered a comparable tissue area like our images. HPA-derived images showed round shaped tissue cores surrounded by background, as exemplified for images of nucleophosmin staining (cf. Figures 1C and 1D).
In contrast to our own nucleophosmin stainings (cf. Figure 1A and 1B) no protein staining was found in the nuclei of the tumor tissue (cf. Figures 1C and 1D). Instead, brown color staining of cytosol regions was dominating. Most of the tumor cells and the connective tissue showed unspecific blue background staining. In agreement with our control samples, only unspecifically stained connective tissue (light blue), cell nuclei from cells building the milk ducts (darker blue) and unstained fat tissue (white) were found in the images of the control (gland) tissues stored in the HPA.
It should be mentioned that the images from the HPA are originally stored in TIF format as well, but are only available for download as JPEG files of unknown compression rate. The image compression factor cannot be accessed by the user, but in our image analyses this issue turned out to be neglectable. Both, our own immunohistochemistry images as well as images from the HPA were validated to be representative for each type of tissue and for sufficient immunoreactivity in each image through a pathologist before conducting computer-assisted image analysis.
Quantitative image analysis of IHC-stained tissue slices
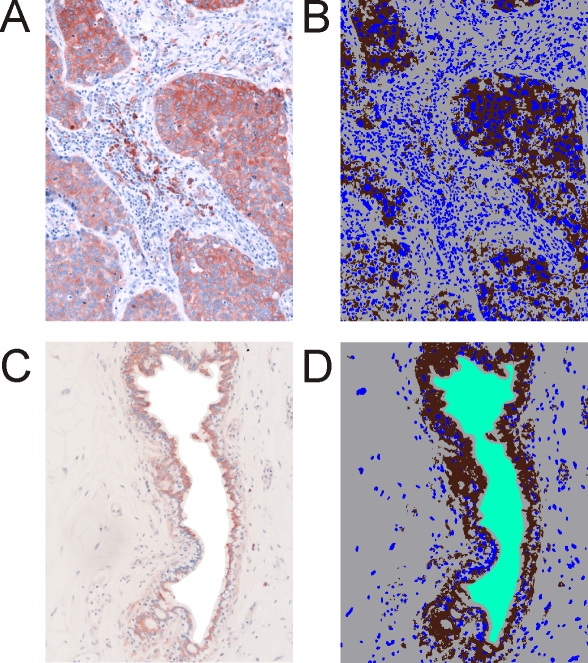
For computer-assisted quantification of the protein expression on the IHC/Hx-stained tissue slices we used algorithms that were provided by the Definiens Developer XD 1.1. Including unstained background, four colors were needed to characterize each image followed by quantitative analysis. Hence, each tissue slice was represented by a false color classification image in which brown pixels corresponded to protein staining, blue pixels to cell staining, and gray pixels represented unstained tissues (Figures 2A and 2B), as is exemplified for NDKA/B staining.
Figure 2.
Immunohistochemical staining of the protein nucleoside diphosphate kinase A (P15531). A. Staining on paraffin embedded tissue of invasive ductal breast carcinoma. B. Classification image for the tumor sample. C. Healthy breast tissue (gland). D. Classification image for the control sample. Cells are stained with hematoxylin. 20-fold magnification. Brown: protein staining, blue: cell staining, grey: unstained tissue, and green: tissue-free background.
NDKA/B is localized to the cytosol resulting in images with blue stained nuclei surrounded by brown protein staining in the cytosol when tumor regions of the tumor-containing images were analyzed. The tissues that surrounded the tumorous areas were negative for NDKA/B staining in the cytosol (white/light blue color) as well as in nuclei of the respective cells. In the control images, only endothelial cells of the milk ducts showed NDKA/B staining, again localized in the cytosol. The surrounding connective and fat tissues were negative for both, protein and background staining.
In some cases of our self-made images some areas were not covered with tissue, for example due to defects in the tissue slices or because of the presence of rather large milk ducts in studied cross sections. In these areas without tissue, the original pixels were replaced manually by white pixels (Figure 2C). The white pixels were added to the class of background pixels (Figure 2D) which were represented as green pixels (tissue-free background) in the false-color representation. This was necessary to avoid that these background areas were erroneously classified as unstained tissue by the computer program. Subsequently, the ruleset of the computer program classified each image by assigning the four different colors to the pixels of the micrographs.
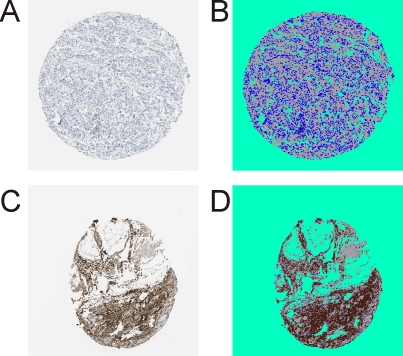
Such manual pixel assignment was not necessary for the HPA-derived images because the tissue cores were in all cases surrounded by background and the software learned to differentiate tissue cores from background automatically. In all cases the background color from the tissue-surrounding areas was also assigned to similarly colored areas within the tissue areas, as is shown for immunohistochemically stained mimecan/osteoinductive factor (Figure 3). Intense mimecan/osteoinductive factor staining was only found in the cytosol of cells from the control (gland) tissue, whereas cells from the tumor tissues were largely unstained. The tumor images showed equally shaped, undifferentiated tumor cells with blue stained cell nuclei. Surrounding connective tissue was stained light blue or white.
Figure 3.
Immunohistochemical staining (HPA013132) of the protein mimecan/osteoinductive factor (P20774) from the Human Protein Atlas. A. Staining on paraffin embedded tissue of invasive ductal breast carcinoma. B. Classification image for the tumor sample. C. Healthy breast tissue (gland). D. Classification image for the control sample. Cells are stained with hematoxylin. 20-fold magnification. Brown: protein staining, blue: cell staining, grey: unstained tissue, and green: tissue-free background.
After batch process driven image analysis, the classification images and the pixel numbers for all four color classes were stored for each tissue slice. The sum of all pixels per image not being background pixels were defined as the total tissue area. The expression amount of the protein of interest was defined as the normalized proportion of the area with brown pixels (representing the stained protein) from the total tissue area, i.e. we calculated the quotient of brown (protein)/total tissue (sum of brown, blue and gray) pixels for each image. The mean values of these (dimensionless) quotients for all tumor images of one protein staining and all control images of that same protein staining were calculated separately for each antibody (Table 3). In order to determine the differential expression of a protein we compared the so obtained mean values of the tumor groups to the mean values of those from the control (gland) groups.
Table 3.
Results from protein expression analyses
| Ace. No. | Protein name | Gene name | Experiment | Differential expression | ||||
|---|---|---|---|---|---|---|---|---|
| mean T | n T d | mean G | n G d | p-value e | ||||
| P15531 | Nucleoside diphosphate kinase A | NME1/NME2 | IHC anti nm23 a) | 0.340 | 9 | 0.076 | 9 | < 0.05 |
| IHC HPA CAB002169 a) | 0.216 | 3 | 0.062 | 3 | 0.32 | |||
| IHC HPA HPA008467 a) | 0.410 | 6 | 0.018 | 2 | < 0.01 | |||
| LC-MS b) | 0.0003 | 9 | not found | 9 | not determined | |||
| 2DE c) | 357822 | 12 | 68049 | 12 | < 0.01 | |||
| P14618 | Pyruvate kinase isozym M2 | PKM2 | IHC anti PKM2 a) | 0.402 | 6 | 0.165 | 7 | < 0.05 |
| IHC HPA CAB019421 a) | 0.149 | 8 | 0.010 | 3 | < 0.05 | |||
| IHC HPA HPA029501 a) | 0.324 | 12 | 0.151 | 2 | < 0.05 | |||
| LC-MS b) | 0.0025 | 9 | not found | 9 | not determined | |||
| 2DE c) | 100592 | 12 | 18628 | 12 | < 0.05 | |||
| P06748 | Nucleophosmin | NPM1 | IHC anti NCP a) | 0.168 | 9 | 0.102 | 13 | 0.12 |
| IHC HPA CAB012983 a) | 0.336 | 8 | 0.053 | 3 | < 0.01 | |||
| IHC HPA HPA011384 a) | 0.243 | 8 | 0.026 | 2 | < 0.01 | |||
| LC-MS b) | 0.001 | 9 | not found | 9 | not determined | |||
| 2DE c) | 2247279 | 12 | 256551 | 12 | < 0.01 | |||
| P20774 | Mimecan/osteoinductive factor | OGN | IHC HPA HPA013132 a) | 0.010 | 8 | 0.447 | 2 | < 0.01 |
| LC-MS b) | 0.001 | 9 | 0.007 | 9 | < 0.01 | |||
| 2DE c) | 321566 | 12 | 1995002 | 12 | < 0.01 | |||
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | IHC HPA CAB005197 a) | 0.192 | 5 | 0.013 | 2 | < 0.05 |
| IHC HPA CAB016392 a) | 0.389 | 8 | 0.078 | 2 | < 0.01 | |||
| LC-MS b) | 0.007 | 9 | 0.002 | 9 | < 0.01 | |||
| 2DE c) | 1349792 | 12 | 106121 | 12 | < 0.01 | |||
proportion of brown pixels per tissue pixels in images from tumor tissue (mean T) and control tissue (gland; mean G) after image analysis of immunohistochemically stained tissue slices
normalized protein expression amounts in tumor tissue (mean T) and control tissue (gland; mean G) using quantitative label-free LC-MS data [12]
normalized protein expression amounts in tumor tissue (mean T) and control tissue (gland; mean G) using densitometric data from 2D gels [11]
number of technical replicates for the tumor group (n T) and control group (gland; n G), respectively
p-values were determined using the Welch's t-testfor IHC data and the Student's t-testfor LC-MS and 2-DE data, respectively
In total we investigated differential expressions of five proteins from our predefined proteome signature for invasive ductal breast carcinoma (nucleoside diphosphate kinase A, pyruvate kinase M2, nucleophosmin, glyceraldehydes-3-phosphate dehydrogenase and mimecan/ osteoinductive factor) using immunohistochemically stained tissue slices from either own stains (53 images) or from the HPA source (87 images). For determining differential expression between both groups for each antibody staining Welch's t-tests for two populations was performed (Table 3).
Nucleoside diphosphate kinase A (NDKA) was found in larger areas (mean: 0.34) in the tumor group compared to the control (gland) group (mean: 0.076) and, hence, can be assigned as significantly up-regulated (p-value < 0.05) in the immunohistochemically stained samples from our patients (Figure 4A). When analyzing the images of the stainings with the antibody used by the HPA group (HPA008467) a similar result was obtained. Mean of the NDKA stain in the tumor group (0.41) was higher than that of the control group (0.018) and a p-value < 0.01 was obtained (Figure 4B). The same tendency of differential NDKA expression was found for the second HPA antibody CAB002169 (Table 3), however without reaching significance (p = 0.32).
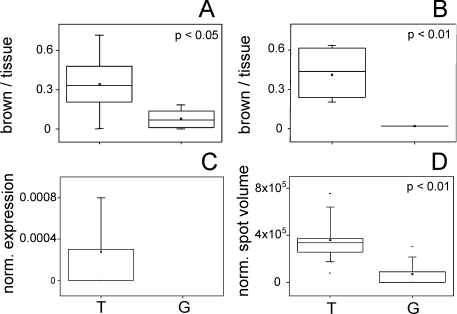
Figure 4.
Box and whisker plots for expression data of nucleoside diphosphate kinase A (P15531). A. Expression amount on proprietary immunohistochemically stained tissue slices (anti nm23) represented by the proportion of brown pixels per tissue pixels. Replicates: n = 9 for T, n = 9 for G. B. Expression amount on immunohistochemically stained (HPA008467) tissue slices from the Human Protein Atlas represented by the proportion of brown pixels per tissue pixels. Replicates: n = 6 for T, n = 2 for G. C. Normalized protein expression amounts using quantitative label-free LC-MS data. Replicates: n = 9 for T, protein not found in G. D. Normalized protein expression amounts using densitometric data (normalized spot volumes) from 2-D gels. Replicates: n = 12 for T, n = 12 for G. Tumor samples (T) are depicted on the left and control (gland) samples (G) on the right, p-values are given.
Pyruvate kinase M2 was also found to be significantly up-regulated in the tumor samples (mean: 0.402) compared to the control (gland) samples (mean: 0.165) in the IHC analysis of our images. The protein expression difference was statistically significant (p-value < 0.05). Both antibody stains from the HPA (CAB019421 and HPA029501) support this tendency of differential expression (p-value < 0.05 in both cases; Table 3). Similarly, nucleophosmin was stained in larger areas in the tumor group (mean: 0.168) as compared to the controls (mean: 0.102) when analyzing the IHC images of our patients (Table 3) and both antibody stains from the HPA (CAB012983 and HPA011384) showed a significant up-regulation in the tumor images as compared to the control (gland) images (p-value < 0.01 in both cases).
Immunohistochemical stains from our own patient cohort were neither available to us for glyceraldehyde-3-phosphate dehydrogenase nor for mime-can/osteo-inductive factor. Instead, images from the HPA were analyzed for differential expression of these proteins. The glyceraldehyde-3-phosphate dehydrogenase IHC staining with the HPA antibody CAB016392 (Table 3) showed a stronger expression of this protein in the tumor samples (mean: 0.389) compared to that in the control (gland) samples (mean: 0.078) with statistical significance (p-value < 0.01). The matching tendency was found for the second HPA antibody CAB005197 (p-value < 0.05).
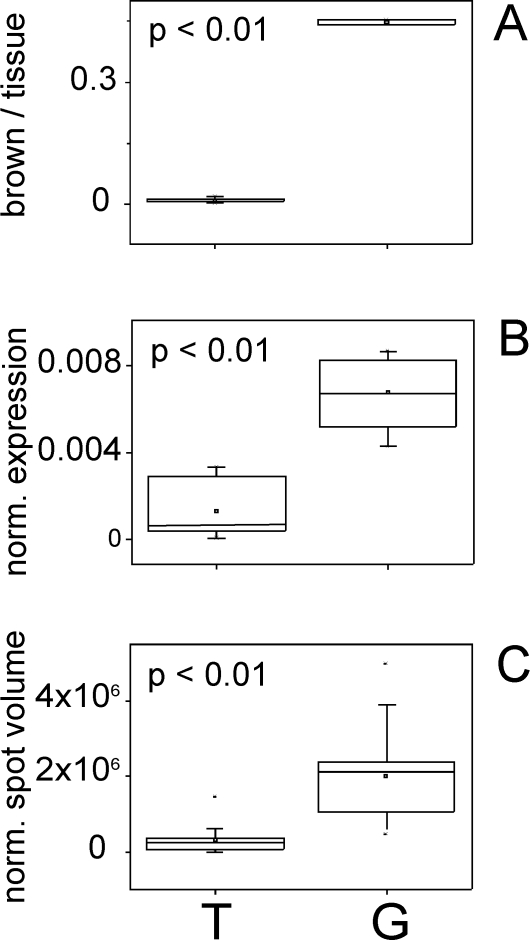
In contrast to the four above mentioned proteins mimecan/osteoinductive factor was significantly down-regulated (p-value < 0.01) in the tumor group (mean: 0.01) compared to the control (gland) group (mean: 0.447) when analyzing the IHC staining of the HPA antibody HPA013132 (Figure 5A).
Figure 5.
Box and whisker plots for expression data of mimecan/osteoinductive factor (P20774). A. Expression amount on immunohistochemically stained (HPA013132) tissue slices from the Human Protein Atlas represented by the proportion of brown pixels per tissue pixels. Replicates: n = 8 for T, n = 2 for G. B. Normalized protein expression amounts using quantitative label-free LC-MS data. Replicates: n = 9 for T, n = 9 for G. C. Normalized protein expression amounts using densitometric data (normalized spot volumes) from 2-D gels. Replicates: n = 12 for T, n = 12 for G. Tumor samples (T) are depicted on the left and control (gland) samples (G) on the right, p-values are given.
The high consistence of differential protein expression between the two IHC image sources suggests that the IHC image analysis procedure, as described here, enables validation of quantitative proteome analyses with publicly available IHC data.
Comparison of immunohistochemistry-based analysis with quantitative proteome analysis
In order to estimate whether publicly available IHC data may function as a suitable reference for proteome analysis results, we compared the IHC quantitation results with previous expression difference analyses using nano-LC ESI-MSE and 2-D gel electrophoresis, respectively, of invasive ductal breast carcinoma tissue and control (gland) tissue. Quantitative data from nano-LC ESI-MSE experiments, termed normalized expression, showed for NDKA that this protein was only found in samples from the tumor group but not in the control (gland) group (Table 3, Figure 4C). In the follow-up study using 2-D gel analysis, again NDKA was found to be up-regulated in the tumor samples compared to the control (gland) samples (p-value < 0.01) as derived from the normalized spot volumes (Table 3, Figure 4D). Hence, the differential expression of NDKA is confirmed with a fairly large group of patients from two completely different sources.
Similarly, in 2-D gel-based analyses pyruvate kinase M2 was found up-regulated in the tumor group (p-value < 0.05), whereas in the nano-LC ESI-MSE experiments the protein was found in the tumor group only (Table 3). Again, all sets of completely independent experiments confirm each other. Also, findings on nucleophosmin expression as determined by IHC image analysis again validated the results from the previous experiments where nucleophosmin was only found in the tumor samples by nano-LC ESI-MSEexperiments and was determined as significantly up-regulated in the tumor samples (p-value < 0.01) upon 2-D gel analysis (Table 3). Both, the nano-LC ESI- MSEexperiments and the 2-D gel analyses corresponded again to the here described results obtained with IHC image analysis of glyceraldehydes-3-phosphate dehydrogenase (Table 3) which was found to be significantly up-regulated (p-values < 0.01). Finally, mimecan/osteoinductive factor expression as determined by IHC image analysis also correlated to the results from the nano-LC ESI- MSE and the 2-D gel experiments. A significant down-regulation of mimecan/osteoinductive factor (p-values < 0.01) was detected in proteome analyses (Figures 5B and 5C), corresponding well to the here described results.
In conclusion, we successfully analyzed IHC images of self-made stains as well as images provided by the Human Protein Atlas, cross-referencing the quantitative proteome signature with the most important clinically relevant technology for protein topology analysis in cancer. Hence, due to the combination of results, with IHC image analysis, protein location in the tissue is added to the quantitative expression data derived from quantitative proteomics.
Discussion
In the case of breast cancer investigations, several groups have shown that using a combination of IHC markers, molecular subgroups of breast carcinoma [18] can be identified, and clinical outcome of ER positive breast cancer patients [19] can be predicted. Similarly, clinically relevant prognostic subgroups in patient cohorts hosting a primary breast cancer with involvement of four or more axillary lymph nodes can be identified [20] when monitoring the IHC-stainings with the defined set of antibodies. Comprehensive immunohistochemistry with multiple overlayed protein localization analyses was termed toponomics - and for projects which were mainly subjected to diagnostic purposes the term toponostics was suggested [21, 22]. In parallel to IHC-based investigations, we and others investigated protein expression differences in patients suffering from invasive ductal breast cancer using a proteome research approach. We defined a proteome signature for invasive ductal breast cancer that contained twenty proteins and enabled clear separation of both groups using principal component analysis and hierarchical clustering [12]. Further, detailed protein structure investigations enabled to refine our proteome signature, opening the way to relational structure-function-studies with respect to disease processes and/or therapeutic intervention [11]. Joining toponomics and proteomics allows to access new aspects of protein research beyond validation of quantitative protein expression differences for exemplary proteins. Combining protein toponomics and proteomics will enable to tackle systems biology approaches, where quantitative and time-resolved data are needed, for a defined compartment, i.e. specific cell types within a tissue of an organ in even greater quality (Figure 6).
Figure 6.
Schematic presentation of key developments in proteome and toponome research. Both research areas are regarded as corner stones of “Toponostics” for systems biology approaches. Quantitative and time-resolved data together with precise cellular localization information are needed for pathway analyses, to understand tumor development, and to enable modeling of tissue processes.
Similarly, for clinical studies it becomes clear that the dissection of tissue into its specific cell components is needed before protein (or RNA) expression differences can be advanced to pathway analyses that may be of interest for understanding the principle of function for a given therapeutic agent. In such a combined approach it becomes more and more important to consider the role of antibody specificity and cross-reactivity for each protein target during toponome analysis. In particular the epitope sequences against which antibodies of interest are directed ought to be known and should be provided by the manufacturers whenever possible. Otherwise the antibody-binding regions on a protein target need to be determined experimentally using epitope mapping approaches, e.g. using mass spectrometry [23-25]. Taken together we showed that despite the fact that the principle applicability of immunohistochemistry for protein quantitation has been questioned [26, 27], e.g. because of the influence of fixation on IHC results [3, 28] as well as because of other issues, computer-assisted IHC image analysis turned out to provide reliable results. Particularly when the determined protein expression differences were supported by quantitative proteome signatures which provided lead informations on the differential expression of the proteins of interest. Obviously, determining quantitative protein expression differences with high accuracy in clinical tissues is a stronghold of proteome analysis [22, 29] whereas visualizing the spatial location of a protein of interest with high resolution is currently reached best by microscopic methods [30, 31].
In the near future the emerging methods of MALDI-MS Imaging promise fast and accurate localizations of proteins from a predefined proteome signature, even when suitable antibodies are not readily available [32, 33]. Hence, particularly for clinical studies MALDI-MS Imaging seems to be perfectly accompanying proteome research for guiding pathologists' investigations working with antibody-based immunohistochemistry on above mentioned tissues. More and more established biobanks in the clinics will serve as a repository for joint investigations in which proteome analysis, pathology, and clinical investigations will be working together to untangle complex pathways and disease-related protein network disturbances. Ultimately, it can be envisaged that the combination of quantitative information from protein expression profiling with data on spatial distribution of protein abundances from histology/pathology enables the identification of candidate proteins serving as key-markers for diagnostics and/or prognostics, or as future therapeutic targets for the treatment of breast cancer.
Acknowledgments
We thank Michael Kreutzer and Manuela Sieb for excellent technical assistance and Dr. Michael Hecker for supporting information on bio-statistics. We like to express our thanks to Dr. Falk Pommerenke from the “Partnerschaft der Fachärzte für Pathologie” at the Südstadt Clinical Center, Rostock, for supplying histological data on the individual samples and Dr. Christian George for clinical information on the individual tissue samples.
Abbreviations
- DAB
Diaminobenzidine
- FFPE
formalin-fixed and paraffin-embedded
- G
Gland
- HPA
Human Protein Atlas
- Hx
Hematoxylin
- IHC
Immunohistochemistry
- T
Tumor
References
- 1.Decaestecker C, Moles Lopez X, D'Haene N, Roland I, Guendouz S, Duponchelle C, Alix B, Debeir O, Salmon I. Requirements for the valid quantification of immunostains on tissue microarray materials using image analysis. Proteomics. 2009;9:4478–4494. doi: 10.1002/pmic.200800936. [DOI] [PubMed] [Google Scholar]
- 2.Pontén F, Jirstrom K, Uhlen M. The Human Protein Atlas-a tool for pathology. J Pathol. 2008;216:387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 3.Warford A, Howat W, McCafferty J. Expression profiling by high-throughput immunohistochemistry. J Immunol Methods. 2004;290:81–92. doi: 10.1016/j.jim.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Taylor CR, Levenson RM. Quantification of immunohistochemistry-issues concerning methods, utility and semiquantitative assessment II. Histopathology. 2006;49:411–424. doi: 10.1111/j.1365-2559.2006.02513.x. [DOI] [PubMed] [Google Scholar]
- 5.Nakhleh RE. Error Reduction in Surgical Pathology. Arch. Pathol. Lab. Med. 2006;130:630–632. doi: 10.5858/2006-130-630-ERISP. [DOI] [PubMed] [Google Scholar]
- 6.Takikita M, Chung J-Y, Hewitt SM. Tissue microarrays enabling high-throughput molecular pathology. Curr Opin Biotechnol. 2007;18:318–325. doi: 10.1016/j.copbio.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Tuominen VJ, Ruotoistenmäki S, Viitanen A, Jumppanen M, Isola J. ImmunoRation: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res. 2010;12:R56. doi: 10.1186/bcr2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rojo MG, Bueno G, Slodkowska J. Review of imaging solutions for integrated quantitative immunohistochemistry in the Pathology daily practice. Folia Histochem Cytobiol. 2009;47:349–354. doi: 10.2478/v10042-008-0114-4. [DOI] [PubMed] [Google Scholar]
- 9.Strömberg S, Björklund MG, Asplund C, Sköllermo A, Persson A, Wester K, Kampf C, Nilsson P, Andersson A-C, Uhlen M, Kononen J, Pontén F, Asplund A. A high-throughput strategy for protein profiling in cell microarrays using automated image analysis. Proteomics. 2007;7:2142–2150. doi: 10.1002/pmic.200700199. [DOI] [PubMed] [Google Scholar]
- 10.Gloriam DE, Orchard S, Bertinetti D, Björling E, Bongcam-Rudloff E, Borrebaeck CAK, Bourbeillon J, Bradbury ARM, de Daruvar A, Dübel S, Frank R, Gibson TJ, Gold L, Haslam N, Herberg FW, Hiltke T, Hoheisel JD, Kerrien S, Koegl M, Konthur Z, Korn B, Landegren U, Montecchi-Palazzi L, Palcy S, Rodriguez H, Schweinsberg S, Sievert V, Stoevesandt O, Taussig MJ, Ueffing M, Uhlén M, van der Maarel S, Wingren C, Woollard P, Sherman DJ, Hermjakob H. A Community Standard Format for the Representation of Protein Affinity Reagents. Mol Cell Proteomics. 2009;9:1–10. doi: 10.1074/mcp.M900185-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Röwer C, Koy C, Hecker M, Reimer T, Gerber B, Thiesen H-J, Glocker MO. Mass Spectrometric Characterization of Protein Structure Details Refines the Proteome Signature for Invasive Ductal Breast Carcinoma. J Am Soc Mass Spectrom. 2011;22:440–456. doi: 10.1007/s13361-010-0031-6. [DOI] [PubMed] [Google Scholar]
- 12.Röwer C, Vissers JPC, Koy C, Kipping M, Hecker M, Reimer T, Gerber B, Thiesen H-J, Glocker MO. Towards a proteome signature for invasive ductal breast carcinoma derived from label-free nanoscale LC-MS protein expression profiling of tumorous and glandular tissue. Anal Bioanal Chem. 2009;395:2443–2456. doi: 10.1007/s00216-009-3187-9. [DOI] [PubMed] [Google Scholar]
- 13.Uhlen M, Björling E, Agaton C, Al-Khalili Szigyarto C, Amini B, Andersen E, Andersson A-C, Angelidou P, Asplund A, Asplund C, Berglund L, Bergström K, Brumer H, Cerjan D, Ekström M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J, Forsberg M, Björklund MG, Gumbel K, Halimi A, Hallin I, Hamsten C, Hansson M, Hedhammar M, Hercules G, Kampf C, Larsson K, Lindskog M, Lodewyckx W, Lund J, Lundeberg J, Magnusson K, Malm E, Nilsson P, Ödling J, Oksvold P, Olsson I, Öster E, Ottosson J, Paavilainen L, Persson A, Rimini R, Rockberg J, Runeson M, Sivertsson A, Sköllermo A, Steen J, Stenvall M, Sterky F, Strömberg S, Sundberg M, Tegel H, Tourle S, Wahlund E, Waldén A, Wan J, Wernérus H, Westberg J, Wester K, Wrethagen U, Xu LL, Hober S, Pontén F. A Human Protein Atlas for Normal and Cancer Tissues Based on Antibody Proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Tukey JW. Exploratory Data Analysis. 1977. Addison Wesley.
- 15.Welch BL. The generalization of “Student's” problem when several different population variances are involved. Biometrika. 1947;34:28–35. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]
- 16.Elston CW. Classification and grading of invasive breast carcinoma. Verh. Dtsch Ges Pathol. 2005;89:35–44. [PubMed] [Google Scholar]
- 17.Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe. 1987;8:138–140. [PubMed] [Google Scholar]
- 18.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressier L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M, Perou CM. Immunohistochemical and Clinical Characterization of the Basal-Like Subtype of Invasive Breast Carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 19.Ring BZ, Seitz RS, Beck R, Shasteen WJ, Tarr SM, Cheang MCU, Yoder BJ, Budd GT, Nielsen TO, Hicks DG, Estopinal NC, Ross DT. Novel Prognostic Immunohistochemical Biomarker Panel for Estrogen Receptor-Positive Breast Cancer. J Clin Oncol. 2006;24:3039–3047. doi: 10.1200/JCO.2006.05.6564. [DOI] [PubMed] [Google Scholar]
- 20.Crabb SJ, Bajdik CD, Leung S, Speers CH, Kennecke H, Huntsmann DG, Gelmon KA. Can clinically relevant prognostic subsets of breast cancer patients with four or more involved axillary lymph nodes be identified through immunohistochemical biomarkers? A tissue microarray feasibility study. Breast Cancer Res. 2008;10:R6. doi: 10.1186/bcr1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schubert W. Topological proteomics, toponomics, MELK-technology. Adv Biochem Eng Biotechnol. 2003;83:189–209. doi: 10.1007/3-540-36459-5_8. [DOI] [PubMed] [Google Scholar]
- 22.Glocker MO, Guthke R, Kekow J, Thiesen H-J. Rheumatoid arthritis, a complex multifactorial disease: on the way toward individualized medicine. Med Res Rev. 2006;26:63–87. doi: 10.1002/med.20045. [DOI] [PubMed] [Google Scholar]
- 23.Suckau D, Köhl J, Karwath G, Schneider K, Casaretto M, Bitter-Suermann D, Przybylski M. Molecular epitope identification by limited proteolysis of an immobilized antigen-antibody complex and mass spectrometric peptide mapping. Proc Natl Acad Sci USA. 1990;87:9848–9852. doi: 10.1073/pnas.87.24.9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker CE, Tomer KB. MALDI/MS-based epitope mapping of antigens bound to immobilized antibodies. Mol Biotechnol. 2002;20:49–62. doi: 10.1385/MB:20:1:049. [DOI] [PubMed] [Google Scholar]
- 25.El-Kased RF, Koy C, Lorenz P, Montgomery H, Tanaka K, Thiesen H-J, Glocker MO. A Novel Mass Spectrometric Epitope Mapping Approach without Immobilization of the Antibody. J Proteomics Bioinform. 2011;4:1–9. [Google Scholar]
- 26.Leong AS. Quantitation in immunohistology: fact or fiction? A discussion of variables that influence results. Appl. Immunohistochem. Mol Morphol. 2004;12:1–7. doi: 10.1097/00129039-200403000-00001. [DOI] [PubMed] [Google Scholar]
- 27.Apfeldorfer C, Ulrich K, Jones G, Goodwin D, Collins S, Schenck E, Richard V. Object orientated automated image analysis: quantitative and qualitative estimation of inflammation in mouse lung. Diagn Pathol. 2008;3(Suppl1):S16. doi: 10.1186/1746-1596-3-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paavilainen L, Edvinsson A, Asplund A, Hober S, Kampf C, Pontén F, Wester K. The Impact of Tissue Fixatives on Morphology and Antibody-based Protein Profiling in Tissues and Cells. J Histochem Cytochem. 2010;58:237–246. doi: 10.1369/jhc.2009.954321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hewitt SM, Takikita M, Abedi-Ardekani B, Kris Y, Bexfield K, Braunschweig T, Chung J-Y. Validation of proteomic-based discovery with tissue microarrays. Proteomics Clin Appl. 2008;2:1460–1466. doi: 10.1002/prca.200800003. [DOI] [PubMed] [Google Scholar]
- 30.Zieba A, Wählby C, Hjelm F, Jordan L, Berg J, Landegren U, Pardali K. Bright-Field Microscopy Visualization of Proteins and Protein Complexes by In Situ Proximity Ligation with Peroxidase Detection. Clin Chem. 2010;56:99–110. doi: 10.1373/clinchem.2009.134452. [DOI] [PubMed] [Google Scholar]
- 31.Pierre S, Scholich K. Toponomics: studying protein-protein interactions and protein networks in intact tissue. Mol Biosyst. 2010;6:641–647. doi: 10.1039/b910653g. [DOI] [PubMed] [Google Scholar]
- 32.Schwamborn K, Caprioli RM. Molecular imaging by mass spectrometry-looking beyond classical histology. Nat Rev Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 33.McDonnell LA, Corthals GL, Willems SM, van Remoortere A, van Zeijl RJM, Deelder AM. Peptide and protein imaging mass spectrometry in cancer research. J Proteomics. 2010;73:1921–1944. doi: 10.1016/j.jprot.2010.05.007. [DOI] [PubMed] [Google Scholar]